Abstract
Peptides from natural sources such as milk are shown to have a wide spectrum of biological activities. In this study, three peptides with antioxidant capacity were identified from camel milk protein hydrolysate. Pepsin and pancreatin were used for hydrolysis of milk proteins. Ultrafiltration and reverse-phase high-performance liquid chromatography were used for the concentration and purification of the hydrolysate, respectively. Sequences of the three peptides, which were determined by matrix-assisted laser desorption/ionization time-of-flight spectrophotometry, were LEEQQQTEDEQQDQL [molecular weight (MW): 1860.85 Da, LL-15], YLEELHRLNAGY (MW: 1477.63 Da, YY-11), and RGLHPVPQ (MW: 903.04 Da, RQ-8). The 3-(4,5-dimethylthia-zol-2-yl)-2,5-diphenyltetrazolium bromide assay was used to evaluate the cytotoxicity of these chemically synthesized peptides against HepG2 cells. In vitro analysis showed antioxidant properties and radical scavenging activities of these peptides on 2,2-diphenyl-1-picrylhydrazyl, 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)+, O2−, and OH− free radicals. HepG2 cells were treated with YY-11 peptide for 48 hours, and the expression of superoxide dismutase and catalase genes was examined using real-time polymerase chain reaction. The results revealed a significant increase in the expression of superoxide dismutase and catalase genes in treated HepG2 cells.
Keywords: antioxidant activity, camel milk protein, gene expression, HepG2 cell line, peptide
1. Introduction
Free radicals are produced in a wide range of biological and chemical systems [1]. Reactive oxygen species (ROS) including superoxide anions, hydroxyl radicals, nitric oxide radicals, and peroxyl radicals are various forms of free radicals that are produced as byproducts of cellular respiration in mitochondria [2]. Free radicals have dual functions, in which they can play a role in signaling pathways and defense responses against pathogens, but excessive free radicals can damage biomolecules such as DNA, proteins, and lipids and eventually cause oxidative stress [3]. Under normal conditions, ROS can be neutralized by the enzymatic and nonenzymatic mechanisms in the body; however, increases in the amount of ROS in the body will lead to an imbalance between free radicals and antioxidants, which finally leads to oxidative stress [2]. A variety of diseases such as cancers are in association with oxidative stress [4]. Owing to the harmful effects of free radicals and oxidative stress in the body, prevention of these reactions seems necessary [5]. Body cells have effective strategies to prevent DNA damage induced by free radicals [6]. Antioxidant enzymes such as glutathione peroxidase, superoxide dismutase (SOD), and catalase (CAT) are part of defense mechanisms against oxidative stress and are able to inhibit ROS rapidly [7]. Levels of these enzymes increase in oxidative stress conditions to prevent possible damage; however, in some cases, the amount of endogenous antioxidants is not enough to inhibit free radicals and an external source of antioxidants is required [8]. Some synthetic antioxidant compounds such as butylated hydroxytoluene and butylated hydroxyanisole, despite their use in medicine, have adverse side effects on the body [9]. Therefore, researches have focused on the identification and extraction of antioxidant compounds from natural sources. Peptides as natural antioxidants have some regulatory effects including nutrient uptake, immune defense, and antioxidant properties [10]. Several studies have been performed on the antioxidant capacity of protein hydrolysates or peptides extracted from natural sources such as egg-yolk protein [11], milk kefir and soymilk kefir [12], casein [13], algae protein waste [14], and buckwheat protein [15]. Camel milk is a rich source of proteins, which is suggested to have biological activity including antibacterial, antiviral, and antioxidant activities (PMID: 1319434). A previous finding has shown that due to it antioxidant activity, camel milk can be considered as a potential therapeutic approach for the treatment of autism spectrum disorder (PMID: 24069051, 20175528). The aim of this study was to investigate the antioxidant properties of the three peptides derived from camel milk proteins and also to evaluate the expression of SOD and CAT genes in HepG2 cells treated with the selected peptide YY-11.
2. Materials and methods
2.1. Materials
Enzymes and chemical compounds used in this study including pepsin, pancreatin, 2,2-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), glutathione, butylated hydroxyanisole, potassium persulfate, trichloroacetic acid, 3-(4,5-dimethylthia-zol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), EDTA, and trifluoroacetic acid were obtained from Sigma Chemicals Co. (St. Louis, MO, USA). All cell cultured components were purchased from Gibco, Grand Island, New York, USA. RNA isolation and complementary DNA (cDNA) synthesis kits were purchased from Roche (Mannheim, Germany) and Thermo Fisher Scientific Company (168 Third Avenue Waltham, MA 02451, USA), respectively.
2.2. Preparation of hydrolysate from camel milk using pepsin–pancreatin enzymes
Camel milk was hydrolyzed using a mixture of pepsin–pancreatin enzymes according to Quirós et al [16] with some modifications. For this purpose, first the mixture of pepsin and glycine–HCl buffer (pH 2) was prepared and stirred in special conditions (at 37°C for 2 hours). Then using NaHCO3 (0.9M) and NaOH (1.0M), pH values were adjusted to 5.3 and 7.5, respectively. After reaching the desired pH, pancreatin was added and the mixture was kept at 37°C for 4 hours. In order to inhibit the enzyme activity, the hydrolysate were placed in boiling water for 10 minutes, and then incubated at room temperature (25°C) for 15 minutes and finally centrifuged (at 7000g, 4°C, for 15 minutes). The resulting hydrolysate (supernatant) was lyophilized and maintained at −20°C for later analysis. The resulting hydrolysate was dissolved in distilled water and passed through Millipore ultrafiltration membrane of 3 kDa (Billerica, MA, USA). The filtered solution was collected, lyophilized, and stored at −20°C.
