Abstract
The vast majority of bacteria in the environment have yet to be cultured. Consequently, a major proportion of both genetic diversity within known gene families and an unknown number of novel gene families reside in these uncultured organisms. Isolation of these genes is limited by lack of sequence information. Where such sequence data exist, PCR directed at conserved sequence motifs recovers only partial genes. Here we outline a strategy for recovering complete open reading frames from environmental DNA samples. PCR assays were designed to target the 59-base element family of recombination sites that flank gene cassettes associated with integrons. Using such assays, diverse gene cassettes could be amplified from the vast majority of environmental DNA samples tested. These gene cassettes contained complete open reading frames, the majority of which were associated with ribosome binding sites. Novel genes with clear homologies to phosphotransferase, DNA glycosylase, methyl transferase, and thiotransferase genes were identified. However, the majority of amplified gene cassettes contained open reading frames with no identifiable homologues in databases. Accumulation analysis of the gene cassettes amplified from soil samples showed no signs of saturation, and soil samples taken at 1-m intervals along transects demonstrated different amplification profiles. Taken together, the genetic novelty, steep accumulation curves, and spatial heterogeneity of genes recovered show that this method taps into a vast pool of unexploited genetic diversity. The success of this approach indicates that mobile gene cassettes and, by inference, integrons are widespread in natural environments and are likely to contribute significantly to bacterial diversity.
Over the past decade, it has become clear that the majority of bacteria in environmental samples remain undescribed. Examination of environmental samples with molecular methods or microscopy has revealed a large discrepancy between the relatively few organisms capable of being cultured from such samples and the diversity and numbers of organisms actually present (9, 15, 20). Because the majority of bacteria are yet to be discovered or brought into culture, it is clear that the majority of their genetic diversity is unknown. This unknown diversity is in the form of both undiscovered gene families and undiscovered genetic variation within known gene families. In anticipation of recovering useful genes from this unexplored gene pool, various research groups have designed methods for identifying genes in the yet to be cultured fraction of the microbiota. Two approaches are currently being used: PCR amplification of known gene families (14, 23, 29) and screening of shotgun libraries of large DNA fragments generated from environmental sources (often in bacterial artificial chromosomes) by using mass sequencing, hybridization, or activity assays (11, 12, 25). The shotgun approach is limited by the effort involved in identifying genes within the large sequence fragments. PCR has the potential to overcome this problem, but is limited by the availability of suitable priming sites and does not recover intact genes.
The genomics era has clearly indicated that a large proportion of bacterial genes have been acquired by horizontal gene transfer (19). Thus, there is an opportunity to recover a significant proportion of the “undiscovered” bacterial gene pool, not by targeting gene sequences themselves, but rather by targeting conserved sequences associated with mobile elements. Horizontal gene transfer is facilitated by a number of genetic elements in bacteria, including plasmids, transposons, and integrons. Traditionally, most attention has focused on plasmids and transposons. This is particularly true for environmental microbiology (24). However, since integrons have recently been demonstrated to occur in the chromosomes of diverse bacterial species (22) and integron integrases are recoverable from environmental samples (18), we reasoned that integrons are widespread in natural environments.
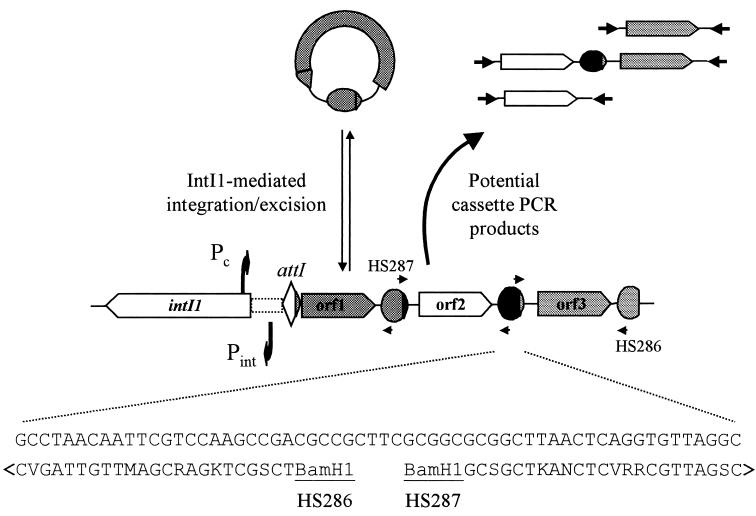
Integrons are gene acquisition and expression systems. The units of DNA captured by integrons, gene cassettes, are the simplest known mobile elements and consist of only a gene and a recombination site known as a 59-be (59-base element) (1, 21). Cassettes are inserted into or excised from integrons by a site-specific recombination reaction catalyzed by the integron integrase, IntI (Fig. 1) (3, 5, 7, 17, 26). Multiple insertion events lead to the formation of multicassette arrays, which in chromosomal integrons may contain over 150 cassettes (2). In such arrays, essentially all cassette-associated genes are flanked by 59-be recombination sites. While these sites are variable in terms of both their sequence and length, they do have a number of common features, including a conserved sequence of about 25 bp at each end that forms imperfect inverted repeats (Fig. 1) (27).
