Abstract
A glucuronan lyase extracted from Sinorhizobium meliloti strain M5N1CS was purified to homogeneity by anion-exchange chromatography. The purified enzyme corresponds to a monomer with a molecular mass of 20 kDa and a pI of 4.9. A specific activity was found only for polyglucuronates leading to the production of 4,5-unsaturated oligoglucuronates. The enzyme activity was optimal at pH 6.5 and 50°C. Zn2+, Cu2+, and Hg2+ (1 mM) inhibited the enzyme activity. No homology of the enzyme N-terminal amino acid sequence was found with any of the previously published protein sequences. This enzyme purified from S. meliloti strain M5N1CS corresponding to a new lyase was classified as an endopolyglucuronate lyase.
Bacterial polysaccharides as well as plant polysaccharides can be degraded by specific glycosidases corresponding either to hydrolases or to lyases. Among the polyanionic polymers, some of them (i.e., alginate) are exclusively degraded by lyases (35, 40, 42). The lyase cleaving mechanism consists of a β-elimination, in which a general base-catalyzed abstraction of the proton at C-5 of a uronic acid occurs. A transfer of electrons from the carboxyl group to form a double bond between C-4 and C-5 results in the elimination of the 4-O-glycosidic bond and in the formation of 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid. This reaction leads to the formation of an unsaturated uronate at the newly generated, nonreducing end. Oligosaccharides from a degree of polymerization of 2 to 3 or 5 can be obtained by polysaccharide degradation with lyases (3, 20, 40). To date, lyase activities on various polymers have been described as alginate (16, 32, 35), gellan (41), hyaluronan (39), ulvan (25), and xanthan (15). The tested lyases were found in many organisms such as marine gastropods (19) or fungi (36, 45) as well as in many bacteria (14, 15, 31) and bacteriophages (1, 4). The enzyme localization in the producing organisms may be in the cytosol (28, 36) and the periplasmic space (22), as for alginate lyase from Azotobacter species, or in extracellular fractions, as for a Bacillus circulans lyase (26). Lyases present possible applications in the medical field, such as, for example, the alginate lyase, which promotes diffusion of antibiotics through extracellular polymers produced by the pathogenic Pseudomonas sp. (17). Due to potential applications, studies concerning polysaccharide lyases have been increasing.
The Sinorhizobium meliloti mutant strain M5N1CS (NCIMB 40472) (7), which induces the formation of effective nodules on alfalfa roots (12), produces a glucuronan (6, 18) corresponding to high-molecular-weight (HMW) and low-molecular-weight (LMW) (1→4)-β-d-polyglucuronic acid partially acetylated at the C-2 and/or C-3 position. The detection of LMW glucuronan containing a 4-5 unsaturated glucuronic residue at the nonreducing end (29, 30) led us to suspect a polymer degradation by a glucuronan lyase.
In order to produce oligoglucuronans, a lyase degrading various acetylated and nonacetylated glucuronans was purified and characterized.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
For glucuronan lyase production, S. meliloti strain M5N1CS (NCIMB 40472) (6) was aerobically cultured in a 2-liter Erlenmeyer flask containing 1 liter of tryptone yeast (TY) medium (pH 7.2) (2) supplemented with sucrose (1% [wt/vol]) (TYS), during 75 h at 30°C on a rotary shaker (100 rpm). The inoculum was 100 ml of a S. meliloti M5N1CS suspension in the same TYS medium, grown 20 h at 30°C under agitation (100 rpm). A 10-ml bacterial suspension grown 20 h at 30°C was used to inoculate the 100-ml TYS medium.
Glucuronan production.
S. meliloti strain M5N1CS was cultivated at 30°C in a 6-liter reactor (LSL Biolaffite, Saint-Germain-en-Laye, France) containing 4.5 liters of Rhizobium Complete medium (5) supplemented with sucrose (1% [wt/vol]) (RCS). The inoculum was 0.5 liter of RCS in a 1-liter Erlenmeyer flask inoculated with S. meliloti M5N1CS. After 20 h of incubation on a rotative shaker (100 rpm) at 30°C, the inoculum was transferred to the reactor and incubated as previously described (29), with or without addition of 0.15% (wt/vol) MgSO4 · 7H2O for the production of mainly 3-O-acetylated (standard) or 2,3-di-O-acetylated (highly acetylated) glucuronans.
Glucuronan isolation, purification, and characterization.
Two HMW glucuronans, one which was standard (mainly 3-O-acetylated) and one which was highly acetylated (mainly 2,3-di-O-acetylated), were produced during two 75-h fermentations. Then the broths were centrifuged (33,900 × g, 40 min, 20°C), and the HMW polysaccharides were concentrated from the cell-free broth by tangential ultrafiltration on a 105 normal-molecular-weight cutoff (NMWCO) polysulfone membrane (0.1 m2) (Sartorius, Göttingen, Germany). The concentrates were diluted with 1 volume of distilled water and ultrafiltered as above; this step was repeated five times, and then the last concentrated products were dried under vacuum. The substitution degree determined by 1H nuclear magnetic resonance (NMR) spectroscopy was one acetate for 1.3 residue and one acetate for 0.7 residue for the standard and highly acetylated glucuronans, respectively (8). A fraction of the standard glucuronan was completely deacetylated by 2 M KOH during 12 h at 50°C (pH 12). The complete deacetylation of glucuronan was confirmed by 1H NMR spectroscopy.
