Abstract
A flow-sorting technique was developed to determine unperturbed metabolic activities of phylogenetically characterized bacterioplankton groups with incorporation rates of [35S]methionine tracer. According to fluorescence in situ hybridization with rRNA targeted oligonucleotide probes, a clade of α-proteobacteria, related to Roseobacter spp., and a Cytophaga-Flavobacterium cluster dominated the different groups. Cytometric characterization revealed both these groups to have high DNA (HNA) content, while the α-proteobacteria exhibited high light scatter (hs) and the Cytophaga-Flavobacterium cluster exhibited low light scatter (ls). A third abundant group with low DNA (LNA) content contained cells from a SAR86 cluster of γ-proteobacteria. Cellular specific activities of the HNA-hs group were 4- and 1.7-fold higher than the activities in the HNA-ls and LNA groups, respectively. However, the higher cellular protein synthesis by the HNA-hs could simply be explained by their maintenance of a larger cellular protein biomass. Similar biomass specific activities of the different groups strongly support the main assumption that underlies the determination of bacterial production: different bacteria in a complex community incorporate amino acids at a rate proportional to their protein synthesis. The fact that the highest growth-specific rates were determined for the smallest cells of the LNA group can explain the dominance of this group in nutrient-limited waters. The metabolic activities of the three groups accounted for almost the total bacterioplankton activity, indicating their key biogeochemical role in the planktonic ecosystem of the Celtic Sea.
A contemporary challenge in microbial ecology is to understand the functional role of phylogenetically defined bacterial populations in natural ecosystems. Extensive biogeochemical studies have shown that bacteria constitute a major component of carbon cycling in aquatic ecosystems as the main consumers of dissolved organic matter (DOM) (see, for example, references 2, 3, 12, and 13). Although the importance of DOM mineralization is well recognized, the relative contributions of the major phylogenetic groups of bacterioplankton to DOM consumption in the sea are still under investigation.
Studies of heterogeneous natural microbial communities have been significantly improved with the introduction of new techniques. One of these uses a combination of microautoradiography and fluorescence in situ hybridization (FISH) to identify individual bacterial cells that have incorporated isotopically labeled tracers (24, 33). A second complementary technique is one that allows quantification of tracer incorporation in flow cytometrically sorted cells (6, 25, 34, 35). Several cellular optical properties can be used to distinguish bacterial groups by flow cytometry, and the groups may be flow sorted to provide concentrated group-specific sample. The sorted cells can be subsequently used for molecular (4, 39) and radioactive tracer analyses.
The ease with which bacterioplankton can be stained for nucleic acid content and analyzed cytometrically has allowed a distinction to be made between a general group of bacteria with low DNA content (LNA) and another with high DNA content (HNA) (8, 16, 26, 30, 31). However, the biological and ecological nature of cytometrically defined groups remains poorly understood. The potential of phylogenetic affiliation of cells within discrete cytometric groups was demonstrated for cyanobacteria (see, for example, references 9, 25, and 38). A recent study of bacterioplankton in the North Sea showed that each cytometric group was dominated by a different phylogenetic group of bacteria (43). However, molecular analyses of sorted nonphototrophic bacteria have demonstrated that cytometric groups could also be comprised of different phylogenetic groups of bacteria (14).
The aim of the present study was to extend these recent developments in bacterial phylogeny by determining the in situ metabolic activities of dominant groups of marine bacterioplankton. Our approach has been to use a radioactively labeled amino acid tracer to quantify cellular activity. We hypothesized that all bacterioplankton cells incorporate the tracer, with incorporation rate indicating cellular metabolic activities, and that these could be related to cellular biomass. To test the hypothesis, we developed a sensitive tracer technique and applied it to natural bacterioplankton communities of the Celtic Sea. We compared the in situ metabolic activities of cells flow sorted from the dominant groups of bacterioplankton collected in water layers with different biogeochemical regimens. The selected stations offered a range of hydrological conditions suitable for revealing general trends by using a pooled data set of bacterioplankton activity.
MATERIALS AND METHODS
Sampling site.
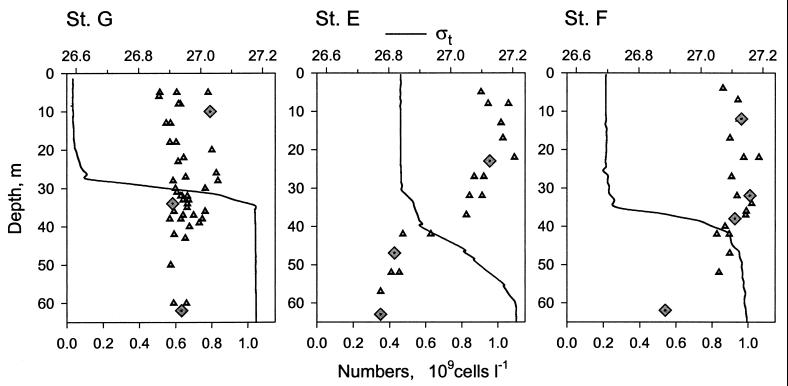
The study was undertaken on cruise D246 on board the R.R.S. Discovery in shelf waters of the Celtic Sea between 22 and 28 May 2000. Three stations with different hydrographic regimens were chosen: one in the vicinity of the Irish coast (station G, 51o30′N, 7o20′W), one close to the shelf break (station E, 50o11′N, 8o36′W), and one in the middle of the Celtic Sea Deep (station F, 51o13′N, 6o23′W). The stations had a pronounced vertical density gradient, i.e., a pycnocline, that separated the surface mixed layer from deeper waters (Fig. 1). Because it was affected by internal waves, the pycnocline spanned 25 to 30 m at station E, while in more coastal waters the pycnocline was sharper at only 5 to 10 m thick. Each station was sampled two to four times at 12-h intervals. Seawater was collected with a rosette of Niskin bottles mounted on a Neil Brown Mk3C conductivity-temperature-depth profiler (General Oceanics). Ten to twelve depths were sampled from the top 65 m of 80- to 130-m water columns with a depth resolution of 1 to 10 m. The pycnocline layer was sampled with the highest depth resolution. Bacterial abundance was determined at all depths sampled. Rates of bacterioplankton incorporation of [3H]leucine and [35S]methionine were measured in water samples collected at six depths on each CTD cast. The samples used for rate determinations were initially collected into acid-washed 1-liter thermos flasks by using acid-soaked silicone tubing and then processed within 1 h of sampling. In addition, 10 selected samples representative of the three distinct water layers, namely, the surface mixed layer, the pycnocline, and the deep-water layer at the three stations, were used for more detailed analyses (diamonds, Fig. 1). The analyses included flow sorting of three main groups of bacterioplankton for FISH and tracer studies.
