Abstract
Natural genetic transformation offers a direct route by which synthetic gene constructs can be placed into the single circular chromosome of Streptococcus pneumoniae. However, the lack of a general negative-selection marker has hampered the introduction of constructs that do not confer a selectable phenotype. A 1.3-kb cassette was constructed comprising a kanamycin (Kn) resistance marker (kan) and a counterselectable rpsL+ marker. The cassette conferred dominant streptomycin (Sm) sensitivity in an Sm-resistant background in S. pneumoniae. It was demonstrated that it could be used in a two-step transformation procedure to place DNA of arbitrary sequence at a chosen target site. The first transformation into an Sm-resistant strain used the cassette to tag a target gene on the chromosome by homologous recombination while conferring Kn resistance but Sm sensitivity on the recombinant. Replacement of the cassette by an arbitrary segment of DNA during a second transformation restored Sm resistance (and Kn sensitivity), allowing construction of silent mutations and deletions or other gene replacements which lack a selectable phenotype. It was also shown that gene conversion occurred between the two rpsL alleles in a process that depended on recA and that was susceptible to correction by mismatch repair.
Streptococcus pneumoniae, a widespread human pathogen associated with high rates of disease and mortality, is being used increasingly as a genetically tractable model pathogen for application of genomics to searches for new drugs and drug targets. Many wild-type strains of the pneumococcus are readily transformable (18), so that effects of disruption of specific genes can be readily assessed in model host systems. To date, these gene disruptions have commonly been made by inserting a drug resistance gene that provides direct selection of rare recombinants. While powerful, this method does have drawbacks. As design of strains with multiple mutations becomes more sophisticated, for example, an accumulation of drug markers in the mutated strains could become cumbersome and possibly compromise interpretations of experimental results. Also, many important categories of gene mutation, such as missense substitutions and in-frame deletions, usually confer no selectable phenotype.
While there are several currently available ways to circumvent or accommodate these limitations, a particularly convenient approach, used in a variety of other bacteria but not yet applied in S. pneumoniae, employs a bicistronic cassette permitting both selection for its acquisition and selection for its loss. One marker allowing the required negative selection is based on a common spontaneous bacterial streptomycin (Sm) resistance mutation in the gene rpsL that causes a lysine replacement in protein S12 of the small ribosomal subunit (21). As this mutation is recessive, an rpsL+ allele has been employed to provide a dominant drug-sensitive phenotype in genetic contexts where it can provide direct selection for deletion, mutation, or replacement events (5, 7, 8, 17, 20, 22–25). Here we describe application of this principle for construction of an rpsL cassette for use in S. pneumoniae that allows use of antibiotics at both selection steps and show that it can be used with natural genetic transformation for gene replacement through negative selection.
MATERIALS AND METHODS
Strains, media, and DNA sources.
S. pneumoniae strains used in this work (Table 1) are derivatives of strains R6 and Rx, whose origins are traced by Tiraby et al. (26). The recessive str41 mutation (4) corresponds to a single base substitution (A to C transversion) converting Lys56 (AAA) to Thr (ACA) (21); the allele str1 confers the same K56T substitution on S12 but also carries two silent transitions (GC to AT), at positions 150 and 405 in the rpsL gene (13). We refer to these Smr alleles here as rpsL41 and rpsL1, respectively. Growth media and culture methods for genetic transformation have been described (10, 12). Unless stated otherwise, donor DNA was used at a final concentration of 100 ng/ml of competent culture. Drug selection was as described (10) using final concentrations in selective agar of 150 μg of Sm per ml, 200 μg of kanamycin (Kn) per ml, and 2.5 μg of novobiocin (Nv) per ml. For transformation, cultures growing in broth were induced with synthetic competence-stimulating pheromone peptide (CSP-1; Chiron Mimitopes, Raleigh, N.C.) as described (6).
TABLE 1.