Preparation of a crude enzyme fraction.
All steps were carried out at 4°C. At the end of the incubation (75 h) the bacterial suspension was centrifuged (13,880 × g, 30 min), and the pellet was collected and suspended in 10 mM Tris-HCl buffer (pH 8.0) (final volume, 80 ml). The bacterial suspension was disrupted with an ultrasonicator (Vibra-Cell model VCX 600 W; Danbury, Conn.) at 20 kHz (13-mm-diameter standard probe) for 30 min, and then the mixture was centrifuged at 50,000 × g for 1 h. The supernatant, called crude enzyme fraction, was collected and stored at −80°C.
Enzyme purification.
The enzyme was purified at room temperature, through a two-step procedure using low-pressure liquid chromatography (ProTeam LC System 210; Isco, Lincoln, Nebr.).
(i) Step 1.
The purification was performed on a ready-to-use anion exchanger Sartobind membrane adsorber (MA) Q100 (Sartorius, Goettingen, Germany), which had first been equilibrated with 10 mM Tris-HCl buffer (pH 8.0). Ten milliliters of crude enzyme fraction was loaded onto the Sartobind cartridge. The Sartobind MA unit was first washed with the equilibration buffer (112 ml) with a flow rate of 4.3 ml/min; then the eluent consisted of a KCl gradient from 0 to 50 mM in 30 s and a 50 mM KCl buffer (69 ml) at the same flow rate as previously. A KCl buffer (at least 100 mM) was used. Fractions (8.6 ml) were collected with a Foxy 200 collector (Isco). Enzymatic activity was tested on each fraction as described below. Fractions presenting a glucuronan lyase activity were pooled and ultrafiltered in a stirred Amicon cell (Beverly, Mass.) through a 104 NMWCO polyethersulfone membrane (Sartorius). The retentate dispersed in the buffer used for step 2 (10 mM Tris-HCl [pH 7.2]) was ultrafiltered as previously (this step was repeated twice) and concentrated to a final volume of 1.3 ml.
(ii) Step 2.
The concentrated enzyme solution from step 1 was applied as described above to the Sartobind MA, which had first been equilibrated with a 10 mM Tris-HCl buffer (pH 7.2). The Sartobind MA unit was then washed with the equilibration buffer (129 ml), and a KCl gradient (ranging from 0 to 30 mM in 30 s) was applied. Proteins were eluted with a 30 mM KCl buffer (77 ml). The flow rate was as above. Fractions presenting a glucuronan lyase activity were pooled, washed, concentrated by ultrafiltration to a final volume of 2 ml, and stored at −80°C.
Enzyme assays.
During the glucuronan lyase purification steps, the enzyme was tested on the reaction mixture consisting of 1 ml of 10 mM Tris-HCl buffer (pH 7.2), 0.5 ml of glucuronan in 10 mM Tris-HCl buffer (pH 7.2) (0.2% [wt/vol]) and 0.5 ml of enzyme solution. A fraction aliquot, which was immediately frozen, (−80°C) corresponded to the standard (t = 0 h). The remaining reaction mixture was incubated 15 h at 30°C on an HS 500 shaker (Jankel & Kunkel, Staufen, Germany). The polysaccharide degradation was determined by quantitative analysis of the reducing sugar, using the 2,2′-bicinchoninate method (43). The lyase activity was confirmed by measuring A235 arising from the double bond in the reaction product, on a Uvikon 930 spectrophotometer (Kontron, Montigny Lebretonneux, France). One unit (U) of the enzyme activity was defined as an increase of 1 unit per h in the absorbance at 235 nm. Specific activity was expressed as units per milligram of protein.
The glucuronan lyase relative activity was determined on 60 μl of the deacetylated glucuronan (1.3 g/liter) in 50 mM KH2PO4 buffer (pH 6.5) mixed to 20 μl of the purified glucuronan lyase. Then, the mixture was incubated at room temperature for 2 h. Analytical chromatographic separation of the oligo-uronides obtained by enzymatic hydrolysis of deacetylated glucuronan was performed on a Millipore Waters Model 510 high-performance liquid chromatography apparatus, using two high-performance gel filtration columns in sequence: an OHpak B-805 (8 by 500 mm) from Shodex (Tokyo, Japan) and a TSKgel G3000PW 10 μm (7.5 by 300 mm) from Interchim (Montluçon, France). Chromatography was run isocratically at room temperature, with 0.1 M NaNO3 as eluent (1 ml/min). Detection was performed online with a R-410 Waters differential refractometric detector. Chromatograms were analyzed and recorded with a C-R4A chromatopac integrator (Shimadzu). Glucuronan lyase relative activity was estimated by comparison of chromatograms obtained with the substrate alone and after enzymic hydrolysis.
Protein determination.
Protein in Sartobind MA fractions was routinely monitored by measuring A280 with a UA-6 UV detector (Isco). The amounts of protein in the crude and purified enzyme fractions were estimated with a bicinchoninic acid protein assay; bovine serum albumin was used as a standard (38).
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in a vertical slab unit (Mini Protean II Slab Cell; Bio-Rad, Richmond, Calif.) according to the procedure described by Laemmli (23). The separating gel (18%) was overlaid with a stacking gel (4%). The samples were incubated in 50 mM Tris-HCl (pH 7.5), with 1% (wt/vol) SDS and 1% (vol/vol) 2-mercaptoethanol. Protein samples (from 28.5 to 107 μg) and a standard protein mixture (from 14,400 to 97,400 Da) (Bio-Rad) were applied to the gel. The electrophoresis was performed for 15 h at room temperature at 80 V. Gels were stained with Coomassie brilliant blue R-250 (Bio-Rad).