FIG. 1.
Vertical distribution of seawater density and total bacterioplankton numbers at three studied stations in the Celtic Sea (St. G, E, and F). Lines show the individual profiles of seawater density index, ςt, triangles show the bacterial numbers at sampled depths, and diamonds indicate the samples chosen for flow-sorting studies. Seawater density (in kilograms decimeter−3) is equal to 1 + (ςt × 10−3).
Flow cytometry.
For flow cytometric analyses, replicated 2-ml samples were fixed with 1% paraformaldehyde (PFA) and incubated at 2°C for 24 h. Subsequently, the samples were frozen and kept at −20°C. The abundance of heterotrophic bacteria was routinely determined with a FACSort flow cytometer (Becton Dickinson, Oxford, United Kingdom) after staining with SYBR Green I DNA stain as described previously (30, 42). Bacterioplankton cellular protein contents were determined with a FACStar Plus flow cytometer (Becton Dickinson, Mountain View, Calif.) after simultaneous staining with Hoechst 33342 DNA stain and SYPRO Red protein stain (41). The SYPRO protein cell fluorescence was calibrated by using four bacterial cultures. Yellow-green microspheres of 0.5 μm (Fluoresbrite Microparticles; Polysciences, Warrington, Pa.) were used in all analyses as an internal standard to normalize samples and to calculate bacterial protein content and bacterial concentration. The absolute concentration of beads in a standard stock suspension was determined by flow cytometric counting of beads in volumes dispensed with an automatic microinjector (KD Scientific). Bacterial biomass was calculated by multiplying bacterial concentration with the mean protein content of bacterial cells.
Identification of flow-sorted bacteria.
Cells, double stained with Hoechst 33342 and SYPRO Red, were sorted by using the FACStar flow cytometer. FISH with sorted cells harvested on 0.2-μm (pore-size) polycarbonate filters was done according to the protocol of Glöckner et al. (17) with some modifications (14), including the use of helper probes (15). The sorted cells were hybridized with a set of probes listed in Table 1. New probes were developed with the ARB program package (O. Strunk, B. Gross, B. Reichel, M. May, S. N. Hermann, et al. [http://www.mikro.biologie.tu-muenchen.de.]). The specificity of probes was ensured by altering the formamide concentration of the hybridization buffer used for FISH with natural samples. The probes were commercially synthesized and labeled with CY3 dye (Interactiva, Ulm, Germany). Cells were viewed by using an Axioplan epifluorescence microscope equipped with a 100′ Plan Neofluar objective (Zeiss, Jena, Germany), and at least 300 cells were counted per sorted sample. Probe-positive cells were presented as fractions of cells stained with the general nucleic acid dye DAPI (4′,6′-diamidino-2-phenylindole).
TABLE 1.
Probes used in the present studya
| Probe | Probe sequence (5′→3′) | Specificity | FA (%) | Source or reference |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | Bacteria | 0 | 1 |
| ALF968 | GGTAAGGTTCTGCGCGTT | α-Proteobacteria | 35 | 18 |
| GAM42a | GCCTTCCCACATCGTTT | γ-Proteobacteria | 35 | 28 |
| CF319a | TGGTCCGTGTCTCAGTAC | CF cluster | 35 | 29 |
| COL189 | AUCCCCCCCUUUGGUCCG | Colwellia clade | 35 | This study |
| COL189h5 | TAGACRTTATGCGGTATTAGC | Helper for COL189 | 35 | This study |
| COL189h3 | CAAATGGCGAGAGGTCCGAAG | Helper for COL189 | 35 | This study |
| SAR86/1245 | TTAGCGTCCGTCTGTAT | SAR86 cluster | 20 | This study |
| SAR86/1245h5 | CCATTGTAGCACGTGTGTAGC | Helper for SAR86/1245 | 20 | This study |
| SAR86/1245h3 | GGATTRGCACCACCTCGCGGC | Helper for SAR86/1245 | 20 | This study |
| RSB67 | CGCTCCACCCGAAGGTAG | Roseobacter clade | 20 | 43 |
| RSB67h5 | CGCTCGACTTGCATGTGT | Helper for RSB67 | 20 | 43 |
| RSB67h3 | CGTTACTAACCCGTCCGC | Helper for RSB67 | 20 | 43 |
Abbreviations: FA, formamide; CF, Cytophaga-Flavobacterium.