FIG. 1.
Exploitation of the integron-gene cassette system for recovery of intact genes by PCR. Gene cassettes form integrated, linear arrays when in association with integrons. Cassettes can integrate at either the integron-associated recombination site (attI) or cassette-associated recombination sites (59-be family). In all cases, the recombination boundary is within a highly conserved GTTRRRY motif as shown. The primers HS286 and HS287 target conserved sequence within the left and right halves of 59-bes, respectively. When used in cassette PCR, these primers recover intact genes or arrays of genes as shown. The example sequence shown is that of the aadB 59-be.
Several features of the integron-gene cassette system suggested to us that it might provide a means by which intact novel genes could be recovered directly from environmental DNA by PCR without prior sequence data. First, integrons appear to be a feature of many and diverse bacterial species (18, 22). Second, many of the cassette arrays associated with chromosomal integrons are very large (2, 22). Third, the structure of multiple cassette arrays means that individual genes are flanked by conserved sequences (59-be sites) that are potential targets for PCR primers. Here we show that the use of PCR primers targeting 59-be sites allows the recovery of complete genes, the vast majority of which are novel and do not encode products that have orthologs in protein databases.
MATERIALS AND METHODS
DNA template isolation.
Soil, sediment, biomass, or water samples were collected from a variety of locations in Australia and Antarctica (Table 1). DNA was isolated from 400 ± 20 mg (500 ml for water samples) of material by a bead-beating procedure (28). Details of sample collection and processing have been previously described for most locations (13, 29). The Yerranderie silver mine samples were collected at 1-m intervals along a linear transect crossing the mine drainage channel. Samples from Cape Denison were taken at multiple points around the perimeter of a penguin colony.
TABLE 1.
Environmental sites sampled for gene cassettes
| Locationa | Description | Sample type | No. of samplesb | No. PCR positivec | No. of clones sequenced |
|---|---|---|---|---|---|
| Balmain, NSW | Abandoned industrial site | Contaminated soil | 6 | 4 | 32 |
| Homebush, NSW | Abandoned industrial site | Contaminated estuarine sediment | 10 | 5 | 20 |
| Macquarie, NSW | Urban eucalypt forest | Soil | 8 | 3 | NDd |
| Lidsdale, NSW | Mixed plantation forest | Soil | 2 | 2 | ND |
| Sturt National Park, NSW | Semidesert | Sandy and stony soils | 9 | 5 | 4 |
| Nullarbor, SA | Aquatic cave | Microbial biofilm | 6 | 2 | ND |
| Shelley Beach, NSW | Ocean beach | Seawater | 1 | 0 | ND |
| Hunter River, NSW | Shallow river | Sediment | 2 | 0 | ND |
| Namoi River, NSW | Shallow river | Sediment | 6 | 6 | ND |
| Yerranderie, NSW | Abandoned silver mine | Soil and sediment, low pH | 29 | 28 | 26 |
| Cape Denison, Antarctica | Penguin colony | Ornithogenic soil | 12 | 10 | 8 |
| Flinders Ranges, SA | Hot spring, 48–63°C | Microbial biofilm and sediment | 9 | 9 | 7 |
| Lomatia Creek, NSW | Urban creek | Microbial biofilm and sediment | 6 | 6 | 6 |
| Jubilee 1, NSW | Abandoned dump site | Contaminated soil | 3 | 3 | 5 |
| Jubilee 2, NSW | Urban creek | Sediment | 1 | 1 | 6 |
Samples used for cloning are represented by the following acronyms: Balmain, Bal; Homebush, HB; Pulgamurtie landform, Sturt National Park, Pu; Yerranderie silver mine, SM; Cape Denison ornithogenic soil, Orn; Flinders ranges hot spring, FR; Lomatia creek sediment, Lom; Jubilee dump, Dum; Jubilee creek sediment, Sed. NSW, New South Wales; SA, South Australia.