IEF.
Analytical isoelectric focusing (IEF) was performed on a 5% IEF polyacrylamide gel plate (Bio-Rad) with a broad-range ampholyte (pI ranging from 3 to 10). A protein calibration mix with pI values from 4.45 to 9.6 was used. The IEF was performed for 1 h at room temperature at 100 V, and then 250 V was applied during 1 h, and at least 500 V was applied during 30 min. Proteins were stained with an IEF gel staining solution (Bio-Rad).
Determination of the N-terminal sequence.
N-terminal amino acid sequence of the purified lyase was determined by Edman degradation (10) with a Procise 494 protein sequencing system (Applied Biosystems, Division of Perkin-Elmer).
Fractionation of oligoglucuronates.
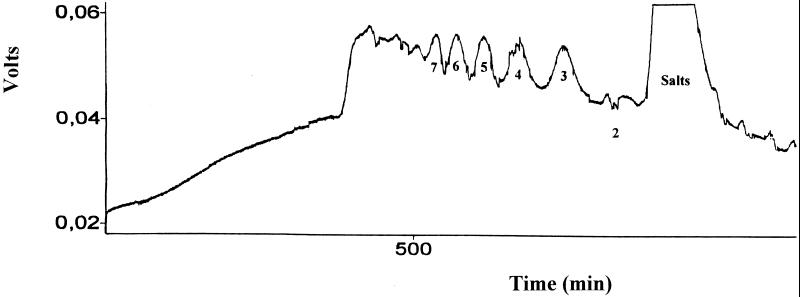
Oligoglucuronates obtained by enzymatic degradation were size fractionated by preparative gel filtration on a glass chromatographic column (2.5 by 100 cm) packed with Bio-Gel P-6 (Bio-Rad), eluted with a 50 mM NaNO3 solution, with a flow rate of 50 ml/h. Detection was achieved with a R-403 Waters differential refractometer. Fractions (10 ml) were collected with a 201–202 model Gilson collector. Fractions belonging to a same peak were combined, concentrated, desalted by high-performance liquid chromatography on a Toyopearl HW 40F/50F (Interchrom) column, and freeze-dried.
NMR.
1H NMR analyses were performed at 85°C with an AC-300 Bruker (Bruker Spectrospin, Wissembourg, France) Fourier transform spectrometer with a 5-mm 1H 13C dual probe. To equilibrate exchangeable protons, the different samples (20 mg) were freeze-dried and then dispersed in 500 μl of D2O. 1H NMR spectra were obtained using a spectral width of 300 MHz, a 16K data-block size, and a pulse sequence of 8 μs; 16 scans with acquisition of 2.73 s were accumulated. The H2O signal was presaturated using the standard Bruker Presat sequence, with a delay of 3 s and a decoupler power of 20 dB at low range. The polymerization degree was estimated by comparison of the integral of the H-1 or H-4 signals of the unsaturated unit to the integral of the other H-1 signals (9).
RESULTS
Glucuronan lyase purification.
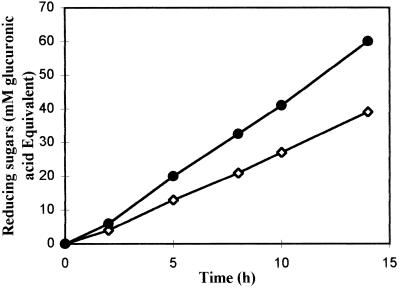
Four liters of a 75-h S. meliloti M5N1CS culture broth was centrifuged (13,880 × g, 30 min). The pellet was suspended in 80 ml of a 10 mM Tris-HCl buffer (pH 8.0) and sonicated. After centrifugation (50,000 × g, 1 h) the supernatant corresponding to the crude enzyme fraction was collected and tested on native and deacetylated glucuronans. Polymer degradation was monitored using the reducing sugar assay (43) (Fig. 1). We noted that the deacetylated glucuronan was the better substrate for lyase activity. As the thiobarbituric acid method (44) routinely used for lyase activity studies leads to the formation of glucuronan precipitates, in this study, the specific lyase activity was determined by UV spectrophotometry (λ = 235 nm). We noted that the specific activity on the crude enzyme fraction was 4 × 10−2 U/mg.
FIG. 1.
Evolution of reducing sugar concentration during incubation of S. meliloti M5N1CS cell extracts with deacetylated (—●—) and native (—◊—) glucuronan fractions (expressed in millimolar glucuronic acid equivalents). Each data point represents the mean of at least three values, which did not vary more than 15% from the mean.
