Abstract
Using a transposon carrying a promoterless lux operon to generate transcriptional fusions by insertional mutagenesis, we have identified a Pseudomonas putida gene with increased expression in the presence of corn root exudates. Expression of the transcriptional fusion, induced by the amino acid lysine, was detected in P. putida in the rhizosphere of plants as well as in response to seed exudates. The mutant was unable to grow on lysine or δ-aminovalerate as carbon sources, which indicates that the affected function is involved in the pathway for lysine catabolism. However, the mutant strain grew with glutaric acid, the product of δ-aminovalerate metabolism via glutaric acid semialdehyde, as a C source. The translated sequence of the interrupted gene showed high levels of similarity with aminotransferases. These sets of data suggest that the product of this gene has δ-aminovalerate aminotransferase activity. This is the first direct genetic evidence correlating a DNA sequence with such activity in Pseudomonadaceae.
The surface of plant roots and the surrounding soil regions (rhizosphere) constitute an environment where nutrients are available for bacterial populations to be established at relatively high cell densities. Root exudates consist of a complex mixture of sugars, amino acids, vitamins, organic acids, and other compounds (15, 16, 18) that provide the necessary elements and energy sources to support bacterial growth in the rhizosphere.
The molecular mechanisms involved in the colonization of plant roots by rhizobacteria are being extensively studied by two different but complementary approaches: (i) a random approach involving the isolation and characterization of mutants with reduced colonization capacities and (ii) a directed design, in which specific functions presumed to be important for colonization are tested and their roles in plant-bacterial interactions are assessed. These two approaches have allowed researchers to establish the importance of elements such as flagella, type IV pili, or chemotactic responses (11, 12, 23, 24). Particular attention has been paid to the so-called plant growth-promoting rhizobacteria and, among them, to Pseudomonas spp. strains that exert beneficial effects on plant health. Pseudomonas spp. genes encoding functions involved in plant root and seed colonization have been identified, including an agglutination factor (8), a site-specific recombinase (9), and a series of proteins involved in adhesion to seeds (13).
In recent years there have been increasing efforts in a third direction: the analysis of bacterial gene expression in the rhizosphere by techniques such as in vivo expression technology. Genes that respond to root exudates or that are preferentially expressed in the rhizosphere have been identified in Rhizobium sp. and Pseudomonas spp. among others (5, 6, 19, 20). Some of the genes identified in Pseudomonas fluorescens are involved in sugar transport and metabolism, amino acid transport, secretion, and oxidative stress response (19).
In the soil bacterium Pseudomonas putida KT2440, which is a very efficient colonizer of the rhizosphere of corn and other agronomically important plants (17), information regarding gene expression in the rhizosphere is still limited. Previous work has shown that the putA and putC genes involved in proline catabolism in this strain are induced in response to corn root exudates, where this amino acid is relatively abundant (25).
To expand these studies we used a transposon carrying a promoterless lux reporter to identify promoters induced in P. putida KT2440 by root exudates. We have isolated and characterized in detail a mutant affected in an aminotransferase involved in lysine metabolism, the expression of which is increased in response to corn root exudates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
P. putida KT2440, a derivative of the P. putida soil isolate mt-2 (14), was used. Escherichia coli strains and plasmids employed for transposon mutagenesis (SM10[λpir] harboring pUT-miniTn5[Km]′luxCDABE and HB101 harboring RK600) have been described before (10, 26). Plasmid pGB1 is a multicopy cloning vector that can replicate in E. coli and Pseudomonas spp. (7). The P. putida KT2440 cosmid library used in this study has been described elsewhere (21).
E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (22). P. putida strains were grown at 30°C either in LB or in minimal medium, which was basal M9 medium (22) supplemented with Fe-citrate, MgSO4, and trace metals, as described previously (1), and with benzoate (15 mM), glucose (0.4% [wt/vol]), or sodium citrate (10 mM) as a carbon source, unless otherwise specified. Where indicated, M9 lacking NH4Cl, which we called M8, was used to test the ability of the different strains to use alternative nitrogen sources. When appropriate, antibiotics were added at the following concentrations (in μg/ml): chloramphenicol, 30; kanamycin, 50; and tetracycline, 15. Chloramphenicol was used in cultures of P. putida KT2440 or its derivatives, since this strain is naturally resistant to this antibiotic.