Amino acid incorporation and bacterioplankton production.
Two amino acids, [3H]leucine and [35S]methionine, were used as precursors. The [3H]leucine incorporation rates estimated production of bacterioplankton biomass (21). The [35S]methionine was added at tracer concentration to determine the in situ rates of amino acid incorporation and therefore the protein synthesis of bacterioplankton. The latter rates were used as an index of cellular metabolic activity.
Triplicate subsamples (1 ml) from samples collected at six depths were inoculated with l-[4,5-3H]leucine (63 Ci mmol−1; ICN Pharmaceuticals, Ltd.) at a 20 nM final concentration or with l-[35S]methionine (>1,000 Ci mmol−1; Amersham Pharmacia Biotech UK, Ltd.) at a <0.3 nM final tracer concentration and then incubated in the dark at the in situ temperature of the surface mixed layer. The whole content of one of three tubes was fixed at 0.5, 1, or 1.5 h by mixing with an equal volume of 10% trichloroacetic acid (TCA). The sample particulate material was harvested onto 0.2-μm (pore-size) nylon filters (Supor; Pall Corporation). Radioactivity incorporated into TCA-insoluble material was counted with a RackBeta 1219 liquid scintillation counter (LKB-Wallac, Turku, Finland). The rate of precursor incorporation was calculated as the slope of the linear regression of radioactivity against incubation time (r2 > 0.99, P < 0.0001). Incorporation rate constants, i.e., the fractions of the added precursor incorporated by bacterioplankton per hour, were used for comparison. Leucine incorporation rates were converted into bacterial biomass production by using a previously determined conversion factor of 640 g of bacterial protein per mol of incorporated leucine (43).
Flow sorting of bacterioplankton cells labeled with [35S]methionine tracer
Methionine incorporation by bacterioplankton was measured as described above with minor modifications, i.e., subsamples were fixed with 1% PFA after a 3-h incubation. Subsamples were stored at 2°C before they were stained with SYBR Green I, and groups were flow sorted by using a FACSort flow cytometer set to single-cell sort mode. Sorted cells were collected onto 0.2-μm (pore-size) nylon filters and washed with 10 ml of ultrapure water (18.2 MΩ resistivity), and the filters were subsequently radioassayed. Three proportional numbers (e.g., 2,000, 4,000, and 6,000) of the cells were sorted, and the cellular activity was determined as the slope of the linear regression of radioactivity against the number of sorted bacteria (r2 > 0.96, P < 0.0001). A mean coefficient of variance for the measured activities of sorted cells was 6%.
RESULTS
Intercalibration of sorting gates of the two flow cytometers used.
Limitations in instrument design prevent the use of a single flow cytometer for all analyses. Accurate cellular DNA and protein measurements were not possible with a single laser FACSort instrument, while flow sorting of radioactively labeled bacteria with the drop sorter of a FACStar instrument was not safe. Therefore, different flow cytometers were used for sorting bacterioplankton groups for FISH and tracer studies.
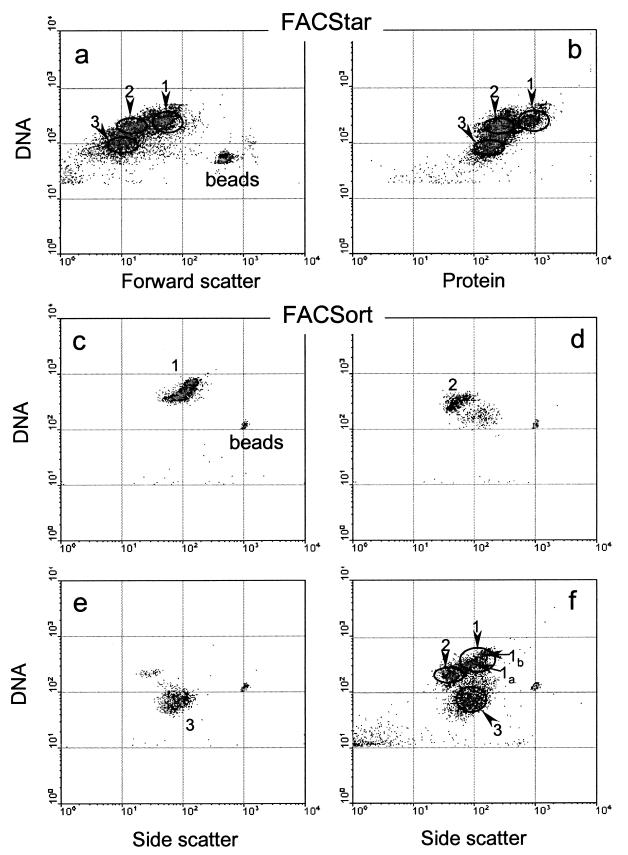
A cross calibration of flow cytometers was undertaken to ensure that the same groups were recognized and sorted by the two instruments. Three dominant groups were visualized on a flow cytometric signature of bacterioplankton double stained for DNA and protein. Sorting gates were drawn around the core of each group, and target cells were sorted by using the FACStar instrument (Fig. 2a and b). Subsequently, the sorted cells were reanalyzed with the FACSort instrument. The sorted cells were seen as tight clusters by the second flow cytometer (Fig. 2c to e). The positions of these clusters matched the positions of the three main groups of bacterioplankton in a natural sample analyzed on the second instrument (Fig. 2f). There was good agreement between the sorting gates on the two instruments, with 95% ± 3% (n = 8) of cells sorted from gate 1 of the FACStar recorded in gate 1 of the FACSort. The level of agreement between the other two gates was slightly lower, with 75% ± 15% (n = 8) and 80% ± 10% (n = 10) of cells sorted on the FACStar instrument recorded on the FACSort instrument in gates 2 and 3, respectively. Thus, we could conclude that the same groups of bacterioplankton were targeted by the two flow cytometers.
