Abstract
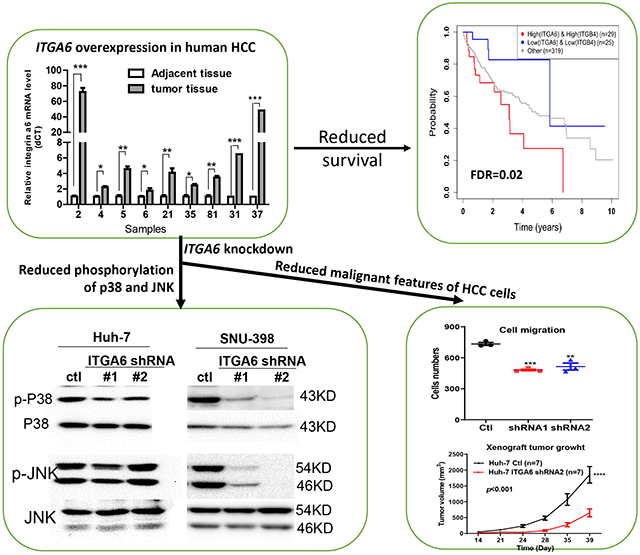
Integrin α6 (ITGA6) forms integrin receptors with either integrin β1 (ITGB1) or integrin β4 (ITGB4). How it functions to regulate hepatocellular carcinoma (HCC) progression is not well-elucidated. We found that ITGA6 RNA and protein expression levels are significantly elevated in human HCC tissues in comparison with paired adjacent non-tumor tissues by RNA sequencing, RT-qPCR, Western blotting and immunofluorescence staining. Stable knockdown of ITGA6 with different ITGA6 shRNA expression lentivectors significantly inhibited proliferation, migration and anchorage-independent growth of HCC cell lines in vitro, and xenograft tumor growth in vivo. The inhibition of anchorage-dependent and -independent growth of HCC cell lines was also confirmed with anti-ITGA6 antibody. ITGA6 knockdown was shown to induce cell-cycle arrest at G0/G1 phase. Immunoprecipitation assay revealed apparent interaction of ITGA6 with ITGB4, but not ITGB1. Expression studies showed that ITGA6 positively regulates the expression of ITGB4 with no or negative regulation of ITGB1 expression. Finally, while high levels of ITGA6 and ITGB4 together were associated with significantly worse survival of HCC patients in TCGA data set, the association was not significant for high levels of ITGA6 and ITGB1. In conclusion, ITGA6 is up-regulated in HCC tumors and has a malignant promoting role in HCC cells through integrin α6β4 complex. Thus, integrin α6β4 may be a therapeutic target for treating patients with HCC.
Keywords: Hepatocellular carcinoma, integrin α6, integrin α6β4, proliferation, migration
Graphical Abstract
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the fourth leading cause of cancer-related mortalityworldwide.1, 2 Although diagnostic techniques and clinical treatment strategies have been improved, the 5-year survival rate for HCC is still less than 17% for all stages combined.3, 4 The death caused by HCC is still rising faster compared to other cancers, having doubled since 1980’s.5, 6 It is crucial to investigate underlying molecular mechanisms that drive hepatic tumorigenesis and progression for enabling the development of novel targeted therapy.
Integrins are a family of heterodimeric transmembrane receptors formed by association of α and β subunits. Until now, 18 α and 8 β subunits have been identified, generating more than 24 αβ complexes.7, 8 Increasing evidence indicates that integrins are essential for cell migration and invasion. Some of them can promote stem cell function and serve as stem cell markers.9-14 Recently, it has been reported that integrin α6 (ITGA6) facilitates tumorigenesis and progression of several cancers.15-19 ITGA6 also known as CD49f, associates with either integrin β1 or integrin β4 to form α6β1 or α6β4 receptors. Previous studies mainly focused on the role of ITGA6 in promoting metastasis and other malignant phenotypes of several cancers.20-24 A few studies 25, 26 and our RNA sequencing data shown below demonstrated that ITGA6 is significantly up-regulated in HCC tumor tissues. Yet, the role and potential mechanism of ITGA6 in the progression of HCC remain largely unknown.
In the present study, we found that ITGA6 RNA and protein levels are elevated in HCC tissues in comparison with paired adjacent non-tumor tissues by RNA sequencing, RT-qPCR, Western blotting and immunofluorescence staining. Knockdown of ITGA6 in HCC cell lines significantly inhibited their migration, anchorage dependent and independent growth in vitro and xenograft growth in vivo. Immunoprecipitation assay was used to analyze whether integrin α6β1 or α6β4 complex plays a major role in HCC cells. Our study demonstrates a tumor oncogenic role of ITGA6 in HCC cells through integrin α6β4 complex.
2. MATERIALS AND METHODS
2.1. Cell cultures
The human HCC cell line SNU-398 (RRID: CVCL 0077) was purchased from ATCC (CRL-2233™) and the human HCC cell line Huh-7 (RRID: CVCL 0336) was a gift from Dr. Robert Lanford at the Texas Biomedical Research Institute in San Antonio, Texas. They were authenticated on June 11, 2021 using short tandem repeat profiling and all experiments were performed with mycoplasma-free cells. The cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin, 1% sodium pyruvate in 5% CO2 atmosphere at 37°C.
2.2. siRNA transfection
SNU-398 and Huh-7 cells were cultured in 60-mm dish and changed to an antibiotic-free medium before transfection. The cells were transiently transfected with 75nM ITGA6 siRNA, ITGB4 siRNA and control siRNA respectively, using lipofectamine 2000 (Invitrogen, Carlsbad, CA, Cat.#2030890) according to the manufacturer’s instruction. After 5h, the medium was replaced with normal culture medium and cells were cultured for another 48h for subsequent analyses.
The ITGA6 siRNA and ITGB4 siRNA were purchased from Sigma-Aldrich Inc (St.Louis, MO). The sequences targeting human ITGA6 and ITGB4 are shown in Supplementary Table 1 and a universal negative control siRNA from Sigma-Aldrich Inc (Cat. # SIC001) were used as the control.
2.3. Expression plasmid and transfection
Lentiviral plasmids of ITGA6 shRNA (TRCN0000296162, TRCN0000289094) and empty control plasmid (SHC201-TRC2-pLKO-puro) were purchased from Sigma-Aldrich Inc (St.Louis, MO). ITGA6 expression plasmid was constructed by inserting ITGA6 cDNA in the pCDH cDNA cloning and expression lentivector (System Biosciences, Mountain View, CA, Cat.# CD511B-1). Each plasmid was co-transfected together with two packaging vectors, psPAX2 and pMD2.G, into 293T cells. Lentiviral particle-containing medium was harvested after 48h or 72h and filtered for infection of HCC cell lines. The infected HCC cell lines were selected with puromycin (1μg/ml for Huh-7 cell and 2μg/ml for SNU-398 cell) for 7 days to establish stably infected cells. The shRNA sequences targeting human ITGA6 are shown in Supplementary Table 1.