Collection of root exudates.
Corn seeds were surface sterilized by washing twice for 15 min with 70% (vol/vol) ethanol and twice with 20% (vol/vol) bleach, followed by thorough rinsing with sterile deionized water. Surface-sterilized seeds were germinated on a petri plate with sterile deionized water at 30°C for 3 days. Seedlings were then transferred to a grid and grown hydroponically in M8 for 3 days, and root exudates were collected and filter sterilized according to the method described by Vílchez et al. (25).
Mutagenesis.
Transposon mutagenesis with a mini-Tn5(Km) derivative carrying ′luxCDABE from Photorhabdus luminescens (26) was performed by triparental mating. The recipient (P. putida KT2440), donor (E. coli SM10[λpir] harboring the suicide vector carrying mini-Tn5), and helper (E. coli HB101 with pRK600) were grown overnight in LB with the appropriate antibiotics. After incubation of the recipient at 42°C for 15 min to temporarily inactivate its restriction systems, 0.7 ml of the recipient was mixed with 0.2 ml of the donor and 0.1 ml of the helper. Cells were collected by centrifugation, suspended in 50 μl of fresh LB, and spotted on an LB plate. After overnight incubation at 30°C, cells were scraped off the plate and resuspended in 1 ml of LB, and serial dilutions were plated on selective minimal medium, which was M9 with benzoate, kanamycin, and chloramphenicol.
Monitoring of bioluminescence.
Bioluminescence in response to seed exudates was detected on soft agar plates to facilitate diffusion of the exudates. In each plate, 100 μl of an overnight culture was mixed with a cooled (42°C) 0.2% (wt/vol) water-agar solution. The mixture was poured onto petri dishes and allowed to solidify. Three surface-sterilized seeds were then placed in different spots on each plate, and the plates were incubated at 30°C for 5 h. Luminescence was detected by exposing the plate to Hyperfilm MP (Amersham) autoradiography film for 1 h.
To monitor luminescence on roots, seeds were incubated in M9 for 1 h with a bacterial suspension (1:1,000 dilution from overnight cultures) and planted in pots containing vermiculite. Plants were kept under greenhouse conditions at 28°C with natural day-night cycles. At different times, the plants were removed, placed on wet filter paper, and covered with clear plastic wrap. Luminescence was detected after overnight exposure on film.
Quantitative measurements of bioluminescence in liquid cultures (three independent experiments) were done with a BioOrbit 1250 luminometer.
Competitive root colonization assays.
For root colonization assays, overnight cultures grown in LB were diluted in M9 salts to a turbidity at 660 nm (optical density at 660 nm) of about 1, and seeds were inoculated with 1:1,000 dilution suspensions of bacterial mixtures (1:1 proportion). After incubation for 1 h, the seeds were washed and planted in pots containing vermiculite or used to determine the number of bacteria attached to the seed (see below). Plants were kept under greenhouse conditions at 28°C with natural day-night cycles for 1 week. At that time the plants were removed, and the roots were cut, weighed, and placed in sterile 50-ml screw-cap tubes containing 20 ml of M9 and 4 g of glass beads (diameter = 3 mm). The tubes were vortexed for 2 min, serial dilutions were plated on selective media (M9-benzoate with the appropriate antibiotics), and the number of CFU per gram of root was determined for each plant. The same process was used with inoculated seeds to determine the number of attached bacteria (i.e., the initial inoculum on the seeds).
DNA techniques.