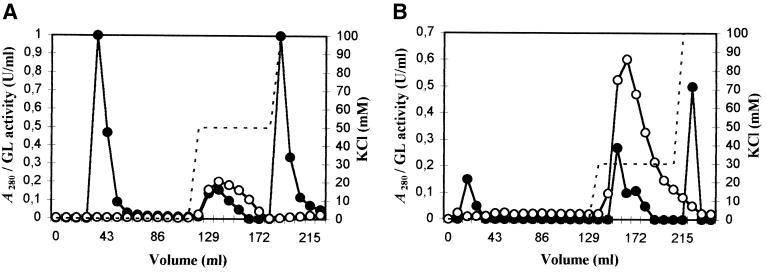
The enzyme purification was performed first with a Sartobind MA Q100 unit by injection of the whole crude enzyme fraction (176 ml) through injections of 10 ml. In the first purification step, the Sartobind MA was washed with 112 ml of the 10 mM Tris-HCl buffer (pH 8.0). Then, the elution was performed with KCl buffers (50 and 100 mM). Three fractions absorbing at 280 nm were identified from the crude enzyme fraction (Fig. 2A): the first one eluted with the equilibration buffer, the second one eluted with the 50 mM KCl buffer, and the third eluted with the 100 mM KCl buffer. The glucuronan lyase activity was detected only in the fraction eluted with 50 mM KCl. The fraction from 120.4 to 137.6 ml was collected, desalted, and concentrated. The activity of the glucuronan lyase chromatographied fraction (noted GLC1) was increased 23-fold.
FIG. 2.
Elution profiles on a Sartobind Q100 MA of the crude enzyme fraction from S. meliloti M5N1CS (step 1) (A) and from GLC1 fraction (step 2) (B). Membranes were equilibrated with 10 mM Tris-HCl buffer at pH 8 (A) and pH 7.2 (B). Elution was carried out first with the equilibration buffer, then with 50 mM KCl buffer for step 1 (A), and then with 30 mM KCl buffer for step 2 (B). Glucuronan lyase (GL) activity (—○—), KCl concentration (–––), and absorbance at 280 nm (—●—).
In order to achieve the glucuronan lyase purification, the GLC1 fraction was applied on the same Sartobind MA unit as above. In the second purification step, the membrane was washed with 129 ml of 10 mM Tris-HCl buffer (pH 7.2) and eluted with 30 and 100 mM KCl buffers (Fig. 2B). The active fraction (eluted with the 30 mM KCl buffer from 146.2 ml to 163.4 ml) was desalted, concentrated by ultrafiltration to a final volume of 2 ml, and stored at −80°C. At this level of purification, the activity of the glucuronan lyase fraction (noted GLC2) was purified 394-fold compared to the crude enzyme fraction, with a yield of 2.5%. We noted that the glucuronan lyase activity remained constant in samples stored at −80°C for several weeks (data not shown).
Basic features of purified glucuronan lyase.
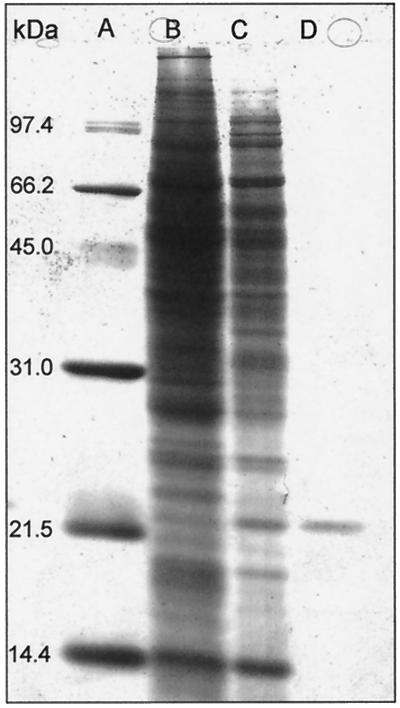
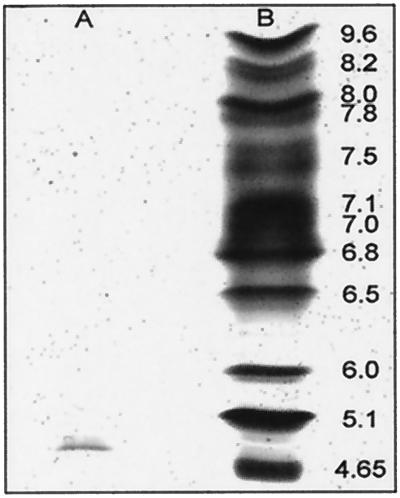
The purified enzyme preparation was examined by SDS-PAGE in denaturing conditions (Fig. 3). The GLC2 fraction was identified as a single protein band (lane D), while the GLC1 fraction (lane C) revealed numerous protein bands. The purification procedures applied to the bacterial extracts revealed to be selective for the purification of a 20-kDa protein exhibiting a glucuronan lyase activity. The GLC2 fraction analyzed by IEF revealed a single band with pI estimated at 4.9 (Fig. 4, lane A).
FIG. 3.
SDS-PAGE of glucuronan lyase fractions at different purification steps, after staining with Coomassie brilliant blue R-250. Lanes: A, molecular mass marker proteins; B, crude enzyme fraction; C, GLC1 fraction; D, GLC2 fraction.
FIG. 4.
Analytical IEF of purified glucuronan lyase from S. meliloti M5N1CS. Lanes: A, purified glucuronan lyase; B, pI marker proteins. The proteins were stained with the IEF gel staining solution.
Effects of environmental conditions.
