Abstract
In this paper a method of using the “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) extraction and gas chromatography coupled to mass spectrometry detection (GC–MS) was developed for the analysis of five frequently applied pesticides in papaya and avocado. The selected pesticides, ametryn, atrazine, carbaryl, carbofuran, and methyl parathion, represent the most commonly used classes (carbamates, organophosphorous, and triazines). Optimum separation achieved the analysis of all pesticides in < 6.5 minutes. Validation using papaya and avocado samples established the proposed method as linear, accurate, and precise. In this sense, the correlation coefficients were > 0.99. The limits of detection (LOD) and quantification (LOQ) in papaya ranged from 0.03 mg/kg to 0.35 mg/kg and from 0.06 mg/kg to 0.75 mg/kg, respectively. Meanwhile for avocado, LOD values varied from 0.14 mg/kg to 0.28 mg/kg and LOQ values ranged from 0.22 mg/kg to 0.40 mg/kg. Recoveries obtained for each pesticide in both matrices ranged between 60.6% and 104.3%. The expanded uncertainty of the method was < 26% for all the pesticides in both fruits. Finally, the method was applied to other fruits.
Keywords: fruits, GC-MS, matrix effect, pesticides, uncertainty
1. Introduction
Pesticides are extensively used to control pests that cause damage to crops. In this way, the application of these compounds intends to ensure the quantity and quality of fruits and vegetables required for consumers. However, this can lead to the bioaccumulation of pesticide residues in them. For this reason and considering the negative effects of the pesticides on human health; such as genotoxicity, inhibition of acetyl cholinesterase activity, hepatic, and renal toxicity [1,2]; pesticide monitoring is important to ensure a minimal exposure to them.
Gas chromatography (GC), coupled with different detectors, is a very useful technique employed for the analysis of volatile pesticides such as organochlorides, organophosphates, and carbamates. Mass spectrometry detector (MS) is the most useful for pesticide residue determination in food matrices when an appropriate sample preparation and cleanup procedures are applied [3].
In this sense, GC coupled to mass spectrometry detection (GC–MS) has been successfully used for the analysis of pesticides in different fruits and vegetables including rice, orange, apple, and spinach [4]; grape [5]; pomegranate, grape, okra, tomato, and onion [6]; banana [7]; orange [8]; rice [9]; apple and tomato [10]; cantaloupe melon, broccoli, sweet potato, and lemon [11]; apple–blueberry sauce, pea, and lime [12]; mango [13]; mango and papaya [14]; turnip, green cabbage, French bean, eggplant, apple, nectarine, and grape [15]; berry fruits [16]; Brazilian melon [17]; apple, orange, carrot, and tomato [18].
Due to the complexity of fruit and vegetable matrices, different extraction procedures have been used for GC analysis of pesticide residues. Established 3500 and 3600 series Environmental Protection Agency methods are widely used for this task [19,20]. However, the current trend in pesticide analysis is to develop more efficient and environmentally friendly methods. These methods involve sample preparation techniques such as microwave assisted extraction (MAE), matrix solid-phase dispersion (MSPD), solid–liquid extraction (SLE) [5], dispersive solid phase extraction (DSPE) [6], solid-phase microextraction (SPME) [10,13,15,21], solid phase extraction (SPE) [16], and “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) method [5–9,11,17,18].
QuEChERS is a procedure which has shown good performance on the difficult task of the extraction of pesticides from food matrices. This sample treatment has been applied for the extraction of a wide range of pesticides with diverse chemical properties in several types of fruits and vegetables, which have different compounds such as sugar, pigments, and high water content [5,9,22]. Owing to the widespread use of this procedure, different versions of QuEChERS method have been developed; among these are the Association Official Analytical Chemists (AOAC; acetate buffering) version and the CEN (citrate-buffering) version. The acetate buffering version had showed higher recoveries for the pH-dependent pesticides, therefore it is more frequently used [12].
Nowadays validation is considered an essential part of the method evaluation; it has the aim of determining if an analytical method is suitable and reliable for its purpose. By using the data produced from method validation the method uncertainty can be estimated. Uncertainty is an important parameter for method evaluation defined as “a parameter associated with the result of a measurement that characterizes the dispersion of the values that could reasonably be attributed to the measurand” [23,24].
The uncertainty may originate from many possible sources, which are related to the different stages of the analytical method. Potential uncertainty sources are sampling, matrix effects, uncertainties of masses and volumetric equipment, reference values, approximations and assumptions incorporated in the measurement method and procedure, and random variation [23,25].
Each uncertainty source should be treated independently to obtain its contribution to the overall uncertainty of an analytical method. In this sense, the contributions of all the uncertainty sources are considered to estimate the combined uncertainty of the method. From the combined uncertainty, an expanded uncertainty is determined. This last term represents an interval within which an analytical result is believed to lie with a high level of confidence [17,23].
In this paper a method using QuEChERS extraction and GC–MS was developed for the analysis of five frequently applied pesticides in papaya and avocado. The studied pesticides were representative of three of the most commonly used classes, which are: triazines (ametryn and atrazine), carbamates (carbaryl and carbofuran), and organophosphorous (methyl parathion). The proposed method was validated according to European guidelines. In addition, the expanded uncertainty was evaluated taking into account the different sources of uncertainty that affect the process.
