Abstract
MicroRNAs are endogenous noncoding small RNAs that posttranscriptionally regulate the expressions of their target genes. Accumulating research shows that miRNAs are crucial regulators of immune cell growth and antitumor immune response. Studies on miRNAs and tumors primarily focus on the tumor itself. At the same time, relatively few studies on the indirect regulatory effects of miRNAs in the development of tumors are achieved by affecting the immune system of tumor hosts and altering their immune responses. This review discusses the influence of miRNAs on the antitumor immune system.
1. Introduction
For a long time, DNA carrying the information and functional proteins in the functioning of living things had been the research hot spot in gene epigenetic regulation. Scientific studies show that only a small portion of genes in the genomes of humans and other advanced eukaryotes encode for proteins; over 97% of the transcriptional products are from non-protein-coding RNAs. These noncoding RNAs were critical in cell growth, proliferation, differentiation, and apoptosis. Lin-4 is the first miRNA identified that could regulate the developmental timing of C. elegans by acting on the mRNA of the crucial developmental protein lin-14 [1, 2]. Additionally, miRNA let-7 was also found to contribute to the development of C. elegans [3]. miRNAs are small noncoding RNAs or ncRNAs, consisting of a small number of nucleotides, usually 21~23 nt in length. miRNAs regulate gene expression at the posttranscriptional or translational levels by acting in a sequence-specific way, that is, on the 3′-untranslated region (3′-UTR) of the target mRNAs [4, 5]. Furthermore, miRNAs have also been involved in gene silencing (protein degradation factors, such as exosomes) at the pretranslational stage (during chromatin remodeling) and even in the cotranslational stage. Briefly, miRNAs are generated in the nucleus by RNA polymerase II from longer primary transcripts (pri-miRNAs). Pri-miRNAs are usually derived from their introns of protein-coding genes or noncoding genes. Pri-miRNAs fold into hairpins, further bound by Drosha and Dicer, two enzymes from the RNase III family. A microprocessor complex is formed when Drosha interacts with DGCR8 in the nucleus and releases the ~70 nt precursor miRNA (pre-miRNA) hairpin. Following its export to the cytoplasm by exportin-5, Dicer further processes the transcript in order to generate mature ∼20 bp miRNA/miRNA duplexes (Figure 1) [6]. In previous studies, miRNAs accounted for about 1% of the human genome. The miRNA families are critical gene families essential to almost every pathological and physiological process in the body. They are critical in the developmental processes of immune cells. At the same time, they also affect the development of tumors by regulating the immune responses [7, 8].
Figure 1.
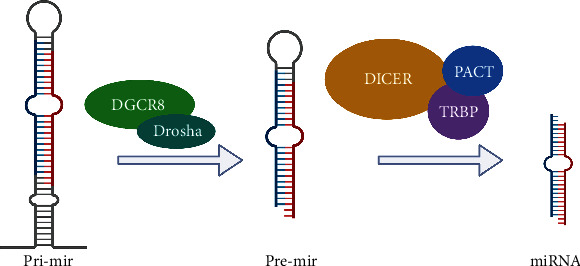
miRNA biogenesis.
2. miRNAs and Tumor Immunity
2.1. miRNAs and Innate Immunity
The innate immune system is the natural immune system, which is gradually formed by organisms during the long-term germline evolution and includes innate immune molecules, innate immune cells (dendritic cells, granulocytes, NK cells, and macrophages), and innate immune barrier. Innate immune molecules and immune cells can rapidly and effectively phagocytose and kill pathogens or “nonself” antigenic foreign bodies in the body. They can recognize antigenic foreign bodies, including pathogens and their products, aging-related injuries, and malformed cells in the body. Nevertheless, cross-reactions can occur if their epitopes are similar enough to those of the original antigen. However, cells in the innate immune system play the dual role of promoting and inhibiting tumor proliferation depending on the surrounding environment [9]. Herein, we mainly focus on the immune cells, including macrophages and dendritic cells (Figure 1).
2.1.1. miRNAs and Macrophages
Depending on their functionally polarized mechanism, macrophages within the tumor microenvironment can either promote or suppress tumor growth. Several miRNAs influence macrophage polarization, which is vital to the progression of cancer, such as extravasation, intravasation, tumor invasion, and premetastatic site formation [10]. For example, miR-21 deficiency induces macrophage polarization to the M1 phenotype in tumor cell environments via the activator of the transcription 1 (STAT1) signaling and the IFN-γ/signaling transducer. By downregulating miR-21, STAT1 signaling can be augmented as well as the expression of programmed death-ligand 1 (PD-L1) in macrophages, which inhibits macrophage antitumor activities [11, 12]. Moreover, miR-127 also enhanced macrophage activation, suppressing the expression of M1 marker genes and promoting the transcription of M2 markers [13]. Induction of elevated miR-127 by LPS decreases the expression of BCL6, which inhibits the expression of the phosphatase Dusp1. By reducing Dusp1, the phosphorylation of JNK is increased, which promotes inflammation and the M1 phenotype [13]. In addition, miRNAs can also affect the M2 polarization of macrophages. Veremeyko et al. [14] observed that IL-13 and IL-4 treatment increased the expression of miR-124 and knockdown of miR-124 activated M1 marker expression (i.e., CD86, TNF, and iNOS) while suppressing M2 markers (i.e., Ym1 and CD206) in M2-polarized macrophages [14]. By modulating macrophage polarization, miRNAs have therapeutic potential for treating inflammation-related diseases (Figure 2).
Figure 2.
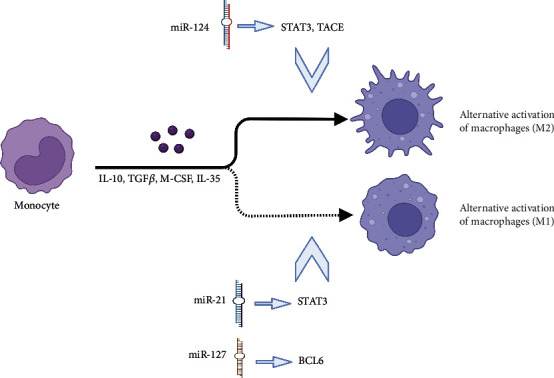
miRNA affects the polarization of macrophages.
Recent studies have shown that miRNAs regulate macrophage growth, mainly when they interact with macrophage precursors, including hematopoietic stem cells (HSCs), thereby regulating macrophage response to cancer. miR-125a can regulate the survival and implantation of HSCs by silencing proapoptotic factors, such as the BCL2 killing factor (BAK1) and BCL2 modifying factor (BMF) and Krueppel-like factor (KLF) 13 [15]. miRNA-126 expressed in HSCs inhibits cell cycle progression and hematopoietic output through the phosphatidylinositol three-kinase (PI3K)/AKT pathway regulation [16]. Therefore, miRNAs can indirectly affect the growth and development of macrophages through HSCs and by regulating immune-related responses (Table 1).