Number of independent samples collected and tested with the cassette PCR.
Number of samples generating PCR products. PCR profiles from the same sample were highly reproducible; independent samples collected at the same location showed high levels of intersample variability.
ND, not determined. Libraries were not constructed from these samples.
PCR amplification.
The primers to conserved sequences in 59-be sites used were HS286 (5′ GGGATCCTCSGCTKGARCGAMTTGTTAGVC 3′) and HS287 (5′ GGGATCCGCSGCTKANCTCVRRCGTTAGSC 3′). These primers target the flanking regions of 59-be sites as shown in Fig. 1. The underlined sequence is a BamHI linker that is not complementary to 59-be sequences. Reaction mixes consisted of approximately 5 ng of template DNA, 100 pmol of each primer, 200 nM deoxynucleoside triphosphate (dNTP) mix, 2 mM MgCl2, and 1 U of Red Hot DNA polymerase (Advanced Biotechnologies) in the reaction buffer supplied with the enzyme. The PCR was carried out by standard techniques with the following cycling program: 94°C for 3 min for 1 cycle, 94°C for 30 s, 55°C for 30 s, 72°C for 2 min 30 s for 35 cycles, and 72°C for 5 min for 1 cycle. All template DNAs gave a positive result in a control 16S ribosomal DNA amplification, performed with the primers f27 and r1492 as previously described (28).
Ligation and transformation.
PCR products were ligated into the pGEM-T Easy vector (Promega, Madison, Wis.) following the manufacturer's instructions. The ligation mixture was transformed by heat shock into Escherichia coli JM109 competent cells (catalog no. L2001; Promega) following the manufacturer's protocol.
Plasmid isolation and sequencing.
Plasmid from clones containing insert was isolated from 3-ml overnight cultures by using the Wizard Plus Miniprep DNA purification system (Promega) as per the manufacturer's instructions. DNA sequencing of cloned inserts was performed at the Macquarie Sequencing Facility (Macquarie University, New South Wales, Australia) with an ABI Prism 377 (PE Biosystems), using primers flanking the insert region pGEMF (5′ CCGACGTCGCATGCTCC 3′) and pGEMR (5′ CTCCCATATGGTCGACCTG 3′). For clones with longer inserts, complete sequence was generated by the use of additional sequencing primers specific for the inserts in question.
Sequence retrieval and analysis.
Sequence analyses were performed with programs available through the BioNavigator package (eBioinformatics Pty., Ltd. [http://www.eBioinformatics.com]) and the GCG package (Genetics Computer Group, Madison, Wis.). Open reading frames (ORFs) were identified by using MAP. Homologs to inferred proteins were searched for by BLASTP and PSI-BLAST. Putative 59-be sites were identified by searching cloned sequences by eye. Identified putative 59-be sites (Fig. 2) fulfilled the following criteria. (i) They possessed the eight invariant residues found within known 59-be sites. (ii) Two putative IntI-like simple sites were present. (iii) They possessed a significant inverted repeat structure that included the two putative simple sites and the sequence located between these sites (27).
FIG. 2.
Comparison of a typical 59-be recovered from environmental DNA with a 59-be from an antibiotic resistance gene cassette. (A) The relatively conserved first 29 and last 28 bases of each element (27) are indicated. The numbers indicate the number of bases in the central region of each 59-be. The aadB 59-be is from the previously described aadB gene cassette (1, 21). The HB14 element is an example of an element from environmental DNA (accession no. AF265263). The sequences are those found in the circular gene cassette. The recombination crossover point for insertion of a circular cassette into an integron array is between the G and first T of site 1R (27). Consequently the last 6 bases of a cassette's 59-be are located at the front of the cassette (Fig. 1). For HB14, these 6 bases are within the binding region of the HS286 primer and are shown in lowercase. Asterisks indicate the eight invariant residues in 59-be sites (7, 27). The left and right simple sites are indicated by the overlined bar. The inverted repeats associated with these simple sites and which comprise putative IntI binding domains are named and indicated by the horizontal arrows and shading (6, 27). (B) The aadB and HB14 elements are shown as foldbacks to emphasize their inverted repeat structure and their structural similarity. The putative IntI binding domains are shaded.