2.4. RNA extraction and sequencing
Ten paired frozen HCC tumor and adjacent non-tumor liver tissues were obtained from the Tissue Biorepository of the Transplant Center at the University of Texas Health Science Center at San Antonio. Non-tumor tissue sections were confirmed to be devoid of tumor cells and HCC tumor sections were verified to have at least 75% tumor cells by a pathologist before they were used for RNA extraction. Total RNA was isolated from the tissues using RNA Mini Spin Column of Enzy Max LLC (Lexington, KY) according to the manufacturer’s instructions. DNase (Thermo Scientific, Rockford, IL, Cat.# EN0521) was added to the first wash solution at 10μg/70μl and incubated for 15 min at room temperature to remove genomic DNA contamination. The quality of RNA samples was analyzed with a Bioanalyzer (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA) by the Mays Cancer Center’s Next Generation Sequencing Shared Resource. Samples with RNA Integrity Number of 7 or greater were used for RNA sequencing library construction using TruSeq Stranded mRNA Library Prep kit according to manufacturer's protocol (cat.#RS-122-2002; Illumina, Inc.). Eight paired samples were sequenced in the Illumina HiSeq 2000 and two paired samples in HiSeq® 3000 system (Illumina, Inc.) using a 100bp paired-end sequencing protocol. All sequence reads were aligned to the UCSC human genome build hg19 using TopHat2, and bam files from the alignment were further processed using HTSeq-count 27 to obtain the read counts per gene in all samples. Sequencing coverage and quality statistics are summarized in Supplementary Table 2. Analysis of the read count data was performed using R version 3.3.3. Raw count data was first filtered to exclude genes that had only one or zero reads across all samples. The data was then transformed for visual inspection using regularized log2 transformation function of R package DESeq2 28, version 1.14.1. Data normalization and differential expression (DE) were performed using R package DESeq2, version 1.14.1. P values of the DE analysis were adjusted for multiple testing using the Benjamini-Hochberg procedure. Statistically and biologically significant DE genes (DEGs) were defined by applying the following stipulations: adjusted p-values <0.05 and absolute log2 fold change ≥1 (corresponding to a minimal two-fold up- or down-regulation).
2.5. RT-PCR
The mRNA level of ITGA6 was measured by quantitative real-time RT-PCR (RT-qPCR). Total RNA (1μg) was extracted from tissues or from a pellet of cultured cells, described above, and was reverse-transcribed to cDNA using random primers and M-MLV reverse transcriptase from Invitrogen Life Technology (Grand Island, NY). Quantitative real-time PCR was performed using SYBR Green PCR Mix from Invitrogen Life Technologies. The cDNA was also used for PCR to detect relative levels of two differentially spliced variants ITGA6A and ITGA6B with the ITGA6 PCR primers, which covers mRNA sequences of the two variants. All primers used in this study were synthesized by Integrated DNA Technologies (Coralville, IA). GAPDH was used as an internal control. The primers used in this study were shown in Supplementary Table 1.
2.6. Western blotting analysis
Protein was extracted from tissues or cell pellets using Laemmli buffer with protease inhibitors. The concentration of protein was quantified by the bicinchoninic acid protein assay (Thermo Scientific, Rockford, IL). Equal amount of total protein was applied to SDS-PAGE electrophoresis and then transferred to nitrocellulose membrane under 100 V. The membrane was blocked with Tris buffered saline with Tween (TBST) containing 5% milk and incubated with primary antibody overnight at 4°C. Washing steps of 10 min for 3 times were applied after primary and appropriate secondary antibody incubation. Proteins were detected with peroxidase-coupled secondary antibody from Sigma-Aldrich, using Electrochemiluminescence (ECL, Thermo Fisher Scientific, Grand Island, NY) and visualized using the FluorChem™ E imager with Alpha View software version 4.1.4 (Protein Simple, San Jose, CA). Antibodies used in the study included Anti-ITGA6 (Abcam, Cambridge, MA Cat.#ab181551), Anti-ITGB4 (Cell Signaling Technology, Danvers, MA, Cat.#14803), Anti-ITGB1 antibody (Cell Signaling Technology, Danvers, MA, Cat.#34971), Anti-phospho-p38(Thr180/Tyr182) (Cell Signaling, Cat.#9211), Anti-p38 (Cell Signaling, Cat.#9212), Anti-phospho-JNK(Thr183/Tyr185) (Cell Signaling, Cat.#4671), Anti-JNK (Cell Signaling, Cat.#9252), Anti-GAPDH (Calbiochem, Billerica, MA, Cat. # 80602-840), Anti-vinculin (Cell Signaling, Cat.#73614), Anti-Mouse IgG (Jackson immunoresearch, West Grove, PA, Cat.#115-035-003), and Anti-Rabbit IgG (Jackson immunoresearch, Cat.#111-035-003).
2.7. Immunofluorescence staining
A small piece of frozen tissues was transferred into a cryomold and then covered with optimal cutting temperature compound (OCT). The covered tissue was put into cold isopropanol to let the OCT solidify. Serial sections (6 μm) were cut with a cryostat, fixed in cold fixation buffer (4% paraformaldehyde, 85 mM Na2HPO4, 75 mM KH2PO4) on ice for 8-10 min and then rehydrated in PBS for 10 min at room temperature. The slides were blocked in PBS containing 0.3% Triton and 10% goat serum for 30 min at room temperature and incubated with an anti-ITGA6 antibody (LS Biologicals, Seattle, WA, Cat.#:LS-A8768, 1:50 dilution) overnight at 4°C followed by 30min incubation with Alexa Fluor® 488 goat anti-rabbit IgG (Abcam, Cambridge, MA Cat.# ab150077, 1:1000 dilution) at room temperature. The mounted sections were counterstained with mounting medium containing DAPI (Santa Cruz Biotechnology, Santa Cruz, CA) and examined under a confocal fluorescence microscope.
2.8. Cell proliferation assay
SNU-398 cells (5,000 cells/well) and Huh-7 cells (4,000 cells/well) were plated in triplicates in a 96-well plate. The viable cells were measured every two days by adding 50 μl of 3-(4,5-Dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide solution (MTT, 2mg/ml in PBS) to each well and incubated at 37°C for 2h. The blue color formazan product was dissolved in 100 μl Dimethyl Sulfoxide (DMSO) on a shaker for 10min at 300 rpm and then measured at 595 nm wavelength for absorbance on a BioTek plate reader. Meanwhile, the confluency of the cultures was measured daily with BioTek CYTATION 5 imager.
2.9. Cell migration assay
Cell migration assays were performed in 24-well transwells with 8 μm pore polycarbonate membranes (BD Biosciences, San Diego, CA). SNU-398 and Huh-7 cells (40,000 cells /well) were seeded in serum-free RPMI medium in the upper insert in triplicates. The lower chamber medium contained 10% FBS as a chemo attractant. After incubating for 16h for SNU-398 cells or 18h for Huh-7 cells, the cells that did not migrate across the membrane were removed with a cotton swab and the migrated cells were fixed and stained using the Hema 3 Stain 18 kit (Fisher Scientific, Waltham, MA) according to the manufacturer’s instruction.
2.10. Colony formation assay in soft agar
Five hundred microliter of 0.8% low-melting agarose (Life Technologies, Carlsbad, CA) in RPMI containing 10% FBS was added in a 24-well tissue culture plate as underlayer. Five thousand cells in the culture medium were mixed with 0.8% soft agarose in equal volume ratio and were plated on the top of the underlayer drop by drop. After 12-14 days of incubation, cell colonies were visualized by overnight staining with 250 μl p-iodonitrotetrazolium violet staining (Sigma, St Louis, MO). The plates were scanned with a scanner and colony number in each well was quantified using Image J.