Bacterial strains and primers used in this study
| Strain or primer | Description | Source or reference | |
|---|---|---|---|
| S. pneumoniae | |||
| CP1200 | Rx derivative; hex mal rpsL1; Hex+ Kns RecA+ Smr | 14 | |
| CP1250 | CP1200 but bgl-1; Kns Smr | 16 | |
| CP1296 | CP1250 but cbp3::kan-rpsL+ (by transformation with PCR construct; see Materials and Methods); Knr Sms | This study | |
| CP1326 | CP1296 but rpsL+/+ by spontaneous conversion; cbp3::kan-rpsL+; Knr Sms | This study | |
| R800 | R6 derivative; Hex+ Kns RecA+ Sms | 11 | |
| R239 | R800 but recA::ermAM (by transformation with a ligation mixture)-deletion ClaI (recA)-ClaI (within RUP, downstream of recA), blunt ended and ligated to a BstUI-ermAM fragment; Eryr | Bernard Martin | |
| R304 | R800 but nov1 rif23 rpsL41; Novr Rifr Smr | 15 | |
| R416 | R800 but rpsL41 (by transformation with R304 chromosomal DNA); Smr | This study | |
| R810 | R800 but comC::pXF520 (comC+) ebg::spc, and carries UP mutation; ComCDEUP Cmr Spcr | 12 | |
| R960 | R416 but cbp3::kan-rpsL+ (by transformation with CP1296 chromosomal DNA); Hex+rpsL41 Knr Sms | This study | |
| R961 | R800 but cbp3::kan-rpsL+ (by transformation with CP1296 chromosomal DNA); Hex+ Knr | This study | |
| R974 | R960 but Smr through spontaneous conversion; cbp3::kan-rpsL41 rpsL41; Knr Smr | This study | |
| R981 | R800 but rpsL1 (by transformation with CP1200 chromosomal DNA); Smr | This study | |
| R989 | R960 but recA::ermAM (by transformation with R239 chromosomal DNA); cbp3::kan-rpsL+ Hex+rpsL41; Eryr Knr Sms | This study | |
| R990 | R981 but cbp3::kan-rpsL+ (by transformation with CP1296 chromosomal DNA); rpsL1; Knr Sms | This study | |
| R1001 | R989 but spontaneous revertant to Smr; cbp3::kan-rpsL0recA::ermAM rpsL41; Eryr Knr Smr | This study | |
| R1004 | R960 but hexA::cat; Cmr Knr Sms | This study | |
| R1005 | R990 but hexA::cat; Cmr Knr Sms | This study | |
| R1029 | R800 but ΔcomC::kan-rpsL+ (by transformation with a SOEing fragment; see Materials and Methods); Knr | This study | |
| R1036 | R981 but ΔcomC::kan-rpsL+ (by transformation with R1029 chromosomal DNA); Knr Sms | This study | |
| Primersa | |||
| B | CATTATCCATTAAAAATCAAACGGAAGCCGGGAAAATTCCCAGC (underlined sequence corresponding to kan cassette; complementary to Kan5, see below); upstream of comC; 679–660; SPU33315 | ||
| BM47 | GATTTGCTAAGTTTGAAATGATTGAG; within orfL, upstream of comCDE; 3–28; SPU33315 | ||
| BM52 | GTCCTCTATCCCTCTCATAC; within comD; 1080–1061; SPU33315 | ||
| BM54 | CATTCCAGCATAATCATGTCG; within comD; 1583–1563; SPU33315 | ||
| BM81 | TCGCGATGACTACTATGAACG; upstream of cbp3; TIGRb | ||
| BM82 | GCTTACAGAAAAGAGCAGGAAAT; downstream of cbp3; TIGR | ||
| C | GGAAAGGGGCCCAGGTCTCTGTAATGAAATAAGGGGAAAGAG (underlined sequence corresponding to rpsL; complementary to primer 7, see below); upstream of comD; 801–822; SPU33315 | ||
| cat3 | AGCCAGTCATTAGGCCTATC; within cat (pC194 plasmid); 1903–1884; L08860 | ||
| DAM301 | CGCGCAAGCTGGGGATCCG; upstream of kan in pR410 | ||
| DAM303 | AAGGGCCCGTTTGATTTTTAATG; positions 7–23 of DAM303 correspond to positions 773–789 upstream of kan; AF411920; TIGR | ||
| DAM313 | AGCTTTCTCGTGGTGTAGAACAAC; upstream of cbp3; TIGR | ||
| DAM314 | ACGAGGATCCGATCCATTTCCTCTGGAATA; within cbp3; TIGR | ||
| DAM315 | AGCAGGGCCCAGGTCTCTGGTAAGTGGTAT; within cbp3; TIGR | ||
| DAM316 | CTCTCAAGGTCGCCCAGCTATG; downstream of cbp3; TIGR | ||
| DAM345 | CAGGAGACATTCCTTCCGTATCTT; within kan; 926–903; AF411920; TIGR | ||
| DAM347 | CCGAATTCTAGGTACTAAAACAATTCATCCAGTAA; downstream of Kan5 in pR410 | ||
| DAM350 | CTGGAATTCACCAAAAATAAAAAAACACAG GAG; upstream of rpsL; TIGR | ||
| DAM351 | CTAGGGCCCCTTTCCTTATGCTTTTGGAC; downsteam of rpsL; TIGR | ||
| Kan5 | CCGTTTGATTTTTAATGGATAATG; positions 3–24 of Kan5 correspond to positions 185–206 of ami; X17337 | ||
| 7 | AGAGACCTGGGCCCCTTTCC; downstream of rpsL; TIGR |
Information given in the order sequence; gene; position within the deposited sequence indicated; and GenBank/EMBL accession number.
TIGR, TIGR website (http://www.tigr.org).
PCR amplification.
Plasmid or genomic DNA (50 ng) and 50 pmol of each primer were used in a total volume of 50 μl of PCR SuperMix (Gibco-BRL); amplification proceeded for 30 cycles as follows: 45 s at 94°C, 45 s at 60°C, and 2 min at 72°C, followed by a 10-min extension cycle. Products were purified using the QIAquick PCR purification kit (Qiagen). Oligonucleotide primers used for various purposes are listed in Table 1.