Nomenclature and classification.
Gene cassettes have few sequence elements in common, but show distinct organizational patterns. All clones obtained here were assigned to groups according to the number of cassettes inferred to be present in the amplicon. Thus, group 1 PCR products are inferred to contain a single gene cassette, group 2 products are inferred to contain two cassettes in an array, and group 3 products are inferred to contain three cassettes. The cassettes within each clone were classified into one of seven distinct types (A to G) according to the number and orientation of ORFs (Table 2). The orientation of an ORF was defined by its associated 59-be if present or from the position of the HS287 primer for group 1 clones. All clones are thus classified according to both the number and type of cassettes they contain. For example, a group 3ABA clone has three cassettes, a type A, followed by type B, and then another type A. Individual clones are designated according to a two- to four-letter code indicating the sample of origin followed by a unique number (or alphanumeric) for that library (Table 1). Identified ORFs within gene cassettes were generally greater than 50 codons, beginning with an ATG, GTG, or TTG. Where more than one ORF matched these criteria, the longest ORF was considered to be the most likely coding sequence.
TABLE 2.
Distribution of structural arrangements in known gene cassette pools
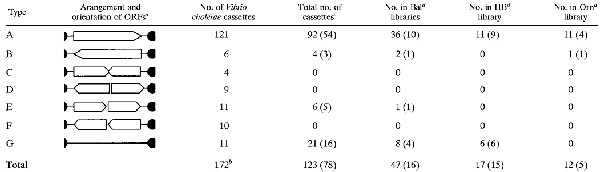 |
Open arrowed boxes denote ORFs and their orientation. No obvious ORFs are present in type G cassettes.
An additional seven cassettes in V. cholerae are atypical and may represent illegitimate recombination or transposition events (10).
Forty-five cassettes were contained within arrays, and the presence of internal 59-be sites within such arrays confirms their identity as gene cassettes. The remaining 78 cassettes were not part of arrays and therefore did not possess an internal 59-be (this includes OrnC5, which comprises two type A cassettes within a single plasmid). The numbers of these single cassettes for each type are indicated in parentheses in this and the remaining columns.
Libraries were constructed from 15 samples. These columns display data from the three most extensively sequenced libraries, Balmain (Bal), Homebush (HB), and Cape Denison ornithogenic soil (Orn).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences described herein are AF265260 to AF265275, AF349046 to AF349111, AF356540, and AF378527 to AF378541.
RESULTS
PCR with 59-be primers is cassette specific.
In an attempt to recover cassette-associated bacterial genes from natural environments, PCR primers to the left- and right-hand conserved regions of 59-be sites were designed. These primers, HS286 and HS287, specific for the left and right halves, respectively, are oriented as shown in Fig. 1. PCR was performed with 113 DNA samples derived from a total of 15 sites (Table 1) in Australia and Antarctica. Sites were selected to represent a diverse range of environments and included marine and freshwater, as well as terrestrial environments that have suffered various levels of anthropogenic disturbance. Multiple, independent samples were collected from each site, and it was found that the vast majority of samples provided DNA that produced cassette PCR products (Table 1). Where present, multiple products were recovered with fragment sizes mostly falling within the range of 300 to 1,000 bp.
Clone libraries were constructed from 15 of the DNA samples (Table 1). A total of 114 clones were sequenced from these libraries. A small number of sequences were found to occur more than once in the libraries, giving a total of 99 distinct clone types (now referred to as clones) in all libraries. The insert sizes of the cloned fragments ranged from 251 to 1,497 bp. These sequences were analyzed to assess the specificity of the PCR for gene cassettes in different environments.
