Abstract
We introduce the TRUST4 open-source algorithm for reconstructing immune receptor repertoires in αβ/γδ T-cells and B-cells from RNA-seq data. Compared to competing methods, TRUST4 supports both FASTQ and BAM formats, is faster and more sensitive in assembling longer, even full-length, receptor repertoires. TRUST4 can also call repertoire sequences from single-cell RNA-seq (scRNA-seq) data without V(D)J enrichment, and is compatible with both SMART-seq and 5’ 10X Genomics platforms.
T cells and B cells can generate diverse receptor (TCR and BCR) repertoires, through somatic V(D)J recombination, to recognize various external antigens or tumor neo-antigens. Upon antigen recognition, BCRs also undergo somatic hypermutations (SHMs) to further improve antigen-binding affinity. Repertoire-sequencing has been increasingly adopted in infectious disease1, allergy2, auto-immune3, tumor immuology4, and cancer immunotherapy5 studies, but it is an expensive assay and consumes valuable tissue samples. Alternatively, RNA-seq data contain expressed TCR and BCR sequences in tissues or peripheral blood mononuclear cells (PBMC). However, repertoire sequences from V(D)J recombination and SHM are different from the germline thus are often eliminated in the read mapping step.
Previously, we developed the TRUST algorithm6–8 to de novo assemble immune receptor repertories directly from tissue or blood RNA-seq data. When applied to The Cancer Genome Atlas tumor RNA-seq data, TRUST revealed significant biological insights on the repertoires of tumor-infiltrating T cells6 and B cells8, as well as their associated tumor immunity. Although less sensitive than TCR-seq and BCR-seq, TRUST is able to identify the abundantly expressed and potentially more clonally expanded TCRs/BCRs in the RNA-seq data which are more likely to be involved in antigen binding9. Recent years also see other computational methods for immune repertoire construction from RNA-seq data, such as V’DJer10, MiXCR11, CATT12 and ImRep13. These methods focus on reconstructing complementary-determining region 3 (CDR3s) with limited ability to assemble the full-length V(D)J receptor sequences, although CDR1/CDR2 on the V sequence still contribute significantly to antigen recognition and binding. For example, five out of six mutations predicted in a recent study to influence antibody affinity in the acidic tumor environment are located in CDR1 and CDR214; four out of nine positions contributing most to the 4A8 antibody binding to the SARS-COV-2 Spike protein are in CDR1 and CDR215. Therefore, algorithms that can infer the full-length immune receptor repertoires can facilitate better receptor-antigen interaction modeling.
With the advance of single-cell RNA sequencing (scRNA-seq) technologies, researchers can study immune cell gene expression and receptor repertoire sequences simultaneously. Several algorithms, such as MiXCR11, BALDR16, BASIC17, and VDJPuzzle18, have been developed to construct full-length paired TCRs or BCRs from the SMART-seq scRNA-seq platform19. In contrast to SMART-seq, droplet-based scRNA-seq platforms such as 10X Genomics20, while yielding sparser transcript coverage per cell, can process orders of magnitude more cells at a lower cost. To analyze immune repertoires using the 10X Genomics platform, currently researchers need to prepare extra libraries to amplify TCR/BCR sequences.
In this study, we redesigned the TRUST algorithm to TRUST4 with significantly enhanced features and improved performance for immune repertoire reconstruction (Figure 1a). First, TRUST4 supports fast extraction of TCR/BCR candidate reads from either FASTQ or BAM files. Second, TRUST4 prioritizes candidate read assembly by abundance and assembles all the candidate reads with partial overlaps against contigs, thus increasing the algorithm speed. Third, TRUST4 explicitly represents highly similar reads in the contig consensus, thus accommodating somatic hypermutations and improving memory efficiency (Online Methods). Fourth, TRUST4 can assemble full-length V(D)J sequences on TCRs and BCRs. Finally, TRUST4 supports repertoire reconstruction from scRNA-seq platforms without requiring the extra 10X V(DJ) amplification steps.
Figure 1.