FIG. 2.
(a and b) Flow-cytometric signatures of natural bacterioplankton stained for nucleic acids and proteins, with superimposed gates used for sorting three distinct groups by the FACStar instrument. (c to f) Flow-cytometric signatures of sorted HNA-hs, HNA-ls, and LNA (1, 2, and 3, respectively) groups (c, d, and e, respectively) reanalyzed by the FACSort instrument and compared with the flow-cytometric signature of the same unsorted bacterioplankton sample (f), with the gates and regions used for sorting on the second instrument indicated.
Phylogenetic affiliation of sorted cells.
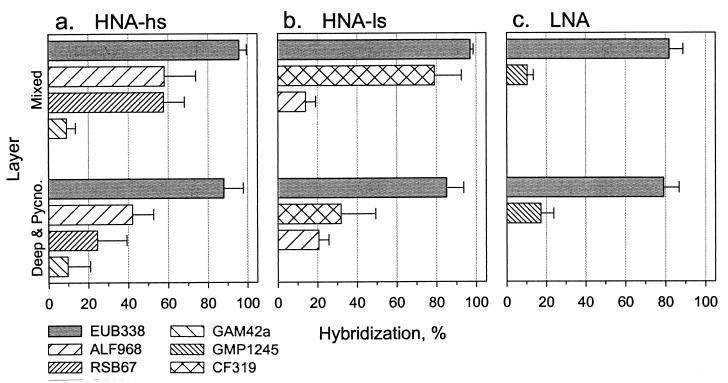
Bacterial cells sorted from the groups were identified by FISH. Because the composition of cells sorted from the bacterioplankton samples at the three stations was similar, the FISH results were pooled (Fig. 3). For the same reason, the FISH results for the pycnocline and deep layer were also combined. In fact, no major differences in bacterioplankton community structure were detected between the surface mixed layer and deeper waters. However, a higher percentage of cells hybridized with oligonucleotide probes in the mixed layer compared to deeper waters.
FIG. 3.
FISH with cells flow sorted from three groups (HNA-hs [a], HNA-ls [b], and LNA [c]) of bacterioplankton, collected in the surface mixed layer (Mixed) and the combined deep and pycnocline layers (Deep & Pycno.). Hybridization results are presented as percentages of probe-positive cells stained with DAPI. Probes: EUB338 for Bacteria, ALF968 for α-proteobacteria, RSB67 for the Roseobacter clade, GAM42a for γ-proteobacteria, SAR86/GMP1245 for the SAR86 cluster of γ-proteobacteria, CF319a for the Cytophaga-Flavobacterium cluster (see Table 1 for details).
Between 79 and 97% of sorted cells belonged to the domain Bacteria (Table 1, Fig. 3). Proteobacteria of the α subdivision numerically dominated the cells sorted from the HNA-high light scatter (hs) group, i.e., 60 and 40% of cells in the mixed layer and the deeper waters, respectively (Fig. 3a). Furthermore, one particular clade related to Roseobacter spp. (43) represented most of these α-proteobacteria. Proteobacteria of the γ subdivision were another relatively abundant group, although they represented ≤10% of all sorted cells. The other phylogenetic groups of bacteria tested (Table 1), i.e., the SAR86 cluster, the Cytophaga-Flavobacterium cluster, and the Colwellia clade, represented even smaller percentages of the total cells. The Cytophaga-Flavobacterium cluster dominated cells sorted from the HNA-low light scatter (ls) group, comprising 80 and 30% of the bacteria in the mixed layer and the deeper waters, respectively (Fig. 3b). The α-proteobacteria comprised 15 to 20% of all sorted cells from the HNA-ls group. The α-proteobacteria detected in this group could possibly be a result of sorting impurities (Fig. 2d). Tiny bacteria from the LNA group were more difficult to identify by FISH. The only probe that gave reproducible hybridization with these cells was the SAR86 probe, and even then only 10 to 20% of cells could be detected and these showed a weak signal (Fig. 3c).
Cellular and biomass specific activities of sorted groups.
The three cytometric groups were also sorted in order to estimate group-specific metabolic activity (Fig. 2f). In fact, two different regions of the HNA-hs group (Fig. 2f, 1a and 1b) were sorted on seven of ten occasions. On average, the cellular activity of bacteria sorted from region 1b was 2.7 ± 0.45 times higher than the cellular activity of bacteria sorted from region 1a. The α-proteobacteria comprised 55% ± 13% and 64% ± 26%, Roseobacter spp. comprised 51% ± 17% and 58% ± 15%, and γ-proteobacteria comprised 7% ± 2.5% and 15% ± 4.5% of cells sorted from regions 1a and 1b, respectively. Because the phylogenetic compositions of bacteria within regions 1a and 1b were similar, the activities of cells for regions 1a and 1b were pooled together to determine the mean activities of bacteria in the HNA-hs group.