Construction of a positive/negative selection cassette, kan-rpsL+.
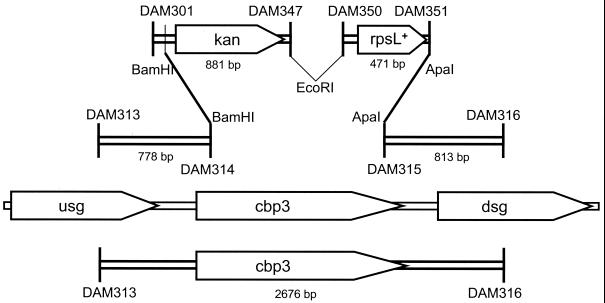
A 435-bp fragment (PCRII) containing rpsL+ was amplified from chromosomal DNA of strain R800 (EcoRI and ApaI termini) using the primer pair DAM350 and DAM351. An 896-bp fragment (PCRIII) containing the kan marker was amplified from plasmid pR410 using DAM301 and DAM347 (BamHI and EcoRI termini). Plasmid pR410 (kindly provided by Marc Prudhommme) carries a synthetic kan cassette derived from the aphIII gene of plasmid pJHI (27) and was designed similarly to the previously described erythromycin and chloramphenicol resistance cassettes (1). Two DNA fragments (PCRI and PCRIV) flanking the dispensable target gene cbp3 (19) were prepared by PCR using chromosomal DNA of strain CP1250 as a template (PCRI, with a BamHI 3′ terminus [DAM313-DAM314] and PCRIV, with an ApaI 5′ terminus [DAM315-DAM316]. PCRI, PCRII, PCRIII, and PCRIV were digested with corresponding restriction nucleases, purified, ligated, and used to transform CP1250, with selection for Knr. After backcrossing one Knr clone to CP1250, one of the resulting Knr transformants, named CP1296, was shown to carry a disruption of the target gene and insertion of the kan and rpsL fragments with the same polarity as cbp3. The predicted sequence of the disrupted cbp3 locus is available (GenBank accession no. AF411920).
The structure of the insertion in CP1296 was verified by PCR with primer pairs DAM313-DAM347, DAM303-DAM351, and DAM350-DAM316; the expected junction fragments of 1.7, 1.3, and 1.2 kb, respectively, were obtained. The sequences of junctions were also verified by reading sequence in the junction fragments with primers DAM345, DAM351, and DAM350. The entire sequence within the rpsL+ gene was read in both directions and matched the published sequence (Z15120). We propose the trivial name Janus for this cassette, which uses both forward and reverse selection to allow formation of junctions and new structures not under selection.
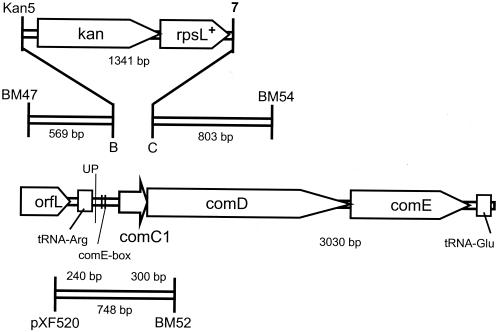
Construction of R1029, a ΔcomC::kan-rpsL+ strain.
A strain harboring a substitution of the comC gene by the kan-rpsL+ cassette was constructed as follows. First, a PCR fragment containing the region immediately upstream of comC was amplified from R800 chromosomal DNA with the primers BM47 and B, and a kan-rpsL fragment was amplified from strain R960 with the Kan5 and 7 primers (Table 1). Following purification through QIAquick columns, the two fragments were mixed and connected to generate fragment A by PCR with the BM47-7 primer pair. A fragment containing part of comD was then amplified with the C-BM54 primer pair (Table 1), purified using a QIAquick column, and mixed with fragment A for assembly into a unique product through PCR amplification with primers BM47 and BM54. Strain R1029 (R800 ΔcomC::kan-rpsL+; Table 1) was obtained following transformation of strain R800 with the final PCR product by selection for Knr transformants. Normal transformant yields obtained with strain R1029 following treatment with synthetic competence pheromone (data not shown) indicated that the synthetic promoter in kan-rpsL+ drove sufficient expression of comDE for pheromone signal transmission and full activation of the competence cascade.
RESULTS
Construction and properties of a dominant Sms cassette.
A bicistronic positive/negative selection cassette was constructed by combining PCR amplicons containing a Kn resistance gene preceded by a constitutive promoter from the amiA (oligopeptide permease) locus (1) and the wild-type S12 gene, rpsL. The cassette was inserted into a dispensable gene coding for a putative choline-binding protein (cbp3) (19) in strain CP1250, which carries the rpsL1 mutation in the “genuine” or chromosomal copy of rpsL, as described in Materials and Methods and illustrated in Fig. 1. PCR with characteristic sets of primers directed to the cassette and to the insertion locus confirmed that junction fragments and an internal kan-rpsL fragment were readily amplified from DNA of CP1296. The detailed organization of the cassette is displayed in AF411920, including the rpsL sequence determined for the copy of the gene obtained from R800 and its context in CP1296. Although CP1250 is Smr, CP1296 was found to be fully sensitive to this drug, as predicted for addition of a dominant Sms allele; the cassette reduced the Sm MIC from >250 to 5 μg/ml, the same as the MIC of R800, the Sms source of rpsL+ (data not shown).