Preparation of plasmid and chromosomal DNA, digestion with restriction enzymes (Roche and New England BioLabs), ligation, electrophoresis, and Southern blotting were done using standard methods (3, 22). Hybridizations were done using the DIG DNA Labeling and Detection kit (Roche) according to the manufacturer's instructions. Plasmid sequencing was done using universal or reverse pUC19/M13 oligonucleotides as primers. The transposon insertion point was identified by PCR amplification with Taq polymerase (Amersham Pharmacia), followed by direct sequencing with primers TNEXT2 (5′-CTTTATTGATTCCATTTTTACACT-3′) and DAVT2 (5′-AGGCGATTTCAGCGAAGCAC-3′). DAVT2 corresponds to the 3′ end (complementary strand) of davT, and TNEXT2 corresponds to the transposon [94 bp from the right end of mini-Tn5(Km)′luxCDABE]. Sequencing was done on an ABI PRISM 310 automated sequencer. Sequences were analyzed with Omiga 2.0 software (Oxford Molecular) and compared with data from the P. putida KT2440 genome, obtained from The Institute for Genomic Research (http://www.tigr.org), and with the GenBank database using BLAST programs (2).
Nucleotide sequence accession number.
Sequences have been deposited in GenBank under accession number AF299291.
RESULTS
Isolation of mutants in root exudate-inducible (rei) genes.
To identify genes that are preferentially expressed in response to corn root exudates, P. putida KT2440 was mutagenized with a mini-Tn5 derivative carrying a promoterless lux operon and a kanamycin resistance gene (26). Insertion of this transposon in the chromosome allows the generation of transcriptional fusions with the lux operon, which can be identified by the production of bioluminescence (26).
One thousand kanamycin-resistant mutants were picked after P. putida KT2440 mutagenesis and transferred to M9-benzoate plates with or without root exudates (100 μl of 3-day root exudates per plate). After overnight incubation, the plates were checked for light emission. Around 14% of the clones were luminescent. Three clones showed increased luminescence in the presence of root exudates, indicating that the transposon insertion had given rise to a transcriptional fusion induced by the root exudates. One of these mutants, which we named rei-2 (for root exudate induced), consistently showed the strongest signal and was chosen for detailed analysis.
Lysine utilization is impaired in mutant rei-2.
As an initial approach to determine which components of the root exudate were responsible for the increased luminescence, rei-2 was grown in minimal medium with different sugars, i.e., glucose, fructose, sucrose, or xylose, and with or without Casamino Acids. We observed increased luminescence only when Casamino Acids were present in the growth medium (data not shown). Similar experiments with the other two rei mutants did not result in an increase in luminescence with any of the substrates tested. To find out whether rei-2 responded to a specific amino acid, the clone was spotted on M9-benzoate plates supplemented with each amino acid separately (40 μg/ml), and light emission was checked after overnight incubation. Luminescence was significantly more intense in the presence of lysine than in the presence of any other amino acid. Similar results were obtained in liquid cultures grown in minimal medium with benzoate supplemented with lysine (Fig. 1).
FIG. 1.
Bioluminescence of rei-2 in response to different amino acids. Cultures were grown overnight in tubes with M9-benzoate (−) or M9-benzoate supplemented with 40-μg/ml concentrations of various amino acids. Tubes were then placed together on a metal rack and exposed to autoradiography film for 2 min.
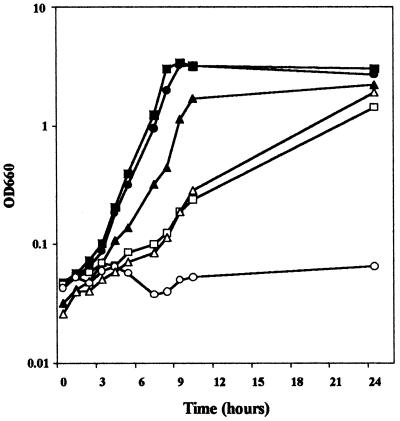
On the basis of these results, we considered the possibility that the mutation in rei-2 affected a gene involved in lysine metabolism. Consequently, we tested whether growth of this mutant was impaired with lysine as a carbon and/or nitrogen source. P. putida KT2440 and rei-2 were streaked on M9 and M8 (which is identical to M9 except that it lacks NH4Cl) supplemented with glucose and lysine or with lysine alone. Both strains grew similarly on M9 supplemented with both glucose and lysine. However, P. putida KT2440 could use lysine as a nitrogen and carbon source (although not very efficiently in the latter case), whereas rei-2 grew poorly on lysine as the only nitrogen source and did not grow at all when the amino acid was the sole carbon or carbon and nitrogen source (results not shown). These results were confirmed in cultures in liquid M8 medium with lysine as the sole carbon and nitrogen source (Fig. 2) and indicated that the transposon had been inserted in a gene involved in lysine catabolism. No difference was observed between the two strains in cultures grown in M9 with glucose as the carbon and energy source (Fig. 2).