2. Methods
2.1. Materials and reagents
Standards of the pesticides carbofuran (CF), carbaryl (CAR), atrazine (ATZ), ametryn (AME), and methyl parathion (MeP), all with purity > 98%, acetic acid, acetonitrile, and methanol, all HPLC grade, were obtained from Sigma-Aldrich (St. Louis, MO, USA). Helium (99.999% purity) was supplied by Praxair (Colima, México). A methanol stock solution of each pesticide at 50 mg/L was weekly prepared. Methanolic working solutions were prepared daily by mixing pesticide stock solutions. These solutions were stored in the dark at 4°C. Working solutions were used for GC–MS method optimization and spiking fruit matrices used for method validation. Other chemicals used in this work were analytical reagent grade.
DisQuE Dispersive Sample Preparation kit containing extraction and clean-up tubes from Waters (Milford, MA, USA) and nylon filters (0.45 μm pore size) from Phenomenex (Torrance, CA, USA) were used for sample preparation.
2.2. Instruments
A Varian 3900 GC coupled to a Saturn-2100T mass spectrometry detector and equipped with a CombiPAL autosampler and MS Workstation version 6.9 software from Varian (Palo Alto, CA, USA) were used for chromatographic analysis. The analytical column Zebron ZB-5MS Crossbond (5% phenyl–95% dimethyl-polysiloxane; 30 m, 0.25 mm internal diameter, 0.25 μm film thickness) from Phenomenex (Torrance, CA, USA) was used in this study.
An NB-101B food processor (Homeland Houseware, Los Angeles, CA, USA), an analytical balance model CX220 (Citizen Scale, Metuchen, NJ, USA), a Vortex-Genie 2 mixer (Scientific Industries, Bohemia, NY, USA), a Sorvall Biofuge Primo R centrifuge (Thermo Electron Corp., Schwerte, Germany), and a 5415D centrifuge (Eppendorf, Hamburg, Germany) were also used for the pesticide extraction procedure.
2.3. GC–MS analysis
The initial oven temperature was set at 140°C, increased to 240°C at a rate of 15°C/min, increased again at a rate of 40°C/min to 280°C, this temperature was maintained for 3 minutes. The total run time was 6.5 minutes. One μL of sample was injected in split mode (2:1). The injector temperature was set at 240°C. The helium carrier gas flow was 1.1 mL/min.
The mass spectrometer was operated in electron impact ionization mode at 70 eV of collision energy with 2 minute solvent delay to prevent damage to the filament of the ion source. The temperature of the ion source, manifold, and transfer line was set at 240°C, 50°C, and 260°C, respectively. For qualitative analysis a full scan from 75 mass/charge (m/z) to 280 mass/charge (m/z) was applied. For quantitative pesticide analysis selected ion monitoring (SIM) mode was used. Table 1 shows the molecular weight and ions used for quantitative analysis.
Table 1.
Selected ions for pesticides detection and retention times.
| Pesticide | MW a | Ions monitoring (m/z) | Retention time (min) |
|---|---|---|---|
| CF | 221.3 | 164, 149 | 2.57, 5.18 |
| CAR | 201.2 | 144, 115 | 3.88 |
| ATZ | 215.7 | 200, 215 | 5.29 |
| MeP | 263.2 | 263, 109 | 6.25 |
| AME | 227.3 | 227, 212 | 6.34 |
AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; MeP = methyl parathion; MW = molecular weight.
Molecular weight.
2.4. Sample preparation
Papaya and avocado fruits were selected as samples for this study. These fruits were chosen due to their economic importance, matrix complexity, and for having different compositions. With regard to these fruits composition, papaya is characterized by 88% water content, moderate carbohydrates content (10%), and low lipids content (0.3%); meanwhile, avocado is characterized by 72% water content, moderate carbohydrates content (8%), and high lipids content (15%) [26]. After washing, peeling, and homogenizing the fruits, a portion of 0.6 kg was stored at −20°C in a dark glass flask for further treatment.
The fruits samples were treated according to the QuECh-ERS methodology (AOAC Official Method 2007.01) [12]. The homogenized sample was thawed, and then 15 g was weighed in a 50-mL conic tube. Afterward, the extraction solvent (15 mL of acetonitrile with 1% acetic acid) was added to the sample and stirred for 5 minutes using a vortex mixer. Next, the sample was poured into the extraction tube and immediately vortexed for 1 minute and centrifuged at 2700 g for 5 minutes. Subsequently, 1 mL of the extract was placed into the clean-up tube, vortex mixed, and centrifuged for 1 minute at 10700 g. Lastly, the supernatant was filtered and discharged in a 2-mL vial. One μL of the extract was injected in the GC–MS system.
For calibration curves, samples were fortificated with all pesticides at 1.0 mg/kg, 1.5 mg/kg, 2.0 mg/kg, 4.0 mg/kg, 6.0 mg/kg, and 8.0 mg/kg. The fortificated fruit samples were allowed to equilibrate for 120 minutes before treatment. Blank samples were also prepared according this procedure.
2.5. Method validation and matrix effect
The proposed method was validated following the European guidelines [27,28]. The parameters assessed were selectivity, linearity, limits of detection, and quantification, precision, and recovery.
Matrix effect was estimated by comparing the slopes of standard curves dissolved in extraction solvent and matrix-matched calibration curves. The matrix effect was calculated as: 100 × [1 – (solvent slope)/(matrix slope)] [24].
2.6. Uncertainty assessment
The measurement uncertainty was estimated according to the procedures recommended by EURACHEM/CITAC and Quantifying Uncertainty in Analytical Measurement [23]. The expanded uncertainty (Ue) at a 95% of confidence level was obtained using the medium level concentration of the linear range (4 mg/kg) and Eq. (1).
| (1) |
where Uc is the combined uncertainty.