Table 1.
The function of miRNAs in adaptive immunity.
| miRNA | Type of immunity | Functions | References |
|---|---|---|---|
| miR-28 | Cellular immunity | Silences PD-1 and attenuates the exhaustion of T cells | [32] |
| miR-155 | Cellular immunity | Activates the expression of PD-L1 | [33] |
| miR-195 | Cellular immunity | Inhibits CD4T cells through CCDC88C | [34] |
| miR-448 | Cellular immunity | Inhibits IDO1 and enhances CD8+ T cell response | [36] |
| miR-181a | Cellular immunity | Control TCR signal | [38] |
| miR-126 | Cellular immunity | Acts on IRS-1, regulates T cell activation | [39] |
| miR-150 | Humoral immunity | Acts on BCR, controls B cell differentiation | [40] |
| miR-34a | Humoral immunity | Affects BCR signaling by the DDR/miR-34a axis | [41] |
2.1.2. miRNAs and NK Cells
miRNAs can directly regulate the proliferation and cell lethality in NK cells, along with the regulation of their physiological functions through the introduction of other proteins. NK cells kill the activating receptors as they recognize the non-MHC class I molecule ligands, which in turn affirm the high expression of receptors on the surface of tumors and cells that are infected with viruses while having low expression in normal cells, thereby achieving the targeted goal of killing tumor cells without damaging the normal cells [17]. After overexpression of miR-152, Zhu et al. found that NK cell-mediated cytolysis increased by RT-PCR and WB. The result suggests that miR-152 may act as an immune promoter by upregulating host-mediated cytolysis in NK cells [18]. NK cells and T cells are inhibited by B7-H3, an immunomodulatory glycoprotein on the surface of cells [19]. Although the transcription of B7-H3 is ubiquitous in normal tissues, its protein is preferentially expressed in solid tumor cells only. Xu [20] used PCR to demonstrate that miR-29 levels are high in normal tissues while miR-29 levels are lower in tumors, including brain tumors and sarcomas. Transfection with miR-29 lowers B7-H3 expression, according to the study. The expression of B7-H3 increases after knocking out miR-29, indicating that the transcription of miR-29 is negatively correlated with the expression of B7-H3 [20]. This study was subsequently reported by Asuthkar's team, which further found that the overexpression of miR-29 in MYC+ medulloblastoma cells mediated the downregulation of B7-H3 and inhibited angiogenesis in medulloblastoma. They hypothesized that this biological process might be associated with JAK/STAT1 signaling [21].
2.1.3. miRNAs and Dendritic Cells
miRNAs play the dual role of regulating dendritic cells' (DCs) antigenic transmission and immunogenicity. DCs are professional antigen-presenting cells; immature DCs exhibit strong migration and antigen uptake abilities, whereas mature DCs can modify T cells' viability effectively [22]. Several miRNAs exert positive effects on the immune responses of DCs. Sun et al. show that miR-142 promotes the increase in the inflammatory cytokine production and enhances the T cell activation [22]. miR155 is essential in promoting DC maturation and migration; it can induce the DCs to activate T cells by secreting effector factors. In the breast cancer tumor microenvironment, miR-155 expression in DCs is downregulated, thereby inhibiting the DC-mediated immune response [23]. miRNAs can promote DC-mediated immune reactions and suppress DC-mediated antitumor immune reactions. For instance, miR-21 and miR-28 inhibit the activation and maturation of DCs as well as the activation of T cell-mediated immune reactions [24]. Thus, miRNAs have dual effects on immune responses in DCs.
2.2. miRNAs and Adaptive Immunity (Table 1)
Adaptive immunity refers to the immune responses that can be recognized and initiated against a specific pathogen after prior contact with this pathogen [25]. The immune system triggers the immune responses through the lymphocyte-mediated recognition of the corresponding specific antigens and their antigenic receptors by the major histocompatibility complexes (MHCs) involved in several immune cells and immune molecules [26]. The signaling pathways associated with T and B cells are important in specific immunity [27].
2.2.1. miRNAs and Cellular Immunity
The specific immune reactions mediated by T cells are critical to antitumor immunity. Cytotoxic T lymphocytes (CTLs) are the central effector cells in antitumor responses, as they specifically kill mutant cells or tumor cells through the granzyme/perforin pathway and death receptor pathway (TNF-TNFR and Fas-Fasl pathways) [28]. CD4+ Th cells not only activate CD8+ CTLs by releasing several cytokines and enhancing the immune efficacies of CTLs but also produce chemokines, IFNs, and TNF, which directly act on the tumor cells [29].
miRNAs are essential in T cell-mediated tumor immunity. Programmed cell death receptor 1 (PD-1) promotes the apoptosis of antigen-specific T cells in the lymph nodes (programmed cell death) [30]. miRNAs impact T cell exhaustion by regulating PD-1 expression [31]. Li [32] observed increased T cell depletion in melanoma mice. Moreover, miRNA microarray and PD-1 exhaustive analyses showed that the levels of 11 miRNAs changed significantly in the melanoma environment. After screening, it was found that miR-28 could bind to multiple insulin receptor substrates (IRs) and subsequently silence PD-1, thereby attenuating and regulating the depletion of T cells and the expression of PD-1. Zheng et al. [33] also show that miR-155 enhances PD-L1 expression in the diffused sizeable B cell lymphoma microenvironment, wherein CD8+ T cells are recruited by PD-1/PD-L1 interaction, and the function of CD8+ T cells is inhibited by phosphorylated AKT and ERK, in such a way that the efficacy of T cell-mediated tumor immune reactions is reduced.
Additionally, miR-195 and miR-448 also play critical roles in antitumor response pathways. In lung adenocarcinoma, miR-195 acts as a suppressor and regulates the response of CD+4 T cells through the CCDC88C [34]. The T cell is controlled by indoleamine 2,3-dioxygenase (IDO1), which is important to immune tolerance; upregulated expression of IDO1 is reported in several types of cancer [35]. Lou et al. show that IDO1 inhibits the activation of CD8+ T cells in colon cancer. miR-448, a tumor suppressor, attenuates the expression of IDO1 and enhances the activation of CD8+ T cells [36]. The mutual characteristics of miRNAs, PD-1, and other tumor-related molecules have essential value in tumor immunotherapy.
During the development of normal human T cells, they need to undergo gene rearrangement of antigen recognition receptor (TCR), express multiple TCRs, and experience positive and negative selection, which are the core procedures in the growth and development of T cells [37]. miR-181a has a primary function in T cell activation, wherein it regulates thymic selection and the activation threshold of antigen recognition receptors (TCRs) by targeting different signaling pathways [38]. YY1, which modulates TCR signaling by upregulating miR-181a and inhibiting the miR-181a-mediated negative feedback loop, is correlated with a decrease in miR-181a expression. Chu et al. report that insulin receptor 1 (IRS-1), a functional target of miR-126, was increased in the miR-126-knockout mice. The activation, proliferation, and interferon expression of CD4+ T cells were higher than those of the wild-type mice [39]. In conclusion, miRNAs are associated with T cells' activation, development, and maturation.