In the strategy employed here, successful amplification relies on the occurrence of multiple cassettes incorporated in an array and could result in the amplification of a single cassette or multiple cassettes (Fig. 1). In the simplest scenario, where primer binding sites flank a single cassette, the amplification products will not include any sequence elements conserved between all gene cassettes (group 1 products [see Materials and Methods]). Therefore, to assess the specificity of our PCR technique, we examined the net pattern of arrangement of sequence features in our amplicons. As shown in Table 2, there are only seven known arrangements for genetic features within gene cassettes with a very strong bias toward arrangement type A.
The most obvious sequence feature in the amplicons is represented by the PCR primers, which show a nonrandom distribution among the amplicons. In 97 of the 99 clones, the HS286 primer is at one end and the HS287 primer is at the other. This indicates that the PCR products reflect a very strong trend for binding sites for these two primers to be linked and that this linkage of primer binding sites is due to their specificity for 59-be sites present in linear gene cassette arrays.
The competitive nature of PCR is expected to favor the amplification of shorter PCR products resulting in a bias toward amplicons representing a single gene cassette. Given the strong bias toward single-ORF, and therefore generally smaller, cassettes (type A [see Materials and Methods]), we anticipated the majority of amplicons would contain a single ORF with its start and stop codons in close proximity to the HS287 and HS286 primers, respectively. Examination of all cloned sequences showed this to be the case for 54 clones, which we consider to represent group 1A amplicons (i.e., one cassette of arrangement type A) (Table 2). The orientation of the ORF with respect to the two primers is equivalent to the specific orientation of cassette ORFs in known integron arrays. This proportion of intact ORFs, in defined orientation, is highly unlikely to be recovered by chance.
The defining feature of a gene cassette is a 59-be recombination site. We anticipated a significant proportion of amplicons to represent multiple gene cassettes and include 59-be sites. All clones were examined for putative 59-be sites (see Materials and Methods). As expected, none of the group1A amplicons contained a putative 59-be, further suggesting that these clones represent a single cassette. A total of 20 clones were found to contain one or more putative 59-be sites (Fig. 2). In 12 of these, the putative 59-be was located in the sequence between two forward ORFs. These clones are considered to represent group 2AA amplicons, derived from two type A cassettes (Table 2) within an array. We also recovered three group 3AAA amplicons, each of which included three ORFs and two putative 59-be sites. In each of these cases, the putative element was located between two adjacent ORFs. The remaining clones included alternate cassette arrangement types in their array. Clone Bal25 represents a group 2BA amplicon, since the ORF in the first cassette is in reverse orientation (Table 2). Bal33 (group 2AG) and SM63A4 (group 2GA) each included one cassette with no apparent ORF. The first cassette in Pu8 (group 3EAA) contained two ORFs in the forward orientation, and none of the cassettes in Bal48 (group 3GGG) contained an ORF.
Taken together, these data demonstrate that the HS286-HS287 primer pair is highly selective for integron-associated cassette arrays. As such, it is probable that the majority of the remaining (24 of 99) clones represent single cassettes of arrangement types B, E, and G. On this basis, we infer that the environmental gene cassette libraries analyzed here contain a total of 123 cassettes (Table 2). Two observations provide additional support for this conclusion. First, the relative proportions of cassette arrangement types in the environmental clones are consistent with the relative proportion of such arrangement types in the fully characterized Vibrio cholerae N16961 integron (Table 2). Second, of the 16 group 1G amplicons, 8 show sequence homology (>60%) to the type G cassette recovered in Bal33 (group 2AG) and 4 show sequence homology to the first cassette of the array in Bal48 (group 3GGG). These type G cassettes appear to constitute relatively common families of unknown function.
Cassettes are abundant in natural environments.
A total of 123 cassettes (Table 2) were identified in the clone libraries. Only 17 of these cassettes were recovered more than once, and where this occurred, identical cassettes were obtained from the same PCR sample. Several features of the recovered cassettes are notable, including the fact that the pool of cassettes in natural environments is very large, suggesting that the number sampled is significantly less than the total available pool of cassettes present in the environments tested. This is true even for the environments most extensively assayed (Table 2). In the case of Balmain, 47 distinct cassettes were recovered from a total sample size of 50.