2.3. High-performance liquid chromatography and amino acid sequencing
High-performance liquid chromatography (Knauer, Berlin, Germany) was used for the purification of camel milk protein hydrolysate. A flow rate of 2.0 mL/min and a linear gradient of 5–65% acetonitrile containing 0.1% trifluoroacetic acid were used for fractionation over 60 minutes. Five hundred microliters of the resulting hydrolysate (at 20 mg/mL) were prepared and filtered through a 0.45-μm filter, and then loaded on a C18 column (Macherey-Nagel, Düren, Germany). Elution of peaks was monitored at 220 nm, and fractions were collected and lyophilized. The collected fractions were further purified using the same gradient of acetonitrile as mentioned above at a flow rate of 1 mL/min. The peptides of interest were analyzed through de novo sequencing using matrix-assisted laser desorption/ionization time-of-flight (at Proteomics International Pty Ltd (Nedlands, Western Australia).
2.4. Evaluation of DPPH free radical scavenging
DPPH assay was used to evaluate the scavenging ability of the peptides according to Binsan et al [17] with some modifications. Briefly, a mixture containing 0.5 mL of sample (0–1 mg/ mL, final concentration) and 0.5 mL of DPPH (0.15mM in 95% ethanol) was prepared. The mixture was incubated for 30 minutes in the dark at room temperature (25°C).
Sample absorbance (ASample) was measured at 570 nm using a spectrophotometer (Epoch, Winooski, Vermont, USA). Free radical scavenging ability of the peptides was assessed by the following formula [17]:
A mixture containing 0.5 mL water and 0.5 mL DPPH in ethanol was used as a control.
2.5. Evaluation of ABTS free radical scavenging
ABTS radical cation solution was first produced by mixing ABTS with potassium persulfate and incubating the mixture in the dark at room temperature (25°C) for 16 hours [17]. To prepare the working solution, the mixture was diluted with phosphate buffered saline to reach the absorbance of 0.70 ± 0.02 at 734 nm. Thereupon, 0.5 mL of the working solution with 0.5 mL of various concentrations of peptides (0–1 mg/mL) were mixed and incubated at 37°C for 1 hour. Finally, the absorption was measured at 734 nm. Free radical scavenging capacity of the peptides was measured using the following equation [17]:
A mixture made up of phosphate buffered saline (0.5 mL) and the working solution (0.5 mL) was used as a control.
2.6. Evaluation of hydroxyl free radical scavenging
Hydroxyl free radical scavenging ability of the peptides was evaluated by the method of Li et al [18] with slight modifications. A mixture of phenanthroline (0.75mM) and FeSO4 (0.75mM) in phosphate buffer (pH 7.4) was prepared, and after adding 0.01% H2O2 and the peptide sample, the mixture was incubated at 37°C for 1 hour. Ultimately, the absorbance was evaluated at 536 nm. Hydroxyl free radical scavenging capacity of the peptides was evaluated by the following equation:
where AS is the absorbance of the sample; A1 is the absorbance of the control containing 1,10-phenanthroline, FeSO4, and H2O2; and A0 is the absorbance of the blank containing 1,10-phenanthroline and FeSO4.
2.7. Evaluation of superoxide free radical scavenging
This method is based on the ability of a desired compound to scavenge O−2 through the production of a chromophoric compound during the course of the reaction. For this purpose, 80 μL of hydrolysate along with 80 μL of Tris-HCl buffer (0.05M, pH 8.3) was placed in a 96-well microplate; thereupon, 40 μL of pyrogallol solution (1.5mM) was added to each well of the plate. Finally, absorption of the mixture was measured at 320 nm for 5 minutes at 25°C. Butylated hydroxytoluene (at 0–1 mg/mL, final concentration) was used as a positive control (AC) to evaluate the superoxide free radical scavenging capacity of the peptide sample (AS) [18,19]:
2.8. Cell culture
HepG2 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) containing 10% fetal bovine serum supplemented with L-glutamine and 1% penicillin/streptomycin at 37°C in a humidified, 5% CO2 incubator. Cell treatment was performed when the cells were in the logarithmic phase of the growth (with 80% confluence).
2.9. Viability assay
Viability of HepG2 cells treated with various concentrations of peptides (from 0 mg/mL to 1 mg/mL) was evaluated by MTT assay. For this purpose, HepG2 cells were seeded and incubated for 24 hours, and then the cells were treated with different concentrations of peptides for 24 hours, 48 hours, and 72 hours. After the incubation time, MTT solution was added and the mixture was then incubated in dark at 37°C for 4 hours. Finally, Dimethyl sulfoxide (DMSO) was added to each well and the optical absorbance of the sample was recorded at 570 nm using a spectrophotometer.