Using the determined activities and proportions of the total number of cells in each of the three groups, we calculated the activity of an average bacterioplankton cell in each analyzed sample and compared them with the estimated mean activities of all bacterioplankton cells. The latter values were calculated by dividing the measured rates of [35S]methionine tracer incorporation per milliliter by the total number of bacterioplankton cells per milliliter. The sorted cells accounted for 93% of the total bacterioplankton activity. The different fixation procedures used for bulk measurements (5% TCA) and bacterioplankton sorting (1% PFA) did not affect the determined rates. This is in close agreement with earlier studies, wherein oceanic bacterioplankton, labeled with either [3H]thymidine or [14C]leucine and harvested unfixed and fixed (5% TCA) at the end of incubations, had very similar radioactivity independent of fixation (40).
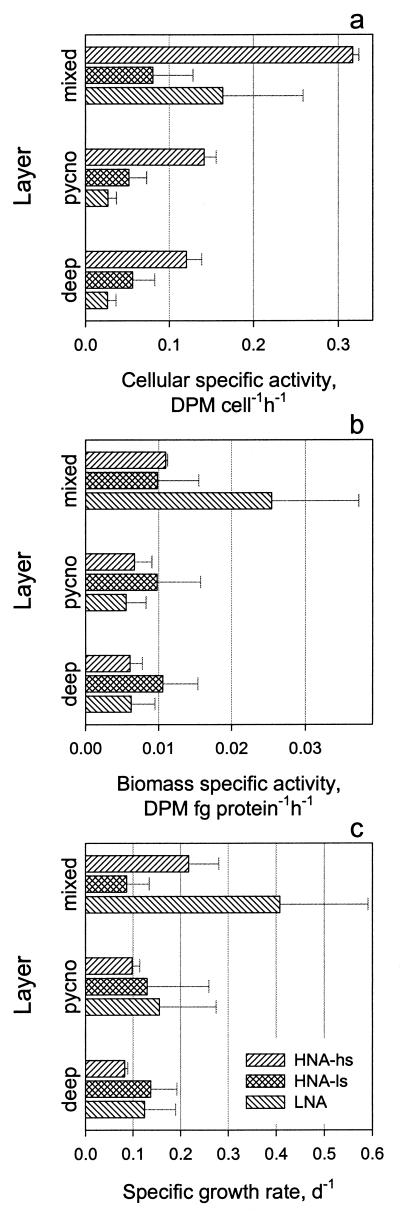
The rates of [35S]methionine tracer incorporation by sorted cells from the HNA-hs group were approximately twice as high in the surface mixed layer as in either the pycnocline or the deep-water layers (Fig. 4a). Also the activities of cells from the HNA-hs group were considerably higher than the activities of cells from the HNA-ls and LNA groups. The activities of cells from the HNA-ls group were less variable between water layers. Activities of cells from the LNA group reached high values in the surface mixed layer and were comparable with activities of cells in the HNA-hs group. This was somewhat surprising because the mean protein content of cells from the LNA group was only ca. 20% of the protein content of cells from the HNA-hs group.
FIG. 4.
Comparison of cellular specific activities (a), corresponding biomass specific activities (b), and specific growth rates (c) of cells sorted from the bacterioplankton groups (HNA-hs, HNA-ls, and LNA) from the surface mixed layers (mixed), pycnocline (pycno), and deep-water layers (deep) sampled. Error bars indicate a single standard deviation of specific activities determined at three studied stations.
Cellular protein contents, averaged for all stations, were significantly higher (t test, α = 5%) in the surface mixed layer than in deeper waters: 31 ± 4 versus 22 ± 8 fg of protein cell−1, 7.4 ± 1.3 versus 5.7 ± 1.3 fg of protein cell−1, and 6.3 ± 0.9 versus 4.7 ± 0.7 fg of protein cell−1 for the HNA-hs, HNA-ls, and LNA groups, respectively. Consequently, in the surface mixed layer the LNA group had 2.5 times higher biomass specific activity than the HNA groups (Fig. 4b). The biomass specific activity of the HNA-ls group remained constant throughout the water column, while the activities of the other two groups decreased in deeper waters. The biomass specific activities of the LNA and HNA-hs groups were remarkably similar in the pycnocline and deep layers despite a nearly fivefold difference in protein content.
Contributions of groups to total bacterioplankton biomass and activity.
Despite the sharp pycnocline, the vertical distribution of bacterioplankton at station G was uniform at ca. 0.6 × 109 cells liter−1 throughout the water column, while the surface mixed layer was enriched with bacterioplankton at the other two stations (Fig. 1). The concentrations of total bacterioplankton decreased from ca. 0.9 × 109 cells liter−1 in the surface mixed layer to 0.45 × 109 cells liter−1 in the deep layer at stations E and F. Cells from the HNA-ls group were the most numerous in the surface layer, constituting ca. 45% of the bacterioplankton. Their contribution to the total numbers was lower at depth, representing ca. 30%. The percentage of cells from the HNA-hs group gradually decreased with depth from ca. 40 to 15%, while the percentage of cells from the LNA group increased with depth from 10 to 60%. The latter numerically dominated the deep-water layer. The total biomass of bacterioplankton, 10 to 15 μg of protein liter−1, was significantly higher in the surface mixed layer than in the deep layer, i.e., 2 to 4 μg of protein liter−1. The HNA-hs group dominated the bacterioplankton biomass in the water column, particularly in the surface layer, where it accounted for almost 80% of biomass (Fig. 5). The HNA-ls group contributed ca. 20% to biomass in all layers, and the contribution of the LNA group rose with depth from <10 to 25%.