2.11. Cell cycle analysis
Cells were trypsinized after reaching 70~80% confluence and washed with cold PBS twice. One million cells were transferred into a new tube and spun down. The pellet was resuspended very gently with about 100 μl cold PBS, fixed by adding 3ml cold 70% ethanol drop by drop while vortexing, and then kept at room temperature for 20min. The cells were washed 3 times with cold PBS and stained with 300 μl DNA staining solution (TritonX-100 (0.5%), Propidium iodide (PI) (500μg/ml), RNase A (10μg/ml) in PBS) for 30min in the dark. Then cells were analyzed for fluorescence intensity using a FACScan flow cytometer (Becton Dickinson, San Diego, CA).
2.12. Mouse xenograft study
Animal experiments were conducted following appropriate guidelines and the animal health and behavior was monitored daily. Six-week-old female nude mice were used for our study. Huh-7 and SNU-398 control cells and ITGA6 knockdown cells (2x106 cells/100 μl/mouse) were suspended in cold PBS containing 50% Matrigel and injected subcutaneously into the right flank of each mouse. After tumor cell inoculation for 1-2 weeks, the size of growing tumors was measured and recorded twice a week. Tumor volumes were calculated with the following formula: volume (mm3) = length × width × width / 2. The mice were euthanized at the end of the study and the excised tumors were weighed.
2.13. Immunoprecipitation
Total protein extraction and measurement of concentration were performed as described under Western blotting method. One thousand micrograms of total protein were incubated with 10 μl of Anti-ITGA6 antibody conjugated with agarose (Santa Cruz Biotechnology, Santa Cruz, CA, Cat.# sc-374057) overnight at 4°C. The pellet was collected by centrifugation at 3,000rpm for 30s and washed 3 times with cold PBS. Then the pellet was resuspended in electrophoresis sample buffer and applied to SDS-PAGE electrophoresis as described under Western blotting method.
2.14. Survival analysis
We used the expression data of ITGA6, ITGB1, and ITGB4 of HCC cases in The Cancer Genome Atlas (TCGA) data set to determine whether expression of the pair of ITGA6 and ITGB4 is associated with patient survival. We repeated the same analysis for ITGA6 and ITGB1. Following our approach in Zainulabdeen et.al.29 for cases for each of these three genes, we divided the range of expression values from 373 TCGA HCC cases into 20 bins in a way that the number of cases in each bin is roughly similar. We examined two arbitrary cutoff values for each gene and considered the cases with an expression level below the smaller cutoff low expressing, and the cases with an expression level above the larger cutoff high expressing. We categorized cases into three groups: 1) high-high cases with high expression levels of both ITGA6 and ITGB4, 2) low-low cases with low expression of both ITGA6 and ITGB4, and 3) the rest of cases, who were not included in the following concordance analysis. 30, 31 We examined all possible cutoff values aligning with bins that led to groups with at least 18 (5%) cases. Using survival package Version 2.44-1, 32 we computed concordance, which measures how well the grouping correlates with overall survival data. If a set of cutoff values for two genes led to a concordance value within half standard deviation from the best concordance (Fig. S5), we tested the null hypothesis that the survival of the low-low group is not different from the survival of the high-high group using a log rank test. We selected the best set of cutoffs based on the resulting p-value and computed false discovery rate (FDR) to adjust for multiple testing (Fig. S6).
2.15. Statistical analysis
Two-tailed t-test was used to determine statistical difference between two means. One-way ANOVA followed with multiple comparisons was used to determine statistical difference between means of control versus various treatment groups. Two-way repeated measures ANOVA was used to determine statistical difference between two growth curves. We used R 33 to study the effect of expression of ITGA6 combined with ITGB4 or ITGB1 on survival. Other statistical analyses were performed using Prism 8.
3. RESULTS
3.1. Increased expression of ITGA6 in hepatocellular carcinoma
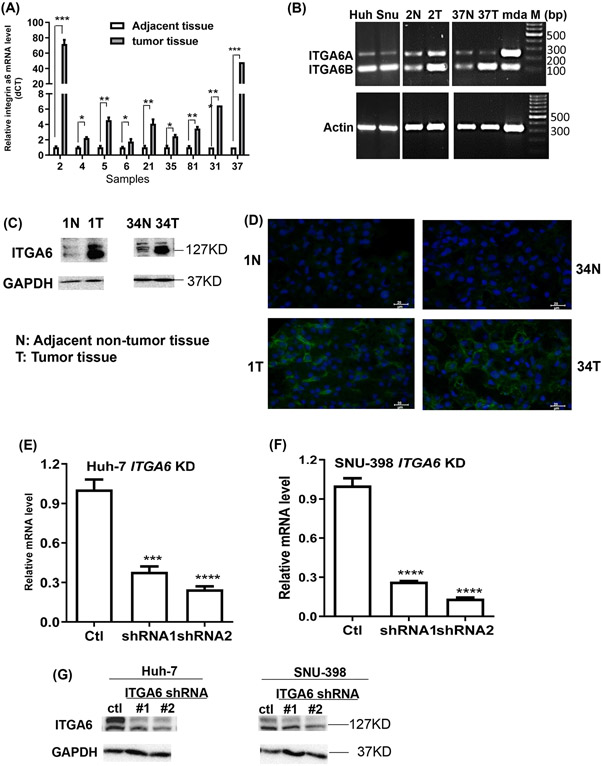
Whole transcriptome RNA sequencing in paired HCC tumor and adjacent non-tumor liver tissue samples from ten local Hispanic patients was performed. The average expression of ITGA6 was found increased by over 3.5-fold in HCC tumor tissues in comparison with their adjacent non-tumor tissues with an adjusted P value of 1.73x10−9 from DESeq2 analysis. Wilcoxon test of the ITGA6 gene reads also showed significant difference between the ten paired samples (Figure S1A). We further examined mRNA and protein levels of ITGA6 in paired human HCC tumor tissues and adjacent non-tumor tissues. ITGA6 was significantly up-regulated in HCC tumor tissues by RT-qPCR (Figure 1A). Interestingly, while the transcripts of the two splice variants ITGA6A and ITGA6B showed a similar level in two non-tumor tissue samples, ITGA6B RNA appears higher than ITGA6A RNA in the two human HCC cell lines and the two tumor samples (Figure 1B). The upregulation of ITGA6 in tumor samples was also confirmed at the protein level by Western blotting (Figure 1C) and by immunofluorescence staining (Figure 1D). To confirm our results, the RNA sequencing data of 50 adjacent non-tumor tissues, their paired HCC tumors, and 373 HCC tumors in TCGA dataset was downloaded and analyzed. We found that ITGA6 was also significantly up-regulated in HCC tumor tissues (Figure S1B), and the up-regulation was across all tumor stages and grades (Figure S1C and D).
Figure1.