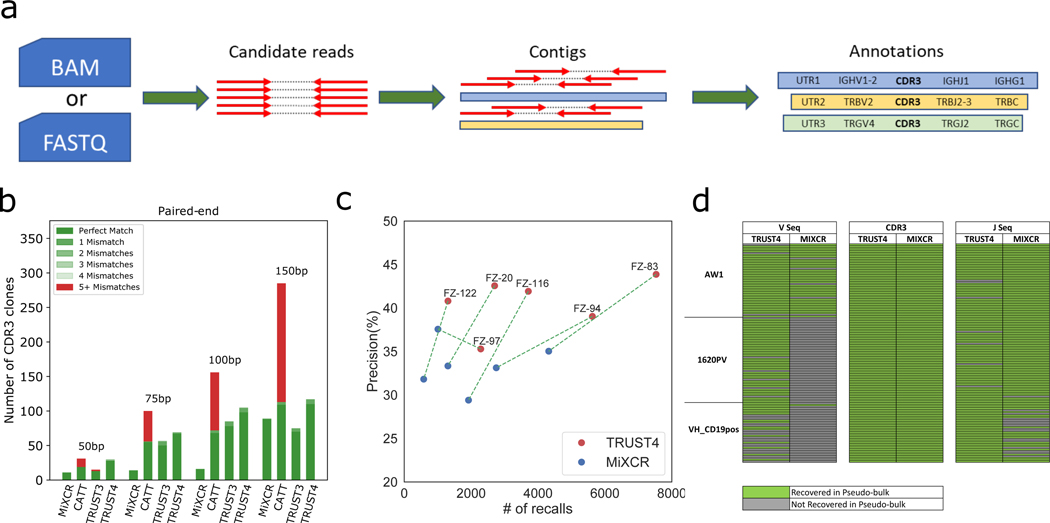
TRUST4 on bulk RNA-seq data (a) Overview of TRUST4 (b) Number of TRB CDR3s reported by MiXCR, CATT, TRUST3 and TRUST4 from the in silico RNA-seq data (c) Precision and recall number of six bulk RNA-seq samples using BCR-seq’s results as the gold standard. (d) Evaluation of the full-length V, CDR3 and J sequences assembled by TRUST4 and MiXCR on the pseudo-bulk RNA-seq by grouping SMART-seq data. Each row represents whether the cell’s sequences were recovered in the pseudo-bulk data.
We evaluated TRUST4 performance on TCR/BCR reconstruction from bulk RNA-seq using three different approaches. First, for TCR evaluation, we used the in silico RNA-seq datasets with known TRB sequences from an earlier study11. On average, TRUST4 called 281% more CDR3s than MiXCR, 22.9% more than CATT, 57.8% more than TRUST3, and maintained a zero false positive rate across different read lengths (Figure 1b, more parameter settings in Supplementary Figure 1a). Second, for BCR evaluation, we used six tumor RNA-seq samples of ~100M pairs of 150bp reads with corresponding immunoglobulin heavy chain (IGH) BCR-seq as the gold standard8. Since BCRs also have somatic hypermutation and isotype switch during clonal expansion, we required the algorithm call to match CDR3 and V, J, C (isotype) gene assignments as BCR-seq. TRUST4 showed better precision (> 18%) and sensitivity (> 74%) than MiXCR in five out of six samples (Figure 1c, more parameter settings in Supplementary Figure 2a). On the sixth sample, TRUST4 only lost 6% precision with twice the sensitivity than MiXCR (FZ-97). We note that BCR-seq and RNA-seq were conducted on different slices of the same tumor. Even two technical replicates of repertoire sequencing on the same DNA/RNA could not achieve 100% precision and sensitivity, so the performance metrics are likely to be under estimations. TRUST4 consistently assembled more IGHs across different abundance ranges reported in BCR-seq (Supplementary Figure 2b), and found twice as much as IGHs with a single mRNA copy than MiXCR. In addition, TRUST4 only used 20–25% of the time it took MiXCR to process these samples on average (Supplementary Table 1), at < 6GB memory usage on an 8-thread processor. Furthermore, TRUST4 ran directly on the FASTQ files was significantly faster than read mapping to generate BAM files followed by TRUST4 on the BAM files. Third, for base-level full-length assembly evaluation, we created a pseudo-bulk RNA-seq data by randomly picking 25M read pairs from 137 SMART-seq B cells as a testing case. To establish a gold-standard of the BCR calls, we used the 128 IGH assemblies that were consistently called by BALDR and BASIC at the single-cell level (Supplementary Figure 3a). TRUST4 and MiXCR correctly identified all the 128 CDR3s, and TRUST4 reconstructed 93 full-length IGH sequences while MiXCR only found 39 (Figure 1d). TRUST4 was able to call some BCRs with only 5,000 randomly sampled read pairs in the SMART-seq data set (18 read pairs/chain), and showed higher sensitivity than MiXCR across all abundance ranges (Supplementary Figure 3b). The high efficiency of TRUST4 allowed us to characterize immune repertoire in tumor samples, and we identified an association IgA1 B cell clonal expansion with poor prognosis in colon adenocarcinoma from The Cancer Genome Atlas (TCGA) RNA-seq samples (Online Methods, Supplementary Figure 4). We noted that IGHA1 over expression is not associated with survival, suggesting that immune repertoire analysis provides additional insights onto tumor immunity.