FIG. 2.
Growth of P. putida KT2440 and its mutant derivative rei-2 with and without plasmid pLYS24. Wild-type P. putida KT2440 (squares), rei-2 (circles), and rei-2(pLYS24) (triangles) were grown on M9 minimal medium with 20 mM glucose (closed symbols) or on M8 with 25 mM lysine (open symbols) as the only carbon and nitrogen source. Growth was determined by measuring turbidity at 660 nm at the indicated time (hours after inoculation).
The gene interrupted in rei-2 encodes a putative aminotransferase.
To identify the gene affected in rei-2, a cosmid library of P. putida KT2440 (21) was used to complement the mutation. After transfer of the library to the mutant by conjugation, we selected a clone that could grow on M8 with lysine as the sole carbon and nitrogen source. The cosmid that allowed complementation of the growth deficiency was isolated and digested with PstI, and the resulting fragments were subcloned in the pGB1 shuttle vector. After transformation of rei-2, eight clones able to grow on lysine as the sole carbon and nitrogen source were analyzed. In all cases, the plasmid contained a 2.1-kb PstI fragment. One of these was chosen and named pLYS24. This plasmid complemented the growth deficiency of rei-2 in both solid and liquid media with lysine as the carbon and nitrogen source (Fig. 2). However, rei-2 harboring pLYS24 grew slightly more slowly in M9 with glucose, probably as the result of the metabolic load imposed by the multicopy plasmid.
To confirm that the cloned fragment corresponded to the locus where the insertional mutation had occurred, the 2.1-kb PstI fragment was isolated, labeled with digoxigenin, and used as a probe on a Southern hybridization with chromosomal DNA isolated from P. putida KT2440 and rei-2 digested with various restriction enzymes. In all cases, a band that hybridized with the probe showed less mobility in the mutant than in the wild-type strain (not shown), indicating that the 2.1-kb fragment did correspond to the locus where the transposon had been inserted.
Plasmid pLYS24 was also used for partial sequencing with the M13 universal and reverse primers. The sequence obtained was compared with the available data from the P. putida genome obtained from The Institute for Genomic Research. The sequence could be unambiguously identified in contig 10746 with the exact size of the PstI fragment (deduced from the genome sequence data) being 2,123 bp. Restriction analysis of pLYS24 and comparison of the results with those expected from the genome sequence data confirmed that this fragment was indeed the one cloned in pLYS24.
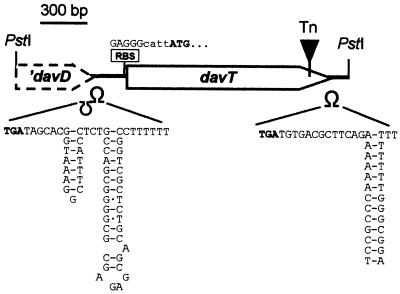
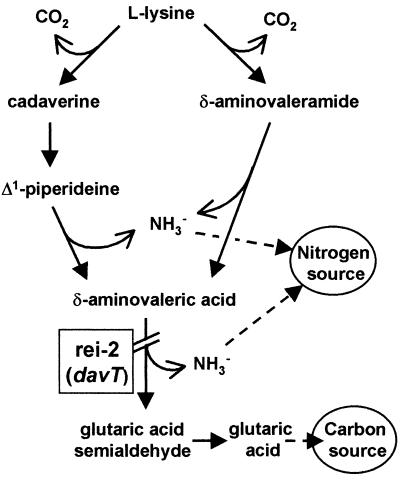
Analysis of the genomic sequence revealed a 1,275-bp open reading frame preceded by a potential ribosome binding site (GAGGG) (Fig. 3). Downstream of the stop codon, a stem-loop structure that might correspond to a rho-independent terminator was also identified (Fig. 3). Similarity searches were then performed with the databases using BLAST programs (2). The 1,275-bp open reading frame showed a high degree of similarity with genes coding for aminotransferases. In particular, the deduced amino acid sequence was 73% identical to the product of the E. coli gabT gene, γ-aminobutyrate (GABA) aminotransferase (EC 2.6.1.19), the enzyme that catalyzes the conversion of GABA into succinic semialdehyde (4). This similarity, and the fact that rei-2 can use lysine as a nitrogen source to a certain extent, suggested that the