Expression of ITGA6 in HCC tumor and paired adjacent non-tumor tissues and knockdown of ITGA6 in HCC cell lines. (A) ITGA6 mRNA levels in 9 cases by RT-qPCR. Data presented are mean±SEM of triplicate measurements. Statistical significance of the difference was assessed by two-tailed t-test: *, P< 0.05, **, P< 0.01, ***, P< 0.001. (B) Relative mRNA levels of ITGA6A and ITGA6B variants are shown by agarose gel electrophoresis of PCR products amplified for 30 cycles with a pair of primers covering both variant RNA sequences. Huh and Snu stand for human HCC Huh-7 and SNU-398 cell lines, respectively. mda stands for human breast cancer MDA-MB-231 cell line, which is known to express the two ITGA6 variants and used as a positive control. M: molecular marker in base pair. (C) ITGA6 protein levels in 2 cases by Western blotting assay. GAPDH levels were used to verify equal sample loading. (D) Increased ITGA6 protein levels in tumors as compared with adjacent non-tumor tissues in 2 cases by immunofluorescence staining. ITGA6 was shown in green, and DAPI was used to stain cell nuclei in blue. The scale bar=20 μm. (E & F) Two ITGA6 shRNA expression vectors and a control vector were transfected into HCC cell lines via lentiviral infection. ITGA6 stable knockdown (KD) in Huh-7 (E) and SNU-398 (F) cells at its mRNA level was confirmed with RT-qPCR. Data presented are mean±SEM of triplicate measurements. Statistical significance of the differences was assessed by one-way ANOVA: ***, P< 0.001, ****, P<0.0001. (G) ITGA6 stable knockdown was also confirmed at its protein level with Western blotting. GAPDH protein levels were used to verify equal sample loading.
3.2. Knockdown of ITGA6 inhibits cell proliferation and migration
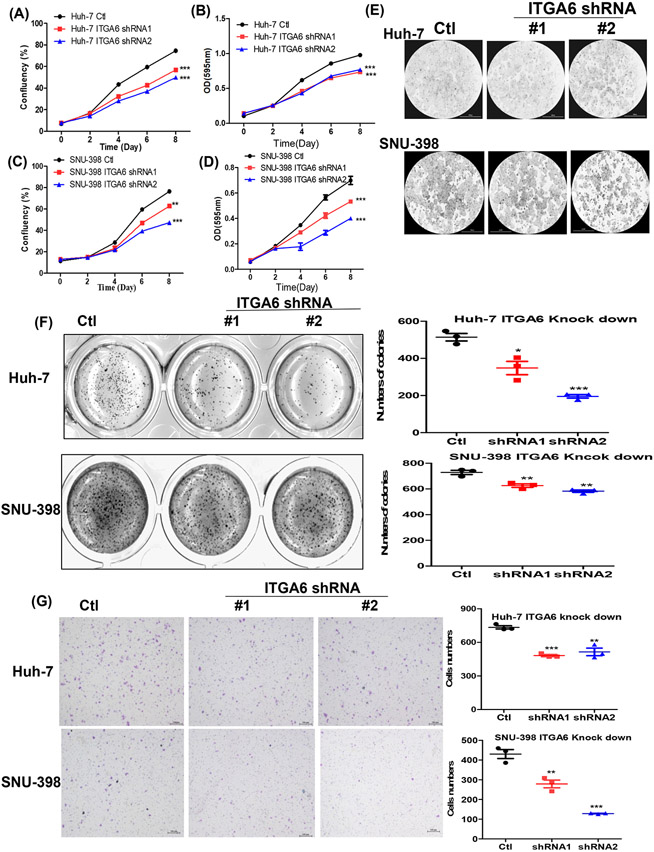
We used two ITGA6-shRNA lentiviral plasmids and their corresponding control shRNA (Ctl) lentivector to stably knockdown ITGA6 expression in human HCC Huh-7 and SNU-398 cell lines. The decreased expression of ITGA6 was confirmed at mRNA level by RT-qPCR (Figure 1E and 1F) and protein level by Western blotting (Figure 1G) after stable lentiviral transfection. The two bands in the Western blots showing reduced levels of ITG6A in the two knockdown cells were likely the heavy and light chains of ITG6A protein according to published study. 34 We then examined whether ITGA6 is necessary for malignant features of HCC cells, and found that knockdown of ITGA6 significantly inhibited the growth of Huh-7 and SNU-398 cells as measured over 8 days by culture confluency (Figure 2A, C and E) and by MTT assay (Figure 2B and D). The malignant-promoting role of ITGA6 was further demonstrated with soft agar colony formation assay in SNU-398 and Huh-7 cells (Figure 2F). Additionally, ITGA6 knockdown also significantly inhibited the migration in these two HCC cell lines (Figure 2G). These results are consistent with those initially obtained in our preliminary experiments with two ITGA6 siRNAs. Transient knockdown of ITGA6 with the siRNAs also reduced ITGA6 expression in the two HCC cell lines (Figure S2A-D) and inhibited their growth on plastic (Figure S2E) and in soft agar (Figure S2F) as well as their migration (Figure S2G).
Figure 2.
Knockdown of ITGA6 inhibited the growth and migration of HCC cells. Anchorage-dependent growth of the paired cell lines was assessed with a confluence assay (A and C) with representative confluence images (E) and an MTT assay (B and D). Data presented are mean±SEM of four replicate measurements. Statistical significance of the differences was assessed by two-way ANOVA test: **, P< 0.01, ***, P<0.001. Anchorage-independent growth of the paired HCC cell lines was assessed with a soft agar assay (F), and their migration in a transwell assay (G, scale bar=200 μm). Data presented are mean±SEM of triplicate measurements. Statistical significance of the differences is assessed by one-way ANOVA: *, P< 0.05, **, P< 0.01, ***, P< 0.001.
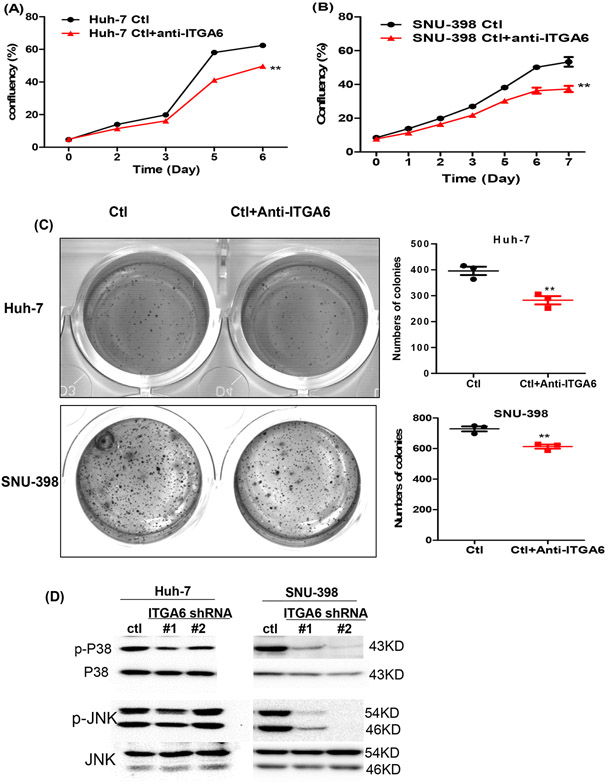
To exclude the possibility of non-specific targeting of ITGA6 siRNAs and shRNAs, we also confirmed the role of ITGA6 using an anti-ITGA6 blocking antibody (GoH3 clone, Biolegend, San Diego, CA, Cat.# 313614). Our results demonstrated that blocking ITGA6 function with the anti-ITGA6 antibody also significantly inhibited anchorage-dependent and -independent growth of HCC cell lines (Figure 3A-C).