Next, we evaluated TRUST4 performance on the 5’ 10X Genomics scRNA-seq data on peripheral blood mononuclear cells (PBMC). For this dataset, the two separately processed T cell and B cell 10X V(D)J libraries served as the gold standards. When considering the single cells that passed the Seurat21 cell-level QC, TRUST4 made 5,091 T cell and 1,318 B cell calls respectively (Figure 2a and Supplementary Figure 5a). Among the CDR3s reported by 10X V(D)J, TRUST4 recovered 48.1% (6035/12558) of TCR CDR3s and 78.0% (1946/2494) of BCR CDR3s. The higher sensitivity of TRUST4 on BCR is due to the higher expression level of BCR in B cells (Supplementary Figure 5b). For precision, 94.6% of TCR CDR3s and 98.2% of BCR CDR3s from TRUST4 were identical with 10X V(D)J (Figure 2b). Although CellRanger_VDJ was designed for 10X V(D)J data, we tested it on 5’ 10X scRNA-seq data which has the same data format. TRUST4 found 78% more TCR CDR3s and 16% more BCR CDR3s in the cells that passed QC (Supplementary Figure 5c). In addition, TRUST4 was over 10 times faster and over twice more memory efficient than CellRanger_VDJ. Furthermore, TRUST4 also reported 83 γδT cells, which 10X V(D)J currently does not have a kit to profile. In this data, Seurat did not annotate any γδT cells, instead called 71 out of the 83 TRUST4-annotated γδT cells as CD8 T cells. Close examination of gene expression in these 83 cells revealed that they have higher δV and δC gene expression but lower CD8A or CD8B expression (Supplementary Figure 5d), supporting TRUST4’s annotation of these cells as γδT cells.
Figure 2.
TRUST4 on 5’ 10X Genomics scRNA-seq data (a) UMAP of the 5’ 10X Genomics PBMC data. (b) Number of CDR3s matched with 10X Genomics V(D)J enriched library from the cells annotated by Seurat. (c) TRUST4’s assembled V gene similarities against the reference germline V gene sequences from paired full-length IGH and IGK/IGL assemblies of the 5’ 10X Genomics NSCLC data. Each dot represents one cell.
We further tested TRUST4 on a 10X Genomics non-small cell lung cancer (NSCLC) dataset. In this case, TRUST4 called 1,241 T cells and 2,478 B cells (Supplementary Figure 6). TRUST4 assembled 142 IGH CDR3s out of the Seurat-annotated 144 plasma B cells, while 10X V(D)J only found 131. For these plasma B cells, TRUST4 also reconstructed full-length paired BCRs for 104 cells, in which we observed a high correlation on the SHM rate between IGHs and IGK/IGLs in these cells (Figure 2d, Pearson r=0.67, p-value=8e-15), suggesting coordinated SHMs on two chains during B cell division. Furthermore, TRUST4 found more somatic hypermutations on IGH than on IGK/IGL (p-value<1e-10, two-sided Wilcoxon signed-rank test), supporting the more important role of antibody heavy chain on antigen binding affinity.