product of this gene could act as a δ-aminovalerate aminotransferase, the enzyme responsible for the conversion of δ-aminovalerate (an intermediate in lysine metabolism) (Fig. 4) into glutaric acid semialdehyde (18). To test this hypothesis, P. putida KT2440, rei-2, and rei-2(pLYS24) were inoculated on plates containing 20 mM δ-aminovalerate as a carbon and/or nitrogen source or 20 mM glutarate as a carbon source. As with lysine, rei-2 was unable to grow with δ-aminovalerate as a carbon source, and the defect was reversed by pLYS24. All three strains grew well on glutarate. Also, δ-aminovalerate acted as a strong effector of the transcriptional fusion in rei-2 (see below). Taken together, these results suggest that δ-aminovalerate is indeed the substrate for the aminotransferase encoded by the gene interrupted in rei-2 (Fig. 4). We therefore named this gene davT (for delta-aminovalerate transaminase).
FIG. 3.
Structure of the 2.1-kb fragment cloned in pLYS24. The ribosome binding site (RBS) of davT, the putative terminators (Ω), and the transposon insertion point (Tn) are indicated. Start and stop codons are shown in bold.
FIG. 4.
Lysine catabolism pathway in P. putida. The diagram is based on that of Phillips (18) and indicates the step interrupted in mutant rei-2.
The point of insertion of the mini-Tn5 was precisely defined by PCR amplification with an oligonucleotide reading outward from the transposon and an oligonucleotide corresponding to the complementary strand of davT at the 3′ end of the gene (see Materials and Methods). The amplified product was isolated from an agarose gel and used for sequencing with either of the two oligonucleotides as primers. In this way, we located the insertion 125 bp upstream of the stop codon of davT (Fig. 3).
Upstream of davT, the 3′ end of a truncated open reading frame was found within the 2.1-kb PstI fragment also followed by a potential terminator (Fig. 3). Comparison of this sequence with the databases revealed high homology with succinic semialdehyde dehydrogenases (EC 1.2.1.16). The 169-amino-acid sequence was 80% identical to the last 169 amino acids of succinic semialdehyde dehydrogenase of E. coli, the enzyme encoded by gabD and responsible for the conversion of succinic semialdehyde into succinate (4). The fact that the enzymes in this family are around 480 amino acids long suggested that this gene was indeed truncated in the fragment cloned in pLYS24. This was confirmed by analyzing the sequence upstream of the PstI site in the P. putida genome. Its similarity to semialdehyde dehydrogenases and its proximity to davT suggested that this gene, which we tentatively named davD, encodes glutaric semialdehyde dehydrogenase. A similar organization appears in gabD and gabT of E. coli (4). However, in E. coli these genes appear in a cluster with two other genes, gabC and gabP (a regulator and a GABA transporter, respectively), whereas in P. putida, no similar genes were found upstream or downstream from davD and davT in the genome sequence.
Expression of davT.
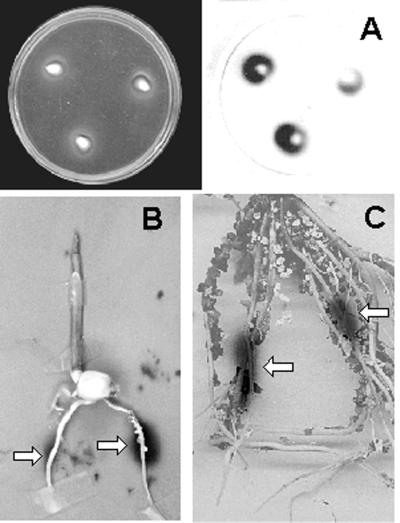
The response of rei-2 to corn exudates was assayed at different stages of seed germination and root development. Examples of these experiments are shown in Fig. 5. An initial assay was performed on soft agar plates with corn seeds, followed by detection of luminescence as described in Materials and Methods. After 5 h of incubation, intense luminescence was detected in the area adjacent to the seeds (Fig. 5A).