Figure 3.
Blockade or knockdown ITGA6 inhibited the growth of HCC cells and phosphorylation of p38 and JNK. Anchorage-dependent growth of control antibody (Ctl) or anti-ITGA6-treated cell lines (anti-ITGA6) was assessed with confluence assay (A and B). Data presented are mean±SEM of four replicate measurements. Statistical significance of the differences was assessed by two-way ANOVA test: **, P< 0.01. Anchorage-independent growth of the Ctl and anti-ITGA6-treated HCC cell lines was assessed with a soft agar assay (C). Data presented are mean±SEM of triplicate measurements. Statistical significance of the differences is assessed by two-tailed t-test: **, P< 0.01. (D) Knockdown of ITGA6 reduced the levels of phosphor-p38 (p-p38) and p-JNK relative to the levels of total p38 and JNK in Huh-7 and SNU-398 cells by Western blotting.
3.3. Knockdown of ITGA6 reduces phosphorylation of p38 and JNK
Integrins including Integrin α6β4 are known to interact with growth factor receptors resulting in the modulation of the activity of their downstream targets including the mitogen-activated protein kinases (MAPKs). 35 The MAPKs (ERK, p38, JNK) are well known to play a major role in regulating cell proliferation as well as migration. 36,37 To gain an insight to how ITGA6 knockdown may affect the proliferation of HCC cells, we compared the levels of phosphorylated/activated ERK, p38 and JNK between the control and ITGA6 knockdown cells. While the level of phospho-ERK at Thr202/Tyr 204 was not altered after ITGA6 knockdown (data not shown), the level of phospho-p38 (p-p38) at Thr180/Tyr182 was reduced after ITGA6 knockdown in both HCC cell lines (Figure 3D). The level of p-JNK at Thr183/Tyr185 was modestly reduced by ITGA6 shRNA #1 in Huh-7 cells and more obviously reduced by both ITGA6 shRNAs in SNU-398 cells.
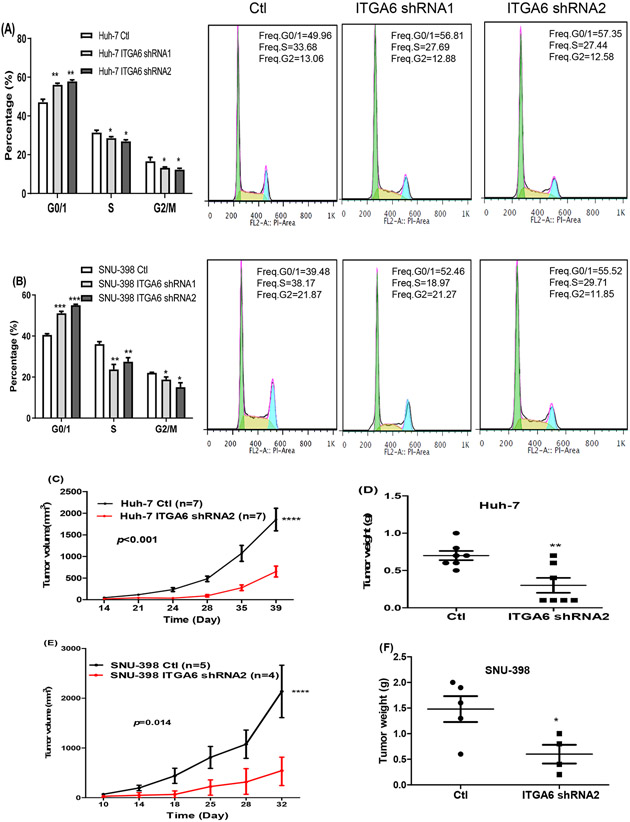
3.4. Knockdown of ITGA6 induces cell cycle arrest at G0/G1 phase
The above results indicated that ITGA6 may facilitate cell division and consequently increase the proliferative capacity of HCC cells. To extend the finding, we performed cell cycle and apoptosis analysis by flow cytometry. Our results revealed that knockdown of ITGA6 increased the percentage of cells in the G0/G1 phase and decreased the percentage of cells in the S and G2/M phase (Figure 4A, B). However, knockdown of ITGA6 did not affect apoptosis in HCC cell lines (Data not shown). Thus, knockdown of ITGA6 inhibited HCC cell growth by inducing G0/G1 cell cycle arrest but not apoptosis.
Figure 4.
Knockdown of ITGA6 induced cell-cycle arrest at G0/G1 phase and inhibited xenograft tumor growth. Knockdown of ITGA6 induced cell cycle arrest at G0/G1 in Huh-7 cells (A) and SNU-398 cell (B). Data presented are mean±SEM of triplicate measurements. Statistical significance of the differences of each cycle phase was assessed by one-way ANOVA: *, P< 0.05, **, P< 0.01, ***, P< 0.001. Representative flowcytometry histograms of the cell distribution in the three phases of cell cycle are also provided for the control and ITGA6 knockdown cells. Tumor growth curve and weight of tumors formed by Huh-7 (C, D) and SNU-398 (E, F) Ctl and matched ITGA6-shRNA cells in nude mice. The tumor volume was calculated using the formula: v=length × width × width / 2. Statistical significance of the differences of survival curve was assessed by two-way ANOVA test: ****, P< 0.0001. Individual tumor weight from two group mice was collected at the end of experiment. Statistical significance of the differences was assessed by two-tailed t-test: *, P< 0.05, **, P< 0.01.
3.5. Knockdown of ITGA6 inhibits tumor growth
To further verify the malignancy-promoting role of ITGA6 in HCC, we determined whether ITGA6 was necessary for promoting tumor growth in vivo. We inoculated Huh-7 and SNU-398 ITGA6 knockdown cells and their matched control (Ctl) cells subcutaneously in nude mice. Seven of 11 mice inoculated with Huh-7 cells produced xenograft tumors. Out of 9 mice inoculated with SNU-398 cells, there were 5 tumors formed by the control cells and 4 formed by the ITGA6 knockdown cells. As shown in Figure 4C and 4E, knockdown of ITGA6 resulted in significantly slower tumor growth by both Huh-7 and SNU-398 cells. The average weight of ITGA6 knockdown cell-formed tumors was also significantly lower than that of Ctl cell-formed tumors (Figure 4D and 4F).
3.6. ITGA6 interacts with ITGB4 and stimulates ITGB4 expression in HCC cells
Since ITGA6 can form complex with either ITGB4 or ITGB1, we next examined which β subunit of the two was the major partner of ITGA6 in HCC cells by immunoprecipitation assay. Proteins of the two HCC cell lines (Huh-7 and SNU-398) were pulled down by an Anti-ITGA6antibody conjugated with agarose and then subjected to SDS-PAGE electrophoresis to detect ITGB4 and ITGB1. The results indicated that while both ITGB4 and ITGB1 were detected in the input, we only detected ITGB4, but not ITGB1, in the proteins pulled down by anti-ITGA6 antibody in HCC cell lines (Figure 5A).
Figure 5.