In summary, TRUST4 is an effective method to infer TCR and BCR repertoires from bulk RNA-seq or scRNA-seq data. TRUST4 not only has high efficiency, sensitivity, and precision on reconstructing CDR3s, but can also assemble full-length immune receptor sequences from bulk RNA-seq data. Furthermore, TRUST4 can reconstruct immune receptor sequences at single-cell level, including γδT cells, directly from 5’ 10X Genomics scRNA-seq data without specific 10X V(D)J enrichment libraries. Our results support the advantage of the 5’ 10X Genomics scRNA-seq platform, which not only provides gene expression information but also enables computational calling of immune repertoires. TRUST4 is available open source at https://github.com/liulab-dfci/TRUST4, and can be an important method for tumor immunity and immunotherapy studies.
Methods
Methods, evaluation details and data availability are available in the online method section.
Online methods
Algorithm overview
TRUST4 reconstructs the immune repertoire in three stages: candidate reads extraction, de novo assembly, and annotation (Figure 1a).
Candidate reads extraction
TRUST4 can find the candidate TCR and BCR reads from either raw sequence files or the alignment file produced by aligners, such as STAR22, HISAT23. When the input is an alignment file, if a read or its mate aligns on the V, J, C loci, this read is added to the candidate read set. If a read is unmapped and is not a candidate based on the mate information, TRUST4 will test whether this read has a significant overlap with V, J, C genes. If so, this read and its mate are also candidate reads. When the input is raw sequence files, TRUST4 applies the significant overlap criterion to every read or read pair to find candidate reads. To identify whether a read has significant overlap with one of the V, J, C genes, TRUST4 first locates the receptor gene with the most number of k-mer hits (default: k=9) from the read. TRUST4 then computes the longest chain from these k-mers to filter incompatible hits. Lastly, if the union bases of the k-mers in the longest chain reached the threshold, TRUST4 will claim the read has a significant overlap with the gene. The threshold is max(21, read_length/5+1), so data with shorter reads has less stringent criterion. Since TRUST4 avoids alignment in the candidate reads extraction algorithm, this stage is fast even if the input data is raw sequence files.
If the data has barcode information, such as 10X Genomics scRNA-seq data, TRUST4 also corrects erroneous barcode for each candidate reads when given the whitelist. TRUST4 first builds the barcode usage distribution from the first 2 million reads before correcting. Then, for each input barcode that is not in the whitelist, TRUST4 finds all the neighbor barcode within one hamming distance in the whitelist (at most 4*barcode_length) and report the one that is the most frequent barcode in the usage distribution. If there are multiple valid neighbor barcode with the same frequency in the usage distribution, TRUST4 will correct on the base with the lowest FASTQ quality.
De novo assembly
When assembling the candidate reads into immune receptor sequences, TRUST4 adopts the read overlap scheme. Cells like plasma B cells can generate thousands of reads for each recombined receptor gene, so comparing every pair of reads to construct the overlap graph as in previous versions of TRUST is inefficient. TRUST4 implements a greedy extension approach by aligning the candidate read to existing contigs one by one. To perform alignment, TRUST4 builds an index for all the k-mers in the contigs and applies the seed-extension paradigm to identify the alignments. TRUST4 deems a read overlaps with a contig if they have a highly similar (90% for BCR, 95% for TCR) alignment block containing at least 31bp exact matches and the unaligned bases of the read are outside of the contig. Based on the overlaps, TRUST4 will update the contigs with following rules: (1) If a read partially overlaps with one contig, TRUST4 extends this contig; (2) if a read partially overlaps several contigs, TRUST4 merges corresponding contigs; (3) If a read does not overlap with any existing contigs, TRUST4 creates a new contig with this read’s sequence. When processing the reads, TRUST4 prioritizes the reads coming from highly expressed TCRs/BCRs. To achieve this, TRUST4 first counts the frequency of k-mers (21-mer by default) across all the candidate reads. If a read comes from a highly expressed receptor sequence, all of its k-mers would have high frequencies in the data. Therefore, the minimum frequency of a read’s k-mers can roughly indicate the abundance of the gene. TRUST4 then sorts the read based on the minimum k-mer frequency rule. The ordering of reads is equivalent to picking the most frequent k-mer as the start point in the de Bruijn graph-based transcriptome assembler Trinity24.
