Abstract
The use of isiA expression to monitor the iron status of cyanobacteria was investigated. Studies of laboratory cultures of the cyanobacterium Synechocystis sp. strain PCC 6803 showed that isiA expression is dependent on the organism's response to iron deficiency; isiA expression starts as soon as a decline in the rate of growth begins. isiA expression is switched on at concentrations of iron citrate of less than 0.7 μM. A PCR method was developed for the specific amplification of the iron-regulated isiA gene from a variety of cyanobacteria. After we developed degenerate primers, 15 new internal isiA fragments (840 bp) were amplified, cloned, and sequenced from strains obtained from algal collections, from new isolates, and from enriched field samples. Furthermore, isiA expression could be detected by means of reverse transcription-PCR when enriched field samples were exposed to restricted iron availability. These results imply that determining the level of iron-regulated isiA expression can serve to indicate iron deficiency in cyanobacterial samples of differing origins from the field.
Iron as an important micronutrient of phytoplankton had been neglected for a long period before Martin and coworkers (28–30) revived its consideration in their approach to defining the basic pattern of plankton productivity in the open oceans. Subsequent research dealt with iron as the crucial nutrient in high-nitrate, low-chlorophyll regions of the open oceans and its ecological importance (1, 12, 16, 22, 43). Some evidence of iron-limited phytoplankton development also exists for coastal upwelling regions (13). Owing to the complex chemical behavior of iron, the bioavailable iron concentration cannot easily be manipulated in oxygen-rich waters by iron enrichment and iron depletion procedures. Trace-metal precipitation and adsorption to particles as well as ion-exchange processes (for a review, see reference 39) are only a few of the potential effects of these procedures that can hamper the desirable effects of fertilization and depletion experiments. Although these are methods that provide support for the characterization of the concentration of bioavailable iron in water (42), the great number and variety of variously efficient mechanisms of organismic iron sequestration (6, 14, 43) limit the interpretation of the corresponding analyses. Thus, the use of molecular markers to detect the physiological state of iron limitation in microalgae without the use of artificial manipulations of water chemistry would represent an important achievement in proving iron limitation in parts of phytoplankton.
Besides producing results of general interest, the use of molecular approaches became necessary to go beyond the limits of conventional methodology in aquatic ecology. Scanlan et al. (36) introduced the phosphate-binding protein PstS as a potential diagnostic marker for investigating phosphate stress in photosynthetic picoplankton. For iron limitation, flavodoxin accumulation was used as a biochemical marker and its detection in single cells provided some insight into the physiological adaptation of natural phytoplankton on the molecular level (22). However, variability in levels of flavodoxin expression has been found and strains of coastal origin, among them the only tested cyanobacterial representative, showed no flavodoxin expression under iron-depleted conditions at all (8). In addition, the study of iron limitation in cyanobacterial field populations using molecular markers is more difficult than in eukaryotic algae (23).
Cyanobacteria have a significantly higher demand for iron than eukaryotic algae (4, 44). Investigations of cyanobacterial iron accumulation in the field are rare. Cyanobacterial responses to iron stress resemble those of heterotrophic bacteria (for a review, see reference 44), but some filamentous species express flavodoxin constitutively after N deprivation in heterocysts (35; U. Geiß et al., unpublished data). In coccoid cyanobacteria, the flavodoxin-encoding gene isiB is cotranscribed in a dicistronic operon with isiA. Under iron-limiting conditions, transcription is highly induced but the dicistronic message is 10-fold less abundant than the monocistronic isiA transcript (25, 40), which is iron regulated as well. IsiA (also known as CP43′) is a chlorophyll a-binding protein similar to CP43 (PsbC) of photosystem II and the Pcb protein of prochlorophytes (21). Its exact function in iron-starved cells is still a matter of discussion. It has been hypothesized to serve as a chlorophyll a storage system or an extra light-absorbing system to protect the photosystems against excess light under iron-depleted conditions (5, 9, 11, 32). Four isiA genes, which reveal a high degree of homology, are known so far (24–26).