ITGA6 interacts with ITGB4 and stimulates ITGB4 expression in HCC cells. (A) Anti-ITGA6 antibody and matched control Anti-IgG were used to pull down proteins by immunoprecipitation assay in HCC cells. ITGB4 was detected in proteins pulled by the anti-ITGA6 antibody by Western blotting assay. The Western blots of Input shows equal amounts of ITGA6, ITGB4, ITGB1 and GAPDH proteins were subjected to pulldown by the control or anti-ITGA6 antibody. (B) Knockdown of ITGA6 significantly reduced ITGB4 mRNA levels in Huh-7 and SNU-398 cells. Data presented are mean±SEM of triplicate measurements with RT-qPCR assay. Statistical significance of the differences of each gene between the Ctl and two knockdown cells was assessed by one-way ANOVA: P< 0.01, ***, P< 0.001. (C) Knockdown of ITGA6 reduced protein levels of ITGB4, but not ITGB1, in Huh-7 and SNU-398 cells. (D and E) Over-expression of ITGA6 (OE) in HCC cells resulted in an increase of ITGB4 mRNA and protein, but had no effect or even decreased the expression of ITGB1. Data presented are mean±SEM of triplicate measurements in RT-qPCR assay. Statistical significance of the differences of each gene between the Ctl and OE cells was assessed by two-tailed t-test: P< 0.01, ***, P< 0.001.
When we examined the expression of ITGB4 and ITGB1 in HCC cells with ITGA6 knockdown or ITGA6 over-expression, we found that knockdown of ITGA6 resulted in a significant decrease of ITGB4 in both mRNA and protein levels but had either no effect or even increased the expression of ITGB1 (Figure 5B, C). Conversely, overexpression of ITGA6 resulted in an increase of ITGB4 but had no effect or even decreased the expression of ITGB1 (Figure 5D, E). Similar to the knockdown of ITGA6, knockdown of ITGB4 also led to increased cell numbers in G0/G1 phase (Figure S3A, B) and significantly inhibited the migration of the HCC cells (Figure S4A-C). These results indicate that ITGA6 appears to promote the malignancy of HCC cells by stimulating the expression of and physically interacting with ITGB4.
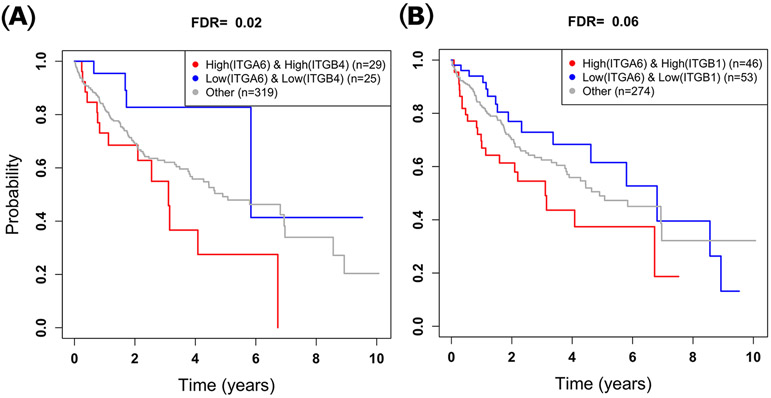
3.7. High ITGA6 and ITGB4 expression is associated with worse survival of HCC patients
Using the expression data of 373 HCC cases in TCGA data set, we grouped the patients into three risk categories to determine whether expression of the ITGA6/ITGB4 pair is associated with patient overall survival. Kaplan-Meier 38 plots of ITGA6 and ITGB4 showed that the group of patients we identified as high–risk had significantly shorter overall survival compared to the low–risk group (FDR=0.022, Figure 6A). In contrast, for the ITGA6/ITGB1 gene pair, the difference between the overall survival of the two risk-groups was insignificant after correcting for multiple testing (FDR=0.060, Figure 6B). Also, the overall survival of the high-risk group of the ITGA6/ITGB4 pair (≤6.5 years) was worse compared to that of the ITGA6/ITGB1 pair (≥7.0 years) even though the difference is not statistically significant.
Figure 6.
Association of expression levels of ITGA6 and ITGB4 pair orITGA6 and ITGB1 pair with HCC patient survival. (A) Patients with high expression levels of ITGA6 and ITGB4 have significantly worse overall survival compared to those with low expression of levels of ITGA6 and ITGB4. (B) The difference in overall survival time between patients who have high vs. low expression levels of ITGA6 and ITGB1 is insignificant.
4. DISCUSSION
Integrins are heterodimeric transmembrane receptors that directly bind components of the extracellular matrix (ECM) proteins and provide the traction necessary for cell motility and invasion.8, 39 It has been reported that integrins are involved in several carcinogenic processes including the initiation, proliferation, survival, and metastasis of solid tumors. The importance of integrins in affecting tumor progression has made them targets for cancer therapy.40, 41 As a member of the integrin family, integrin α6 (ITGA6) has been shown to play crucial roles in several cancers.8, 16, 22, 23, 42 In the present study, we found that ITGA6 was significantly up-regulated in HCC tumor tissues at mRNA and protein levels from our local Hispanic patients. We also found significant upregulation of ITGA6 in HCC tumors in the TCGA data set, which is consistent with a previous study.43 Furthermore, HCC cells and tumors appear to express higher levels of ITGA6 splice variants ITGA6B than ITGA6A. The former was shown to confer cancer stem cell properties and higher malignacy in complex with integrin β1 than the latter in breast cancer models. 14 These findings led us to investigate whether ITGA6 plays a tumor promoting role in HCC.
Indeed, knockdown of ITGA6 or blockade of ITGA6 function significantly inhibited the growth of HCC cells under anchorage-dependent and independent conditions. The growth-promoting activity of ITGA6 was apparently due to its positive effect on cell cycle progression. Jiang et al 44 demonstrated that exosome-derived ENO1 promoted the growth and metastasis of HCC cells by upregulating integrin α6β4 expression and activating the FAK/Src-p38MAPK pathway. We found that knockdown of ITGA6 reduced the levels of active p38 and JNK, which likely led to the reduced proliferation in the knockdown cells. Consistently, we found that down-regulation of ITGA6 inhibited the growth of xenograft tumors in nude mice demonstrating the malignancy-promoting role of ITGA6 in vivo. Given its prominent role in promoting cell-ECM adhesion and cell motility, ITGA6 down-regulation also significantly inhibited the migration of HCC cells, which are necessary for metastatic dissemination of tumors. Indeed, reports by Lv et al 25 and Ke et al 26 showed that a high level of ITGA6 is associated with metastasis and early recurrence in HCC patients.