To estimate the Uc of the method six main uncertainty sources (ui) were identified, as shown in Eq. (2). These sources are associated with: standard solution preparation (u1), calibration curve preparation (u2), sample preparation treatment (u3), precision (u4), accuracy/bias (u5), and the linear least square fitting (u6).
| (2) |
where ui is a source standard uncertainty.
The standard uncertainty of each source takes into account the contribution of factors associated with it. In this case, the following equation is used.
| (3) |
where p, q, and r are the different factors contributing to the standard uncertainty of each source.
The assessment of each uncertainty source identified is presented in the following paragraphs.
For the evaluation of u1 three uncertainty factors were considered: (1) purity of standards; (2) weighing of standards; and (3) flask volume measurement. For standard purity u(P), the standard deviation (SD) equals 0.02 and a rectangular distribution (d = 3) was considered (Eq. 4). With regards to the weighing of standards u(W) was considered 0.1 mg because this value corresponds to the SD provided by the manufacturer. In case of flask volume measurement, two factors were considered: flask calibration and temperature. The standard uncertainty of the flask calibration, u(Cal), is calculated considering the SD = 0.02 mL and a triangular distribution (d = 6) in Eq. (4). Due to the flask having been calibrated at a temperature different to the working one, the uncertainty from temperature, u(T), was calculated using Eq. (5).
| (4) |
where SD is the standard deviation specified by the manufacturer and d is the type of distribution.
| (5) |
where V (10 mL) is the flask volume, ΔT (5°C) is the difference between the working and calibration temperature, and α (0.0015°C−1) is the volume expansion coefficient of the MeOH.
For the evaluation of u2, the volume measurement uncertainty of: micropipette and flask was considered. In case of the micropipette, the standard uncertainty was calculated according Eq. (4) using the SD = 0.5 μL and a triangular distribution (d = 6). For the flask volume measurement uncertainty the calculus applied for u1 was considered.
For the evaluation of u3, the three uncertainty factors of the QuEChERS procedure were considered: weighing of the sample and volume measurement using micropipette and flask. The uncertainty of weighing of the sample, u(W), was calculated as described in u1. Meanwhile, volume measurement using flask and micropipette was estimated in same way as u1 and u2, respectively.
For the evaluation of u4, Eq. (6) was used, where SD is the standard deviation of experimental data obtained by sample analysis in different days and n is the number of assays considered.
| (6) |
For the evaluation of u5, Eq. (6) was also used. In this case, SD is the standard deviation of the recoveries obtained of various assays and n is the number of these assays.
For the evaluation of u6, the experimental data was obtained from the matrix-matched calibration curves of papaya and avocado. The following equations were used for this evaluation.
| (7) |
where S is the residual standard deviation (calculated with Eq. 8), B1 is the calibration curve slope, c0 is the assessed concentration, p is the number of repetitions performed to determine c0, n is the total number of measurements of the standards used in the calibration curve, and c is the average concentration of the calibration standards.
| (8) |
where Aj is the signal obtained for each calibration standard, B0 is the intercept of the calibration curve, and cj is the concentration of each used calibration standard.
2.7. Method feasibility
For method feasibility evaluation, the developed and validated method was applied to lime, mango, melon, and banana samples. These fruits were selected because of their economic importance and high production level, as well as their characteristic composition. Lime was selected as a food with high acidity, mango and melon as high water content commodities, and banana as high carbohydrates content commodity.
Blanks samples were prepared using the procedure described in Section 2.4. The chromatograms obtained from the injections of these blanks samples and fortificated papaya and avocado matrices at 8 mg/kg were compared. This comparison was performed to identify matrix interferences and to determine which matrix-match calibration curve is more appropriate for quantification of these compounds in lime, mango, melon, and/or banana. The presence of matrix interferences from these fruits were checked by monitoring the specific ions for each pesticide at the retention time interval expected for their elution (Table 1).
3. Results and discussion
3.1. Optimization of separation conditions using GC–MS
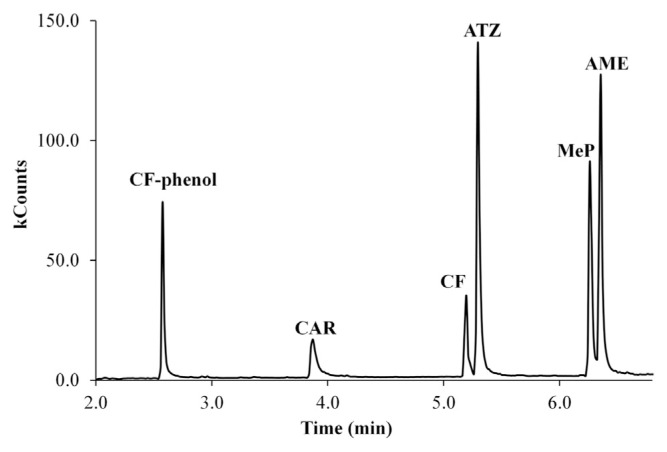
The temperature gradient for the GC–MS analysis of the pesticides studied was optimized with the purpose to obtain the highest resolution between peaks and the shortest analysis time. The optimization was performed in full scan mode using a 50-mg/L pesticide standard mixture. The optimized gradient is described in Section 2.3. Resolution values > 1.5 between peaks calculated with peak width at half-height are considered base-line resolution. Therefore, all pesticides were baseline resolved in a total analysis time close to 6.5 minutes. Table 1 shows the retention time of each pesticide under optimal separation conditions. Figure 1 shows the chromatogram obtained from the pesticide standard mixture.
Figure 1.
Total ion chromatogram of standard solution mixture of pesticides. AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; MeP = methyl parathion.