In the present study, the reliability of isiA expression with respect to iron depletion was investigated by means of a model cyanobacterium. The relation of growth rate to isiA expression as well as the induction of the isiAB promoter was examined. Additionally, a PCR method was developed for amplifying specifically isiA genes from diverse cyanobacterial sources. Application of this method extended the sequence information on isiA genes from miscellaneous cyanobacterial samples and provided a tool for a first isiA expression study using enriched cultures originating from brackish phytoplankton. Besides the integrated flavodoxin approach (7, 8), detection of isiA expression may support investigations into the iron supply available to cyanobacterium-dominated coastal phytoplankton.
MATERIALS AND METHODS
Organisms and culture conditions.
The strains listed in Table 1 were derived from the Sammlung von Algenkulturen (SAG), Universität Göttingen, Göttingen, Germany, and the Pasteur Culture Collection. The cyanobacterial isolates were kindly provided by the Pharmaceutical Institute, University of Greifswald. Field samples were taken on 30 July 2000 at a water depth of 0.3 m from the Darss-Zingst estuary (Bodstedter Bodden, pier of Zingst, Germany), which is characterized by salinities of 5 to 10‰. Fifty-milliliter phytoplankton samples were concentrated by centrifugation and plated on 1% Kobe agar (Roth) in medium C (17). After growth on petri dishes in daylight, the algal consortia were transferred into the synthetic growth medium BG11 (33), leading to the growth of mixtures of cyanobacteria, green algae, and bacteria.
TABLE 1.
Organisms examined in PCR studies
| Sectiona | Tested strain or sample | Strain or culturef | Presence or absence of isiA gene |
|---|---|---|---|
| Strains | |||
| I | Chamaesiphon polonicus | SAG 32.87 | − |
| Cyanothece sp. | PCC 9224 | − | |
| Gloeobacter violaceus | SAG 7.82 | − | |
| Microcystis aeruginosab | BM Mi/5 | + | |
| Synechocystis sp. | PCC 6803 | + | |
| Synechococcus sp. | PCC 7942 | + | |
| PCC 7002 | + | ||
| SAG 14.02-1 | + | ||
| II | Chroococcidiopsis thermalis | SAG 42.79 | − |
| Dermocarpella sp. | SAG 29.84 | − | |
| Myxosarcina sp. | SAG 30.84 | − | |
| Stanieria sp. | SAG 27.84 | + | |
| Xenococcus sp. | SAG 28.84 | − | |
| III | Lyngbya sp. | SAG 36.91 | − |
| Lyngbya lagerheimii | SAG 24.99 | + | |
| Microcoleus chthonoplastes | SAG 31.92 | − | |
| Oscillatorialesc | 99-2/6.2.2 | + | |
| 99-3/5.3.1 | + | ||
| IV | Anabaena sp. | PCC 7120 | + |
| Anabaena torulosa | SAG 26.79 | + | |
| Anabaenopsis elenkinii | SAG 252.80 | + | |
| Microchaete sp. | SAG 47.93 | − | |
| Tolypothrix sp. | PCC 7601 | + | |
| V | Fischerella sp. | PCC 73103 | + |
| Chlorogloeopsis fritschii | SAG 1411-1a | + | |
| Field samples | |||
| Nodularia bloomd | R | + | |
| Enrichment culturesc | BB 1 | + | |
| BB 2 | + | ||
| BB 3 | + | ||
| BB 4 | − | ||
| BB 5 | − | ||
| BB 6 | − | ||
| BB 7 | + |
Taxonomic sections are as described by Rippka (33).
From the Max-Planck-Institute for Limnology, Plön, Germany.
From the Pharmaceutical Institute, University of Greifswald, Greifswald, Germany.
From the Baltic Sea, Rerik, Mecklenburg-Vorpommern, Germany.
From the Darss-Zingst estuary, Bodstedter Bodden, Mecklenburg-Vorpommern.