FIG. 1.
Construction of Janus cassette in the S. pneumoniae cbp3 locus. Pentagons marked rpsL and kan, modules of the Janus cassette. Primers used to amplify two cassette modules and two targeting fragments are indicated at the termini of those PCR products. After synthesis, restriction enzyme digestion, and purification, the product of a single ligation reaction was screened directly by transformation of CP1250 to obtain correctly linked modules giving the Knr phenotype. Top, PCR fragment used to construct strain CP1296 (cbp3::kan-rpsL+). Middle, cbp3 chromosomal region. Bottom, a 2,676-bp fragment containing cbp3 amplified from chromosomal DNA of strain CP1250 with primers DAM313 and DAM316 and used for reintroduction of cbp3 to replace the Janus cassette.
kan-rpsL+ cassette can be deleted by targeted recombination.
Although Rimini et al. (19) reported transformation deficiency for an insertion-duplication mutation at cbp3, CP1296 was transformed as readily as its parent, CP1250. In contrast to a minority of spontaneous Smr revertants found in CP1296 cultures (see below), deliberate transformation of this strain with DNA from strain R800, which is Sms but carries an intact cbp3 gene, generated a large number of Smr clones which had also become Kns (Table 2). In accord with the design of this cassette, we interpret these DNA-dependent recombinants as arising from excisional recombination exchange replacing the entire kan-rpsL cassette in a single event directed by flanking homology at the cbp3 locus. As expected, transformation with a pure amplicon containing the cbp3 gene (Fig. 1) was also effective at generating Kns Smr recombinants (data not shown). To verify this interpretation of the genetic results, several putative excisional transformants were examined by growing colonies from single Smr colonies; Kns clones were found only when a cbp3-containing donor DNA had been used for the cross (data not shown).
TABLE 2.
Excisional transformation replaces kan-rpsL cassette with cbp3a
| Donor DNA | Induction by CSP | Fold increaseb in Smr | Growth (CFU/ml) with selection on:
|
|
|---|---|---|---|---|
| Sm | Sm and Kn | |||
| None | + | 1.1 | 1,000 | 830 |
| − | 1 | 910 | 820 | |
| R800 (Sms) | + | 28 | 22,000 | 1,000 |
| − | 1 | 780 | 710 | |
Strain CP1296 was grown to an optical density of 0.05, diluted 10-fold, and treated as indicated with CSP-1 (100 ng/ml) and DNA (10 ng/ml) for 60 min. Cell types are reported among 10 to 20 million cells per ml in the transformed population.
Ratio of Smr in CSP-treated culture to that in untreated control.
Replacement of the entire kan-rpsL cassette via transformation with a donor DNA lacking the cassette is thus readily recognized, both because such excisional Smr transformants can occur in higher numbers than the spontaneous revertants and because they become Kns (Table 2).
Spontaneous Smr clones in rpsL heterozygotes.
The kan-rpsL+ cassette was not completely stable in the heterozygous state. This was indicated by the composition of clones arising by introduction of the cassette into an Smr strain by transformation: cultures grown from single Knr colonies had a variable minority of Knr Smr cells (0.1 to 10 per 10,000). On subculturing single cells, all Smr subclones tested were Knr, while other Knr subclones were Sms, but again contained a variable but nonzero minority of Smr (data not shown). As Knr Smr arose as well in comA (noncompetent) cultures, the generation of the Smr minority did not depend on transformation but apparently arose by intracellular recombination events between the Sms and Smr rpsL alleles to convert Sms to Smr (“genuine → cassette” gene conversion) while leaving kan in place. Since such spontaneous Smr subclones would represent false-positives during the practical applications envisaged for this cassette, it is important to understand their source. It would be especially valuable to know how to minimize their occurrence when applying the method to mutate virulent isolates, in which the level of competence for genetic transformation may not be as high as in standard laboratory stocks.
Spontaneous Smr clones result from gene conversion.
A frequency of 2.4 Smr clones per 10,000 Sms cells was observed through plating of several independent cultures of strain R960 (Table 1). Chromosomal DNA from Knr derivatives of strain R800 obtained through transformation with R974 DNA (Table 3) readily transformed strain R800 to Smr (data not shown), demonstrating linkage of Smr to Knr and strongly suggesting that the rpsL41 allele had replaced the cassette-linked rpsL+ copy. Consistent with this interpretation, introduction of the kan-linked rpsL41 gene of R974 into strain R416 did not abolish its resistance to Sm (Table 3). R974 is therefore a candidate prototype for genuine → cassette spontaneous conversion of rpsL sequences. Other explanations, such as spontaneous mutation to Smr of the kan-linked rpsL copy, appeared unlikely inasmuch as the rate of mutation to Smr is very low in a wild-type background (13). In line with this interpretation, no spontaneous Smr could be obtained in strain R961, which harbors two rpsL+ alleles, indicating that the presence of a resident rpsL41 mutant allele was required for the production of Smr clones.