FIG. 5.
Bioluminescence of rei-2 in response to corn exudates in planta in the presence of corn seeds (A) and in the rhizosphere after 6 (B) or 15 (C) days of inoculation. Luminescence was detected by exposure on film as described in Materials and Methods. The images have been overlaid in panels B and C to show the root areas where luminescence was more intense (arrows).
To detect expression of the davT::lux fusion in the rhizosphere, hydrated seeds were inoculated with a suspension of rei-2 and planted on vermiculite. At different times, plants were removed and the roots were exposed to film. Luminescence was detected around the central areas of the root at all of the times tested. Examples are shown in Fig. 5B (6 days) and C (15 days). No luminescence was observed in samples of vermiculite taken from areas that had not been in contact with the root (not shown).
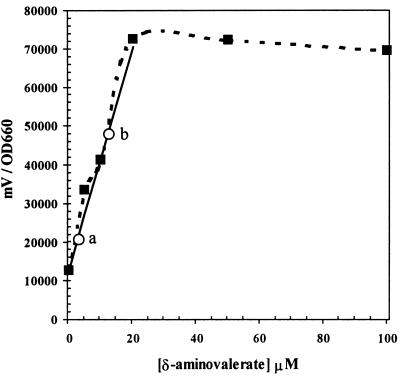
Expression of the davT::lux fusion was also measured in liquid cultures with different concentrations of δ-aminovaleric acid as well as with corn root exudates. An overnight culture of rei-2 was diluted 1:1,000 in 2 ml of fresh M9-citrate medium supplemented with increasing concentrations of δ-aminovalerate or with dilutions of 3-day corn root exudates. Cultures were allowed to grow for 5 h at 30°C, and at that time, culture turbidity and luminescence were determined (Fig. 6). Maximal luminescence was obtained with 20 μM δ-aminovalerate. Higher concentrations did not result in any increase in luminescence. The least-squares line of the linear portion of the curve was adjusted in order to extrapolate the data obtained with root exudates. Two of these data are shown in Fig. 6. Extrapolation resulted in calculated concentrations of ∼12 μM for inducer(s) in cultures to which 50 μl of root exudate had been added and of ∼3.7 μM for those to which 20 μl of root exudate had been added. We were therefore able to estimate the concentration of inducer(s) in root exudates collected after 3 days to be ∼0.42 mM.
FIG. 6.
Quantitative measurement of luminescence relative to optical density at 660 nm in liquid cultures in the presence of 0, 5, 10, 20, 50, or 100 μM δ-aminovalerate. Squares and broken line indicate actual values. The continuous line corresponds to the least-squares adjustment of the linear portion of the curve. Circles indicate the values obtained in cultures supplemented with different dilutions of root exudates. Two data points are shown: 20 (a) and 50 (b) μl of root exudates. Experiments were done in triplicate, and the results for a representative experiment are shown (the induction pattern and deduced concentration of inducers in root exudates were the same in all cases).
Relevance of davT in root colonization.
Competitive root colonization assays were performed to determine the potential importance of lysine catabolism in root colonization by P. putida KT2440. Overnight cultures of KT2440 and rei-2 were diluted in M9 and mixed at a proportion of 1:1. Hydrated corn seeds were then inoculated with this mixture and sown in pots containing vermiculite. After 7 days, plants were removed and the number of bacteria present on the root was determined as described in Materials and Methods. As shown in Table 1, the number of CFU per gram of root of KT2440 was twice as high as that of rei-2. Similar results were obtained when rei-2 was tested in colonization assays with rei-2(pLYS24) instead of KT2440, whereas there were no differences in colonization when KT2440 was tested in competition with the complemented mutant (Table 1). These differences were consistently observed (with less than 12% deviation) in spite of some variability in the total number of cells recovered from plant to plant. Thus, the mutation in davT resulted in a slight but not very significant reduction in the colonization capacity of KT2440.