PCC, Pasteur Culture Collection; SAG, Sammlung von Algenkulturen, Göttingen, Germany.
Cyanobacterial strains were grown at 29°C under constant illumination (40 μmol of photons m−2 s−1) for physiological investigations and mRNA expression studies using BG11 medium (33). All other strains were grown in medium as recommended by the culture collections. For all iron depletion experiments, media were prepared without an iron source. Only significant sources of iron contamination (water, NaNO3 stock solution) were purified in a Chelex 100 (Bio-Rad) column in order to minimize secondary iron contaminations of less abundant ultrapure salts owing to extensive handling of solutions and glassware. Acid-rinsed glassware was used throughout the experiments. At the onset of iron limitation experiments, cells were washed three times with iron-deficient medium. During incubation, cells were transferred every second day into fresh medium. Strains were harvested when cultures showed a blue shift of chlorophyll a absorbance from approximately 680 to 673 nm, indicating iron deficiency (11). Owing to the presence of other algae, this indicator could not be applied to enriched field samples that were harvested after about 12 days of cultivation, when slight chlorosis could be observed.
For investigations on growth rate and induction of the isiAB promotor, Synechocystis strain MpIGisi (18) was used. This mutant is a derivative of Synechocystis sp. strain PCC 6803T with a chromosomally integrated fusion product of the isiAB promotor with the gfp gene (18). MpIGisi cells were cultivated semicontinuously (daily medium exchange and adjustment of cell density) in the presence of kanamycin (50 μg ml−1) at 29°C under constant illumination (175 μmol of photons m−2 s−1) and continuous aeration (2% [vol/vol] CO2) using a nitrate-containing mineral medium (2).
Cells of Escherichia coli strain TG1 were used for routine DNA manipulations after cultivation in Luria broth at 37°C (34).
DNA and RNA techniques.
All DNA techniques such as plasmid isolation, transformation of E. coli, and ligation were performed according to standard procedures (34). Chromosomal DNA was extracted from cyanobacterial cells by phenol and chloroform treatment (41). Nucleotide sequences of known isiA genes from the strains Synechocystis sp. strain PCC 6803, Synechococcus sp. strain PCC 7942, Synechococcus sp. strain PCC 7002, and Anabaena sp. strain PCC 7120 were aligned using the software package ALIGN (Align Plus, version 2.0, copyright 1989, 1992; Scientific & Educational Software) to design the degenerate primers isiAfw (5′-AAD TAY GAH TGG TGG GC-3′ [bp 28 to 44 of the Synechocystis sp. strain PCC 6803 isiA gene]) and isiArev (5′-CGT TTC GGC AAA RTA RGG-3′ [bp 849 to 866 of the Synechocystis sp. strain PCC 6803 isiA gene]). A predicted 840-bp-long fragment was obtained in the PCR performed with Taq PCR Master Mix (Qiagen). For the combined amplification of psbC, pcb, and isiA fragments, the reverse primer HLWHA-1 (5′-GCG TGC CAS AGR TGA CC-3′ [bp 948 to 964 of the Synechocystis sp. strain PCC 6803 isiA gene]) was used together with the isiAfw primer mentioned above. The PCR program consisted of an initial 94°C denaturation step for 5 min and 1 min each of denaturation (94°C), annealing (52°C), and polymerization (72°C) for 33 cycles before the final elongation step (10 min). PCR fragments were evaluated by Southern hybridization by applying digoxigenin-labeled isiA probes from Synechocystis, Synechococcus strain PCC 7002, and a Fischerella sp., which were obtained using a PCR digoxigenin probe synthesis kit (Roche Biochemicals). All PCR products were separated on 0.8% agarose gels, and the bands were excised and eluted using a Qiaex kit (Qiagen). The obtained DNA fragments were cloned into plasmid pGEM-T (Promega). Both strands of these fragments were sequenced using the dideoxy chain termination method by applying a Thermo Sequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Life Science). Universal primers, which were fluorescently labeled with an infrared dye, IRD 800 (MWG Biotech), were used for sequencing.