2.10. SOD and CAT gene expression
To evaluate the SOD and CAT gene expression, total RNA was isolated from HepG2 cells (treated and untreated) using a high pure RNA isolation kit according to the company’s instructions. The extracted RNA (2 μL) was used to synthesize cDNA using 1 μL Oligo (dT) primer, 4 μL reaction buffer, 1 μL RiboLock RNase inhibitor, 2 μL dNTP, 1 μL RevertAid RT, and 9 μL nuclease-free water, and then incubated at 42°C for 60 minutes and 70°C for 5 minutes in a thermal cycler (Biotech, London, England). The Bio-Rad CFX96 detection system was utilized for real-time polymerase chain reaction. Reaction solution was composed of 2 μL cDNA, 10 μL SYBR Green Master Mix, 2 μL primers, and 6 μL distilled water.
Afterward, levels of gene expression in cells treated with various concentrations of YY-11 peptide (125 μg/mL, 250 μg/ mL, and 250 μg/mL) were compared with those in untreated control cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Sequences of primers used are presented in Table 1.
Table 1.
Sequences of primers used in real-time PCR.
| Genes | Forward 5′ → 3′ | Reverse 5′ → 3′ |
|---|---|---|
| GAPDH | 5′-TGTGTCCGTCGTGGATCTGA-3′ | 5′TTGCTGTTGAAGTCGCAGGAG-3′ |
| SOD | 5′ TCTGGATGGGTGTGGCTTGCTCT 3′ | 5′ GCATGCTCCCAAACATCGATC 3′ |
| CAT | 5′ CGTGCTGAATGAGGAACAGA3′ | 5′ AGTCAGGGTGGACCTCAGTG3′ |
| GAPDH = | Glyceraldehyde 3-phosphate dehydrogenase; CAT = catalase; PCR = | polymerase chain reaction; SOD = superoxide dismutase. |
2.11. Data analysis
Statistical analyses were performed using SPSS software (version 16). One-way analysis of variance, and Least Significant Difference (LSD) test were used for data analysis, and the results were expressed as mean ± standard deviation. A p value of <0.05 was considered as a significant level.
3. Results
3.1. Hydrolysis of camel milk proteins
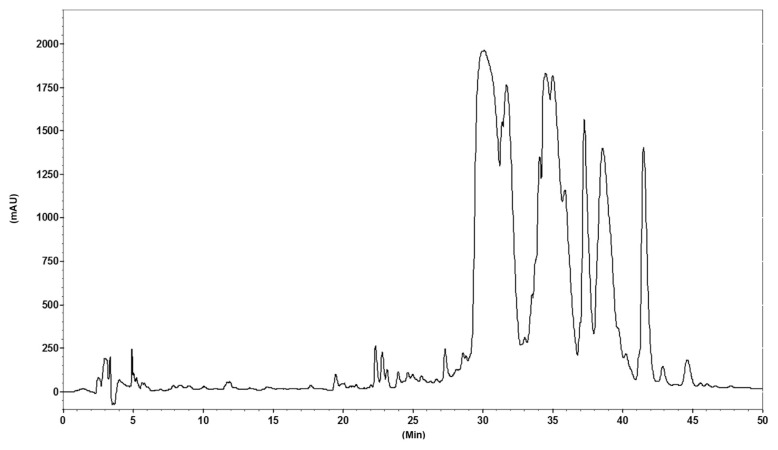
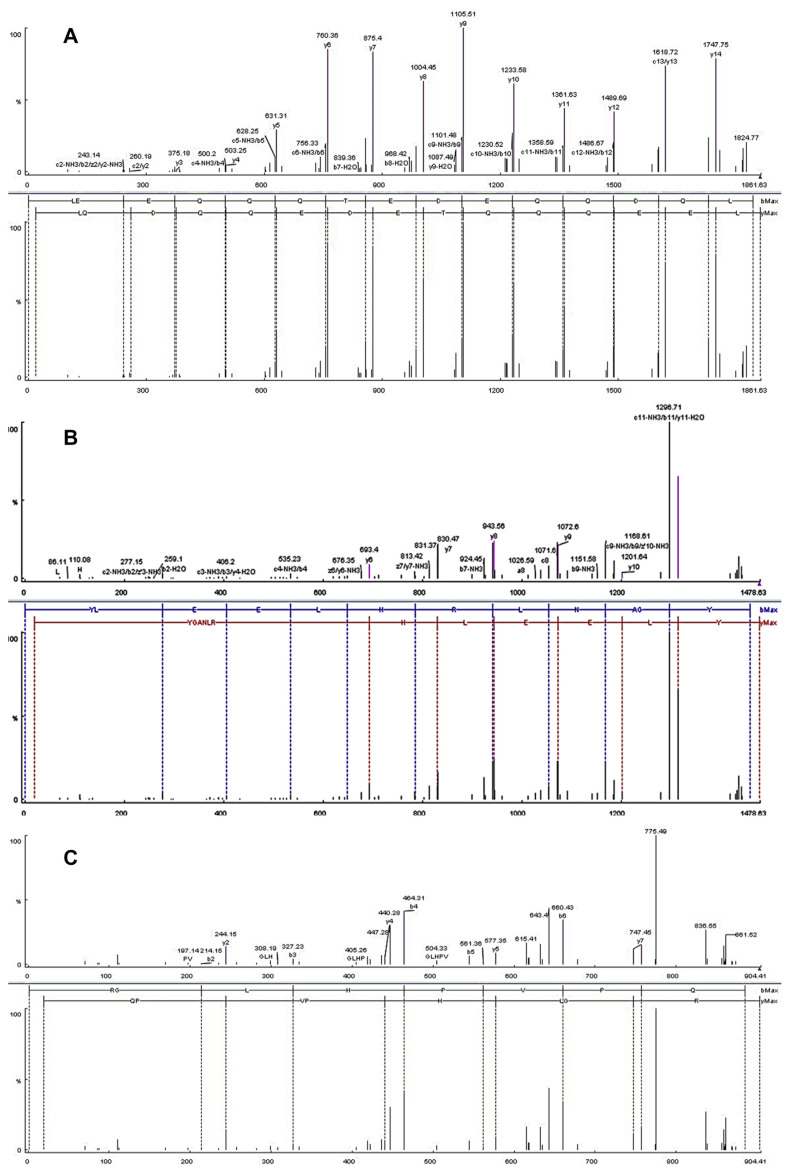