TABLE 1.
Competitive root colonization assaysa
| Strain | Initial population on seeds | Population on corn root after 1 wk |
|---|---|---|
| KT2440 | 1.5 × 104 | 3 × 106 |
| rei-2 | 1.9 × 104 | 1.4 × 106 |
| rei-2(pLYS24) | 2 × 104 | 3.4 × 106 |
| rei-2 | 1.1 × 104 | 1.1 × 106 |
| KT2440 | 5.5 × 104 | 6.3 × 106 |
| rei-2(pLYS24) | 3 × 104 | 4.7 × 106 |
Results are reported in CFU per gram of root or seed. Details for culture of bacteria and seed inoculation are given in Materials and Methods. Data are the averages of three independent assays done in triplicate. Standard deviations were below 12% of the given values.
DISCUSSION
The study of bacterial gene expression in response to root exudates is beginning to shed light on the physiological state of bacteria in the rhizosphere. In this work we used transposon mutagenesis with a reporter lux operon to identify genes induced in the presence of corn root exudates. Three out of 1,000 clones showed induction of bioluminescence in the presence of root exudates, which might seem like a surprisingly small proportion (0.3%). The number of mutants screened so far is not sufficient for a full survey; however, it should be noted that only 14% of the clones were luminescent (i.e., the transposon insertion had given rise to a functional fusion with the reporter). Thus, 3 out of 140 transcriptional fusions were induced by root exudates, which gives an estimate of around 2% of the genes responding to exudates. The selection method used here is perhaps less appropriate for mass screening of rhizosphere-inducible genes than other systems such as in vivo expression technology, but it has the advantage of allowing easy in situ detection and quantitative measurement of expression, as shown in Fig. 5 and 6, respectively.
We characterized in detail one of the genes which encodes an enzyme with δ-aminovalerate aminotransferase activity involved in lysine catabolism. To our knowledge, this is the first direct genetic evidence linking the δ-aminovalerate transaminase activity in pseudomonads with a specific DNA sequence. Expression of a davT::lux fusion is induced by lysine and δ-aminovaleric acid and can be detected in bacteria that colonize the root system of corn plants. The strong signal obtained in response to seed exudates probably reflects the abundance of at least certain amino acids in the early steps of seed germination and plant development. This is consistent with the relatively high concentration of inducers of davT expression estimated to be present in exudates (0.42 mM). Given the similarity of DavT to other aminotransferases, we cannot rule out the possibility that other compounds structurally similar to lysine or δ-aminovalerate also act as inducers.
In root colonization assays, the signal detected with the davT::lux transcriptional fusion was localized mainly in the central parts of the root. It has been proposed that exudates are different in composition in different root areas, with sugars being more abundant in the root tips and amino acids being released mainly by older parts (15). Our data seem to support this idea.
Many of the genes previously found in P. fluorescens to be induced in the rhizosphere correspond to functions involved in nutrient transport and utilization (19). The results presented here support the relevance of nutrient utilization pathways for bacterial life in the rhizosphere. A P. putida mutant unable to use lysine as a carbon source was slightly less competitive in root colonization assays than its parental strain. The nature of root exudates, a complex mixture in which various compounds are present in small amounts, probably makes it important for rhizobacteria to be able to utilize many different carbon and energy sources in order to efficiently compete and establish in this ecological niche. Yet at the same time, the availability of various alternative nutrients in the rhizosphere may compensate for the defect in lysine utilization, and this mechanism may explain why the differences observed between the wild type and the davT mutant in their ability to colonize the root system were not very significant.
ACKNOWLEDGMENTS
We are grateful to S. Swift for strains and plasmids. We thank A. Salido for technical assistance, A. Hurtado for DNA sequencing, and M. I. Ramos-González for advice on the use of the P. putida KT2440 cosmid library. We thank K. Shashok for editing the revised version of the manuscript.