During the chromatographic analysis, degradation of the two carbamate pesticides (CF and CAR) was observed. This conclusion was based on the fact that the molecular ion was not recorded. Degradation compounds were identified and differentiated from the parent compounds taking into account fragmentation patterns. In this sense, CAR was completely degraded to 1-naphthol which corresponds to the peak at 3.88 minutes. Meanwhile, CF is partially degraded into two peaks; both peaks were integrated as a single signal to quantify this pesticide. The first peak 7-phenol carbofuran (CF-phenol) is observed at 2.57 minutes and the second peak corresponds to the parent CF compound at 5.18 minutes. As can be seen in Figure 1, the degradation product of CF has a retention time lower than the parent compound. This degradation is called McLafferty rearrangement, and it is due to CF and CAR having an aromatic ring and a hydrogen atom in the γ-position with respect to their aromatic ring that makes them thermally unstable. As can be seen in Figure 2 both CF and CAR decompose to give the corresponding phenol and methyl-isocynate.
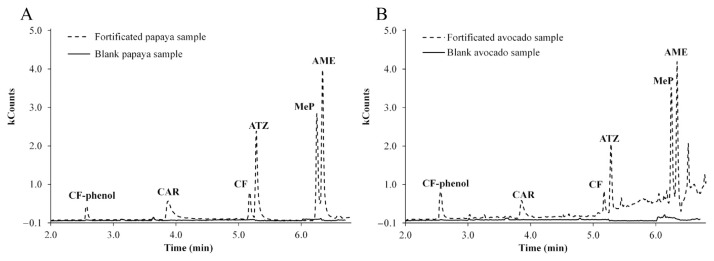
Figure 2.
Chromatograms (SIM mode) obtained for injection of blank (solid line) and fortified (dotted line) samples of (A) papaya and (B) avocado for checking matrix interferences. Pesticide concentration was 8 mg/L. AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; MeP = methyl parathion; SIM = selected ion monitoring.
In this paper, experiments were performed to estimate carbamates degradation using different injector temperatures (120°C, 240°C, and 250°C) and different initial gradient temperatures (80°C, 100°C, 120°C, and 140°C). The obtained result showed no significant reduction of the degradation; however, the further degradation of the compounds was observed at the highest temperature. Also, the influence of the chromatographic column age was evaluated, it was found that this parameter has a greater impact on the CF and CAR degradation, because in the chromatograms using a new column only the parents compounds were observed, meanwhile in the chromatograms using the same column after 6 months of use the degradation compounds were observed. This behavior is consistent with previous studies, Carabias-Martínez et al [21] reported that carbamates are not completely degraded in the injector, but it also occurs during elution along the chromatographic column. This behavior could be due to a gradual degradation of stationary phase (e.g., phenyl or dimethyl-polysiloxane groups) that could interact with the carbamates causing their degradation.
It is important to highlight that the degradation of carbamates was reproducible under the analytical conditions described in the experimental section; this is demonstrated with the precision of the method.
3.2. Method validation and matrix effect
Selectivity was evaluated by checking the presence of coextracted interferences in the chromatograms from blank samples. Hence, chromatograms of blank and fortificated (8 mg/kg) samples of papaya and avocado were compared (Figure 2). The presence of matrix interferences were examined by monitoring the SIM chromatograms for each pesticide at the retention time window expected for each compound (Table 1). Although there are coextracted compounds from the fruit matrices during the pesticides extraction, the signal of these inferences is minimized by using SIM detection mode. This is due to the most abundant ions produced from the fragmentation of each pesticide are employed for its quantification. It can be observed that there are no extracted matrix interferences for the determination of the pesticides studied in both fruits; therefore this method can be considered as selective.
Linearity was studied by means of calibration curves in the 1.0–8.0 mg/L range in extraction solvent and in the 1.0–8.0 mg/kg range in matrix-matched of papaya and avocado; both ranges are equivalent. Correlation coefficient and slope values from pesticides in extraction solvent calibration curves and matrix-matched calibration curves are shown in Table 2. As can be seen, correlation coefficients were > 0.99.
Table 2.
Matrix effect and linearity parameters in solvent extraction and fruit matrices.
| Pesticide | Solvent extraction | Matrix match papaya | Matrix match avocado | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Slope | r | Slope | r | Matrix effect (%) | Slope | r | Matrix effect (%) | |
| CF | 4196 | 0.998 | 9060 | 0.999 | 53.7 | 9369 | 0.997 | 55.2 |
| CAR | 5302 | 0.998 | 7056 | 0.999 | 24.9 | 6558 | 0.999 | 19.2 |
| ATZ | 6119 | 0.997 | 5041 | 0.995 | −21.4 | 5740 | 0.995 | −6.6 |
| MeP | 2681 | 0.996 | 4198 | 0.995 | 36.1 | 5619 | 0.999 | 52.3 |
| AME | 8322 | 0.997 | 8195 | 0.997 | −1.5 | 8348 | 0.996 | 0.3 |
AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; MeP = methyl parathion.
Limits of detection (LOD) and quantification (LOQ) were calculated using the equation X × SDb + Yb, where SDb is the standard deviation of 20 blank samples, Yb the blank sample signal and X = 3 for LOD or X = 10 for LOQ. The calculated limits are shown in Table 3. In the case of papaya, the LOD values ranged from 0.03 mg/kg to 0.35 mg/kg and LOQ values varied between 0.06 mg/kg and 0.75 mg/kg; whereas for avocado, LOD, and LOQ values varied from 0.14 mg/kg to 0.28 mg/kg, 0.22 mg/kg, and 0.40 mg/kg, respectively. The baseline (basal signal or noise) in blank samples at the time window in which each pesticide is expected is different due to the quantification being made using different ions. For this reason, the LOD and LOQ calculation are different for each compound. From these results it can be concluded that the method has good sensibility for papaya and avocado fruits because only one pesticide has LOD values < 0.40 mg/kg and the rest of the pesticides have LOD values < 0.2 mg/kg. Also, only one pesticide in the papaya sample has an LOQ value > 0.50 mg/kg, the rest of the pesticides in papaya and avocado samples have LOD values < 0.50 mg/kg.