After the fractionation of the hydrolysate derived through enzymatic digestion of camel milk protein by ultrafiltration equipped with a membrane [3 kD molecular weight (MW) cutoff] and reverse-phase high-performance liquid chromatography, three fractions (labeled as 1, 2 and 3) were collected (Figure 1). After rechromatography by analytical reverse-phase high-performance liquid chromatography, the peptides were freeze dried and their sequences were analyzed by the matrix-assisted laser desorption/ionization time-of-flight technique. As shown in Figure 2, sequences of the three identified peptides were as follows: Peak 1: LEEQQQTE-DEQQDQL (MW: 1198.37 Da, LL-15); Peak 2: YLEELHRLNAGY (MW: 1477.67 Da, YY-11); and Peak 3: RGLHPVPQ (MW: 903.4 Da, RQ-8).
Figure 1.
Purification of camel milk protein hydrolysate on a C8 semipreparative column (10 × 250 mm2). The eluted fractions were numbered as indicated on HPLC chromatogram. Fractions named 1, 2, and 3 were further characterized. HPLC = high-performance liquid chromatography.
Figure 2.
Peptide sequencing of (A) LL-15, (B) YY-11, and (C) RQ-8.
3.2. Free radical scavenging activity
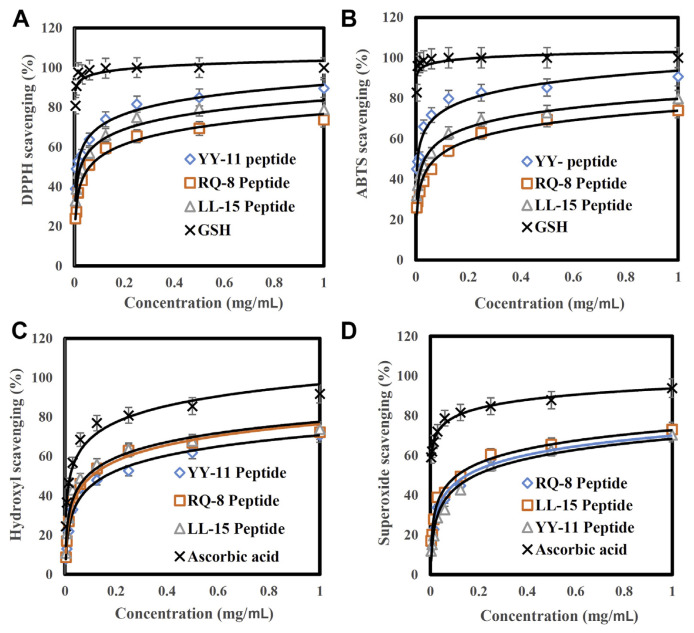
Figure 3 illustrates the DPPH free radical scavenging activity of the three peptides in comparison with glutathione as a standard. As shown in Figure 3, YY-11 peptide (The half maximal inhibitory concentration (IC50) about 0.01 mg/mL and <0.01 mg/mL for DPPH and ABTS, respectively) effectively inhibited DPPH free radicals more than RQ-8 (IC50 about 0.06 mg/mL and 0.07 mg/mL for DPPH and ABTS, respectively) and LL-15 peptides (IC50 about 0.03 mg/mL for DPPH and ABTS). The peptide YY-11 showed potent radical scavenging activity that was attributed to the presence of His and Tyr. By contrast, DPPH free radical scavenging activity of all the peptides showed an increase with increasing peptide concentration. DPPH and ABTS radical scavenging potency of the peptides was as follows: YY-11 > LL-15> RQ-8 (Figures 3A and 3B); hydroxyl free radical scavenging potency of the extracted peptides was as follows: LL-15 (IC50 about 0.08 mg/mL) > RQ-8 (IC50 about 0.1 mg/mL) > YY-11 (IC50 about 0.15 mg/mL) (Figure 3C); and their superoxide free radical scavenging efficacy was as follows: LQ (IC50 about 0.11 mg/mL) > RQ-8 (IC50 about 0.15 mg/mL) > YY-11 (IC50 about 0.19 mg/mL) (Figure 3D).