The enzyme activity was investigated under different buffer conditions at room temperature. The lyase activity tested in KH2PO4 buffers (pH 6.5), ranging from 10 to 200 mM (data not shown), was quite similar to those obtained in glycine-sodium hydroxyde and potassium phosphate buffers at pH ranging from 7 to 8. The 50 mM KH2PO4 buffer at pH 6.5 was used in all further experiments. The glucuronan lyase activity tested between 6 and 79°C in a 50 mM KH2PO4 buffer revealed a maximal relative activity at 50°C. This activity was reduced by one-half at 52°C. Lyase activity tested on heated enzyme fractions in the 50 mM KH2PO4 buffer indicated that the enzyme was stable at up to 50°C and decreased at higher temperatures. The enzyme was completely inactivated at 67°C. The lyase activity was tested on enzyme samples (in 50 mM KH2PO4 buffer at pH 6.5) incubated previously with 1 mM metal salts for 2 h at 50°C. Under the conditions tested, no effect was observed with magnesium (MgSO4), whereas sodium (NaN3), cadmium [Cd(NO3)2], and lithium (LiCl) reduced activity by 26 to 29%, and similar reductions were observed with manganese (MnSO4), EDTA, and calcium (CaCl2) (21 to 30%). However, the enzyme proved to be very sensitive to zinc (ZnCl2) (59% reduction), copper (CuCl2) (85%), mercury (HgCl2) (94%), and silver (AgNO3) (100% reduction).
N-terminal amino acid sequence.
From the purified enzyme, the sequence of 17 amino acids at the N-terminal site was identified as A-E-I-K-D-P-E-N-T-I-L-M-E-T-T-K-G. A comparison of the N-terminal amino acid sequence from the S. meliloti M5N1CS lyase to protein sequences of the GenBank coding sequence data (398,151 sequences searched) revealed no homology with any previously published protein sequences.
Substrate specificity of glucuronan lyase.
Three glucuronan substrates (0.4% [wt/vol] in the incubation buffer), corresponding to a deacetylated glucuronan, a mainly 3-O-acetylated glucuronan, and a mainly 2,3-di-O-acetylated glucuronan, were incubated at 50°C overnight with the enzyme preparation. The polymer degradation was monitored by size exclusion chromatography of the reaction mixture. As described in Table 1, the enzyme was highly specific to deacetylated glucuronan; a smaller activity was noted with the mainly 3-O-acetylated glucuronan (standard), while the mainly 2,3-di-O-acetylated glucuronan (highly acetylated) was not cleaved.
TABLE 1.
Substrate specificity of the glucuronan lyase
| Substratea | Main linkages | Relative activity (%)b |
|---|---|---|
| Deacetylated glucuronan | β-(1→4) | 100.0 |
| 3-O-Acetylated glucuronan | β-(1→4) | 84.4 |
| 2,3-Di-O-acetylated glucuronan | β-(1→4) | NDd |
| Glucoglucuronanc | β-(1→3), β-(1→4) | ND |
| β-d-Mannuronates blocks | β-(1→4) | ND |
| α-l-Guluronates blocks | α-(1→4) | ND |
| Hyaluronate | β-(1→3), β-(1→4) | ND |
| Polygalacturonate | α-(1→4) | ND |
| Polygalacturonate methyl ester | α-(1→4) | ND |
Four g/liter in 50 mM KPB (pH 6.5).
Relative activity was calculated using deacetylated glucuronan as the reference.
Glucoglucuronan repeating unit, β-(1→3)-d-GlcpA-β-(1→4)-d-Glcp. The glucoglucuronan was from our lab (13).
ND, not detectable.
Degradation products from deacetylated glucuronan.
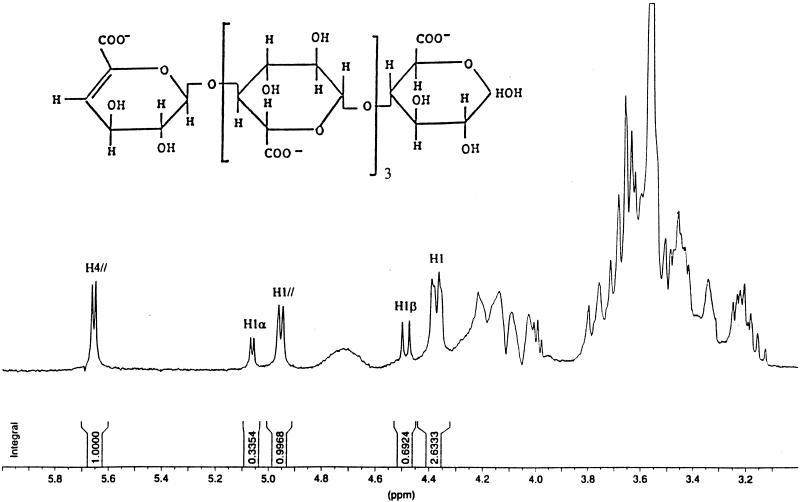
A deacetylated glucuronan solution (2% [wt/vol]) was incubated with 0.46 U of enzyme. After 15 h of incubation the solution was chromatographed on a Bio-Gel P-6 column, and the elution profile was compared to those obtained with a sample corresponding to a deacetylated glucuronan solution as above, incubated without enzyme. No oligosaccharides were detected with the enzyme-free sample, while oligoglucuronates with degrees of polymerization (dp) ranging from 2 to >7 were separated (Fig. 5). 1H NMR spectra from each oligoglucuronate fractions obtained after enzymatic degradation of the glucuronan sample (Fig. 6) revealed doublets at 5.68 ppm, characteristic of H-4 of an unsaturated unit, and signals at 4.96 and 4.32 ppm assigned to H-1 of an unsaturated residue and to H-1 of the repeating unit. The pure oligoglucuronans obtained (dp, 2 to 7), containing an unsaturated residue at the non-reducing end, were significative of a randomly cleaving enzyme.
FIG. 5.