Table 3.
Parameters of validation for pesticides.
| Pesticides | Papaya | Avocado | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| LOD a | LOQ b | Precision (%RSD) c | Recovery (%) | LOD a | LOQ b | Precision (%RSD) c | Recovery (%) | |||||
|
|
|
|
|
|||||||||
| 1 mg/kg | 8 mg/kg | 1 mg/kg | 8 mg/kg | 1 mg/kg | 8 mg/kg | 1 mg/kg | 8 mg/kg | |||||
| CF | 0.03 | 0.06 | 13.9 | 17.2 | 93.9 | 98.1 | 0.19 | 0.36 | 17.6 | 15.0 | 69.7 | 97.7 |
| CAR | 0.17 | 0.31 | 12.5 | 18.3 | 90.1 | 100.0 | 0.14 | 0.22 | 27.3 | 14.7 | 92.3 | 100.7 |
| ATZ | 0.18 | 0.42 | 5.4 | 11.8 | 72.0 | 97.3 | 0.14 | 0.29 | 13.3 | 17.2 | 60.6 | 99.0 |
| MeP | 0.35 | 0.75 | 15.3 | 17.4 | 76.8 | 96.7 | 0.16 | 0.24 | 11.0 | 13.5 | 104.3 | 100.1 |
| AME | 0.11 | 0.22 | 14.1 | 17.1 | 91.3 | 97.5 | 0.28 | 0.40 | 7.0 | 12.4 | 79.7 | 100.1 |
AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; LOD = limit of detection; LOQ = limit of quantification; MeP = methyl parathion; RSD = relative standard deviation.
Limit of detection (mg/kg).
Limit of quantification (mg/kg).
Calculated at 1 mg/kg and 8 mg/kg in fortificated papaya and avocado matrix.
Recovery was determined at two concentration values; these levels correspond to the lowest and highest concentration of the linear range. The recovery results are shown in Table 3. Recoveries obtained for each pesticide in both matrices ranged between 72.0% and 104.3%, except for the concentration at 1.0 mg/kg of CF and ATZ in avocado (69.7% and 60.6%, respectively). This range is considered acceptable for this parameter according to the SANCO/12495/2011 guideline [27].
Precision was expressed as relative standard deviation (RSD) and evaluated in different days (see Table 3). For this purpose, five fortificated matrices at two levels of concentration (1.0 mg/kg and 8.0 mg/kg) were treated and analyzed in five different days. The RSD values calculated were > 20% (27.3%) for CAR in avocado at 1.0 mg/kg. For the other compounds, at both concentration levels evaluated in papaya and avocado, the RSD values were < 20%. The RSD values obtained are considered satisfactory according to the SANCO/12495/2011 guideline [27].
Matrix effect (ME) results are shown in Table 2. As can be seen, ME for carbofuran in papaya and avocado samples presents a strong signal enhance (> 50%). For carbaryl, ME is considered medium signal enhance in papaya and mild signal enhance in avocado. For methyl parathion, ME is considered medium signal enhance in papaya and strong signal enhance in avocado. Ametryn showed mild signal enhance in avocado and mild signal suppression in papaya; whereas the ME for atrazine was mild suppression signal in papaya and medium suppression signal in avocado. It can be observed that ametryn showed the lowest ME.
In previous studies, it has been reported that signal enhance caused by matrix effect is produced for interactions of analyte functional groups such as hydroxyl, amino, and phosphate, with the active surfaces in the GC–MS (injector, column, detector) system [4]. It is important to highlight that the pesticides studied in this work contain at least one of these functional groups. During the matrix effect evaluation, a discrepancy was observed in the signal detection between pesticides (standards) and pesticides extracted from matrices (papaya and avocado). Therefore, to avoid this matrix effect and ensure reliable results it is necessary to use matrix match calibration curves.
3.3. Uncertainty assessment
The results of method uncertainty assessment considering six sources are presented in Table 4. For papaya samples the overall contributions to Uc of the method of the uncertainties associated with u1 (standard solution preparation), u2 (calibration curve preparation), u3 (sample preparation treatment), u4 (precision), and u5 (accuracy/bias) ranged from 6.7% to 11.7% and u6 (linear least square fitting) ranged from 88.3% to 93.3%. In the same way, for avocado, the overall contributions of the uncertainties associated with u1, u2, u3, u4, and u5 varied from 5.8% to 13.0% and u6 ranged from 87.0% to 94.2%. In the light of these results, the uncertainty associated with u6 presents the highest contribution for all the pesticides under study in papaya and avocado.
Table 4.