Figure 3.
(A) DPPH, (B) ABTS, (C) hydroxyl, and (D) superoxide free radical scavenging capacity of peptides compared with GSH and ascorbic acid as positive controls. ABTS = 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid); DPPH = 2,2-diphenyl-1-picrylhydrazyl; GSH = glutathione.
3.3. Cytotoxic effect of RQ-8, LQ-10, and YY-11 peptides on HepG2 cells
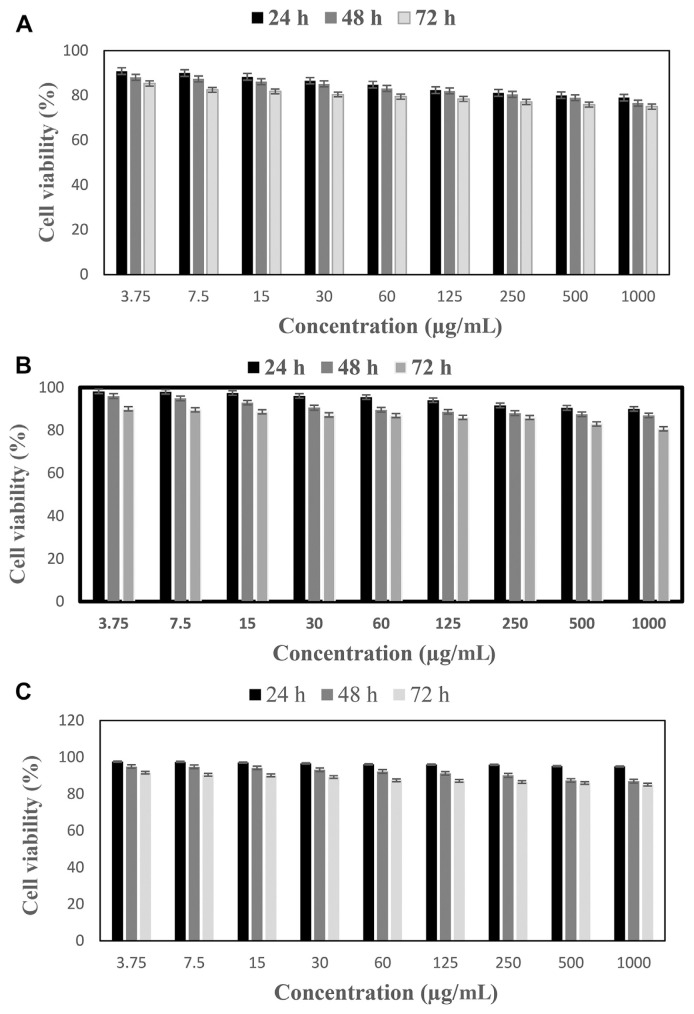
Cytotoxic effects of different concentrations of RQ-8, LL-15, and YY-11 peptides on HepG2 cells after 24 hours, 48 hours, and 72 hours were evaluated by MTT assay, and the results are presented in Figure 4. Growth inhibition of HepG2 cells treated with peptides was about 20% at the highest concentration tested (1 mg/mL). This suggests that all three peptides have low toxicity to HepG2 cells.
Figure 4.
Effects of (A) RQ-8, (B) LL-15 peptide, and (C) YY-11 peptide on HepG2 cell viability compared with untreated cells. Treatment of the cells with RQ-8, LL-15, and YY-11 peptides resulted in a dose- and time-dependent decrease in cell viability.
3.4. SOD and CAT gene expression in HepG2 cells
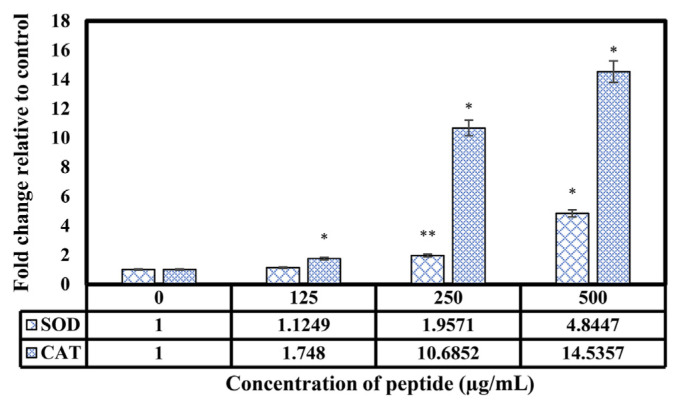
Figure 5 shows the expression alterations of SOD and CAT in HepG2 cells after 48 hours of treatment with peptides. Expression analysis revealed a notable increase (* p < 0.05, ** p < 0.01, *** p < 0.001) at gene expression levels of SOD and CAT in HepG2 cells exposed to peptides at concentrations of 125 μg/ mL, 250 μg/mL, and 500 μg/mL in comparison with untreated cells.
Figure 5.