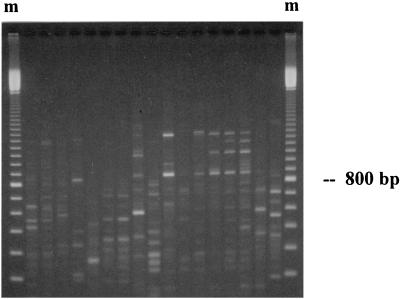
To further assess the diversity of the cassette gene pool, we examined spatial variation in the cassette-PCR product profile. At three locations, Yerranderie, Cape Denison, and Sturt National Park, multiple samples were collected along transects. The electrophoretic profile for independent amplification reactions of the same DNA sample was highly reproducible at all sites (data not shown). In contrast, independent samples produced different patterns of PCR products (Fig. 3). This included adjacent sample sites separated by as little as 1 m. Although we cannot extrapolate from the number of observed bands to the number of cassettes, these data clearly indicate a high level of spatial variability in the cassette pool. This is likely to be due to differences in both cassette composition and relative abundance between sample points.
FIG. 3.
Amplification of gene cassettes along a soil transect. Soil samples were collected at 1-m intervals along a transect spanning a drainage channel at an abandoned silver mine in Yerranderie (Table 1). DNA extracted from each sample was amplified with the primers HS286 and HS287. Products were separated by electrophoresis on 2% agarose and stained with ethidium bromide. Adjacent lanes represent samples separated by 1 m. Note the diversity of amplification products generated and their spatial heterogeneity. Only a few adjacent samples exhibit similar amplification profiles. Each marker lane (m) contains a 100-bp ladder.
Cassette-associated genes are novel.
In total, 107 ORFs were found in cassettes of types A, B, and E (Table 2). For none of these ORFs could an obvious promoter be identified. Additionally, there was no room for a promoter in approximately 80% of cases, since the cassette boundary is less than 40 bases from the putative start codon in these clones. However, this arrangement is typical for most cassette-associated genes, because, where it has been examined, transcription is driven by the integron promoter Pc (4, 16). For 57 of the 107 ORFs, a putative ribosome binding site was identified.
Database searches were carried out with the predicted products of all 107 ORFs. All were found to be previously undescribed, with only 13 displaying a significant relationship to proteins present in sequence databases (Table 3). Of these, eight matched to hypothetical proteins found in a range of Proteobacteria and, in one case, to a bacteriophage-encoded protein. The predicted protein from orf90_Pu8 (Table 3), also showed a significant match to a hypothetical protein (encoded by vca0332) located in a cassette in the V. cholerae integron (10). Protein families that had homologues in environmental cassettes were a hygromycin phosphotransferase, a putative toxin antidote protein, a PemK-like plasmid maintenance protein, an RNA methyl transferase, a thiosulfate thiotransferase, and a pyrimidine dimer DNA glycosylase.
TABLE 3.
Cassette gene products with database matches
| Gene product | Top database hit | Function of matching protein |
|---|---|---|
| orf297_Bal40 | Bacillus subtilis (CAB15191) (P = 1.3e–15) | Conserved hypothetical protein |
| orf101_FR60F2 | Streptomyces coelicolor (CAA22728) (P = 8.5e–21) | Conserved hypothetical protein |
| orf117_HB2 | Bacteriophage 933W (AAD25429) (P = 7.5e–31) | Hypothetical protein |
| orf346_Pu5 | Streptomyces sp. (KYYB_STRHY) (P = 0.002) | Aminoglycoside phosphotransferase |
| orf271_Pu11 | Streptomyces peucetius (CAA06603) (P = 1.3e–5) | Possible thiosulfate thiotransferase |
| orf81_SedF7 | Pyrococcus horikoshii (BAA29861) (P = 0.00056) | Possible toxin antidote protein |
| orf113_SedF7 | Staphylococcus aureus (CAA71064) (P = 1e–21) | PemK family, toxin part of plasmid maintenance system |
| orf132_Bal2-28 | Pseudomonas aeruginosa (AAG07752) (P = 1e–15) | Hypothetical protein |
| orf208_Bal31 | Thermus thermophilus (BAB17605) (P = 8e–17) | RNA methyl transferase |
| orf133_Bal32 | Mycobacterium tuberculosis (Y034_MYCTU) (P = 1.8e–18) | Hypothetical protein |
| orf90_Pu8 | Synechocystis sp. strain PCC6803 (BAA17409) (P = 1.3e–27) | Conserved hypothetical protein |
| orf105_Pu8 | Mycobacterium tuberculosis (YM71_MYCTU) (P = 2.6e–16) | Hypothetical protein |
| orf147_SM63E3 | Micrococcus luteus (BAA11346) (P = 6.4e–48) | Pyrimidine dimer DNA glycosylase |
DISCUSSION
It is increasingly clear that integrons are a common feature of bacterial genomes, and the number of integron classes recovered from natural environments now exceeds those from clinical environments (18, 22). The range of genera now known to host integrons is also very broad and includes Vibrio, Shewanella, Geobacter, Treponema, and Nitrosomonas (18, 22), and this list will undoubtedly continue to grow. This diversity of species hosting integrons means that platforms suitable for the storage, acquisition, rearrangement, and expression of gene cassettes may be widespread in nature. In this context, the data discussed below are particularly significant, since they strongly support the hypothesis presented above that integrons are very common, if not ubiquitous, in natural bacterial populations.