Prior to RNA isolation, cells were broken by freezing them in liquid nitrogen and crushing them with a pestle. After an additional preextraction step with phenol (Aqua-Roti-Phenol; Roth) for 10 min (pH 4.5 to 5; 65°C), total RNA was isolated using a High Pure RNA isolation kit (Roche Biochemicals). The separation of the RNA and the transfer onto nylon membranes were carried out as described previously (40). DNA probes for Northern blot experiments were synthesized by PCR using the above-mentioned degenerate isiA primers. A 16S rRNA-specific probe was generated using the universal primers for the eubacterial 16S rRNAs 27f and 1525r (20). The PCR products were purified on agarose gels, and specific bands were excised and labeled with [α-32P]dATP (Amersham) using a random primer labeling kit (MBI-Fermentas). Quantification of the signals obtained in the Northern blot experiments was done with a phosphorimager (BAS-1000; Fuji). To avoid differences caused by improper gel loading, the quantitative data were calculated on the basis of signals obtained after rehybridization of the same filters with the 16S rRNA-specific probe, with slight variations due to potential growth rate-dependent variations of rRNA content being accepted. Reverse transcription (RT)-PCR was done by applying SUPERSCRIPT II RNase H reverse transcriptase (Gibco BRL, Life Technologies) with the degenerate isiArev primer before regular PCR.
Computer analysis.
We searched for sequence similarities in databases with the assistance of the BLAST software program (3). Sequence alignments were performed with the software Align Plus (version 2.0; Scientific & Educational Software). The phylogram was constructed by means of a multiple sequence alignment of protein sequences using the software program PAUP (phylogenetic analysis using parsimony, beta version 4.0; David Swofford, Laboratory of Molecular Systematics, Smithsonian Institution).
Nucleotide sequence accession numbers.
The nucleotide sequences of the isiA/psbC gene fragments of Synechococcus sp. strain PCC 7942 (EBI accession no. P15347), Synechococcus sp. strain PCC 7002 (EBI accession no. P31157), Synechocystis sp. strain PCC 6803 (EBI accession no. P73884), and Anabaena sp. strain PCC 7120 (isiA, EBI accession no. S42648; psbC, EBI accession no. S42647) were extracted from databases. Complete and partial sequences of cyanobacterial isiA genes from Fischerella muscicola PCC 73103 (EBI accession no. AJ296146; for a previous study, see reference 10), Stanieria sp. strain SAG 27.84 (EBI accession no. AJ311682), Oscillatoriales sp. strain 99-2/6.2.2 (EBI accession no. AJ311683), Oscillatoriales sp. strain 99-3/5.3.1 (EBI accession no. AJ311684), Lyngbya lagerheimii SAG 24.99 (EBI accession no. AJ311685). Anabaena torulosa (EBI accession no. AJ311686), Anabaenopsis elenkinii SAG 252.80 (EBI accession no. AJ311687), BB 1 clone 1 (EBI accession no. AJ311688), BB 1 clone 2 (EBI accession no. AJ311698), BB 1 clone 5 (EBI accession no. AJ311690), BB 2 clone 4 (EBI accession no. AJ311691), BB 3 clone 4 (EBI accession no. AJ311692), BB 7 clone 3 (EBI accession no. AJ311693), R3 (EBI accession no. AJ311694), and R10 (EBI accession no. AJ311695) were obtained during this study and were submitted to the database.
RESULTS
Iron-dependent alterations of growth and isiA expression in Synechocystis sp. strain PCC 6803.