Integrin α6 makes one part of two proteins known as integrin α6β4 and integrin α6β1. The integrin β1 subunit can pair with α1-α11 subunits and αV subunit in addition to α6 subunit. In contrast, integrin α6 is the only possible α subunit partner of integrin β4.45, 46 It has been reported that integrin α6β4 was associated with increased invasive and metastatic potential of various carcinomas.47-49 In agreement with other studies, our immunoprecipitation assay showed that ITGB4, but not ITGB1, was pulled down by anti-ITGA6 antibody in HCC cell lines. Furthermore, silencing either ITGA6 or ITGB4 caused cell cycle arrest at G1 phase and decreased their migration. Interestingly, silencing and over-expressing ITGA6 in HCC cells resulted in the decreased or increased expression of ITGB4 respectively. Furthermore, high levels of ITGA6 and ITGB4 were associated with significantly worse survival in TCGA HCC patients. These results indicated that ITGA6 appears to perform its malignant role through the integrin α6β4 complex in HCC. Given that HCC cells and tumors appear to express increased levels of ITGA6B variant than ITGA6A variant in comparison to the non-tumor liver tissues, it is likely that the malignancy-promoting activities observed in our study was mainly carried out by α6Bβ4, which remains to be confirmed in future studies. On the other hand, previous studies also demonstrated an oncogenic role of integrin α6β1 in HCC cells. Ke et al 26 showed that CD151 formed a complex with integrin α6β1, and a high level of CD151/α6 expression promoted invasion and metastasis of HCCLM3 and HepG2 cells via PI3K pathway. Thus, while it is clear that ITGA6 promotes HCC progression, whether it acts by forming complex with ITGB1 or ITGB4 may be contextual and dependent on model systems.
In summary, our study demonstrated a tumor promoting role of ITGA6 since knockdown of ITGA6 significantly inhibited the growth of HCC cells in vitro and in vivo and also inhibited the migration of HCC cells. Additionally, ITGA6 appears to perform its malignant role through integrin α6β4 complex in the HCC cells, which appears to be an important molecular mechanism that drive HCC progression and may be targeted for novel therapeutic intervention of HCC.
Supplementary Material
Novelty & Impact Statements.
ITGA6 forms integrin receptors with either ITGB1 or ITGB4. How it regulates progression of hepatocellular carcinoma (HCC) is not well-understood. We report that ITGA6 is significantly up-regulated in human HCC tumors. ITGA6 increases ITGB4 expression and promotes malignancy of HCC cells in complex with ITGB4 not ITGB1. High levels of ITGA6 and ITGB4 were associated with reduced patient survival. Thus, integrin α6β4 may be a therapeutic target for treating patients with HCC.
ACKNOWLEDGEMENTS
This work was in part supported by a grant from Clayton Foundation for Research to FGC and L-Z S, and by National Cancer Institute Cancer Center Support Grant P30 CA054174 to the Shared Resources of Mays Cancer Center’s Flowcytometry, Optical Imaging, and Next Generation Sequencing. Next Generation Sequencing Shared Resources is also supported by NIH Shared Instrument grant 1S10OD021805-01 (S10 grant), and CPRIT Core Facility Award (RP160732).GZ was supported in part by a scholarship from China Scholarship Council. CRZ was supported in part by an institutional training grant from NIH T32CA148724 and by an individual training grant from NIH F32CA228435.The authors thank Dr. Arthur M. Mercurio for the lentiviral expression vector pCDH-copGFP with and without human ITGA6 cDNA.
Abbreviations
- ATCC
American Type Culture Collection
- DE
differential expression
- DMSO
Dimethyl Sulfoxide
- ECM
extracellular matrix
- FDR
false discovery rate
- HCC
hepatocellular carcinoma
- ITGA6
integrin α6
- ITGB1
integrin β1
- ITGB4
integrin β4
- OCT
optimal cutting temperature compound
- PI
propidium iodide
- RT-qPCR
reverse transcription and quantitative polymerase chain reaction
- STR
short tandem repeats
- TBST
Tris buffered saline with Tween
- TCGA
The Cancer Genome Atlas
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
ETHICS STATEMENT
Human liver tissue samples used for RNA extraction and sequencing were obtained from the biorepository of the Transplant Center of the University of Texas Health Science Center at San Antonio (UTHSCSA) and approved by the Institutional Review Board of UTHSCSA. A written informed consent was obtained from each patient. All mouse experiments were approved by the Institutional Animal Care and Use Committee and monitored by the Department of Laboratory Animal Resources at the University of Texas Health Science Center at San Antonio.
DATA AVAILABILITY STATEMENT
The RNA-seq data have been deposited in GEO database (Accession #: GSE202853). Other data that supports the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017;152: 745–61. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365: 1118–27. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Mo L, Wang X, Chen B, Hua Y, Gong L, Yang F, Li Y, Chen F, Zhu G, Ni W, Zhang C, et al. TPGS-1000 exhibits potent anticancer activity for hepatocellular carcinoma in vitro and in vivo. Aging (Albany NY) 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen S, Peng H, Wang Y, Xu M, Lin M, Xie X, Peng B, Kuang M. Screening for immune-potentiating antigens from hepatocellular carcinoma patients after radiofrequency ablation by serum proteomic analysis. BMC Cancer 2018;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monsour HP Jr, Asham E, McFadden RS, Victor III DW, Muthuswamy B, Zaheer I. . Hepatocellular carcinoma: the rising tide from east to west—a review of epidemiology, screening and tumor markers. Translational Cancer Research 2013;2: 492–506. [Google Scholar]
- 7.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 2010;12: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Xu J, Chen B, Ma D, Chen Z, Li JC, Zhu C. Elevated integrin alpha6 expression is involved in the occurrence and development of lung adenocarcinoma, and predicts a poor prognosis: a study based on immunohistochemical analysis and bioinformatics. J Cancer Res Clin Oncol 2019;145: 1681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viquez OM, Yazlovitskaya EM, Tu T, Mernaugh G, Secades P, McKee KK, Georges-Labouesse E, De Arcangelis A, Quaranta V, Yurchenco P, Gewin LC, Sonnenberg A, et al. Integrin alpha6 maintains the structural integrity of the kidney collecting system. Matrix Biol 2017;57-58: 244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohaqiq M, Movahedin M, Mokhtari Dizaji M, Mazaheri Z. Upregulation of Integrin-alpha6 and Integrin-beta1 Gene Expressions in Mouse Spermatogonial Stem Cells after Continues and Pulsed Low Intensity Ultrasound Stimulation. Cell J 2018;19: 634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SH, Sud N, Lee N, Subramaniyam S, Chung CY. Regulation of Integrin alpha6 Recycling by Calcium-independent Phospholipase A2 (iPLA2) to Promote Microglia Chemotaxis on Laminin. J Biol Chem 2016;291: 23645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krebsbach PH, Villa-Diaz LG. The Role of Integrin alpha6 (CD49f) in Stem Cells: More than a Conserved Biomarker. Stem Cells Dev 2017;26: 1090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogervorst F KI, van Kessel AG, Sonnenberg A. Molecular cloning of the human alpha 6 integrin subunit. Alternative splicing of alpha 6 mRNA and chromosomal localization of the alpha 6 and beta 4 genes. Eur J Biochem 1991;199: 425–33. [DOI] [PubMed] [Google Scholar]