TABLE 3.
Analysis of cotransfer of Knr and Smr to characterize spontaneous Smr revertants
| Recipient strain | Donor DNAa | Growth (CFU/ml) with selection on:
|
Smr frequency per 100 Knr cells | |
|---|---|---|---|---|
| Kn | Sm | |||
| R800 | R974 | 2.99 × 106 | 3.6 × 104 | 1.20 |
| R989 | 4.18 × 106 | <10 | <0.0002 | |
| R1001 | 2.86 × 106 | <10 | <0.0003 | |
| R416 | R974 | 7.76 × 106 | Rb | |
| R989 | 7.76 × 106 | Sb | ||
| R1001 | 7.76 × 106 | Rb | ||
DNA from Knr transformant of R800 after transformation with the indicated strain.
Cells from two Knr colonies were plated in the presence or absence of 200 μg of Sm per ml to assess their resistance (R) or susceptibility (S).
To characterize the conversions more precisely, we sequenced the cassette rpsL genes in spontaneous Smr revertants arising in cultures of CP1296, which are described above. As reported previously (13), the rpsL1 allele of rpsL differs from the R6 wild-type gene sequence (Z15120) at three positions, although two of those are synonymous substitutions. Sequencing of the cassette copy of rpsL from four Smr revertants obtained from CP1296 revealed in each case that all three nucleotide substitutions characteristic of the distinctive rpsL1 allele were present in the kan-rpsL cassette. Thus, the cassette rpsL+ had been replaced by a copy of the genuine Smr rpsL1 allele precisely, establishing the origin of spontaneous Smr as gene conversion, not mutation.
Smr spontaneous convertants were also observed in cultures of Knr derivatives of strain R800 obtained through transformation with R974 DNA (data not shown), indicating that the rpsL41 allele linked to kan could be donated to the rpsL+ gene. This observation suggested that cassette → genuine conversion could occur as readily as genuine → cassette conversion. Such a conversion would account for the recovery of CP1326, a spontaneous derivative of CP1296 that was stably Sms (Table 1). Indeed, sequencing both copies of rpsL in CP1326 revealed a wild-type allele at both sites, showing that all three divergent bases of the rpsL1 allele had been replaced by R800 (rpsL+) sequence, and confirmed the apparent conversion of the rpsL1 gene to the sequence of the Sms allele carried in the kan-rpsL+ cassette.
Spontaneous conversion is RecA dependent.
To establish whether spontaneous conversion events occurred through homologous recombination, a recA derivative of the rpsL+/rpsL41 heterozygous strain R960 was constructed (strain R989; Table 1). To improve growth of the recA derivative, colonies and liquid cultures were grown in the absence of oxygen. The frequency of appearance of spontaneous Smr revertants was diminished 39-fold in this strain compared to the isogenic recA+ parent similarly grown under anaerobic conditions (Table 4). Interestingly, the spontaneous conversion rate of the wild-type parent itself was diminished fivefold compared to the rate under aerobic conditions (Table 4). This observation suggests that oxygen could induce DNA lesions, possibly leading to chromosome breaks and subsequent conversion events during repair.
TABLE 4.
Genuine → cassette gene conversion is RecA dependent
| Strain | Genotype | Smr frequency per 10,000 cellsa | Fold reductionb |
|---|---|---|---|
| R960c | rpsL41 cbp3::kan-rpsL+ hex+ | 0.47 ± 0.17 | 5.3 |
| R989c | R960 but recA | 0.012 ± 0.0057 | 207.5 (39.2d) |
| R960 | rpsL41 cbp3::kan-rpsL+hex+ | 2.49 ± 0.62 | 1.0 |
| R1004 | R960 but hexA::cat | 2.40 ± 0.63 | 1.0 |
| R1005 | R990 but hexA::cat | 1.95 ± 0.93 | 1.3 |
Calculated from four to five independent cultures grown from individual colonies resuspended in 2 ml of Todd-Hewitt broth supplemented with 0.5% yeast extract and grown to an OD550 of 0.5 to 0.7 before plating.
Compared to the frequency of spontaneous conversion with the rpsL41-rpsL+ combination (hex+ background) for cultures grown in the presence of O2 (last three lines; results taken from Table 5).
Colonies and cultures grown in the absence of O2.
Fold reduction compared to wild type cultures grown in the absence of O2.
To understand the residual source of Smr clones in the recA background, a representative spontaneous Smr derivative (R1001) of the recA mutant strain was retained for analysis. Transformation of the kan-rpsL cassette of R1001 into strain R416 did not abolish its resistance to Sm (Table 3), showing the lack of the rpsL+ allele in the cassette. However, the cassette also did not carry an Smr gene, as no transformation of strain R800 to Smr could be obtained with chromosomal DNA from a Knr derivative of strain R800 generated through transformation with R1001 DNA (Table 3). Together, these results indicate that the kan-linked rpsL copy in strain R1001 was inactivated by mutation. We conclude that it is likely that all Smr revertants produced in a recombination-deficient recA background occur by this mechanism.