This work was supported by grant BIO4-CT98-0283 from the European Union and grant 99178 from the U.S.-Spain Joint Commission for Scientific and Technological Cooperation.
REFERENCES
- 1.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Bartsch K, von Johnn-Marteville A, Schulz A. Molecular analysis of two genes of the Escherichia coli gab cluster: sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD) J Bacteriol. 1990;172:7035–7042. doi: 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayliss C, Bent E, Culham D E, MacLellan S, Clarke A J, Brown G L, Wood J M. Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3-canola mutualism: identification of an exudate-inducible sugar transporter. Can J Microbiol. 1997;43:809–818. doi: 10.1139/m97-118. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat A A, Keister D L. Identification and cloning of Bradyrhizobium japonicum genes expressed strain selectively in soil and rhizosphere. Appl Environ Microbiol. 1992;58:1490–1495. doi: 10.1128/aem.58.5.1490-1495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buell C R, Anderson A J. Genetic analysis of the aggA locus involved in agglutination and adherence of Pseudomonas putida, a beneficial fluorescent pseudomonad. Mol Plant-Microbe Interact. 1992;5:154–162. doi: 10.1094/mpmi-5-154. [DOI] [PubMed] [Google Scholar]
- 9.Dekkers L, Phoelich C C, Van der Fits L, Lugtenberg B J J. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc Natl Acad Sci USA. 1998;95:7051–7056. doi: 10.1073/pnas.95.12.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Weger L A, Van der Vlugt C I M, Wijfjes A H M, Bakker P A H M, Schippers B, Lugtenberg B J J. Flagella of a plant growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol. 1987;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dörr J, Hurek T, Reinhold-Hurek B. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol. 1998;30:7–17. doi: 10.1046/j.1365-2958.1998.01010.x. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa-Urgel M, Salido A, Ramos J L. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;182:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaeger C H, III, Lindow S E, Miller W, Clark E, Firestone M K. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol. 1999;65:2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugtenberg B J J, Kravchenko L V, Simons M. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol. 1999;1:439–446. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- 17.Molina L, Ramos C, Duque E, Ronchel M C, García J M, Wyke L, Ramos J L. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem. 2000;32:315–321. [Google Scholar]
- 18.Phillips A T. Biosynthetic and catabolic features of amino acid metabolism in Pseudomonas. In: Sokatch J R, editor. The bacteria. A treatise on structure and function, vol. X. The biology of Pseudomonas. Orlando, Fla: Academic Press; 1986. pp. 385–438. [Google Scholar]
- 19.Rainey P B. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol. 1999;1:243–257. doi: 10.1046/j.1462-2920.1999.00040.x. [DOI] [PubMed] [Google Scholar]
- 20.Rodelas B, Lithgow J K, Wisniewski-Dye F, Hardman A, Wilkinson A, Economou A, Williams P, Downie J A. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J Bacteriol. 1999;181:3816–3823. doi: 10.1128/jb.181.12.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Herva J J, Ramos-González M I, Ramos J L. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell envelope. J Bacteriol. 1996;178:1699–1706. doi: 10.1128/jb.178.6.1699-1706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Smit G, Swart S, Lugtenberg B J J, Kijne J W. Molecular mechanisms of attachment of Rhizobium bacteria to plant roots. Mol Microbiol. 1992;6:2897–2903. doi: 10.1111/j.1365-2958.1992.tb01748.x. [DOI] [PubMed] [Google Scholar]
- 24.Vande Broek A, Lambrecht M, Vanderleyden J. Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasilense. Microbiology. 1998;144:2599–2606. doi: 10.1099/00221287-144-9-2599. [DOI] [PubMed] [Google Scholar]
- 25.Vílchez S, Molina L, Ramos C, Ramos J L. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J Bacteriol. 2000;182:91–99. doi: 10.1128/jb.182.1.91-99.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winson M K, Swift S, Hill P J, Sims C M, Griesmayr G, Bycroft B W, Williams P, Stewart G S A B. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]