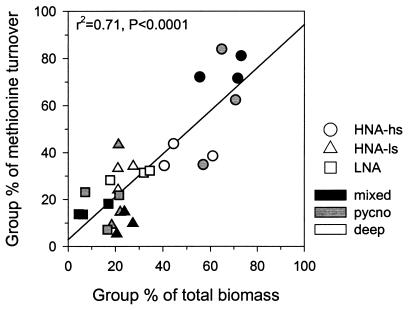
FIG. 5.
Comparison of sorted group (HNA-hs, HNA-ls, and LNA) contribution to total bacterioplankton biomass, with corresponding group contributions to total metabolic activities. A solid line shows statistically significant linear regression; r2 is the corresponding regression coefficient.
Methionine was rapidly incorporated by bacteria at a rate of 4 to 7% h−1 (1 to 1.7 times day−1) and 0.5 to 2% h−1 (0.1 to 0.5 times day−1) in the mixed layer and deeper layers, respectively. It was apparent that the HNA-hs group was the major contributor (70 to 80%) to bacterioplankton incorporation of methionine in the surface mixed layer and pycnocline layer, but the contributions of all three groups was comparable in the deep layer (Fig. 5). There was a significant positive relationship between the contribution to bacterioplankton biomass and to bacterioplankton metabolic activity by each group (Fig. 5). Additionally, the bacterial production determined as the rate of leucine incorporation was strongly correlated with the abundance of the HNA-hs group (r2 = 0.84, n = 43, P < 0.0001) but was not significantly related to the abundance of either of the other two groups (r2 ≤ 0.3) or to total bacterioplankton abundance (r2 = 0.5).
Correlation between incorporation rates of methionine and bacterioplankton production.
Bacterial production was determined by adding 20 nM [3H]leucine that deliberately elevated ambient amino acid concentration. Rates of [35S]methionine incorporation (fraction hour−1) strongly correlated with the corresponding estimates of bacterial production (micrograms of protein liter−1 day−1) (slope, 62.3 ± 1.8; r2 = 0.88; n = 48; P < 0.0001). The strong correlation between [35S]methionine (<0.3 nM) and [3H]leucine (20 nM) incorporation rates by bacterioplankton in the Celtic Sea agrees with the observations of limnic bacterioplankton in summer (22). The strong relationship between the two parameters allowed the estimation of group-specific growth rates (Fig. 4c). The methionine incorporation rate constants of the groups were converted into leucine incorporation rates by using the regression calculation described above. Group-specific growth rates were calculated from the values of group-specific production and group-specific protein biomass of each bacterioplankton group. The group-specific growth rates were within the range of values reported for oceanic bacterioplankton (see, for example, reference 42). The LNA group had a growth rate of 0.4 day−1 in the surface mixed layer two and four times higher than the growth rates of HNA-hs and HNA-ls groups, respectively. In the pycnocline and deeper waters, all groups had relatively similar growth rates of ca. 0.12 day−1 (Fig. 4c).
DISCUSSION
Determination of in situ metabolic activity of flow-sorted bacterioplankton.
Microbial activity is often measured as the rate of incorporation of isotopically labeled compounds into microbial biomass. Flow cytometric sorting has been employed successfully for determining ultraphytoplankton group-specific assimilation of CO2 (25, 34) and inorganic nitrogen (27). Specific activities of bacterial groups have also been determined by using 3H-labeled amino acids (mixed) (6, 7) or [3H]leucine (35). In these experiments, labeled amino acids were added at concentrations up to 80 nM and, as a result, elevated the ambient concentrations of amino acids in seawater (see, for example, reference 20). In such amino acid enrichment experiments, the activities of some groups of bacteria could be overestimated because of their ability to use the added compound as an alternative nutrient. It was also recently shown that α-proteobacteria could differ in their preferences for amino acids compared to bacteria from the Cytophaga-Flavobacterium cluster (10).
By using nonperturbing, tracer concentrations of [35S]methionine (<0.3 nM), we could monitor natural levels of protein synthesis in cells sorted from targeted groups. In addition to the advantages of a tracer study, the high specific activity of [35S]methionine allowed us to sort fewer cells (500 to 20,000) from selected groups for reliable determination of their activity. In previous studies, experiments were not replicated presumably because of the long time required for sorting a sufficient number (50,000 to 500,000) of labeled cells (5, 23).
The disadvantage of flow cytometry is that it is limited to separating cells based on DNA, protein contents, and light scatter, and a cytometric group can be phylogenetically diverse. The advantage of using FISH microautoradiography is in enumerating cells in a specific phylogenetic group that are incorporating a particular compound. However, the flow-sorting technique provides more biogeochemically relevant information, such as what fraction of total incorporation can be attributed to a cytometric group, which in certain cases can be dominated by one phylogenetic group or even a single clade of bacteria (Fig. 3a and b). Accurate measurements of cellular activity can be done by using the latter technique, and the measured activity is a mean value for a number of sorted cells. The sorting technique also allows analysis of variability within groups, e.g., in the same HNA-hs group larger cells that were richer in protein were more metabolically active than smaller cells.
The other noteworthy result of the present study was that the activities of the sorted groups accounted for 93% of the total bacterioplankton activity. This implies that the studied groups were ecologically very important, and the contribution of bacteria growing on suspended detrital particles or algae was insignificant. However, it is important to note that we measured total activity in relatively small volumes of seawater, and it is possible that large detrital particles, which can be hot spots of bacterial activity (2), could be underrepresented.
Phylogenetic composition of cytometric groups of marine bacterioplankton.