2.2.2. miRNAs and Humoral Immunity
Multiple genes control B cells during the different stages of development. Previous studies have confirmed that miRNAs are crucial in the growth of normal B cells and the destruction of B cell malignancies; they are expected to provide new markers for clinical diagnosis. It was found that miR-150 has an essential association with c-Myb. miR-150 regulates the differentiation of B cells by targeting c-Myb [40]. In HSCs, there is a high expression of BCR. B cells recognize antigens through BCR, receive antigenic stimulation, and eventually initiate a humoral immune response. Cerna et al. [41] show that DNA damage response (DDR) can activate FOXP1, a positive regulator of BCR; thus, it can be used as an indicator of disease worsening for predicting different clinical courses of chronic lymphocytic leukemia (CCL). Through the DDR/miR-34a axis, B cells can restrict BCR signaling during DDR by downregulating FOXP1. BCR signaling is limited, and antigen delivery and antibody activation are downregulated, indirectly affecting the mechanism of antibody tumor immunity.
3. Effects of miRNAs on Tumor Immune Microenvironment
In 1989, the theory of “seed and soil” proposed by Paget explained how tumors survive in their environment; this is the so-called tumor microenvironment (TME) [42]. The tumor microenvironment includes extracellular matrix, tumor cells, peripheral blood vessels, and other nonmalignant cells, along with signaling molecules [43]. Simultaneously, characterized by low oxygen, low pH, high pressure, and nutritional deficiencies, the inhibitory effects of immune molecules and immune cells on the tumor are substantially reduced in the TME; the sensitivity of malignant tumor cells toward chemotherapeutic drugs also reduces significantly [44]. It is becoming increasingly apparent that miRNAs are crucial in the development of tumors by regulating the TME.
Abnormal transduction of signaling pathways in the TME can induce tumor development and enhance the body's immune responses against tumors. In the TME, cancer-associated fibroblasts (CAFs) supply oncogenic signals that support tumor growth, metastasis, and progression. Shen et al. show that miR-206, miR-31, and miR-1can promote the development of lung cancer by regulating FOXO3a/VEGF/CCL2, thereby converting normal fibroblasts into tumor-associated fibroblasts (CAFs) [45].
Exosomes are 20~100 nm small membrane vesicles. After budding from the nucleus, the cell membrane fuses with the small membrane vesicles and then secretes their contents outside the cell [46]. Exosomes contain several functional proteins, mRNAs, and miRNAs [47]. In tumor cells, tumor-derived exosomes (TEXs) are abundantly secreted. They regulate communications between tumor cells and TME, the proliferation of tumor cells, and metastasis of tumor cells [48–50]. Immunosuppressive activity of myeloid-derived suppressor cells (MDSCs) is observed in the TME of immature myeloid cells [51, 52]. MDSCs protect tumor cells from attack by the immune system and thus are tolerant to immunotherapy, posing a significant obstacle to immunotherapy [53]. In recent studies, TEXs have been shown to have essential functions, including activating and expanding MDSCs and suppressing their immune responses [54]. Therefore, TEXs are a vital element of the TME and play a crucial role in tumor development [55]. The miRNA sequencing data from Li et al. show that the expression of miR-21 is high in hypoxic TEXs [56]; the presence of TEXs containing miR-21 in cancer cells can also stimulate MDSCs to suppress the immune system. Existing literature shows that miR-21 overexpression can enhance the amplification and function of MDSCs [57]. Li et al. showed that miR-21 and miR-155 synergistically downregulate the expression of PTEN and SHIP-1, causing t the STAT3 pathway activation, which induced the expansion of MDSCs [57]. Their research provides new evidence for the miR-21-mediated immune escape of MDSCs from the TME. The transmission of anti-VEGFA/CCL2/pre-miR-1, anti-miR-31, and pre-miR-206 inhibits tumor growth, tumor angiogenesis, and lung metastasis when administered systemically [45]. miRs are transported by extracellular vesicles (EVs) and play a critical role in the communication between cells in the TME. Exosomal miRs are involved in reciprocal interaction between the neuroblastoma (NBL) cells and TME components. Thus, it can induce proinflammatory reaction in monocytes and further promote NBL cell chemotherapy resistance [58]. In addition, exosomes from NK and activated NK cells, which contain miRs (mir-186) and cytotoxic proteins (granzymes A and B, granulysin, and perforin), show anticancer activity. By capturing these exosomes, NBL cells inhibit tumor cell growth and migration, preventing tumor cells from escaping from the NK cell-mediated cytotoxic response and inducing apoptosis [59, 60].
4. miRNAs and Tumor Immunotherapy
In the last few decades, people have made breakthroughs in understanding how cancer cells escape from the immune system, which in turn has promoted a new insight for preventing the immune escape of tumors. miRNAs have an important impact on tumor development, and similarly, tumor proliferation also counteracts the expression of some miRNA [61, 62].
4.1. miRNAs and Immune Checkpoints (ICPs) [63]
As an immunosuppressive pathway, immune checkpoints (ICPs) are critical in maintaining autoimmune tolerance and regulating the duration and range of immune responses in peripheral tissues [64]. miRNAs that regulate the immune system through ICPs are receiving great research attention. ICPs in humans include the B7, the TNF, the CD28, and the TNFR families of proteins, which are popular targets in clinical drug therapy. According to the effect on T cells, ICPs could be classified as cosuppressor proteins (CTLA-4, B7-H4 PD-1, and PD-L1) and costimulatory proteins (ICOS, CD28, B7-H3, B7-H2, CD40, CD40L, CD70, and CD27) [65].
4.1.1. miRNAs and Cosuppressor Proteins
As a member of ICPs, PD-1/PD-L1-related signaling pathways are a research hotspot of tumor immunity due to their effect on the immune escape of cancer cells [66]. Li et al. transfected miR-28 mimics and inhibitors into the B16F10 mouse T cells and evaluated the expression of PD-1 using RT-qPCR. The dual-luciferase report assay of PD-1 3′-UTR indicated that miR-28 could silence PD-1 by combining with PD-1 3′-UTR. At the same time, the miR-28 mimic could also reduce PD-1 expression and silence the 3′-UTR of PD-1 (Table 2) [32]. Wei et al. used RNA22 and miRanda to check whether miR-138 could bind the key ICPs, CLAT-4, and PD-1. After CD4+ T cells were transfected with miR-138, PD-1 and CTLA-4 were downregulated [67]. In a comparative experiment using the glioma mouse model, C57BL/6J, Wei et al. found that the gliomas disappeared gradually in mice transfected with miR-138, while untransfected mice showed continued glioma invasion. Huang et al. transfected the PD-L1 Mut luciferase reporter and PD-L1 WT vectors and miR-374b into T and NK cells; 48 hours after transfection, the PD-L1 WT luciferase activity was significantly lower in the miR-374b group than in the other groups, which confirmed that PD-1 is associated with miR-374b. In the liver cancer model, miR-374b was found to downregulate PD-1 expression and reduce tumor size [68]. miR-374b, miR-138, and miR-28 can aim at the 3′-UTR of PD-L1 mRNA, thereby regulating PD-L1 expression to affect the immunosuppression [64].