Method uncertainties estimated for different sources and, combined (Uc) and expanded (Ue) uncertainty.
| Fruit | ui a (mg/kg (%)) | Pesticide | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| CF | CAR | ATZ | MeP | AME | ||
| Papaya | u 1 | 0.048 (2.3) | 0.048 (2.1) | 0.048 (3.5) | 0.048 (2.0) | 0.048 (1.9) |
| u 2 | 0.034 (1.2) | 0.034 (1.0) | 0.034 (1.7) | 0.034 (1.0) | 0.034 (0.9) | |
| u 3 | 0.032 (1.0) | 0.032 (0.9) | 0.032 (1.6) | 0.032 (0.9) | 0.032 (0.8) | |
| u 4 | 0.024 (0.6) | 0.074 (4.9) | 0.009 (0.1) | 0.009 (0.1) | 0.070 (4.0) | |
| u 5 | 0.042 (1.8) | 0.037 (1.2) | 0.031 (1.5) | 0.056 (2.7) | 0.071 (4.1) | |
| u 6 | 0.303 (93.1) | 0.317 (89.9) | 0.246 (91.6) | 0.329 (93.3) | 0.331 (88.3) | |
| Uc (mg/kg) | 0.315 | 0.334 | 0.257 | 0.341 | 0.352 | |
| Ue b (mg/kg) | 0.629 | 0.669 | 0.514 | 0.682 | 0.704 | |
| Ue (%) | 15.7 | 16.7 | 12.8 | 17.1 | 17.6 | |
| Avocado | u 1 | 0.048 (1.6) | 0.048 (2.1) | 0.048 (0.9) | 0.048 (2.1) | 0.048 (1.6) |
| u 2 | 0.034 (0.8) | 0.034 (1.0) | 0.034 (0.4) | 0.034 (1.1) | 0.034 (0.8) | |
| u 3 | 0.032 (0.7) | 0.032 (0.9) | 0.032 (0.4) | 0.032 (1.0) | 0.032 (0.7) | |
| u 4 | 0.051 (1.8) | 0.035 (1.1) | 0.077 (2.3) | 0.070 (4.5) | 0.098 (6.4) | |
| u 5 | 0.065 (2.9) | 0.045 (1.8) | 0.069 (1.8) | 0.061 (3.4) | 0.072 (3.5) | |
| u 6 | 0.369 (92.2) | 0.323 (93.1) | 0.493 (94.2) | 0.309 (87.9) | 0.359 (87.0) | |
| Uc | 0.384 | 0.334 | 0.508 | 0.329 | 0.385 | |
| Ueb (mg/kg) | 0.768 | 0.669 | 1.016 | 0.658 | 0.770 | |
| Ue (%) | 19.2 | 16.7 | 25.4 | 16.5 | 19.2 | |
AME = ametryn; ATZ = atrazine; CAR = carbaryl; CF = carbofuran; MeP = methyl parathion Uc = combined uncertainty.
ui are the contributions of each uncertainty source to Uc of the method.
Ue is the expanded uncertainty for a 4 mg/kg estimated for a level of confidence of 95%.
As can be seen in Table 4, Ue values at 95% confidence level for papaya ranged from 12.8% to 17.6% and for avocado from 16.5% to 25.4%. In papaya CF, ATZ, and AME present lower values than for avocado; however, for MeP the value is higher. In the case of CAR, for both fruits Ue is the same value. These results are similar to others reported for methods using GC–MS and the QuEChERS procedure. For example, Walorczyk [24] reported Ue values for blackcurrant samples ranging from 7.0% to 53.0%, where the majority of the pesticides had uncertainties < 30.0%. Also da Silva Souse et al. [17] reported Ue values between 7.9% and 36.1%, for melon samples.
At first sight, the results of this method seem to be high, however, according to SANCO/12495/2011 guideline [27] the uncertainty acceptance criteria is when the value is < 50.0%. Even though the use of the internal standard is recommended to minimize uncertainties, in the case of the proposed method the expanded uncertainties indicated it is not necessary.
3.4. Method feasibility
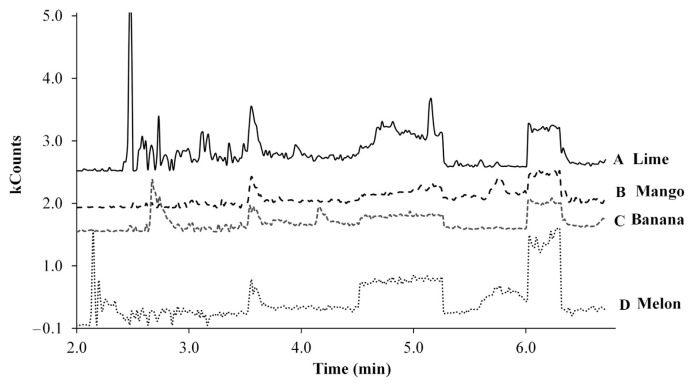
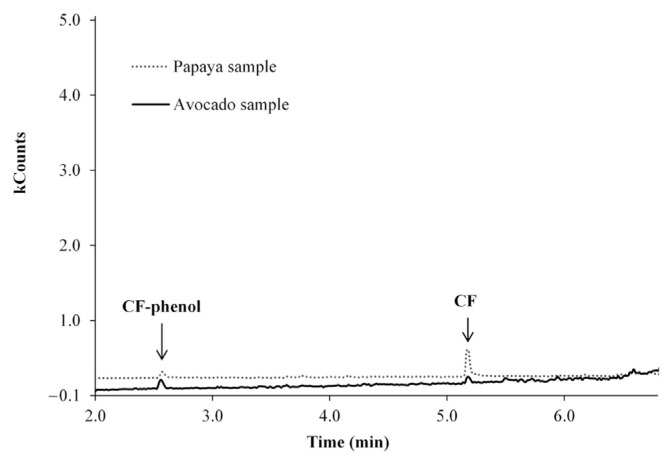
Method feasibility for lime, mango, melon, and banana fruits was carried out as indicated in Section 2.7. The comparison of chromatograms showed that interferences were not observed for any of the pesticides in blank mango and banana samples. However, in chromatograms of blank lime samples interferences for CF and CAR were observed, whereas for ATZ, MeP, and AME interferences were not observed. In melon, interference for CF, CAR, and MeP were observed, whereas for ATZ and AME no interference was observed. As can be seen in Figure 3, the blank profile of lime, mango, and banana is similar to the fortificated avocado sample. This suggests that possibly the matrix-match calibration curve of avocado is more appropriate to be used for pesticide quantification in lime, mango, and banana. In the same way, the blank profile of melon is similar to the fortificated papaya sample; therefore it could be more appropriate for the use of papaya matrix-match calibration curve for quantification purposes in melon. The method validated in this paper also was applied to real samples of papaya and avocado. An example of a chromatogram of papaya and avocado samples with positive analysis is shown in Figure 4.