Spontaneous Smr can also arise in the wild-type background by inactivation of rpsL. One exceptional Smr revertant of CP1296, clone CP1296A4, did not contain rpsL1 in the kan-rpsL cassette. Instead, sequencing of the cassette copy of rpsL in this exceptional revertant showed that it carried a single-base deletion at nucleotide (nt) 99 in rpsL, changing the sequence T(A)6GTT to T(A)5GTT. The frameshift would be expected to inactivate the dominant rpsL+ allele, explaining the loss of the dominant Sms phenotype in this revertant by gene inactivation.
Strategies to reduce spontaneous conversion frequency.
Inasmuch as spontaneous conversion is RecA dependent (see above), it is likely to involve the formation of a transient heteroduplex structure between the two rpsL copies. We reasoned that inclusion of a mismatch normally recognized by the Hex mismatch repair system (2) within the heteroduplex structure leading to conversion would provoke Hex-dependent abortion of the recombination intermediate and therefore would reduce spontaneous conversion. This hypothesis was tested by comparing Smr frequencies in cultures of strain R960 (kan-rpsL+/rpsL41, Table 1) to those in cultures of strain R990 (kan-rpsL+/rpsL1, Table 1).
The rpsL1 allele (transferred from strain CP1200 to the R800 background) contains two silent transitions in addition to the same single base change as that causing Smr in rpsL41 (13). While the rpsL41/rpsL+ mismatch is not efficiently recognized by Hex, transition mismatches are known to be corrected at the heteroduplex stage in transformation (2). The kan-rpsL+/rpsL1 combination was found to reduce the frequency of conversion to Smr 18-fold (Table 5), and this reduction depended on the Hex phenotype of the strain (compare strains R990 and R1005, Table 5). This result further supports the interpretation that spontaneous Smr strains arise by gene conversion and shows directly that the combined use of a hex+ recipient together with an rpsL+/rpsL1 mismatched heterozygous configuration significantly reduces the spontaneous conversion frequencies and can help to lower the background of Smr clones if and when necessary.
TABLE 5.
Mismatch repair-dependent reduction of conversion frequency using the rpsL1-rpsL+ allele combination and a hex+ genetic background
| Strain | Genotype | Hex | Mismatcha | Smr frequency per 10,000 cellsb | Fold reductionc |
|---|---|---|---|---|---|
| R960 | rpsL41 cbp3::kan-rpsL+hex+ | + | HE | 2.49 ± 0.62 | 1.0 |
| R990d | rpsL1 cbp3::kan-rpsL+hex+ | + | LE | 0.14 ± 0.020 | 18 |
| + | LE | 0.11 ± 0.027 | 23 | ||
| R1004 | R960 but hexA::cat | − | HE | 2.40 ± 0.63 | 1.0 |
| R1005 | R990 but hexA::cat | − | LE | 1.95 ± 0.93 | 1.3 |
HE, essentially not recognized or corrected by Hex; LE, transition mismatch recognized or corrected out by Hex.
Calculated from four or five independent cultures grown from individual colonies resuspended in 2 ml of broth and grown to an OD550 of 0.5 to 0.7 before plating.
Compared to the rpsL41-rpsL+ combination (hex+ background).
Duplicate experiments on two different occasions are listed for R990.
Interestingly, the Smr revertants may be enriched during colonial culture growth; pure Sms populations of CP1296 grown in liquid culture exhibited a lower frequency of revertants (7.5 × 10−6) than parallel colonial cultures (1.5 × 10−4) (data not shown), offering a possible alternative to the Hex-dependent strategy for reducing the rate of gene conversion.
Use of Janus to place a regulatory mutation at the comCDE chromosomal locus.
To demonstrate use of Janus for the transfer of a silent mutation not conferring a directly selectable phenotype, an insertion of the cassette was prepared at the comC (ΔcomC::kan-rpsL+; see Materials and Methods) gene to form strain R1036. The three genes, comC, comD, and comE, encode the competence-regulating peptide signal (6), its receptor, and the cognate response regulator, respectively (16). For the excisional transformation step, a 748-bp region overlapping comC and carrying the UP mutation previously characterized as a single nucleotide change within the terminator of the tRNAArg located upstream of comC (12) was amplified from the comCDE chromosomal region of strain R810 with the cat3-BM52 primer pair (Table 1). This fragment was used in transformation of strain R1036 (ΔcomC::kan-rpsL+ rpsL1 hex+; Table 1) to replace the kan-rpsL cassette while reintroducing the comC gene at its normal chromosomal location together with the silent UP mutation (Fig. 2 and Table 6). As the lengths of homologous region allowing this exchange were not large (240 bp in the region upstream of comC and 300 bp downstream of it) (Fig. 2), the Hex strategy was used to reduce the background rate of gene conversion to a level comparable to the transformation frequency of this small donor fragment. With this strategy, transfer of the UP mutation was readily obtained (see Table 6 footnote b).