TRUST4 clusters the reads with somatic hypermutations into the same contig by representing a contig as the consensus of assembled reads. Each position in the contig records four weights as how many reads have the corresponding nucleotide for that position. The consensus means the nucleotide for a position is the nucleotide with highest weight, and the index for alignments stores the k-mers of the consensuses. The read alignment takes the weights into account to tolerate the somatic hypermutations in BCRs. For example, for a position, if nucleotides A and T on the contig have high weights, it is a match if the read has nucleotide A or T. Therefore, reads with different somatic hypermutations can align to the same contig, which avoids creating redundant contigs.
If the input data is paired-end, TRUST4 will use the mate-pair information to extend the contigs. In the first round of contig assembly, due to the read sorting and greedy extension, a contig for the abundant recombined gene attracts all the reads from the same V, J and C genes even though these reads come from different recombination. The mate-pair information fixes this issue by reassigning the reads to the appropriate contigs. The reassignment will extend the contigs and update the position weights in the affected consensus. When the input data is SMART-seq, since there is no need to perfect the assemblies for low abundant sequences in a cell, TRUST4 can skip the extension to reduce running time.
When the input contains barcode information, TRUST4 will assign the read barcode to the contig when creating a new contig, and a read can only align to the contigs with the read’s barcode. As a result, two identical reads with different barcodes will change different sets of contigs. Furthermore, the read-contig overlap criterion is relaxed and requires 17bp instead of 31bp exact matches in the alignment.
Annotation
TRUST4 aligns the assembled contigs to the sequences from the international ImmunoGeneTics (IMGT) database25 to identify the V, J, C genes. IMGT database curates the sequences for V, D, J and constant genes, and is widely used to annotate BCR and TCR sequences, such as in previous TRUST versions and MiXCR. Besides the sequences, IMGT also annotates the start position of CDR3 in the V gene (104th amino acids of the V gene in IMGT coordination). IMGT also defines the end position of CDR3 as amino acid W/F in the amino acid motif W/FGxG in the J gene. TRUST4 determines the CDR3 coordinate based on these IMGT conventions after identifying V and J genes. If the contig is too short to identify the V gene, TRUST4 locates the CDR3 start position as amino acid C in the motif YYC by testing all the open reading frames.
In the final step of annotation, TRUST4 retrieves the somatic hypermutated CDR3s and estimates the CDR3 abundances. If a read fully covers the CDR3 on a contig and the CDR3 sequence from the read is different from the consensus, TRUST4 will report the CDR3 from the read. If there is no such read, TRUST4 directly reports the consensus CDR3 sequence. In the abundance estimation, if reads partially overlap with the CDR3, it could be compatible with several different CDR3 sequences. Therefore, TRUST4 applies the Expectation-Maximization algorithm26 similar in RSEM27 to distribute reads’ count to their compatible CDR3s iteratively. For TCR, TRUST4 filters the CDR3 whose abundance is less than 5% of the most abundant CDR3 from the same contig to avoid sequencing errors.
For the CDR3s that have only start or end positions determined in the contigs, TRUST4 reports them as a partial CDR3s and tries to extend the partial TCR CDR3s as in MiXCR. For an example of missing start position, the extendable partial CDR3 must overlap with the identified V gene in the contig but could not reach the start position. This scenario could happen when the V gene is identified through mate-pair information. TRUST4 then fills the missing sequences with germline sequences of the V gene to complete the partial CDR3. In scRNA-seq, TRUST4 also utilizes information across all cells to extend partial TCR CDR3s. For two cells A, B with the same V and J genes on both chains, cell B can extend its partial CDR3 if B has a complete CDR3 identical to A’s corresponding complete CDR3 and B’s partial CDR3 is a substring of A’s corresponding complete CDR3.
Sequence data
We tested TRUST4 on in silico and real data. The in silico bulk RNA-seq data for evaluating TCRs was generated using the scripts from MiXCR11, where repseqio (https://github.com/repseqio/repseqio) and ART28 generated the simulated TRB and RNA-seq data. As a result, each of the in silico RNA-seq samples contained 1,000 read fragments randomly from 1,000 recombined TRBs. To evaluate BCR reconstruction, we used six lung cancer RNA-seq data and its pairing BCR-seq data from our previous manuscript8. iRepertoire processed the BCR-seq data, and the results were the gold standard for the evaluation. For SMART-seq evaluation, we used three SMART-seq datasets from BALDR: AW1, 1620PV (AW2-AW3), VH_CD19pos. For the pseudo-bulk RNA-seq data, we firstly added a pseudo-mate for the 1620PV’s single-end data with a sequence of one nucleotide N. We then randomly selected 25 million read pairs across all the cells of these three samples to create the pseudo-bulk RNA-seq. In the end, 56%, 33% and 11% of the pseudo-bulk RNA-seq data were from AW1, 1620PV and VH_CD19pos respectively. We applied the same procedure to generate psuedo-bulk samples with fewer read pairs (12million, 6million, …, 2.5K, 1K). The 10X Genomics scRNA-seq data and 10X V(D)J data were downloaded from 10X Genomics website.