The cassette-associated gene pool is very large. Where products were obtained, the PCR protocol used here was highly selective for gene cassettes in all environments tested. Despite this, it is apparent that the 123 cassettes recovered are not representative of the diversity of cassettes in these environments. Indeed only one cassette, that of clone HB5, was sampled more than twice. The extent to which the sequenced clones undersample the diversity present in the PCR products is highlighted by calculation of sampling efficiency. Coverage is a statistical measure of the fraction of an infinite sample set that is included in an actual sample set (8). Calculation of the coverage with respect to cassettes indicates that the sequenced clones would represent only 25% of an infinitely large clone library. This statistic does not give any information on the diversity remaining in the unsampled portion. Consequently, it is not possible to estimate the upper limit for the number of cassette-associated genes recoverable by the present PCR protocol.
The existence of a very large cassette gene pool is also supported by the intersample variability of the PCR. The spatial variability (Fig. 3) reflects variation in cassette composition between microbial populations across relatively small distances. Spatial variation in cassette composition could be due to the presence of distinct integrons in diverse species (18) or differences in cassette profiles between the same integron in closely related individuals (2, 22). In either event, it is clear that the presence of a very large and diverse array of mobile gene cassettes reinforces the observation that integrons are present in many diverse genera. Indeed, the recovery of gene cassettes from all of the environment types tested suggests that these mobile elements, and by implication integrons, have a very wide phylogenetic distribution. This in turn has widespread ramifications for bacterial gene flow within natural populations (19).
A further point should be considered in evaluating the extent of the cassette gene pool sampled by PCR. PCR is capable of introducing considerable sample bias. Two factors are likely to be particularly important in this instance. The first is bias toward smaller amplicons, and the second is primer bias. The primer pair used here was designed against a database primarily composed of 59-be sites from antibiotic resistance gene cassettes found in class 1 integrons. As the database of 59-be sequences has expanded, it has become evident that the primer set does not encompass the sequence diversity in this family of recombination sites. Indeed, several 59-be sites identified within group 2 and 3 clones have diverged significantly from the primer sequences and were only recovered as a consequence of being located between cassettes that are flanked by 59-be sites conforming to the consensus sequences (data not shown). Furthermore, it has recently been suggested that 59-be sites comprise sequence homology groups related to their origin in chromosomal integrons (22). Even if this hypothesis is only applicable to a subset of integrons, it implies that the HS286-HS287 primer set will systematically undersample gene cassette pools. We anticipate that as more integron-gene cassette systems are described, new sets of primers favoring the recovery of distinct cassette pools may be designed.
One observation arising from this study is that the recovered cassettes include a diverse range of genes, the vast majority of which have no known homologues in the databases. Since integrons are widepread features of bacterial populations, it is clear that this pool of novel mobile genes represents a previously unrecognized genomic resource for bacteria. Collectively these data give cause to reconsider our ideas of bacterial genome flexibility and the diversity of proteins likely to be found in even well-known bacterial species. The cassette-associated gene pool of a bacterial community contains a minimum of hundreds of novel genes and is conceivably several orders of magnitude higher. In contrast to the well-known plasmid and transposon systems, these genes are contained in elements capable of facilitating rapid mobilization, reshuffling, and expression of either individual genes or combinations thereof. The rapid emergence of multiple antibiotic resistance in mobile (plasmid and transposon associated) integrons reflects the efficiency of this system in exploiting a vast gene pool.