We found that the percentage of cells detected by FISH with specific probes was higher in the surface layer than in deeper waters, presumably because of lower cellular activities of bacteria in deeper waters (Fig. 4a). Although a range of side scatter and DNA obviously occurs within each group, phylogeny seems to be constant, e.g., groups 1a and 1b. Two cytometric groups were dominated by particular phylogenetic groups of bacteria (Fig. 3). The HNA-hs group was dominated by the Roseobacter-related clade of α-proteobacteria. Interestingly, this clade seems to be widespread since it also dominated the bacterioplankton in the North Sea (43). The HNA-ls group was dominated by the Cytophaga-Flavobacterium cluster. About 80% of the smallest cells sorted from the LNA group hybridized with general bacterial probe Eub338, but only 10 to 20% of cells could be identified by one of the probes tested, the one specific to the SAR86 cluster (32). The difference may possibly be explained by additional diversity not covered by the set of probes used (Table 1). The complete characterization of the phylogenetic diversity of the LNA group is beyond the scope of this study and will be addressed in forthcoming work.
Although the Cytophaga-Flavobacterium cluster often numerically dominates marine bacterioplankton (see, for example, references 11, 18, 37, and 43), these bacteria had very low growth rates (<0.1 day−1) in the surface mixed layer of the Celtic Sea (Fig. 4c). It was the clade of α-proteobacteria that seemed to dominate bacterioplankton biomass and production during the early summer (see, for example, reference 43 and the present study). Possibly, the efficient utilization of seasonally abundant dissolved organic compound or compounds, produced by proliferating phytoplankton, e.g., dimethylsulfoniopropionate, can give these α-proteobacteria an edge in competition with other bacterioplankton (43). However, these bacteria showed similar or lower biomass specific activities than other groups (Fig. 4b), indicating that the HNA-hs group is vulnerable to low DOM in the deeper waters, where its contribution to bacterioplankton activity is considerably decreased (Fig. 5).
Keeping in mind the limitation of the FISH methodology, we can cautiously conclude that the structure of the bacterioplankton community was generally similar in the water column at the three studied stations in the Celtic Sea and was comparable to the bacterioplankton community structure in the North Sea in early summer (43).
Specific activities of discriminated groups.
The fact that biomass specific activities of different groups were relatively uniform, except for high activity of LNA in the mixed layer (Fig. 4b), has several interesting and important ramifications. First, higher cellular protein synthesis by HNA-hs could be explained by maintaining a larger cellular protein biomass. Second, the relatively similar biomass specific activities of different groups strongly supports the validity of the main assumptions that underlie bacterioplankton production determination. Various bacteria in a complex planktonic community incorporate amino acids at a rate proportional to their protein synthesis (21, 36), and consequently a single conversion factor can be employed to translate the former rate into the latter. Third, the fact that the highest biomass specific activity and growth-specific rates (Fig. 4c) were determined for the small cells of the LNA group can explain dominance of this group in oligotrophic oceanic waters (M. V. Zubkov, unpublished data).
Discrimination between bacterial groups with high and low DNA content has been extensively discussed in the literature (see, for example, references 8, 16, 26, 30, and 31). The HNA-hs and HNA-ls groups pooled together were equivalent to the previously identified single group of HNA cells. In the present study this pooled HNA group accounted for 70 to 90% of the total bacterioplankton activity (Fig. 5). More interestingly, according to established views, LNA cells are either inactive or even dead cells or cell fragments (see, for example, references 16, 19, and 23). The present measurements disagree with this view, because cells in the LNA group were at least as active as other members of the community (Fig. 4). Comparable cellular activities for HNA and LNA bacteria were also reported for freshwater bacterioplankton (7). However, the contribution of LNA bacteria to the total bacterioplankton activity in the surface mixed layer did not exceed 15% (Fig. 5) and was even less in more-productive waters (e.g., see reference 23). In deeper waters, the contribution by LNA was more significant, representing up to 30% of total bacterial activity.
In the present study and in the North Sea (43), a significant correlation between bacterial production and concentration of the HNA-hs group was found. Similar correlations between the pooled HNA group and total bacterioplankton activity were found by other researchers (23). The number of HNA bacteria has been suggested as an index for estimating bacterioplankton productivity (16). However, since the HNA-hs do not numerically dominate the HNA, the idea may be misleading, and we caution against relying on numerical dominance as an indicator of group importance in the community. The HNA-ls group dominated by the Cytophaga-Flavobacterium cluster was the most abundant in the surface mixed layer, although its growth-specific activity was the lowest.
Thus, the flow-sorting technique is a promising tool for studying competition for dissolved organic and inorganic compounds between phylogenetically distinct groups within microbial communities. Quantification of these competitive interactions will help to explain mechanisms of microbial control of biogeochemical cycles in aquatic ecosystems.
ACKNOWLEDGMENTS
M.V.Z. thanks Stephen Archer for many stimulating discussions. We thank the anonymous reviewers for helpful critical comments.
This study is a part of a multidisciplinary program, Production and Physical Interactions in the Euphotic Zone (PROPHEZE), of the Plymouth Marine Laboratory and was supported by the U.K. Natural Environment Research Council (NERC) and the Max-Planck Society. This study is part of the NERC Marine and Freshwater Microbial Biodiversity (M&FMB) thematic programme (NER/T/S/2000/00635). The research of M.V.Z. was supported by an NERC postdoctoral research fellowship (GT5/98/16/MSTB).