Performance evaluation
All the methods were tested with their default parameters without explicit clarification. The BAM files as the input for TRUST4 were generated by STAR v2.5.3a. We used MiXCR v3.0.12, CATT with GitHub commit id 0e7b462, TRUST3 v3.0.3, BALDR with GitHub commit id e865b45 and BASIC v1.5.1 in this study. All the evaluations in this work were at the nucleotide level. For example, the match of CDR3 of TRUST4 and BCR-seq gold standard meant their nucleotide sequences were identical. TRUST4 could report both partial and complete CDR3, and we only considered complete CDR3s in the evaluations.
In the TCR evaluation with in silico RNA-seq data, the evaluation criteria were based on the scripts from MiXCR’s manuscript11, We added read length 150bp and ran MiXCR with default parameters. In MiXCR’s original manuscript, the authors used the option “--badQualityThreshold 0” for higher sensitivity (MiXCR_0), and TRUST4 still found about 8% more CDR3s than MiXCR_0 on average (Supplementary Figure 1a). Furthermore, TRUST4 with input from FASTQ and BAM files showed almost identical results, which demonstrated the efficiency of the candidate extraction method. TRUST4 was also the most or one of the most sensitive methods on assembling CDR3s for the TRB chains with different number of reads (Supplementary Figure 1b).
In the bulk RNA-seq data, we mainly evaluated the performance of reconstructing BCR heavy chains, including V, J, C gene assignments and CDR3 sequences. We considered gene assignments in addition to CDR3 sequences in the evaluation because that IGHs had different C genes as isotypes, such as IgM, IgG1, IgA1, and were critical in determining antibodies’ functions. Since CATT could not report C gene and TRUST3 only focused on CDR3 assembly, we omitted CATT and TRUST3 in this evaluation. For the match of V, J genes, we ignored the allele id. For example, if the V gene was annotated as IGHV1–18*01, we regarded it as IGHV1–18. In the evaluation, we excluded the assemblies missing V, J or C gene from TRUST4 and MiXCR. The IGH abundances reported by TRUST4 had a better correlation with the corresponding abundances in BCR-seq than MiXCR’s (Pearson r=0.57 vs. 0.53 on average, Supplementary Figure 2a). We further checked the precision-recall curve by ranking inferred IGHs by abundances (top 100, 500, 1000, …), and TRUST4 consistently outperformed MiXCR across different thresholds (Supplementary Figure 2a). On this real data, MiXCR_0 did not outperform MiXCR as in the in silico data, suggesting the parameter was not effective on real data. TRUST4 with FASTQ and BAM input still showed identical performance across six samples in this real data evaluation. We further evaluated the performance only on CDR3 sequences, which included the results from TRUST3 and CATT. TRUST4 showed the highest sensitivity consistently across all 6 samples, and reported 11% more correct CDR3s than MiXCR_0, the second most sensitive method, with similar precision on average.
The evaluations with SMART-seq data focused on whether the methods could reconstruct all the nucleotides in the variable domain. If the assembled V, J sequences were shorter than the genes’ lengths in the IMGT database, we would regard it as unreconstructed. The match of V or J sequences means the nucleotide bases were the same for the regions annotated as V or J genes. In other words, we ignored the bases before or after V, J genes. In addition to the psuedo-bulk RNA-seq data from the three samples, we ran TRUST4 on original cell level data and compared with BALDR and BASIC on all three samples. We picked the top abundant heavy chain and light chain from TRUST4, and they were identical with at least one of the BALDR and BASIC on 272 out of 274 chains (Supplementary Figure 3). The comparison result indicated that TRUST4 could effectively reconstruct the immune repertoire from SMART-seq scRNA-seq data.