The mRNA expression levels of CAT and SOD genes in HepG2 cells treated with YY-11 peptide (0 μg/mL, 125 μg/mL, 250 μg/mL, and 500 μg/mL) compared with untreated cells. Data were expressed as mean ± SD. * p < 0.001, significant difference. ** p < 0.01, significant difference. CAT = catalase; SD = standard deviation; SOD = superoxide dismutase.
4. Discussion
Camel milk proteins were hydrolyzed using pancreatin and pepsin enzymes, and the toxicity and antioxidant properties of resulting peptides were evaluated. Several studies have shown that proteins extracted from milk have some biological properties such as antioxidant, antimicrobial, and anti-cancer properties [20]. The identified peptides were synthesized using a solid-phase procedure and purified up to 98% on a C8 semipreparative column by the linear gradient of acetonitrile containing 0.1% trifluoroacetic acid. The obtained fractions were freeze dried and safely stored for further analysis.
Several techniques are ordinarily utilized to evaluate antioxidant properties of compounds. In this study, antioxidant activity of the peptides was evaluated by several antioxidant assay methods including DPPH, ABTS, hydroxyl, and superoxide free radicals [21].
Camel milk has powerful antioxidant properties [22]. Jrad et al [23] in 2014 assessed the antioxidant properties of camel milk casein before and after enzymatic digestion by pepsin and pancreatin. ABTS method was used to evaluate the free radical scavenging. The results confirmed the antioxidant effect of this peptide and showed that free radical scavenging activity of the casein peptide increased after enzymatic digestion.
In a study conducted in 2014, the amount of antioxidant gene expression in the livers of rats (intoxicated with CCL4) treated with camel milk was compared with that of untreated samples. The results showed that the expression of antioxidant genes including CAT, SOD, glutathione peroxidase, and glutathione-S transferase in rats treated with camel milk shows a significant increase compared with untreated sample [24]. In a study carried out by El-Said et al [25], the effects of camel milk were examined compared with those of insulin in diabetic rabbits. The study was conducted in four groups of rabbits; the first group included normal rabbits and the next three groups had diabetic rabbits. After the treatment period, blood and tissues, including the liver, kidney, and pancreas, were collected to evaluate the antioxidant enzymes (SOD and CAT). The results showed that camel milk helps improve the oxidative stress caused by diabetes. In addition to proteins, high levels of camel milk minerals, such as iron, sodium, copper, potassium, and magnesium, and a high amount of vitamin C can act as antioxidant agents that are capable of inhibiting free radicals [26]. In a survey conducted in 2014, oxidative stress biomarkers (glutathione, SOD, and myeloperoxidase) were evaluated in children with autism after they took camel milk for 2 weeks. Data analysis showed that all parameters were significantly increased, and it was suggested that camel milk has an important role in reducing oxidative stress through alteration of antioxidant enzymes and nonenzymatic antioxidant molecule levels [27]. The abovementioned studies indicated that camel milk, and peptides and its derivatives have antioxidant effects and can inhibit different free radicals.
Based on the structure and amino acid composition, peptides have different cytotoxic effects. It is shown that brevinin-2R, an antimicrobial peptide isolated from the skin of Rana ridibunda, has no significant toxic effect on A549 cancer cells [28]. Some peptides derived from casein of camel milk [29], peptides extracted from beef sarcoplasmic proteins [30], and peptides isolated from tuna dark muscle [31] have selective cytotoxic effect on certain tumor cell lines. Similar to brevinin-2R, the three isolated peptides derived from camel milk did not show any cytotoxic effect against HepG2 cells.
Anticancer activity of products derived from milk or urine of camels in traditional medicine is accepted, and some studies in this field have proved the anticancer effects of these compounds. Thus, in modern medicine, the effect of these compounds against cancer cells has been reviewed and ultimately approved, leading to the discovery of anticancer drugs. One study has shown that both urine and camel milk are able to inhibit angiogenesis, which is one of the important strategies for cancer inhibition [32].
Although the function and nature of urine and camel milk as anticancer agents are unknown to date, some evidence suggests that a probable reason for this performance could be the presence of iron-binding or lactoferrin as a multifunctional protein [33], which can reduce cancer cell growth by 56% [34].
Studies show that camel milk, due to the presence of protective proteins such as lactoferrin, IgG and IgA, lysozyme, and lactoperoxidase, is effective in the treatment of many diseases and has many therapeutic effects including antioxidant, anticancer, immunological, and antitumor effects [35,36].
Investigations show that proteins in camel milk such as lactoferrin are able to inhibit cancer cells and repair DNA defects [34]. In a study in 2012, the mechanism of anticancer effects of camel milk on MCF7 and HepG2 cancer cells was investigated, and the results showed that camel milk is able to exert its anticancer effects through induction of cell death and oxidative-stress-mediated processes [37]. In some studies, other mechanisms for the anticancer function of camel milk have been reported, for example, Dr Fatin Khorshid has shown that camel milk inhibits the growth of cancer cells in a wide range of cells including the colon, liver, lung, etc., and suggested that the mechanism of this inhibition is related to toxicity and antiangiogenic effects of this compound. Studies show that camel milk, because of its antioxidant and antimicrobial properties, is able to reduce lung inflammation and is effective in the treatment of lung cancer [34].