Table 2.
Cell types and corresponding functions in the main miRNA interactions.
| miRNA | Cell types | Functions | References |
|---|---|---|---|
| miR-21 | Macrophages | Changes in the polarity of macrophages | [11] |
| miR-125a | HSCs | Regulates the survival of HSCs | [15] |
| miR-126 | HSCs | Inhibits the cell cycle | [16] |
| miR-127 | Macrophages | Changes in the polarity of macrophages | [13] |
| miR-124 | Macrophages | Promotes the differentiation of M1 macrophages | [14] |
| miR-152 | NK cell | Upregulates the activity of NK cells | [8] |
| miR-29 | NK cell | Regulates NK cells indirectly through B7-H3 | [19] |
| miR-142 | DC | Enhances the activation of inflammatory factors | [22] |
| miR-155 | DC | Suppresses DC-mediated immune response | [23] |
| miR-28 | T cell | 3′-UTR binds directly to PD-1 | [32] |
| miR-195 | T cell | Regulates CD+4 T cell activation by CCDC88C | [34] |
| miR-448 | T cell | Inhibits IDO1 | [36] |
| miR-181a | T cell | Control TCR signal | [38] |
| miR-126 | T cell | Acts on IRS-1, regulates T cell activation | [39] |
| miR-150 | B cell | Acts on BCR, controls B cell differentiation | [40] |
| miR-1 | CAF | Regulates FOXO3a/VEGF/CCL2 | [45] |
| miR-206 | CAF | Regulates FOXO3a/VEGF/CCL2 | [45] |
| miR-31 | CAF | Regulates FOXO3a/VEGF/CCL2 | [45] |
| miR-186 | NK cell | Suppresses immune escape | [59] |
| miR-142-5p | T cell | Overexpression inhibits the PD-1/PD-L1 pathway | [107] |
| miR-21 | MDSC | Activates MDSC amplification | [57] |
| miR-138 | T cell | Inhibits the expression of CTLA-4 and PD-1 | [67] |
| miR-374b | T cell | Downregulates PD-1 | [68] |
| miR-155 | T cell | Inhibits PD-1 and CTLA-4 | [71] |
| miR-1207-5p | CHO cell | Binding to B7-H4 3′-UTR allele | [77] |
| miR-24 | CHO cell | Associated with B7-H2 | [87] |
| miR-172-92 | CAR-T cell | Enhances CAR-T cell antigenicity | [96] |
| miR-153 | CAR-T cell | Inhibits IDO1, enhances cell lethality | [104] |
CTLA-4 can mediate immunosuppressive pathways and downregulate the activity of T cells [69]. Researchers are trying to enhance the efficacies of tumor vaccines to induce antitumor immune responses by clearing suppressive cells, including Tregs, or blocking immunosuppressive pathways using the CTLA-4 antibodies [70]. Currently, the regulation of the CD28 family by miRNAs remains unclear. By targeting the 3′-UTR of CTLA-4, miR-155 can reduce the production of CTLA-4. At the same time, CD4+ T cell proliferation is significantly enhanced [71]. In miR-155T cell-specific knockout mouse models, ICP blocking treatment against PD-1or CTLA-4 can restore antitumor immune responses [71], which implies that the antibodies of anti-CTLA-4 or anti-PD-1 may be associated with miR-155 overlap. Still, the specific regulatory mechanism needs further investigation.
B7-H4 is expressed chiefly on T cells, B cells, DCs, and macrophages [72]. It is upregulated in colorectal tumor cells [73]. B7-H4 participates in peripheral immune responses and negatively regulates T cell proliferation and immune responses [74]. miRNA-related single-nucleotide polymorphisms (miR-SNPs) are a general term used for a class of functional SNPs that lead to aberrant or lacking miRNA gene regulatory functions [75], which are associated with many complex diseases, including tumors [76]. Wu et al. identified 12 miR-SNPs related to the 3′-UTR of the B7/CD28 family from the NCBI dbSNP BUILED129 and ENSEMBL v58 databases [77]. The constructed related pGL3 vectors of miR-SNPs were cotransfected with the corresponding miRNAs into Chinese hamster ovary cells (CHO cells). miR-1207-5p could significantly inhibit B7-H4 3′-UTR allele expression [77], implying that miR-1207-5p could bond with the 3′-UTR of B7-H4 and inhibit its expression, thereby indirectly regulating the immune response.
4.1.2. miRNAs and Costimulatory Proteins
As T cell-specific molecules, inducible costimulatory molecules (ICOSs) bind specifically to the ligand B7-H2, which leads to ICOS/B7-H2 signaling to stimulate the antitumor response of T cells [78]. T cells in the peripheral blood of tumor patients (such as melanoma and ovarian tumors) express a large amount of ICOSs [79, 80]. ICOS molecules are abundantly expressed in the mouse model tumor treated with anti-CTLA-4 [81], suggesting that ICOS may improve the efficacy of immunotherapy. In a mouse model of colorectal cancer, Wu et al. examined miR-SNPs in the 3′-UTR of the ICOS genes and found that miR-SNPs could substantially weaken the interaction between miRNAs and ICOS, resulting in the upregulation of ICOS expression in T cells [77]. miR-SNPs in ICOS interfere with miR-1279 and miR-2117, inhibit miR-186-5p for ICOS expression, and promote the antitumor responses of T cells. As an immunostimulatory molecule, B7-H2 is primarily expressed in macrophages, DCs, and B cells [82, 83]. It is downregulated in gastric and colorectal cancers [84, 85]. Immune escape of cancer cells in tumors is correlated with B7-H2, but the specific mechanism remains confusing [86]. At present, the only miRNA known to target B7-H2 is miR-24. If the regulatory effect of miR-24 on B7-H2 is disturbed by B7-H2 SNPs, it will increase gastric cancer incidence [87].