For the evaluation with 10X Genomics data, we used TCR library and IG library results from 10X Genomics Immune profiling (10X V(D)J) as the gold standard. Since the computational software CellRanger_VDJ might report multiple CDR3s for a cell, we regarded the most abundant CDR3 as the true CDR3 for a chain, and the less abundant CDR3s as secondary. TRUST4 took the BAM file generated by CellRanger as input, which included the barcode information in the field “CB”. TRUST4 also took FASTQ files as input, and corrected the erroneous barcode based on the whitelist provided in CellRanger package. TRUST4 with FASTQ input reported almost identical results as with BAM input (Supplmentary Figure 5c). Even though CellRanger_VDJ (v3.1.0) was designed for 10X V(D)J data, we ran it on the 10X 5’ scRNA-seq data using IMGT sequences as reference with 8 cores. In our analysis for full-length assemblies, the somatic hypermutation rate was represented by the proportion of matched bases (similarity) between the assembled V genes and the germline sequences (Figure 2c). If there were many somatic hypermutations, the similarity would be low. Besides the 5’ scRNA-seq data, we also evaluated TRUST4 on the 3’ 10X Genomics PBMC data and only 335 cells had reconstructed CDR3s (Supplementary Figure 7). We used the LM22 marker genes from CIBERSORT29 to determine the cell types.
TRUST4 on TCGA-COAD RNA-seq samples
We explored the immune repertoire features on the 466 colon adenocarcinoma (COAD) RNA-seq samples in The Cancer Genome Atlas (TCGA) cohorts. To reduce the effects of somatic hypermutated CDR3s, we first clustered highly similar CDR3 nucleotide sequences of the same length and with the same V, J gene assignments reported from TRUST4. We picked the similarity cutoff as 0.8 by comparing the similarities distribution among the pairs of CDR3s within (intra-patient) and between (inter-patient) samples, where the inter-patient distribution can be regarded as the background random CDR3 pair similarity (Supplementary Figure 4a). Therefore, we defined the clonotype for TCR as the CDR3 sequence and the clonotype for BCR as the cluster with the same V, J gene assignments and similar CDR3 sequences. Although TRB and IGH clonalities were positively correlated with their expression respectively (Spearman r=0.346 for TRB, r=0.085 for IGH), they contained additional information on TCR and BCR clonal expansion (Supplementary Figure 4b). The expression for a chain is computed by the sum of transcripts per millions (TPM) obtained from TCGA cohorts on the constant genes of a chain. We defined clonality as 1-(normalized Shannon entropy) based on the clonotype definition above.
We identified that IgA1 antibody clonal expansion was related to patient survival in colon adenocarcinoma. Unlike in melanoma, where IgG1 and IgA expressions and abundance fractions were positive and negative associated with survival time respectively11, we did not observe such association of survival time in COAD (Supplementary Figure 4c). However, higher clonality of IgA1 B cells was correlated with significantly shorter survival time (p-value=8.1e-5, hazard ratio 9.14 by Cox proportional hazards regression corrected by age), supporting the immunosuppressive property of IgA antibodies30. We hypothesize that the clonal expansion of IgA1 B cells could be related to gut microbiota31, and future work is needed to elucidate the mechanisms.
Data availability
The original scripts for generating and evaluating the in silico RNA-seq data are available at https://github.com/milaboratory/mixcr-rna-seq-paper.
The six bulk RNA-seq samples for BCR evaluation are available in the SRA repository PRJNA492301, and their matched iRepertoire data are available at https://bitbucket.org/liulab/ng-bcr-validate/src/master/iRep.
SMART-seq data is available in the SRA repository SRP126429.
10X Genomics scRNA-seq data is available at https://support.10xgenomics.com/single-cell-vdj/datasets/3.1.0/vdj_nextgem_hs_pbmc3, https://support.10xgenomics.com/single-cell-vdj/datasets/2.2.0/vdj_v1_hs_nsclc_5gex and https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.1.0/5k_pbmc_protein_v3_nextgem.
Code availability
TRUST4’s source code is available at https://github.com/liulab-dfci/TRUST4.