FIG. 2.
Use of Janus at the comCDE chromosomal locus. Top, locations of primers used to generate a 2,669-bp-long PCR product (see Materials and Methods) for the construction of strain R1029. R1029 harbors a substitution of the comC gene by the kan-rpsL+ cassette. Middle, map of the comCDE chromosomal region (16) showing limits of PCR fragments used and the site of the UP mutation previously characterized as a single nucleotide change within the terminator of the tRNAArg located upstream of comC (12). Bottom, limits of the homologous segment carried by the cat3-BM52 PCR fragment and used to transfer the UP mutation into strain R1029. pXF520 refers to the limit of the pneumococcal insert in the nonreplicative plasmid pXF520 (16), which is carried in strain R810 as an insertion in comC.
TABLE 6.
Transfer of a silent point mutation at the comCDE locus: use of Janus-containing strain R1036
| Donor DNA | Growth (CFU/ml) with selection on:
|
Smr frequency per 10,000 cells | Fold increase in Smr frequency | ||
|---|---|---|---|---|---|
| None | Sm | Rifa | |||
| R304 chromosomal DNA | 1.56 × 108 | 2.44 × 106 | 5.8 × 105 | 156 | 246.5 |
| cat3-BM52 (UP) PCR fragment | 2.02 × 104 | 1.3 | 2.04b | ||
| None | 9.9 × 103 | 0.63c | 1 | ||
The ratio Rifr/Smr is normally close to 0.15 in a hex+ recipient because Hex-dependent mismatch repair affects integration of the rif23 marker more severely than of rpsL41. The value (0.24) obtained here was expected, as Smr transformants do not result from substitution of the genuine copy of rpsL by the rpsL41 allele but from excisional replacement of the kan-rpsL cassette by comC+.
Four Smr clones were further analyzed by PCR. Three corresponded to substitution of the kan-rpsL+ cassette by the comC+ segment (excision events) and harbored the UP mutation (see Fig. 2); the fourth clone resulted from a spontaneous conversion event and had retained the kan-rpsL cassette.
A lower spontaneous conversion frequency (0.12 per 10,000 cells) was measured in four independent cultures of strain R1036. This value is totally consistent with spontaneous conversion frequencies measured at the cbp3 locus using the rpsL1-rpsL+ mismatched combination and a hex+ recipient (see Table 5).
DISCUSSION
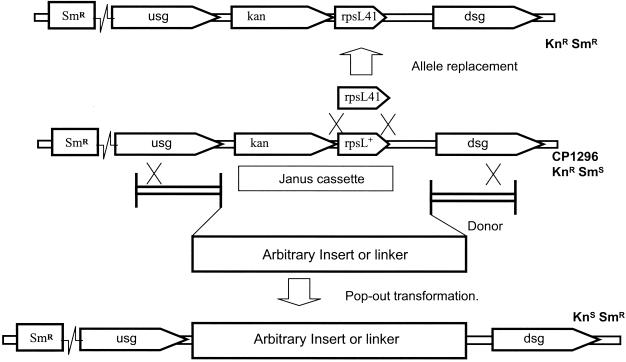
To construct mutant strains of S. pneumoniae without introducing new antibiotic resistance markers, the Janus cassette can be used in a two-step transformation procedure. The first transformation into an Smr strain using the cassette disrupts or tags a target gene on the chromosome by homologous recombination targeted by flanking DNA homology. This Janus strain can subsequently become Smr by two principal routes, as illustrated in Fig. 3. In a second transformation, deletion of the cassette by homologous recombination will restore Sm resistance, allowing construction of mutations without a selectable phenotype. Alternatively, gene conversion eliminating the dominant Sms allele can lead to spontaneous accumulation of Smr clones in a culture of a Janus heterozygote. This conversion is specific, depends on RecA, and involves an intermediate subject to Hex correction. Because Smr revertants accumulate during growth of Smr/s heterozygotes, it may be useful to minimize their frequency in cultures of the intermediate Sms strain by choosing a low-efficiency allele of the Smr locus in a hex+ strain or by controlled subculturing. Since Sm selection after the second transformation step is simply selecting for loss of the cassette, the design of the donor DNA used in that step is broadly unrestricted as long as it includes terminal segments of homology flanking the cassette. Deletions of additional sequences adjacent to the cassette are possible, for example, and virtually any gene(s) (or none) could be inserted in its place.
FIG. 3.
Two fates of Janus. Possible recombination mechanisms for generation of Smr derivatives are illustrated, dependent on (top) gene conversion or (bottom) transformation by exogenous DNA. Crosses show limits of possible single-strand integration or gene conversion events.
The kan-rpsL cassette described here may be useful in applications other than the “drop-in/pop-out” mutagenesis strategy for which we designed it. For example, it is interesting that Sms insertions create strains that could detect and “extract” the wild-type alleles of specific genes from any pneumococcal DNA source, with production of Smr transformants. Also, as illustrated by the experiments reported here with hex and recA, rpsL heterozygotes permit sensitive monitoring of recombination rates independent of the process of genetic transformation.