In addition to providing a means of tracking integron-mediated gene transfer in the environment, the PCR strategy presented here represents a unique opportunity to prospect for new genes of biotechnological importance by culture-independent means. The “floating genome” of the integron-gene cassette system is evidently extensive, exists across multiple species and environments, and includes highly diverse genes. In conjunction with the selective constraints on mobile genes, these features suggest a high probability of biotechnologically useful genes being present within this pool. If this gene pool is accessed by culture-independent cassette PCR, there are a number of corollary benefits to the screening process and subsequent manipulation of the genes. Most notably, identification of gene boundaries and location in a sequence fragment is greatly simplified, the orientation of reading frames is highly predictable, and genes are prepackaged in a form amenable to manipulation by site-specific recombination. Consequently, the PCR strategy outlined here provides rapid access to a significant genetic resource in a way that is independent of prior gene sequence knowledge and recovers the gene in a form ready for direct analysis.
ACKNOWLEDGMENT
This work was supported by a Research Innovation Fund grant from Macquarie University.
REFERENCES
- 1.Cameron F H, Groot Obbink D J, Ackerman V P, Hall R M. Nucleotide sequence of the AAD(2") aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986;14:8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark C A, Purins L, Kaewrakon P, Focareta T, Manning P A. The Vibrio cholerae O1 chromosomal integron. Microbiology. 2000;146:2605–2612. doi: 10.1099/00221287-146-10-2605. [DOI] [PubMed] [Google Scholar]
- 3.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis C M, Grammaticopoulos G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 6.Collis C M, Kim M-J, Stokes H W, Hall R M. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol Microbiol. 1998;29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 7.Collis C M, Recchia G D, Kim M-J, Stokes H W, Hall R M. Efficiency of recombination reactions catalysed by the class 1 integron integrase IntI1. J Bacteriol. 2001;183:2535–2542. doi: 10.1128/JB.183.8.2535-2542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Good I J. The population frequencies of species and the estimation of the population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 9.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 10.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Doson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henne A, Daniel R, Schmitz R A, Gottschalk G. Construction of environmental DNA libraries in Escherichia coli and screening for presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol. 1999;65:3901–3907. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henne A, Schmitz R A, Bomeke M, Gottschalk G, Daniel R. Screening environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol. 2000;66:3113–3116. doi: 10.1128/aem.66.7.3113-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes A J, Bowyer J, Holley M P, O'Donoghue M, Montgomery M, Gillings M R. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol Ecol. 2000;33:111–120. doi: 10.1111/j.1574-6941.2000.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 14.Holmes A J, Roslev P, McDonald I R, Ivesen N, Henricksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 17.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nield B S, Holmes A J, Gillings M R, Recchia G D, Mabbutt B C, Nevalainen K M H, Stokes H W. Recovery of new integron classes from environmental DNA. FEMS Microbiol Lett. 2001;195:59–65. doi: 10.1111/j.1574-6968.2001.tb10498.x. [DOI] [PubMed] [Google Scholar]
- 19.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 20.Pace N R. A molecular view of microbial biodiversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 21.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 22.Rowe-Magnus D A, Guerot A M, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci USA. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seow K-T, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson C R, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smalla K, Krögerrecklenfort E, Heuer H, Dejonghe W, Top E, Osborn M, Niewint J, Tebbe C, Barr M, Bailey M, Gretaed A, Thomas C, Turner S, Young P, Nikolakopoulou D, Karagouni A, Wolters A, van Elsas J D, Dronen K, Sandaa R, Borin S, Brabhu J, Grohmann E, Sobecky P. PCR-based detection of mobile genetic elements in total community DNA. Microbiology. 2000;146:1256–1257. doi: 10.1099/00221287-146-6-1256. [DOI] [PubMed] [Google Scholar]
- 25.Stein J, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 27.Stokes H W, O'Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 28.Yeates C, Gillings M R. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett Appl Microbiol. 1998;27:49–53. [Google Scholar]
- 29.Yeates C, Holmes A J, Gillings M R. Novel forms of ring-hydroxylating dioxygenases are widespread in pristine and contaminated soils. Environ Microbiol. 2001;2:644–653. doi: 10.1046/j.1462-2920.2000.00147.x. [DOI] [PubMed] [Google Scholar]