As the above studies have shown, camel milk proteins are important factors for disease treatment and they inhibit the growth of cancer cells via different mechanisms such as apoptosis, antiangiogenesis, cytotoxicity, and antioxidant effects. The results showed that three isolated peptides (RQ-8, LL-15, and YY-11) derived from camel milk have no significant inhibitory effects on the growth of HepG2 cancer cells, whereas they scavenge DPPH, ABTS, hydroxyl, and superoxide free radicals. SOD, CAT, and glutathione peroxidase are key enzymes that are involved in the inhibition of oxidative stress, and the expression of these enzymes may vary depending on the influence of the compounds. The results of the present study showed that YY-11 peptide was able to exert its antioxidant effect by increasing SOD and CAT gene expression.
Acknowledgments
This work was supported by Science and Research Branch, Islamic Azad University, Mashhad, Iran (Grant number: 75.304.92439) and Ferdowsi University of Mashhad, Mashhad, Iran, which are appreciated by the authors.
Funding Statement
This work was supported by Science and Research Branch, Islamic Azad University, Mashhad, Iran (Grant number: 75.304.92439) and Ferdowsi University of Mashhad, Mashhad, Iran, which are appreciated by the authors.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
REFERENCES
- 1. Gutowski M, Kowalczyk S. A study of free radical chemistry: their role and pathophysiological significance. Acta Biochim Pol. 2013;60:1–16. [PubMed] [Google Scholar]
- 2. Kirkinezos IG, Moraes CT. Reactive oxygen species and mitochondrial diseases. Seminars in Cell and Developmental Biology. 2001;12(6):449–57. doi: 10.1006/scdb.2001.0282. [DOI] [PubMed] [Google Scholar]
- 3. Soltani M, Baharara J. Antioxidant and antiproliferative capacity of dichloromethane extract of Holoturia leucospilota sea cucumber. Int J Data Min Bioinform. 2014;2014:1–9. [Google Scholar]
- 4. Zhu L, Chen J, Tang X, Xiong YL. Reducing, radical scavenging, and chelation properties of in vitro digests of alcalase-treated zein hydrolysate. J Agric Food Chem. 2008;56:2714–21. doi: 10.1021/jf703697e. [DOI] [PubMed] [Google Scholar]
- 5. Zhong S, Ma C, Lin YC, Luo Y. Antioxidant properties of peptide fractions from silver carp (Hypophthalmichthys molitrix) processing by-product protein hydrolysates evaluated by electron spin resonance spectrometry. Food Chem. 2011;126:1636–42. doi: 10.1016/j.foodchem.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 6. Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohankumar K, Ramasamy P. White spot syndrome virus infection decreases the activity of antioxidant enzymes in Fenneropenaeus indicus. Virus Res. 2006;115:69–75. doi: 10.1016/j.virusres.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 8. Crawford EL, Khuder SA, Durham SJ, Frampton M, Utell M, Thilly WG, Weaver DA, Ferencak WJ, Jennings CA, Hammersley JR, Olson DA, Willey JC. Normal bronchial epithelial cell expression of glutathione transferase P1, glutathione transferase M3, and glutathione peroxidase is low in subjects with bronchogenic carcinoma. Cancer Res. 2000;60:1609–18. [PubMed] [Google Scholar]
- 9. Shahidi F, Zhong Y. Novel antioxidants in food quality preservation and health promotion. Eur J Lipid Sci Technol. 2010;112:930–40. [Google Scholar]
- 10. Pihlanto-Leppala A. Bioactive peptides derived from bovine whey proteins: opioid and ace-inhibitory peptides. Trends Food Sci Technol. 2000;11:347–56. [Google Scholar]
- 11. Sakanaka S, Tachibana Y. Active oxygen scavenging activity of egg-yolk protein hydrolysates and their effects on lipid oxidation in beef and tuna homogenates. Food Chem. 2006;95:243–9. [Google Scholar]
- 12. Liu J-R, Chen M-J, Lin C-W. Antimutagenic and antioxidant properties of milk-kefir and soymilk-kefir. J Agric Food Chem. 2005;53:2467–74. doi: 10.1021/jf048934k. [DOI] [PubMed] [Google Scholar]
- 13. Suetsuna K, Ukeda H, Ochi H. Isolation and characterization of free radical scavenging activities peptides derived from casein. J Nutr Biochem. 2000;11:128–31. doi: 10.1016/s0955-2863(99)00083-2. [DOI] [PubMed] [Google Scholar]
- 14. Sheih I-C, Wu T-K, Fang TJ. Antioxidant properties of a new antioxidative peptide from algae protein waste hydrolysate in different oxidation systems. Bioresour Technol. 2009;100:3419–25. doi: 10.1016/j.biortech.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 15. Tang C-H, Peng J, Zhen D-W, Chen Z. Physicochemical and antioxidant properties of buckwheat (Fagopyrum esculentum Moench) protein hydrolysates. Food Chem. 2009;115:672–8. [Google Scholar]