4.2. miRNAs and CAR-T Cells
Chimeric antigen receptor T cell (CAR-T) is a T cell type having a recombinant receptor against tumor-associated antigens. It combines the affinity of antibodies and the killing activity of T cells, bonds with tumor antigens in an antigen-dependent manner, initiates an activation cascade, and exerts a specific killing effect [88–90]. However, many challenges remain to be addressed in the practical utility of CAR-T therapy as follows [91]: (1) in the treatment using CD19 CAR-T cells, antigen escape frequently occurs, resulting in tumor immune tolerance [92]; (2) CAR-T recognizes healthy tissues and produces nontumor toxicity, which can be life-threatening in severe cases [93]; and (3) due to the complexity of TME of solid tumors, the efficacy of CAR-T is still not promising [94]. Zhang et al. show that miRNA expression levels change in their patient samples after CAR-T treatment. The miRNA-TF (transcription factor) gene network shows that miR-148-3a and miR-375 affect the efficacy of CAT-T by regulating the genes encoding transcription factors and participating in the cross-action CAT-T histone treatment; however, the specific mechanism needs further study [95]. Ohno et al. found that CAT-T cells cotransfected with miR-172-92, as compared to CAT-T without miR-172-92 cotransfection, exhibited better antigenicity in malignant gliomas, which implied that CD19 CAR-T therapy combined with mi-172-92 could improve the problem of antigen escape and enhance the efficacy of CAR-T [96].
Although CD19 CAR-T therapy has entered clinical trials and has been successful in chronic lymphocytic leukemia, its utility is restricted because of the single antibody, rare tumor-specific single antigen, and susceptibility to cross-reactivity, resulting in damage to healthy cells [97]. In 2016, O'Rourke et al. proposed dual-receptor AND-gate T cells. By constructing a T cell circuit that requires the activation of two antigens, only the tumor cells can be targeted and killed by T cells, as they express both antigens simultaneously [98]. Currently, the most commonly used single antigen is CD19CAR-T [99]. Researchers are also studying dual-antigen CAR-T systems such as CD19/CD123 [100] and CD19/CD20 [101]. Immune homeostasis, autoimmunity, and self-antigen regulation are influenced by miRNAs [102]. For example, a lack of miR-146a can deregulate autoantigens and cause autoimmune diseases [103]. The research on the relationship between miRNAs, antigens, and the construction of the double-antigen CAR-T system remains unclear. However, through studies on miRNAs and antigens, the selection of different tumor antigens in double-antigen CAR-T systems, the monitoring of the efficacy of double-antigen CAR-T systems, and the construction of double-antigen CAR-T systems are of utmost significance.
CAR-T cells have made breakthroughs in hematological cancer therapy. However, obstacles remain in CAR-T therapy for solid tumors due to several factors, such as CAR-T cells constrained by the complex microenvironment, diverse immune escape mechanisms, and complex tumor antigen targets in solid tumors. Indoleamine 2,3-dioxygenase 1 (IDO1) may be a target for cancer immunotherapy because it is a standard endogenous mechanism of acquired peripheral immunity. It is an essential indicator for predicting tumor prognosis [104]. In immunodeficient mice, Huang et al. found that miR-153 combined with CAR-T cells effectively improved CAR-T cells' tumor cell killing ability by inhibiting IDO1 [104]. Using CAR-T cells and miRNAs to treat solid tumors opens up an entirely new avenue for immunotherapy. Because of miRNA's oncogenic and tumor suppressor functions, its clinical application has been developed, and relevant novel miRNA drugs have already entered clinical trials. As a phase I clinical trial (NCT02369198i) drug, mesomiR-1 delivers miRNA mimics by targeting bacterial minicells. By incorporating miRNA mimics of the tumor suppressor miR-16 in minicells covered with epidermal growth factor receptor (EGFR) antibodies, MesomiR-1 could target tumor cells accurately [105]. In this study, 26 patients with non-small-cell lung cancer (NSCLC) or malignant pleural mesothelioma who tolerated standard treatment were evaluated for the safety of mesomiR-1. The results suggested that mesomiR-1 side effects were within acceptable limits and showed antitumor efficacy in 73% of patients [105]. Thus, this trial provides robust evidence for further studies on the antitumor activity based on miR-16 associated with immune checkpoint inhibitors [106].
5. Conclusion
As a class of highly conserved noncoding small RNAs, miRNAs are important in regulating human tumor immunity, including tumor development, progression, invasion, metastasis and antigen presentation, immune-related molecules, and inhibition of cell activation of the entire immune response process (Table 2 and Figure 3). Previous studies on miRNAs and tumors have investigated the relationship between high or low expressions of miRNAs and tumor proliferation and metastasis or the downstream pathways regulated by miRNAs that affect tumor growth. Research has recently shifted to miRNAs and tumor-related immune responses, wherein miRNAs have been found to modulate tumor immune responses. These include the activation of antigen receptor signaling (miR-181a [38]), antigen-presenting cells (miR-28 [32]), cellular differentiation (miR-124 [14] and miR-21 [11]), and immune escape (miR-21 [24]) or immune surveillance (miR-186 [59]). miRNAs are essential in rapidly developing cancer immunotherapy and have good application prospects. miRNAs serve as biomarkers for tumor diagnosis, treatment, and prognosis and critical targets for tumor immunotherapy. This feature may provide a much-warranted breakthrough for tumor immunotherapy and widen its prospects for treating more malignant tumor types, thereby benefitting more patients.
Figure 3.
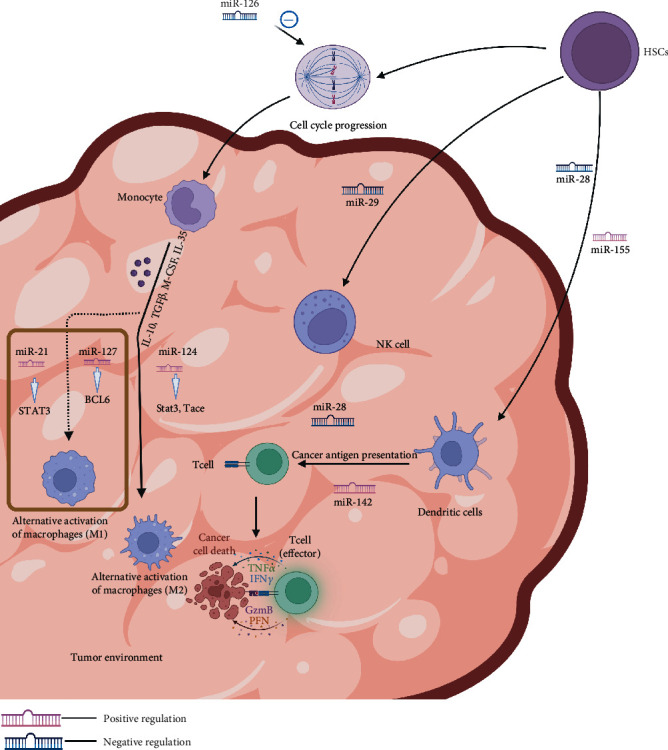
miRNAs participate in the immune responses.