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 3.Azam F, Fenchel T, Feild J G, Gray J S, Meyer-Reil L-A, Thingstad F. The ecological role of water column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 4.Bernard L, Schäfer H, Joux F, Courties C, Muyzer G, Lebaron P. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat Microb Ecol. 2000;23:1–11. [Google Scholar]
- 5.Bernard L, Courties C, Servais P, Troussellier M, Petit M, Lebaron P. Relationships among bacterial cell size, productivity, and genetic diversity in aquatic environments using cell sorting and flow cytometry. Microb Ecol. 2000;40:148–158. doi: 10.1007/s002480000046. [DOI] [PubMed] [Google Scholar]
- 6.Button D K, Robertson B R. Use of high-resolution flow cytometry to determine the activity and distribution of aquatic bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 163–173. [Google Scholar]
- 7.Button D K, Robertson B R. Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol Oceanogr. 2000;45:499–505. [Google Scholar]
- 8.Button D K, Robertson B R, Jüttner F. Microflora of a subalpine lake: bacterial populations, size and DNA distributions, and their dependence on phosphate. FEMS Microbiol Ecol. 1996;21:87–101. [Google Scholar]
- 9.Chisholm S W, Olson R J, Zettler E R, Goericke R, Waterbury J B, Welschmeyer N A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- 10.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell M T, Kirchman D L. Community composition of marine bacterioplankton determined by 16S rDNA clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol. 2000;66:5116–5122. doi: 10.1128/aem.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducklow H. Bacterial production and biomass in the oceans. In: Kirchman D L, editor. Microbial ecology of the oceans. New York, N.Y: Wiley-Liss; 2000. pp. 85–120. [Google Scholar]
- 13.Ducklow H, Carlson C A. Oceanic bacterial production. Adv Microb Ecol. 1992;12:113–181. [Google Scholar]
- 14.Fuchs B M, Zubkov M V, Sahm K, Burkill P H, Amann R. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ Microbiol. 2000;2:191–201. doi: 10.1046/j.1462-2920.2000.00092.x. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs B M, Glöckner F O, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasol J M, Zweifel U L, Peters F, Fuhrman J A, Hagström Å. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol. 1999;65:4475–4483. doi: 10.1128/aem.65.10.4475-4483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 18.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton composition in lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jellett J F, Li W K W, Dickie P M, Boraie A, Kepkay P E. Metabolic activity of bacterioplankton communities assessed by flow cytometry and single carbon substrate utilization. Mar Ecol Prog Ser. 1996;136:213–225. [Google Scholar]
- 20.Kiel R G, Kirchman D L. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquatic Microb Ecol. 1999;18:293–300. [Google Scholar]
- 21.Kirchman D L, K'nees E, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural waters. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchman D L, Newell S Y, Hodson R. Incorporation versus biosynthesis of leucine: implications for measuring rates of protein synthesis and biomass production by bacteria in marine systems. Mar Ecol Prog Ser. 1986;32:47–59. [Google Scholar]
- 23.Lebaron P, Servais P, Agogué H, Courties C, Joux F. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl Environ Microbiol. 2001;67:1775–1782. doi: 10.1128/AEM.67.4.1775-1782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W K W. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 26.Li W K W, Jellett J F, Dickie P M. DNA distribution in planktonic bacteria stained with TOTO or TO-PRO. Limnol Oceanogr. 1995;40:1485–1495. [Google Scholar]
- 27.Lipschultz F. Nitrogen-specific uptake rates of marine phytoplankton isolated from natural populations of particles by flow cytometry. Mar Ecol Prog Ser. 1995;123:245–258. [Google Scholar]
- 28.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 30.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monger B C, Landry M R. Flow cytometric analysis of marine bacteria with Hoechst 33342. Appl Environ Microbiol. 1993;59:905–911. doi: 10.1128/aem.59.3.905-911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 33.Ouverney C C, Fuhrman J A. Combined microautoradiography: 16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivkin R B, Phinney D A, Yentsch C M. Effects of flow cytometric analysis and cell sorting on photosynthetic carbon uptake by phytoplankton in cultures and from natural populations. Appl Environ Microbiol. 1986;52:935–938. doi: 10.1128/aem.52.4.935-938.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Servais P, Courties C, Lebaron P, Troussellier M. Coupling bacterial activity measurements with cell sorting by flow cytometry. Microb Ecol. 1999;38:180–189. doi: 10.1007/s002489900160. [DOI] [PubMed] [Google Scholar]
- 36.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 37.Simon M, Glöckner F O, Amann R. Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat Microb Ecol. 1999;18:275–284. [Google Scholar]
- 38.Vaulot D, Marie D, Olson R J, Chisholm S W. Growth of Prochlorococcus, a photosynthetic prokaryote, in the Equatorial Pacific Ocean. Science. 1995;268:1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- 39.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zubkov M V, Sleigh M A, Burkill P H. Measurements of bacterivory by protists in open ocean waters. FEMS Microbiol Ecol. 1998;27:85–102. [Google Scholar]
- 41.Zubkov M V, Fuchs B M, Eilers H, Burkill P H, Amann R. Determination of total protein content of bacterial cells using SYPRO staining and flow cytometry. Appl Environ Microbiol. 1999;65:3251–3257. doi: 10.1128/aem.65.7.3251-3257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zubkov M V, Sleigh M A, Burkill P H, Leakey R J G. Bacterial growth and grazing loss in contrasting areas of North and South Atlantic. J Plankton Res. 2000;22:685–711. [Google Scholar]
- 43.Zubkov M V, Fuchs B M, Archer S D, Kiene R P, Amann R, Burkill P H. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ Microbiol. 2001;3:304–311. doi: 10.1046/j.1462-2920.2001.00196.x. [DOI] [PubMed] [Google Scholar]