Gel permeation analysis of deacetylated glucuronan incubated with S. meliloti M5N1CS glucuronan lyase, under the following conditions: by Bio-Gel P-6 column, 100 by 2.5 cm; flow rate, 50 ml/h; eluent, 50 mM NaNO3; refractive index detection. The degrees of polymerization are indicated by numbers under the peaks.
FIG. 6.
1H NMR spectrum of deacetylated oligoglucuronan (dp 5) in D2O (300 MHz; 50°C). H4//, H4 of the unsaturated non-reducing terminus; H1β, H1β of the reducing end unit; H1α, H1α of the reducing end unit; H1, H1 of the non-terminus units; H1//, H1 of the unsaturated non-reducing terminus.
DISCUSSION
S. meliloti mutant strain M5N1CS (NCIMB 40472) produces during growth a homopolymer of (1→4)-β-d-glucuronic acid partially acetylated. This exopolysaccharide may have a protecting role, as does EPS II, which is produced by Rhizobium meliloti EFBI (24, 27). In previous works (29), it was noted that the weight-average molecular weight (Mw) of the glucuronan decreased from 7.5 × 105 to 5 × 105 between 27 and 75 h of fermentation. The polymer Mw reduction was correlated with the formation of LMW (from 103 to 5 × 104) glucuronans, whose 1H NMR spectra revealed a doublet at 5.85 ppm characteristic of the H-4 of an unsaturated unit corresponding to the non-reducing terminus unit of the glucuronan. This result was associated to a lyase activity first detected in cell extracts (30). After purification of the enzyme, we determined that the N-terminal amino acid sequence revealed no homology to published protein sequences; the lyase extracted from S. meliloti strain M5N1CS corresponds to a new enzyme. We noted a relative homogeneity between the pI values of specific alginate lyases (from 4.7 to 5.1) (11, 35) and those of the glucuronan lyase (4.9). We noted that addition of magnesium (MgSO4 · 7H2O; 1 mM) to enzyme samples has no effect on the glucuronan degradation, while the addition of magnesium (from 10 to 16 mM MgSO4 · 7H2O) to bacterial suspensions during fermentations leads to glucuronan degradations (29). The polymer degradation may be due to the release of periplasmic proteins as the glucuronan lyase, from bacteria, during magnesium sulfate additions. The glucuronan lyase was tested on uronic acid containing polymers from plants (polygalacturonate and polygalacturonate methyl ester, guluronate, and mannuronate), from animals (hyaluronate), or from bacteria (glucuronan and glucoglucuronan [13]); we noted that under the conditions applied the enzyme cleaved only deacetylated and mainly 3-O-acetylated glucuronans. However, the yield of acetyl residues present on the polymer influences the lyase activity. No degradation was detected when the substrate was a highly acetylated glucuronan (one acetyl for 0.7 residue), while lowly acetylated (mainly 3-O-acetylated) and deacetylated glucuronans were degraded. These results confirm that oligoglucuronans with dp ≥2, containing no 2,3-di-O-acetylated residues, detected in fermentation media of S. meliloti mutant strain M5N1CS correspond to degradation products of unacetylated and monoacetylated glucuronan sequences (33) present on the polymer (8). These results agree with those presented by Skjak-Braek et al. (37), who proposed that acetylation of mannurosyl residues on positions C-2 and C-3 inhibited the alginate lyase activity from different Azotobacter species. Similar results were presented by Kennedy et al. (22) with alginate lyases tested on O-acetylated mannuronate blocks. Pure oligoglucuronan fractions with dp of 2 to 7 were obtained by degradation of a deacetylated glucuronan with the purified enzyme, and unsaturated residues were detected on 1H NMR spectra of each oligoglucuronan fraction. These results lead us to propose that the enzyme cleavage mode was a β-elimination. The presence of oligoglucuronans with dp of 2 to >7 after substrate digestion indicates that the purified lyase was a randomly endolytic enzyme, and these results agree with the literature (40). The glucuronan lyase obtained may be used to produce pure oligoglucuronan fractions unacetylated or partially acetylated (data not shown) in order to test the biological function of the lyase during the life cycle of S. meliloti M5N1CS (NCIMB 40472). On the basis of these results, further studies will be performed in order to clone the glucuronan lyase gene and produce the enzyme. Oligoglucuronans will be produced in large amounts and tested for biological properties as other bacterial oligosaccharides (21, 34).
ACKNOWLEDGMENT
This work was supported by the Pôle Génie des Procédés (Conseil Régional de Picardie).
REFERENCES
- 1.Altman E, Dutton G G S, Stephen A M. Preparative scale depolymerisation of capsular polysaccharide from E. coli K28 using bacteriophage 28-1. S Afr J Sci. 1986;82:45–46. [Google Scholar]
- 2.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 3.Brown B J, Preston J F. l-Guluronan-specific alginate lyase from a marine bacterium associated with Sargassum. Carbohydr Res. 1991;211:91–102. doi: 10.1016/0008-6215(91)84148-8. [DOI] [PubMed] [Google Scholar]
- 4.Clarke B R, Esumeh F, Roberts I S. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J Bacteriol. 2000;182:3761–3766. doi: 10.1128/jb.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courtois B, Hornez J P, Courtois J, Derieux J C. Mise en évidence d'une propriété métabolique de Rhizobium meliloti utilisable pour sa classification. Ann Microbiol. 1983;134:141–147. [PubMed] [Google Scholar]
- 6.Courtois, J., B. Courtois, A. Heyraud, P. Colin-Morel, and M. Rinaudo. 1993. Composés polymères de l'acide glucuronique, procédé de préparation et d'utilisation notamment en tant que moyens gélifiants, épaississants, hydratants, stabilisants, chélatants ou floculants. French patent WO-A. 93/18174.