Evaluation codes in this work are available at https://github.com/liulab-dfci/TRUST4_manuscript_evaluation.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgments
We thank Drs. Bo Li and Chenfei Wang for the helpful discussions. We also acknowledge the funding sources from NIH U01CA226196 and U24CA224316 for supporting this work. The study used some data generated by the TCGA Research Network which is not otherwise cited: https://www.cancer.gov/tcga.
Footnotes
Competing interests
X.S.L. is a cofounder, board member, SAB, and consultant of GV20 Oncotherapy and its subsidiaries, SAB of 3DMedCare, consultant for Genentech, and stockholder of AMGN, JNJ, MRK, PFE, and receives sponsored research funding from Takeda and Sanofi. X.H. conducted the work while being a postdoc fellow and is currently a full time employee of GV20 Oncotherapy.
References
- 1.Lee J. et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22, 1456–1464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiyotani K. et al. Characterization of the B-cell receptor repertoires in peanut allergic subjects undergoing oral immunotherapy. J Hum Genet 63, 239–248 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Liu S. et al. Direct measurement of B-cell receptor repertoire’s composition and variation in systemic lupus erythematosus. Genes Immun 18, 22–27 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Kurtz DM et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 125, 3679–3687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riaz N. et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 171, 934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B. et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet 48, 725–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B. et al. Ultrasensitive detection of TCR hypervariable-region sequences in solid-tissue RNA-seq data. Nat Genet 49, 482–483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X. et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat Genet 51, 560–567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y. et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mose LE et al. Assembly-based inference of B-cell receptor repertoires from short read RNA sequencing data with V’DJer. Bioinformatics 32, 3729–3734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolotin DA et al. Antigen receptor repertoire profiling from RNA-seq data. Nat Biotechnol 35, 908–911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S-Y, Liu C-J, Zhang Q. & Guo A-Y An ultrasensitive T-cell receptor detection method for TCR-Seq and RNA-Seq data. Bioinformatics (2020). [DOI] [PubMed] [Google Scholar]
- 13.Mandric I. et al. Profiling immunoglobulin repertoires across multiple human tissues using RNA sequencing. Nat. Commun. 11, 3126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulea T. et al. Structure-based engineering of pH-dependent antibody binding for selective targeting of solid-tumor microenvironment. MAbs 12, 1682866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi X. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upadhyay AA et al. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med 10, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canzar S, Neu KE, Tang Q, Wilson PC & Khan AA BASIC: BCR assembly from single cells. Bioinformatics 33, 425–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzetto S. et al. B-cell receptor reconstruction from single-cell RNA-seq with VDJPuzzle. Bioinformatics 34, 2846–2847 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hagemann-Jensen M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol 38, 708–714 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Langmead B. & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabherr MG et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefranc M-P IMGT, the International ImMunoGeneTics Information System. Cold Spring Harb Protoc 2011, 595–603 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Dempster AP, Laird NM & Rubin DB Maximum Likelihood from Incomplete Data Via the EM Algorithm. J R Stat Soc Ser. B Stat Methodol 39, 1–22 (1977). [Google Scholar]
- 27.Li B. & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Li L, Myers JR & Marth GT ART: a next-generation sequencing read simulator. Bioinformatics 28, 593–594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV & Chudakov DM B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 20, 294–307 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Bunker JJ & Bendelac A. IgA Responses to Microbiota. Immunity 49, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original scripts for generating and evaluating the in silico RNA-seq data are available at https://github.com/milaboratory/mixcr-rna-seq-paper.
The six bulk RNA-seq samples for BCR evaluation are available in the SRA repository PRJNA492301, and their matched iRepertoire data are available at https://bitbucket.org/liulab/ng-bcr-validate/src/master/iRep.
SMART-seq data is available in the SRA repository SRP126429.
10X Genomics scRNA-seq data is available at https://support.10xgenomics.com/single-cell-vdj/datasets/3.1.0/vdj_nextgem_hs_pbmc3, https://support.10xgenomics.com/single-cell-vdj/datasets/2.2.0/vdj_v1_hs_nsclc_5gex and https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.1.0/5k_pbmc_protein_v3_nextgem.
Code availability
TRUST4’s source code is available at https://github.com/liulab-dfci/TRUST4.
Evaluation codes in this work are available at https://github.com/liulab-dfci/TRUST4_manuscript_evaluation.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.