Acknowledgments
This study was funded by the Natural Science Foundation of Guangdong Province, China (No. 2022A1515012315); the Beijing Science and Technology Medical Development Foundation (No. KC2021-JX-0186-94); the 2021 Special Innovation Project of Department of Education of Guangdong Province (No. 2021KTSCX015); the In-Depth Promotion of the Innovation-Driven Assistance Project in Foshan City (No. 2021043); the 2018 Foshan City Outstanding Young Medical Talent Training Project (No. 600009); the 2020 Shunde District Competition Support Talent Project (no serial number); the Southern Medical University Shunde Hospital Scientific Research Startup Plan (No. SRSP2018001); the Guangdong Medical Science and Technology Research Fund Project (No. A2019302); the Science and Technology Plan Project of Foshan Science and Technology Bureau (No. 2018AB000683); the National Natural Science Foundation of China; the National Natural Science Youth Fund Project (No. 81802879); Southern Medical University Scientific Research Startup Plan (No. PY2018N110); and Foshan City's 13th Five-Year Key Specialty Project (FSGSP2D135051).
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Additional Points
Resources. This study is registered with https://clinicaltrials.gov/ct2/show/NCT02369198.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Authors' Contributions
All authors had made substantial contributions for this manuscript. Yan Lu and Muwen Deng contribute equally to to this work and are co-first authors.
References
- 1.Lee R. C., Feinbaum R. L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell . 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell . 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart B. J., Slack F. J., Basson M., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature . 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell . 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nature Reviews Cancer . 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics . 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 7.Rupaimoole R., Slack F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery . 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 8.Fanini F., Fabbri M. Cancer-derived exosomic microRNAs shape the immune system within the tumor microenvironment: state of the art. Seminars in Cell & Developmental Biology . 2017;67:23–28. doi: 10.1016/j.semcdb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moynihan K. D., Irvine D. J. Roles for innate immunity in combination immunotherapies. Cancer Research . 2017;77(19):5215–5221. doi: 10.1158/0008-5472.CAN-17-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Liu J. M., Luo Y. P. MicroRNAs in tumor immunity: functional regulation in tumor-associated macrophages. Journal of Zhejiang University-Science B . 2020;21(1):12–28. doi: 10.1631/jzus.B1900452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noy R., Pollard J. W. Tumor-associated macrophages: from mechanisms to therapy. Immunity . 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi J., Huang Q., Wang L., et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene . 2018;37(23):3151–3165. doi: 10.1038/s41388-018-0178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying H., Kang Y., Zhang H., et al. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. Journal of immunology (Baltimore, Md: 1950) . 2015;194(3):1239–1251. doi: 10.4049/jimmunol.1402088. [DOI] [PubMed] [Google Scholar]
- 14.Veremeyko T., Siddiqui S., Sotnikov I., Yung A., Ponomarev E. D. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PloS One . 2013;8(12, article e81774) doi: 10.1371/journal.pone.0081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo S., Lu J., Schlanger R., et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proceedings of the National Academy of Sciences of the United States of America . 2010;107(32):14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nucera S., Giustacchini A., Boccalatte F., et al. miRNA-126 orchestrates an oncogenic program in B cell precursor acute lymphoblastic leukemia. Cancer Cell . 2016;29(6):905–921. doi: 10.1016/j.ccell.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Gardiner C. M. NK cell metabolism. Journal of Leukocyte Biology . 2019;105(6):1235–1242. doi: 10.1002/JLB.MR0718-260R. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X. M., Han T., Wang X. H., et al. Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. American journal of obstetrics and Gynecology . 2010;202(6):p. 592.e1-7. doi: 10.1016/j.ajog.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y. H., Martin-Orozco N., Zheng P., et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Research . 2017;27(8):1034–1045. doi: 10.1038/cr.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H., Cheung I. Y., Guo H. F., Cheung N. K. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Research . 2009;69(15):6275–6281. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purvis I. J., Avilala J., Guda M. R., et al. Role of MYC-miR-29-B7-H3 in medulloblastoma growth and angiogenesis. Journal of Clinical Medicine . 2019;8(8):p. 1158. doi: 10.3390/jcm8081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y., Oravecz-Wilson K., Bridges S., et al. miR-142 controls metabolic reprogramming that regulates dendritic cell activation. The Journal of Clinical Investigation . 2019;129(5):2029–2042. doi: 10.1172/JCI123839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X., Wang Y., Yu T., et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro-Oncology . 2017;19(9):1195–1205. doi: 10.1093/neuonc/nox017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth L. A., Boardman D. A., Tung S. L., Lechler R., Lombardi G. MicroRNAs affect dendritic cell function and phenotype. Immunology . 2015;144(2):197–205. doi: 10.1111/imm.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehm T., Swann J. B. Origin and evolution of adaptive immunity. Annual Review of Animal Biosciences . 2014;2(1):259–283. doi: 10.1146/annurev-animal-022513-114201. [DOI] [PubMed] [Google Scholar]
- 26.Bonilla F. A., Oettgen H. C. Adaptive immunity. The Journal of allergy and clinical immunology. . 2010;125(2):S33–S40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Defrance T., Vanbervliet B., Durand I., Briolay J., Banchereau J. Proliferation and differentiation of human CD5+ and CD5- B cell subsets activated through their antigen receptors or CD40 antigens. European Journal of Immunology . 1992;22(11):2831–2839. doi: 10.1002/eji.1830221112. [DOI] [PubMed] [Google Scholar]
- 28.Mami-Chouaib F., Blanc C., Corgnac S., et al. Resident memory T cells, critical components in tumor immunology. Journal for Immunotherapy of Cancer . 2018;6(1):p. 87. doi: 10.1186/s40425-018-0399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borst J., Ahrends T., Bąbała N., Melief C. J. M., Kastenmüller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nature Reviews Immunology . 2018;18(10):635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 30.Xu-Monette Z. Y., Zhou J., Young K. H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood . 2018;131(1):68–83. doi: 10.1182/blood-2017-07-740993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merelli B., Massi D., Cattaneo L., Mandalà M. Targeting the PD1/PD-L1 axis in melanoma: biological rationale, clinical challenges and opportunities. Critical Reviews in Oncology/Hematology . 2014;89(1):140–165. doi: 10.1016/j.critrevonc.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Li Q., Johnston N., Zheng X., et al. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget . 2016;7(33):53735–53750. doi: 10.18632/oncotarget.10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Z., Sun R., Zhao H. J., et al. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Molecular Cancer . 