- 7.Courtois J, Seguin J P, Declomesnil S, Heyraud A, Colin-Morel P, Dantas L, Barbotin J N, Courtois B. A (1→4)-β-d-glucuronan excreted by a mutant of Rhizobium meliloti M5N1 strain. J Carbohydr Chem. 1993;12:441–448. [Google Scholar]
- 8.Courtois J, Seguin J P, Roblot C, Heyraud A, Gey C, Dantas L, Barbotin J N, Courtois B. Exopolysaccharide production by the Rhizobium meliloti M5N1CS strain. Location and quantification of the sites of O-acetylation. Carbohydr Polym. 1994;25:7–12. [Google Scholar]
- 9.Dantas L, Courtois J, Courtois B, Seguin J P, Gey C, Heyraud A. NMR spectroscopic investigation of oligoglucuronates prepared by enzymic hydrolysis of a (1→4)-β-d-glucuronan. Carbohydr Res. 1994;265:303–310. doi: 10.1016/0008-6215(94)00229-0. [DOI] [PubMed] [Google Scholar]
- 10.Edman P, Begg G. A protein sequenator. Eur J Biochem. 1967;1:80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvag H, Erlien F, Skjak-Braek G, Rehm B H A, Valla S. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J Bacteriol. 1998;180:3779–3784. doi: 10.1128/jb.180.15.3779-3784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzales M L, Courtois J, Heyraud A, Colin-Morel P, Michaud P, Barbotin J N, Courtois B. Selection of a succinoglycan deficient Rhizobium meliloti mutant producing a partially acetylated (1→4)-β-d-glucuronan: symbiotic properties analysis. Ann N Y Acad Sci. 1996;782:53–61. [Google Scholar]
- 13.Guentas L, Pheulpin P, Heyraud A, Gey C, Courtois B, Courtois J. Production of a glucoglucuronan by a rhizobia strain infecting alfalfa. Structure of the repeating unit. Int J Biol Macromol. 2000;27:269–277. doi: 10.1016/s0141-8130(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto W, Inose T, Nakajima H, Sato N, Kimura S, Murata K. Purification and characterization of microbial gellan lyase. Appl Environ Microbiol. 1996;62:1475–1477. doi: 10.1128/aem.62.4.1475-1477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto W, Miki H, Tsuchiya N, Nankai H, Murata K. Xanthan lyase of Bacillus sp. strain GL1 liberates pyruvylated mannose from xanthan side chains. Appl Environ Microbiol. 1998;64:3765–3768. doi: 10.1128/aem.64.10.3765-3768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto W, Momma K, Miki H, Mishima Y, Kobayashi E, Miyake O, Kawai S, Nankai H, Mikami B, Murata K. Enzymatic and genetic bases on assimilation, depolymerization, and transport of heteropolysaccharides in bacteria. J Biosci Bioeng. 1999;87:123–136. doi: 10.1016/s1389-1723(99)89001-x. [DOI] [PubMed] [Google Scholar]
- 17.Hatch R A, Schiller N L. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:974–977. doi: 10.1128/aac.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyraud A, Courtois J, Dantas L, Colin-Morel P, Courtois B. Structural characterization and rheological properties of an extracellular glucuronan produced by a Rhizobium meliloti M5N1 mutant strain. Carbohydr Res. 1993;240:71–78. doi: 10.1016/0008-6215(93)84172-3. [DOI] [PubMed] [Google Scholar]
- 19.Heyraud A, Gey C, Leonard C, Rochas C, Girond S, Kloareg B. NMR spectroscopy analysis of oligoguluronates and oligomannuronates prepared by acid or enzymatic hydrolysis of homopolymeric blocks of alginic acid. Application to the determination of the substrate specificity of Haliotis tuberculata alginate lyase. Carbohydr Res. 1996;289:11–23. doi: 10.1016/0008-6215(96)00060-2. [DOI] [PubMed] [Google Scholar]
- 20.Hotchkiss A T, Jr, Revear L G, Hicks K B. Substrate depolymerization pattern of Pseudomonas viridiflava SF-312 pectate lyase. Physiol Mol Plant Pathol. 1996;48:1–9. [Google Scholar]
- 21.Iwasaki K, Matsubara Y. Purification of alginate oligosaccharides with root growth-promoting activity toward lettuce. Biosci Biotechnol Biochem. 2000;64:1067–1070. doi: 10.1271/bbb.64.1067. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy L, McDowell K R, Sutherland I W. Alginases from Azotobacter species. J Gen Microbiol. 1992;138:2465–2471. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lagares A, Caetano-Anolles G, Niehaus K, Lorenzen J, Ljunggren H D, Pühler A, Favelukes G. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J Bacteriol. 1992;174:5941–5952. doi: 10.1128/jb.174.18.5941-5952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahaye M, Brunel M, Bonnin E. Fine chemical structure analysis of oligosaccharides produced by an ulvan-lyase degradation of the water-soluble cell-wall polysaccharides from Ulva sp. (Ulvales, Chlorophyta) Carbohydr Res. 1997;304:325–333. doi: 10.1016/s0008-6215(97)00270-x. [DOI] [PubMed] [Google Scholar]