ACKNOWLEDGMENTS
We thank Chantal Granadel for expert technical assistance and Marc Prudhomme for providing us with plasmid pR410.
This work was supported in part by the U.S. National Science Foundation (grant MCB-9722821 to D.A.M.) and by the European Union (grant QLRK 2000-00543 to J.P.C.). We are indebted to the Institute for Genomic Research (TIGR) for preliminary sequence data that were obtained from its website (http://www.tigr.org) and for permission to use DNA sequence information prior to publication.
REFERENCES
- 1.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 2.Claverys J P, Lacks S A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claverys J P, Lefevre J C, Sicard A M. Transformation of Streptococcus pneumoniae with S. pneumoniae-lambda phage hybrid DNA: induction of deletions. Proc Natl Acad Sci USA. 1980;77:3534–3538. doi: 10.1073/pnas.77.6.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys J P, Roger M, Sicard A M. Excision and repair of mismatched base pairs in transformation of Streptococcus pneumoniae. Mol Gen Genet. 1980;178:191–201. doi: 10.1007/BF00267229. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto-Gotoh T, Tsujimura A, Kuriyama K, Matsuda S. Construction and characterization of new host-vector systems for the enforcement-cloning method. Gene. 1993;137:211–216. doi: 10.1016/0378-1119(93)90008-q. [DOI] [PubMed] [Google Scholar]
- 6.Håvarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosted T J, Baltz R H. Use of rpsL for dominance selection and gene replacement in Streptomyces roseosporus. J Bacteriol. 1997;179:180–186. doi: 10.1128/jb.179.1.180-186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston D M, Cannon J G. Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene. 1999;236:179–184. doi: 10.1016/s0378-1119(99)00238-3. [DOI] [PubMed] [Google Scholar]
- 9.Lataste H, Claverys J P, Sicard A M. Physical and genetic characterization of deletions in Streptococcus pneumoniae. J Bacteriol. 1980;144:422–424. doi: 10.1128/jb.144.1.422-424.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M S, Morrison D A. Identification of a new regulator in Streptococcus pneumoniae linking quorum-sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefevre J C, Claverys J P, Sicard A M. Donor deoxyribonucleic acid length and marker effect in pneumococcal transformation. J Bacteriol. 1979;138:80–86. doi: 10.1128/jb.138.1.80-86.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys J P. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol. 2000;38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 13.Méjean V, Salles C, Bullions L C, Bessman M J, Claverys J P. Characterization of the mutX gene of Streptococcus pneumoniae as a homologue of Escherichia coli mutT, and tentative definition of a catalytic domain of the dGTP pyrophosphohydrolases. Mol Microbiol. 1994;11:323–330. doi: 10.1111/j.1365-2958.1994.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 14.Morrison D A, Trombe M C, Hayden M K, Waszak G A, Chen J D. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta-1. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortier-Barriere I, de Saizieu A, Claverys J P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pnemoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 16.Pestova E V, Håvarstein L S, Morrison D A. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–864. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 17.Prentki P, Binda A, Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991;103:17–23. doi: 10.1016/0378-1119(91)90385-o. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez M, Morrison D A, Tomasz A. Ubiquitous distribution of the competence-related genes comA and comC among isolates of Streptococcus pneumoniae. Microb Drug Res. 1997;3:39–52. doi: 10.1089/mdr.1997.3.39. [DOI] [PubMed] [Google Scholar]
- 19.Rimini R, Jansson B, Feger G, Roberts T C, de Francesco M, Gozzi A, Faggioni F, Domenici E, Wallace D M, Frandsen N, Polissi A. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol Microbiol. 2000;36:1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 20.Russell C B, Dahlquist F W. Exchange of chromosomal and plasmid alleles in Escherichia coli by selection for loss of a dominant antibiotic sensitivity marker. J Bacteriol. 1989;171:2614–2618. doi: 10.1128/jb.171.5.2614-2618.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salles C, Créancier L, Claverys J P, Méjean V. The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 1992;20:6103. doi: 10.1093/nar/20.22.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sander P, Meier A, Bottger E C. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 23.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 24.Skrzypek E, Haddix P L, Plano G V, Straley S C. New suicide vector for gene replacement in yersiniae and other gram-negative bacteria. Plasmid. 1993;29:160–163. doi: 10.1006/plas.1993.1019. [DOI] [PubMed] [Google Scholar]
- 25.Stojiljkovic I, Trgovcevic Z, Salaj-Smic E. Tn5-rpsL: a new derivative of transposon Tn5 useful in plasmid curing. Gene. 1991;99:101–104. doi: 10.1016/0378-1119(91)90039-e. [DOI] [PubMed] [Google Scholar]
- 26.Tiraby G, Fox M S, Bernheimer H. Marker discrimination in DNA-mediated transformation of various Pneumococcus strains. J Bacteriol. 1975;121:608–618. doi: 10.1128/jb.121.2.608-618.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]