2019;18(1):p. 54. doi: 10.1186/s12943-019-0977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan C., Xiang L., Bai R., et al. MiR-195 restrains lung adenocarcinoma by regulating CD4+ T cell activation via the CCDC88C/Wnt signaling pathway: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatic analysis. Annals of Translational Medicine . 2019;7(12):p. 263. doi: 10.21037/atm.2019.05.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhai L., Ladomersky E., Lenzen A., et al. IDO1 in cancer: a Gemini of immune checkpoints. Cellular & Molecular Immunology . 2018;15(5):447–457. doi: 10.1038/cmi.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou Q., Liu R., Yang X., et al. miR-448 targets IDO1 and regulates CD8(+) T cell response in human colon cancer. Journal for Immunotherapy of Cancer . 2019;7(1):p. 210. doi: 10.1186/s40425-019-0691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siska P. J., Rathmell J. C. T cell metabolic fitness in antitumor immunity. Trends in Immunology . 2015;36(4):257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye Z., Li G., Kim C., et al. Regulation of miR-181a expression in T cell aging. Nature cCommunications . 2018;9(1):1–11. doi: 10.1038/s41467-018-05552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu F., Hu Y., Zhou Y., et al. MicroRNA-126 deficiency enhanced the activation and function of CD4+T cells by elevating IRS-1 pathway. Clinical and Experimental Immunology . 2018;191(2):166–179. doi: 10.1111/cei.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao C., Calado D. P., Galler G., et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell . 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 41.Cerna K., Oppelt J., Chochola V., et al. MicroRNA miR-34a downregulates FOXP1 during DNA damage response to limit BCR signalling in chronic lymphocytic leukaemia B cells. Leukemia . 2019;33(2):403–414. doi: 10.1038/s41375-018-0230-x. [DOI] [PubMed] [Google Scholar]
- 42.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Reviews . 1989;8(2):98–101. [PubMed] [Google Scholar]
- 43.Quail D. F., Joyce J. A. Microenvironmental regulation of tumor progression and metastasis. Nature Medicine . 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hui L., Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Letters . 2015;368(1):7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 45.Shen H., Yu X., Yang F., et al. Reprogramming of normal fibroblasts into cancer-associated fibroblasts by miRNAs-mediated CCL2/VEGFA signaling. PLoS Genetics . 2016;12(8, article e1006244) doi: 10.1371/journal.pgen.1006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schorey J. S., Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic (Copenhagen, Denmark) . 2008;9(6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathivanan S., Ji H., Simpson R. J. Exosomes: extracellular organelles important in intercellular communication. Journal of Proteomics . 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 48.King H. W., Michael M. Z., Gleadle J. M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer . 2012;12(1):p. 421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webber J., Steadman R., Mason M. D., Tabi Z., Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Research. . 2010;70(23):9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 50.Hood J. L., San R. S., Wickline S. A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Research . 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 51.Gabrilovich D. I., Bronte V., Chen S. H., et al. The terminology issue for myeloid-derived suppressor cells. Cancer Research . 2007;67(1):p. 425. doi: 10.1158/0008-5472.CAN-06-3037. author reply 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian J., Rui K., Hong Y., et al. Increased GITRL impairs the function of myeloid-derived suppressor cells and exacerbates primary Sjögren syndrome. Journal of Immunology (Baltimore, Md: 1950). . 2019;202(6):1693–1703. doi: 10.4049/jimmunol.1801051. [DOI] [PubMed] [Google Scholar]
- 53.Tesi R. J. MDSC; the most important cell you have never heard of. Trends in Pharmacological Sciences . 2019;40(1):4–7. doi: 10.1016/j.tips.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Whiteside T. L. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Seminars in Immunology . 2018;35:69–79. doi: 10.1016/j.smim.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peinado H., Lavotshkin S., Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Seminars in Cancer Biology . 2011;21(2):139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Li L., Cao B., Liang X., et al. Microenvironmental oxygen pressure orchestrates an anti- and pro-tumoral γδ T cell equilibrium via tumor-derived exosomes. Oncogene . 2019;38(15):2830–2843. doi: 10.1038/s41388-018-0627-z. [DOI] [PubMed] [Google Scholar]
- 57.Li L., Zhang J., Diao W., et al. MicroRNA-155 and microRNA-21 promote the expansion of functional myeloid-derived suppressor cells. Journal of Immunology (Baltimore, Md: 1950) . 2014;192(3):1034–1043. doi: 10.4049/jimmunol.1301309. [DOI] [PubMed] [Google Scholar]
- 58.Challagundla K. B., Wise P. M., Neviani P., et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. Journal of the National Cancer Institute . 2015;107(7) doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neviani P., Wise P. M., Murtadha M., et al. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Research . 2019;79(6):1151–1164. doi: 10.1158/0008-5472.CAN-18-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jong A. Y., Wu C. H., Li J., et al. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. Journal of Extracellular Vesicles . 2017;6(1):p. 1294368. doi: 10.1080/20013078.2017.1294368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams A. H., Liu N., van Rooij E., Olson E. N. MicroRNA control of muscle development and disease. Current Opinion in Cell Biology . 2009;21(3):461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Esau C. C., Monia B. P. Therapeutic potential for microRNAs. Advanced Drug Delivery Reviews . 2007;59(2-3):101–114. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 63.Brunet J. F., Denizot F., Luciani M. F., et al. A new member of the immunoglobulin superfamily--CTLA-4. Nature . 1987;328(6127):267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 64.Yang Q., Cao W., Wang Z., Zhang B., Liu J. Regulation of cancer immune escape: the roles of miRNAs in immune checkpoint proteins. Cancer Letters . 2018;431:73–84. doi: 10.1016/j.canlet.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 65.Abril-Rodriguez G., Ribas A. SnapShot: immune checkpoint inhibitors. Cancer Cell . 2017;31(6):848–848.e1. doi: 10.1016/j.ccell.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 66.Riley J. L., June C. H. The CD28 family: a T-cell rheostat for therapeutic control of T-cell activation. Blood . 2005;105(1):13–21. doi: 10.1182/blood-2004-04-1596. [DOI] [PubMed] [Google Scholar]
- 67.Wei J., Nduom E. K., Kong L. Y., et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro-Oncology . 2016;18(5):639–648. doi: 10.1093/neuonc/nov292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang F., Wang B., Zeng J., Sang S., Lei J., Lu Y. MicroRNA-374b inhibits liver cancer progression via down regulating programmed cell death-1 expression on cytokine-induced killer cells. Oncology Letters . 2018;15(4):4797–4804. doi: 10.3892/ol.2018.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowshanravan B., Halliday N., Sansom D. M. CTLA-4: a moving target in immunotherapy. Blood . 2018;131(1):58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y., Sun E., Li X., et al. miR-155 contributes to Df1-induced asthma by increasing the proliferative response of Th cells via CTLA-4 downregulation. Cellular Immunology . 2017;314:1–9. doi: 10.1016/j.cellimm.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 71.Huffaker T. B., Lee S. H., Tang W. W., et al. Antitumor immunity is defective in T cell-specific microRNA-155–deficient mice and is rescued by immune checkpoint blockade. The Journal of Biological Chemistry . 2017;292(45):18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sica G. L., Choi I. H., Zhu G., et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity . 2003;18(6):849–861. doi: 10.1016/S1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 73.Wang P., Li C., Zhang F., Ma X., Gai X. Clinical value of combined determination of serum B7-H4 with carcinoembryonic antigen, osteopontin, or tissue polypeptide-specific antigen for the diagnosis of colorectal cancer. Disease Markers . 2018;2018:9. doi: 10.1155/2018/4310790.4310790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S., Chen L. Co-signaling molecules of the B7-CD28 family in positive and negative regulation of T lymphocyte responses. Microbes and Infection . 2004;6(8):759–766. doi: 10.1016/j.micinf.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 75.Campayo M., Navarro A., Viñolas N., et al. A dual role for KRT81: a miR-SNP associated with recurrence in non-small-cell lung cancer and a novel marker of squamous cell lung carcinoma. PloS One . 2011;6(7, article e22509) doi: 10.1371/journal.pone.0022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glinsky G. V. Human genome connectivity code links disease-associated SNPs, microRNAs and pyknons. Cell Cycle (Georgetown, Tex) . 2009;8(6):925–930. doi: 10.4161/cc.8.6.7937. [DOI] [PubMed] [Google Scholar]
- 77.Wu D., Tang R., Qi Q., et al. Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cellular Immunology . 2015;293(1):41–48. doi: 10.1016/j.cellimm.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Yao S., Zhu Y., Zhu G., et al. B7-h2 is a co-stimulatory ligand for CD28 in human. Immunity . 2011;34(5):729–740. doi: 10.1016/j.immuni.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liakou C. I., Kamat A., Tang D. N., et al. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proceedings of the National Academy of Sciences of the United States of America . 2008;105(39):14987–14992. doi: 10.1073/pnas.0806075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen H., Liakou C. I., Kamat A., et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both non-malignant and malignant prostate tissues. Proceedings of the National Academy of Sciences of the United States of America. . 2009;106(8):2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan X., Quezada S. A., Sepulveda M. A., Sharma P., Allison J. P. Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. The Journal of Experimental Medicine . 2014;211(4):715–725. doi: 10.1084/jem.20130590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carreno B. M., Collins M. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annual Review of Immunology . 2002;20(1):29–53. doi: 10.1146/annurev.immunol.20.091101.091806. [DOI] [PubMed] [Google Scholar]
- 83.Sharpe A. H., Freeman G. J. The B7-CD28 superfamily. Nature Reviews Immunology . 2002;2(2):116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 84.Lee H., Kim J. H., Yang S. Y., et al. Peripheral blood gene expression of B7 and CD28 family members associated with tumor progression and microscopic lymphovascular invasion in colon cancer patients. Journal of Cancer Research and Clinical Oncology . 2010;136(9):1445–1452. doi: 10.1007/s00432-010-0800-4. [DOI] [PubMed] [Google Scholar]
- 85.Maciejewski R., Radej S., Furmaga J., et al. Evaluation of immature monocyte-derived dendritic cells generated from patients with colorectal cancer. Polski Przeglad Chirurgiczny . 2013;85(12):714–720. doi: 10.2478/pjs-2013-0109. [DOI] [PubMed] [Google Scholar]
- 86.Yao S., Chen L. Adaptive resistance: a tumor strategy to evade immune attack. European Journal of Immunology . 2013;43(3):576–579. doi: 10.1002/eji.201243275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang P., Tang R., Zhu J., et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Molecular Immunology . 2013;56(1-2):98–103. doi: 10.1016/j.molimm.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 88.Topalian S. L., Taube J. M., Anders R. A., Pardoll D. M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nature Reviews Cancer . 2016;16(5):275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han X., Bryson P. D., Zhao Y., et al. Masked chimeric antigen receptor for tumor-specific activation. Molecular therapy: the journal of the American Society of Gene Therapy . 2017;25(1):274–284. doi: 10.1016/j.ymthe.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gill S., Porter D. L. CAR-modified anti-CD19 T cells for the treatment of B-cell malignancies: rules of the road. Expert Opinion on Biological Therapy . 2014;14(1):37–49. doi: 10.1517/14712598.2014.860442. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z., Wu Z., Liu Y., Han W. New development in CAR-T cell therapy. Journal of Hematology & Oncology . 2017;10(1):p. 53. doi: 10.1186/s13045-017-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hamieh M., Dobrin A., Cabriolu A., et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature . 2019;568(7750):112–116. doi: 10.1038/s41586-019-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neelapu S. S. Managing the toxicities of CAR T-cell therapy. Hematological Oncology . 2019;37(Suppl 1):48–52. doi: 10.1002/hon.2595. [DOI] [PubMed] [Google Scholar]
- 94.Newick K., O’Brien S., Moon E., Albelda S. M. CAR T cell therapy for solid tumors. Annual Review of Medicine . 2017;68(1):139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Q., Hu H., Chen S. Y., et al. Transcriptome and regulatory network analyses of CD19-CAR-T immunotherapy for B-ALL. Genomics, Proteomics & Bioinformatics. . 2019;17(2):190–200. doi: 10.1016/j.gpb.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ohno M., Ohkuri T., Kosaka A., et al. Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. Journal for Immunotherapy of Cancer . 2013;1(1):p. 21. doi: 10.1186/2051-1426-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roybal K. T., Rupp L. J., Morsut L., et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell . 2016;164(4):770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Rourke D. M., Nasrallah M. P., Desai A., et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Science Translational Medicine . 2017;9(399):p. eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hirayama A. V., Turtle C. J. Toxicities of CD19 CAR-T cell immunotherapy. American Journal of Hematology . 2019;94(S1):S42–S49. doi: 10.1002/ajh.25445. [DOI] [PubMed] [Google Scholar]
- 100.Ruella M., Barrett D. M., Kenderian S. S., et al. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. The Journal of Clinical Investigation . 2016;126(10):3814–3826. doi: 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schneider D., Xiong Y., Wu D., et al. A tandem CD19/CD20 CAR lentiviral vector drives on-target and off-target antigen modulation in leukemia cell lines. Journal for Immunotherapy of Cancer . 2017;5(1):p. 42. doi: 10.1186/s40425-017-0246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. MicroRNAs: new regulators of immune cell development and function. Nature Immunology . 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 103.Boldin M. P., Taganov K. D., Rao D. S., et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. The Journal of Experimental Medicine . 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang Q., Xia J., Wang L., et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. Journal of Hematology & Oncology . 2018;11(1):1–12. doi: 10.1186/s13045-018-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fuertes T., Ramiro A. R., de Yebenes V. G. miRNA-based therapies in B cell non-Hodgkin lymphoma. Trends in Immunology . 2020;41(10):932–947. doi: 10.1016/j.it.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Witten L., Slack F. J. miR-155 as a novel clinical target for hematological malignancies. Carcinogenesis . 2020;41(1):2–7. doi: 10.1093/carcin/bgz183. [DOI] [PubMed] [Google Scholar]
- 107.Wan J., Ling X., Peng B., Ding G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncology reports. . 2018;40(1):272–282. doi: 10.3892/or.2018.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.