- 26.Larsen B, Hooen K, Ostgaard K. Kinetics and specificity of alginate lyases. Hydrobiologia. 1993;260/261:557–561. [Google Scholar]
- 27.Lloret J, Wulff B B H, Rubio J M, Downie J A, Bonilla I, Rivilla R. Exopolysaccharide II production is regulated by salt in the halotolerant strain Rhizobium meliloti EFB1. Appl Environ Microbiol. 1998;64:1024–1028. doi: 10.1128/aem.64.3.1024-1028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maki H, Mori A, Fujiyama K, Kinoshita S, Yoshida T. Cloning, sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase from Pseudomonas sp. OS-Alg-9. J Gen Microbiol. 1993;139:987–993. doi: 10.1099/00221287-139-5-987. [DOI] [PubMed] [Google Scholar]
- 29.Michaud P, Courtois J, Courtois B, Heyraud A, Colin-Morel P, Seguin J P, Barbotin J N. Physicochemical properties of extracellular (1→4)-β-d-glucuronan produced by the Rhizobium meliloti M5N1CS strain during fermentation: evidence of degradation by an exoenzyme activated by Mg2+ Int J Biol Macromol. 1994;16:301–305. doi: 10.1016/0141-8130(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 30.Michaud P, Pheulpin P, Petit E, Seguin J P, Barbotin J N, Heyraud A, Courtois B, Courtois J. Identification of glucuronan lyase from a mutant strain of Rhizobium meliloti. Int J Biol Macromol. 1997;21:3–9. doi: 10.1016/s0141-8130(97)00033-0. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa A, Ozaki T, Chubachi K, Hosoyama T, Okubo T, Iyobe S, Suzuki T. An effective method for isolating alginate lyase-producing Bacillus sp. ATB-1015 strain and purification and characterization of the lyase. J Appl Microbiol. 1998;84:328–335. doi: 10.1046/j.1365-2672.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- 32.Pecina A, Pascual A, Paneque A. Cloning and expression of the algL gene, encoding the Azotobacter chroococcum alginate lyase: purification and characterization of the enzyme. J Bacteriol. 1999;181:1409–1414. doi: 10.1128/jb.181.5.1409-1414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirlet A S, Pitiot O, Guentas L, Heyraud A, Courtois B, Courtois J, Vijalayakshmi M A. Separation of low-molecular-mass acetylated glucuronans on l-histidine immobilized onto poly(ethylene-vinyl alcohol) hollow-fiber membranes. J Chromatogr. 1998;826:157–166. [Google Scholar]
- 34.Potin P, Bouarab K, Küpper F, Kloareg B. Oligosaccharide recognition signals and defence reactions in marine plant-microbe interactions. Curr Opin Microbiol. 1999;2:276–283. doi: 10.1016/S1369-5274(99)80048-4. [DOI] [PubMed] [Google Scholar]
- 35.Sawabe T, Ohtsuka M, Ezura Y. Novel alginate lyases from marine bacterium Alteromonas sp. strain H-4. Carbohydr Res. 1997;304:69–76. doi: 10.1016/s0008-6215(97)00194-8. [DOI] [PubMed] [Google Scholar]
- 36.Shimokawa T, Yoshida S, Takeuchi T, Murata K, Kobayashi H, Kusakabe I. Purification and characterization of extracellular poly(β-d-1,4-mannuronide) lyase from Dendryphiella salina IFO 32139. Biosci Biotech Biochem. 1997;61:636–640. doi: 10.1271/bbb.61.636. [DOI] [PubMed] [Google Scholar]
- 37.Skjak-Braek G, Grasdalen H, Larsen B. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr Res. 1986;154:239–250. doi: 10.1016/s0008-6215(00)90036-3. [DOI] [PubMed] [Google Scholar]
- 38.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 39.Steiner B, Romero-Steiner S, Cruce D, George R. Cloning and sequencing of the hyaluronate lyase gene from Propionibacterium acnes. Can J Microbiol. 1997;43:315–321. doi: 10.1139/m97-044. [DOI] [PubMed] [Google Scholar]
- 40.Sutherland I W. Polysaccharide lyases. FEMS Microbiol Rev. 1995;16:323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 41.Sutherland I W, Kennedy L. Polysaccharide lyases from gellan-producing Sphingomonas spp. Microbiology. 1996;142:867–872. doi: 10.1099/00221287-142-4-867. [DOI] [PubMed] [Google Scholar]
- 42.Sutherland I W. Polysaccharases for microbial exopolysaccharides. Carbohydr Polym. 1999;38:319–328. [Google Scholar]
- 43.Waffenschmidt S, Jaenicke L. Assay of reducing sugars in the nanomolerange with 2,2′-bicinchoninate. Anal Biochem. 1987;165:337–340. doi: 10.1016/0003-2697(87)90278-8. [DOI] [PubMed] [Google Scholar]
- 44.Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxy heptonic acid in extracts of Escherichia coli B. J Biol Chem. 1958;234:705–712. [PubMed] [Google Scholar]
- 45.Yu S, Christensen T M I E, Kragh K M, Bojsen K, Marcussen J. Efficient purification, characterization and partial amino acid sequencing of two α-1,4-glucan lyases from fungi. Biochim Biophys Acta. 1997;1339:311–320. doi: 10.1016/s0167-4838(97)00014-9. [DOI] [PubMed] [Google Scholar]