Figure 3.
Chromatogram (SIM mode) obtained for blank samples of (A) lime, (B) mango, (C) banana, and (D) melon. SIM = selected ion monitoring.
Figure 4.
Chromatogram (SIM mode) obtained for papaya (dotted line) and avocado (solid line) real samples. SIM = selected ion monitoring.
4. Conclusion
In this paper, an analytical method for the separation of carbofuran, carbaryl, atrazine, methyl parathion, and ametryn in papaya and avocado using QuEChERS method and GC-MS is presented. The optimized conditions allowed the separation of all pesticides with baseline resolution using a Zebron ZB-5MS capillary column (Phenomenex) under a gradient temperature elution. After applying QuEChERS sample treatment, the proposed method was properly validated using papaya and avocado samples. The results indicate that this method is specific, accurate, and reproducible. The expanded uncertainty of the method is acceptable according to SANCO/12495/2011 guideline. The proposed method was also found to be suitable for different kinds of fruits.
Acknowledgments
The authors wish to thank Red Tematica de Toxicologia de Plaguicidas-CONACYT Project No. 253789 for the financial support. Also, N.S.P.-F. wishes to thank CONACYT for the research grant provided.
Funding Statement
The authors wish to thank Red Tematica de Toxicologia de Plaguicidas-CONACYT Project No. 253789 for the financial support. Also, N.S.P.-F. wishes to thank CONACYT for the research grant provided.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
REFERENCES
- 1. Singh S, Kumar V, Thakur S, Banerjee BD, Chandna S, Rautela RS, Grover SS, Rawat DS, Pasha T, Jain SK, Ichhpujani RL, Rai A. DNA damage and cholinesterase activity in occupational workers exposed to pesticides. Environ Toxicol Pharmacol. 2011;31:278–85. doi: 10.1016/j.etap.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 2. Farahi A, Achak M, El Gaini L, El Mhammedi MA, Bakasse M. Electrochemical determination of paraquat in citric fruit based on electrodeposition of silver particles onto carbon paste electrode. J Food Drug Anal. 2015;23:463–71. doi: 10.1016/j.jfda.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.United States Environmental Protection Agency (USEPA) Method USEPA 8270D. Semivolatile organic compounds by gas chromatography/mass spectrometry. Washington, DC: USEPA; 2014. Revision 5. [Google Scholar]
- 4. Kwon H, Lehotay SJ, Geis-Asteggiante L. Variability of matrix effects in liquid and gas chromatography–mass spectrometry analysis of pesticide residues after QuEChERS sample preparation of different food crops. J Chromatogr A. 2012;1270:235–45. doi: 10.1016/j.chroma.2012.10.059. [DOI] [PubMed] [Google Scholar]
- 5. Lagunas-Allué L, Sanz-Asensio J, Martínez-Soria MT. Comparison of four extraction methods for the determination of fungicide residues in grapes through gas chromatography–mass spectrometry. J Chromatogr A. 2012;1270:62–71. doi: 10.1016/j.chroma.2012.10.069. [DOI] [PubMed] [Google Scholar]
- 6. Banerjee K, Utture S, Dasgupta S, Kandaswamy C, Pradhan S, Kulkarni S, Adsule P. Multiresidue determination of 375 organic contaminants including pesticides, polychlorinated biphenyls, and polyaromatic hydrocarbons in fruits and vegetables by gas chromatography–triple quadrupole mass spectrometry with introduction of semiquantification. J Chromatogr A. 2012;1270:283–95. doi: 10.1016/j.chroma.2012.10.066. [DOI] [PubMed] [Google Scholar]
- 7. Hernández-Borges J, Cabrera JC, Rodríguez-Delgado MA, Hernández-Suárez EM, Saúco VG. Analysis of pesticide residues in bananas harvested in the Canary Islands (Spain) Food Chem. 2009;113:313–9. [Google Scholar]
- 8. Herrero A, Ortiz MC, Sarabia LA. D-optimal experimental design coupled with parallel factor analysis 2 decomposition a useful tool in the determination of triazines in oranges by programmed temperature vaporization-gas chromatography–mass spectrometry when using dispersive-solid phase extraction. J Chromatogr A. 2013;1288:111–26. doi: 10.1016/j.chroma.2013.02.088. [DOI] [PubMed] [Google Scholar]
- 9. Hou X, Han M, Dai X, Yang X, Yi S. A multi-residue method for the determination of 124 pesticides in rice by modified QuEChERS extraction and gas chromatography–tandem mass spectrometry. Food Chem. 2013;138:1198–205. doi: 10.1016/j.foodchem.2012.11.089. [DOI] [PubMed] [Google Scholar]
- 10. Jiang Y, Ni Y, Zhu H, Zhu C. Using fast gas chromatography–mass spectrometry with auto-headspace solid-phase microextraction to determine ultra trace residues of organophosphorus pesticides in fruits. J Chromatogr Sci. 2011;49:353–60. doi: 10.1093/chromsci/49.5.353. [DOI] [PubMed] [Google Scholar]
- 11. Koesukwiwat U, Lehotay SJ, Leepipatpiboon N. Fast, low-pressure gas chromatography triple quadrupole tandem mass spectrometry for analysis of 150 pesticide residues in fruits and vegetables. J Chromatogr A. 2011;1218:7039–50. doi: 10.1016/j.chroma.2011.07.094. [DOI] [PubMed] [Google Scholar]