- 16. Quirós A, del Mar Contreras M, Ramos M, Amigo L, Recio I. Stability to gastrointestinal enzymes and structure–activity relationship of β-casein-peptides with antihypertensive properties. Peptides. 2009;30:1848–53. doi: 10.1016/j.peptides.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 17. Binsan W, Benjakul S, Visessanguan W, Roytrakul S, Tanaka M, Kishimura H. Antioxidative activity of Mungoong, an extract paste, from the cephalothorax of white shrimp (Litopenaeus vannamei) Food Chem. 2008;106:185–93. [Google Scholar]
- 18. Li Y, Jiang B, Zhang T, Mu W, Liu J. Antioxidant and free radical-scavenging activities of chickpea protein hydrolysate (CPH) Food Chem. 2008;106:444–50. [Google Scholar]
- 19. Liu C, Hong J, Yang H, Wu J, Ma D, Li D, Lin D, Lai R. Frog skins keep redox homeostasis by antioxidant peptides with rapid radical scavenging ability. Free Radic Biol Med. 2010;48:1173–81. doi: 10.1016/j.freeradbiomed.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 20. Salami M, Moosavi-Movahedi AA, Moosavi-Movahedi F, Ehsani MR, Yousefi R, Farhadi M, Niasari-Naslaji A, Saboury AA, Chobert JM, Haertlé T. Biological activity of camel milk casein following enzymatic digestion. J Dairy Res. 2011;78:471–8. doi: 10.1017/S0022029911000628. [DOI] [PubMed] [Google Scholar]
- 21. Pihlanto A. Antioxidative peptides derived from milk proteins. Int Dairy J. 2006;16:1306–14. [Google Scholar]
- 22. Yadav AK, Kumar R, Priyadarshini L, Singh J. Composition and medicinal properties of camel milk: a review. Asian J Dairy Food Res. 2015;34:83–91. [Google Scholar]
- 23. ad Z, Jr, Girardet J-M, Adt I, Oulahal N, Degraeve P, Khorchani T. Antioxidant activity of camel milk casein before and after in vitro simulated enzymatic digestion. Mljekarstvo. 2014;64:287–94. [Google Scholar]
- 24. El-Bahr S. Camel milk regulates gene expression and activities of hepatic antioxidant enzymes in rats intoxicated with carbon tetrachloride. Asian J Biochem. 2014;9:30–40. [Google Scholar]
- 25.El-Said E, El-Sayed GR, Tantawy E, editors. Effect of camel milk on oxidative stresses in experimentally induced diabetic rabbits. Los Angeles, CA: Veterinary Research Forum, Urmia University, Faculty of Veterinary Medicine; 2010. [Google Scholar]
- 26. Abdalla K. An overview of the therapeutic effects of camel milk in the treatment of type1 diabetes mellitus. J Biomol Res Ther. 20142014 [Google Scholar]
- 27. Kula J. Medicinal values of camel milk. Int J Vet Sci Res. 2016;2:018–25. [Google Scholar]
- 28. Asoodeh A, Haghparast A, Kashef R, Chamani J. Pro-inflammatory cytokine responses of A549 epithelial cells to antimicrobial peptide Brevinin-2R. Int J Peptide Res Ther. 2013;19:157–62. [Google Scholar]
- 29. Osama A, EsmailM E-F, Ehab E-D, TziBun N, Elrashdy MR. Examination of the activity of camel milk casein against hepatitis C virus (genotype-4a) and its apoptotic potential in hepatoma and HeLa cell lines. Hepat Mon. 2011;11:724–30. doi: 10.5812/kowsar.1735143X.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jang A, Jo C, Kang K-S, Lee M. Antimicrobial and human cancer cell cytotoxic effect of synthetic angiotensin-converting enzyme (ACE) inhibitory peptides. Food Chem. 2008;107:327–36. [Google Scholar]
- 31. Hsu K-C, Li-Chan EC, Jao C-L. Antiproliferative activity of peptides prepared from enzymatic hydrolysates of tuna dark muscle on human breast cancer cell line MCF-7. Food Chem. 2011;126:617–22. [Google Scholar]
- 32. Alhaider AA, Gader AGMA, Almeshal N, Saraswati S. Camel urine inhibits inflammatory angiogenesis in murine sponge implant angiogenesis model. Biomed Aging Pathol. 2014;4:9–16. [Google Scholar]
- 33. Gader AGMA, Alhaider AA. The unique medicinal properties of camel products: a review of the scientific evidence. J Taibah Univ Med Sci. 2016;11:98–103. [Google Scholar]
- 34. Kula JT, Tegegne D. Chemical composition and medicinal values of camel milk. Int J Res Stud Biosci. 2016;4:13–25. [Google Scholar]
- 35. Mona E, Ragia O, Abeer A, Mosa T. Biochemical effects of fermented camel milk on diarrhea in rats. N Y Sci J. 2010;3:106–11. [Google Scholar]
- 36. Sharma C, Singh C. Therapeutic value of camel milk—a review. Adv J Pharm Life Sci Res. 2014;2:7–13. [Google Scholar]
- 37. Korashy HM, El Gendy MA, Alhaider AA, El-Kadi AO. Camel milk modulates the expression of aryl hydrocarbon receptor-regulated genesCyp1a1, Nqo1, and Gsta1, in murine hepatoma Hepa 1c1c7 cells. BioMed Res Int. 2012 doi: 10.1155/2012/782642. [DOI] [PMC free article] [PubMed] [Google Scholar]