- 14.Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, Norum JH, Toftgard R, Shaw LM, Mercurio AM. Regulated splicing of the alpha6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep 2014;7: 747–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding YB, Deng B, Huang YS, Xiao WM, Wu J, Zhang YQ, Wang YZ, Wu DC, Lu GT, Wu KY. A high level of integrin alpha6 expression in human intrahepatic cholangiocarcinoma cells is associated with a migratory and invasive phenotype. Dig Dis Sci 2013;58: 1627–35. [DOI] [PubMed] [Google Scholar]
- 16.Yamakawa N, Kaneda K, Saito Y, Ichihara E, Morishita K. The increased expression of integrin alpha6 (ITGA6) enhances drug resistance in EVI1(high) leukemia. PloS one 2012;7: e30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toth RK TJ, Muldong MT, Nollet EA, Schulz VV, Jensen CC, Hazlehurst LA, Corey E, Durden D, Jamieson C, Miranti CK, Warfel NA. Hypoxia-induced PIM kinase and laminin-activated integrin α6 mediate resistance to PI3K inhibitors in bone-metastatic CRPC. Am J Clin Exp Urol 2019. 7: 297–312. [PMC free article] [PubMed] [Google Scholar]
- 18.Ma G JC, Huang F, Li X, Cao X, Liu Z. Integrin α6 promotes esophageal cancer metastasis and is targeted by miR-92b. Oncotarget 2017;8: 6681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groulx JF, Giroux V, Beausejour M, Boudjadi S, Basora N, Carrier JC, Beaulieu JF. Integrin alpha6A splice variant regulates proliferation and the Wnt/beta-catenin pathway in human colorectal cancer cells. Carcinogenesis 2014;35: 1217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan M, Xie F, Xia X, Zhong K, Lian L, Zhang S, Yuan L, Ye J. UNC5Cknockdown enhances the growth and metastasis of breast cancer cells by potentiating the integrin alpha6/beta4 signaling pathway. Int J Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landowski TH, Gard J, Pond E, Pond GD, Nagle RB, Geffre CP, Cress AE. Targeting integrin alpha6 stimulates curative-type bone metastasis lesions in a xenograft model. Mol Cancer Ther 2014;13: 1558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin YP, Hu YP, Wu XS, Wu YS, Ye YY, Li HF, Liu YC, Jiang L, Liu FT, Zhang YJ, Hao YJ, Liu XY, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell death & disease 2018;9: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu T, Zhou R, Zhao Y, Wu G. Integrin alpha6/Akt/Erk signaling is essential for human breast cancer resistance to radiotherapy. Sci Rep 2016;6: 33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groulx JF, Boudjadi S, Beaulieu JF. MYC Regulates alpha6 Integrin Subunit Expression and Splicing Under Its Pro-Proliferative ITGA6A Form in Colorectal Cancer Cells. Cancers (Basel) 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv G, Lv T, Qiao S, Li W, Gao W, Zhao X, Wang J. RNA interference targeting human integrin alpha6 suppresses the metastasis potential of hepatocellular carcinoma cells. Eur J Med Res 2013;18: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke AW, Shi GM, Zhou J, Huang XY, Shi YH, Ding ZB, Wang XY, Devbhandari RP, Fan J. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology 2011;140: 1629–41 e15. [DOI] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2014;31: 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zainulabadeen A, Yao P, Zare H. Underexpression of Specific Interferon Genes Is Associated with Poor Prognosis of Melanoma. PloS one 2017;12: e0170025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steck H KB, Dehing-oberije C, Lambin P, Raykar VC. On ranking in survival analysis: bounds on the concordance index.ed. Proceedings of the Twenty-First Annual Conference on Neural Information Processing Systems (NIPS 2007); 2007 Dec 3-6: Vancouver, BC, Canada. La Jolla (CA): Neural Information Processing Systems Foundation;, 2008. 1209–16. [Google Scholar]
- 31.Kolde R pheatmap: Pretty Heatmaps, ed. R package version 1.0.12. , 2019. [Google Scholar]
- 32.A TT Package for Survival Analysis in R, ed. R package version 2.44–1, 2019. [Google Scholar]
- 33.Team. RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. [Google Scholar]
- 34.Sonnenberg A, Linders CJ, Daams JH, Kennel SJ. The alpha 6 beta 1 (VLA-6) and alpha 6 beta 4 protein complexes: tissue distribution and biochemical properties. J Cell Sci 1990;96 (Pt 2):207–17. [DOI] [PubMed] [Google Scholar]
- 35.Stewart RL, O'Connor KL. Clinical significance of the integrin alpha6beta4 in human malignancies. Lab Invest 2015;95:976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estrada Y, Dong J, Ossowski L. Positive crosstalk between ERK and p38 in melanoma stimulates migration and in vivo proliferation. Pigment Cell Melanoma Res 2009;22:66–76. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, Smalley KS, Mahale A, Eroshkin A, Aaronson S, Ronai Z. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell 2007;11:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan EL MP. Nonparametric estimation from incomplete observations. Journal of the American statistical association 1958;53: 457–81. [Google Scholar]
- 39.Cheresh DA, Stupack DG. Regulation of angiogenesis: apoptotic cues from the ECM. Oncogene 2008;27: 6285–98. [DOI] [PubMed] [Google Scholar]
- 40.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 2010;10: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianconi D, Unseld M, Prager GW. Integrins in the Spotlight of Cancer. International journal of molecular sciences 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz AD, Hall CL, Barney LE, Babbitt CC, Peyton SR. Integrin alpha6 and EGFR signaling converge at mechanosensitive calpain 2. Biomaterials 2018;178: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hass HG, Vogel U, Scheurlen M, Jobst J. Gene-expression Analysis Identifies Specific Patterns of Dysregulated Molecular Pathways and Genetic Subgroups of Human Hepatocellular Carcinoma. Anticancer research 2016;36: 5087–95. [DOI] [PubMed] [Google Scholar]
- 44.Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, Zhang J, Gao Z, Liang R, Wang Q, Wang L. Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell death & disease 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dydensborg AB, Teller IC, Basora N, Groulx JF, Auclair J, Francoeur C, Escaffit F, Pare F, Herring E, Menard D, Beaulieu JF. Differential expression of the integrins alpha6Abeta4 and alpha6Bbeta4 along the crypt-villus axis in the human small intestine. Histochem Cell Biol 2009;131: 531–6. [DOI] [PubMed] [Google Scholar]
- 46.Dydensborg AB TI, Basora N, Groulx JF, Auclair J, Francoeur C, Escaffit F, Paré F, Herring E, Ménard D, Beaulieu JF. Differential expression of the integrins a6A4 and a6B4 along the crypt villus axis in the human small intestine. Histochem Cell Biol 2009;131: 531–6. [DOI] [PubMed] [Google Scholar]
- 47.Meng X, Liu P, Wu Y, Liu X, Huang Y, Yu B, Han J, Jin H, Tan X. Integrin beta 4 (ITGB4) and its tyrosine-1510 phosphorylation promote pancreatic tumorigenesis and regulate the MEK1-ERK1/2 signaling pathway. Bosn J Basic Med Sci 2020;20: 106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan S, Lin M, Zhu Y, Lum L, Thakur A, Jin R, Shao W, Zhang Y, Hu Y, Huang S, Hurt EM, Chang AE, et al. Integrin beta4-Targeted Cancer Immunotherapies Inhibit Tumor Growth and Decrease Metastasis. Cancer Res 2020;80: 771–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li XL, Liu L, Li DD, He YP, Guo LH, Sun LP, Liu LN, Xu HX, Zhang XP. Integrin beta4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Sci Rep 2017;7: 40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in GEO database (Accession #: GSE202853). Other data that supports the findings of this study are available from the corresponding author upon request.