- 12. Lehotay SJ, Son KA, Kwon H, Koesukwiwat U, Fu W, Mastovska K, Hoh E, Leepipatpiboon N. Comparison of QuEChERS sample preparation methods for the analysis of pesticide residues in fruits and vegetables. J Chromatogr A. 2010;1217:2548–60. doi: 10.1016/j.chroma.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 13. Menezes-Filho A, Dos-Santos FN, Pereira PADP. Development, validation, and application of a methodology based on solid-phase micro extraction followed by gas chromatography coupled to mass spectrometry (SPME/GC–MS) for the determination of pesticide residues in mangoes. Talanta. 2010;81:346–54. doi: 10.1016/j.talanta.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 14. Navickiene DSS, Aquino S, Bezerra A. A matrix solid-phase dispersion method for the extraction of seven pesticides from mango and papaya. J. Chromatogr Sci. 2010;48:750–4. doi: 10.1093/chromsci/48.9.750. [DOI] [PubMed] [Google Scholar]
- 15. Sang ZY, Wang YT, Tsoi YK, Leung KSY. CODEX-compliant 11 organophosphorus pesticides screening in multiple commodities using headspace-solid phase microextraction-gas chromatography–mass spectrometry. Food Chem. 2013;136:710–7. doi: 10.1016/j.foodchem.2012.08.060. [DOI] [PubMed] [Google Scholar]
- 16. Yang X, Zhang H, Liu Y, Wang J, Zhang YC, Dong AJ, Zhao HT, Sun CH, Cui J. Multiresidue method for determination of 88 pesticides in berry fruits using solid-phase extraction and gas chromatography–mass spectrometry: determination of 88 pesticides in berries using SPE and GC-MS. Food Chem. 2011;127:855–65. doi: 10.1016/j.foodchem.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 17. da Silva Sousa J, de Castro RC, de Albuquerque Andrade G, Lima CG, Lima LK, Milhome MAL, do Nascimento RF. Evaluation of an analytical methodology using QuEChERS and GC-SQ/MS for the investigation of the level of pesticide residues in Brazilian melons. Food Chem. 2013;141:2675–81. doi: 10.1016/j.foodchem.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 18. Cherta L, Beltran J, López F, Hernández F. Application of fast gas chromatography–mass spectrometry in combination with the QuEChERS method for the determination of pesticide residues in fruits and vegetables. Food Anal Methods. 2012;6:1170–87. [Google Scholar]
- 19.United States Environmental Protection Agency (USEPA) Method USEPA 3500C. Organic extraction and sample preparation. Washington, DC: USEPA; 2007. Revision 3. [Google Scholar]
- 20.United States Environmental Protection Agency (USEPA) Method USEPA 3600C. Cleanup. Washington, DC: USEPA; 1996. Revision 3. [Google Scholar]
- 21. Carabias-Martínez R, García-Hermida C, Rodríguez-Gonzalo E, Ruano-Miguel L. Behaviour of carbamate pesticides in gas chromatography and their determination with solid-phase extraction and solid-phase microextraction as preconcentration steps. J Sep Sci. 2005;28:2130–8. doi: 10.1002/jssc.200400047. [DOI] [PubMed] [Google Scholar]
- 22. Bruzzoniti MC, Checchini L, De-Carlo RM, Orlandini S, Rivoira L, Del-Bubba M. QuEChERS sample preparation for the determination of pesticides and other organic residues in environmental matrices: a critical review. Anal Bioanal Chem. 2014;406:4089–116. doi: 10.1007/s00216-014-7798-4. [DOI] [PubMed] [Google Scholar]
- 23.Ellison SRL, Rosslein M, Williams A, editors. Quantifying uncertainty in analytical measurements. 3rd ed. EURACHEM/CITAC; 2000. [Google Scholar]
- 24. Walorczyk S. Validation and use of a QuEChERS-based gas chromatographic–tandem mass spectrometric method for multiresidue pesticide analysis in blackcurrants including studies of matrix effects and estimation of measurement uncertainty. Talanta. 2014;120:106–13. doi: 10.1016/j.talanta.2013.11.087. [DOI] [PubMed] [Google Scholar]
- 25. Cuadros-Rodrıguez L, Torres MH, Lopez EA, Gonzalez FE, Liebanas FA, Vidal JM. Assessment of uncertainty in pesticide multiresidue analytical methods: main sources and estimation. Anal Chim Acta. 2002;454:297–314. [Google Scholar]
- 26.USDA National Nutrient Database for Standard Reference. National Nutrient Database for Standard Reference; [Accessed 18 May 2015]. Available at: http://ndb.nal.usda.gov/ [Google Scholar]
- 27.European Commission. Document No. SANCO/12495/2011 Method validation and quality control procedures for pesticide residues analysis in food and feed. 2012. Available at: http://www.eurl-pesticides.eu/docs/public/tmplt_article.asp?CntID=727.
- 28.European Commission. 2002/65-7/EC: Commission Decision of 12 August 2002 implementing Council Directive 96/23/EC concerning the performance of analytical methods and the interpretation of results. 2002. Available at: http://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32002D0657.