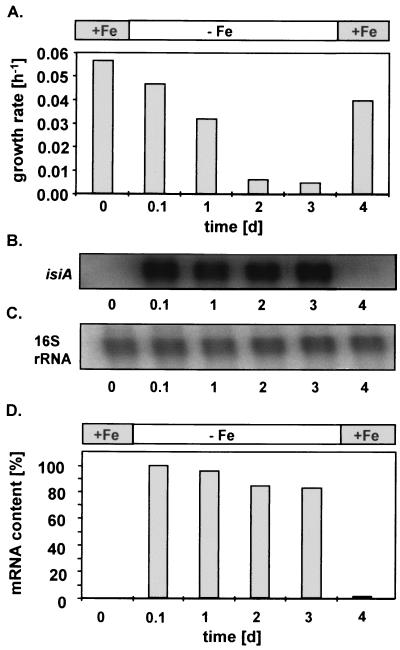
To document isiA induction, we focused on an appropriate model strain to investigate the iron dependency of the growth rate and its relation to isiA expression. During an experiment using aerated Synechocystis cultures, the growth rate and isiA mRNA synthesis were monitored. Slight effects on the growth rate were detected 3 h after the onset of iron-depleted conditions. After 3 days of iron depletion, a >10-fold reduction in the growth rate was observed (Fig. 1A). Additionally, RNA samples were taken each day to measure the content of isiA mRNA in Northern blot experiments with an isiA-specific probe (Fig. 1B to D). When the iron requirements of the cells were met, no transcription of isiA was detectable. However, as soon as 3 h after transfer into iron-depleted medium, isiA transcription was completely induced and the transcript level remained almost constant as long as these conditions were maintained. When cells were incubated under iron-replete conditions again, after as little as 3 h of cultivation, most of the isiA mRNA had disappeared. The monocistronic isiA-specific transcript predominated, whereas the dicistronic isiAB-specific mRNA gave only faint signals in iron-starved cells (not shown).
FIG. 1.
Growth rate and isiA expression of Synechocystis strain MpIGisi and dependence on the starting and stopping of iron-depleted growth in batch cultures with CO2-enriched aeration. +Fe, 19.2 μM Fe(NH4) citrate; −Fe, 0 μM Fe(NH4) citrate. (A) Specific growth rate; (B) Northern blot hybridization signals after application of an isiA-specific probe; (C) Northern blot hybridization signals after application of a 16S rRNA-specific probe; (D) quantitative estimation of isiA mRNA normalized to the 16S rRNA content (highest content corresponds to 100%). d, days.
Induction of the isiAB promoter as a function of the Fe(NH4) citrate concentration.
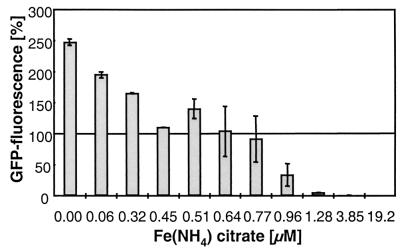
A reporter strain of Synechocystis named MpIGisi (18), carrying a fusion of the isiAB promoter with a promoterless gfp reporter gene, was used to investigate the iron concentration-dependent expression of isiA. The iron requirements of this strain were monitored in BG11 medium by observing the appearance of green fluorescent protein (GFP) fluorescence. In our first attempt, fluorescence-induced cells pregrown under iron-depleted conditions were transferred into medium with Fe(NH4) citrate concentrations ranging from 0 to 19.2 μM. The results shown in Fig. 2 reveal an increase in GFP fluorescence of cells grown with less than 0.77 μM, whereas faint to nondetectable fluorescence was observed at 0.96 to 19.2 μM Fe(NH4) citrate. The highest level of GFP fluorescence was obtained from cells in iron-free medium. These results were corroborated by a second experimental series in which cells with repressed GFP fluorescence after cultivation in iron-replete medium were transferred into media with increasing levels of iron depletion. As found before, iron concentrations above 0.77 to 0.96 μM failed to induce GFP fluorescence in the cells, while increases in fluorescence were induced by further decreases in iron concentrations (data not shown). The kinetics of fluorescence change as well as the level of fluorescence in the different samples reveal a strict dependence of isiAB promoter activity on the extracellular concentration of Fe(NH4) citrate.
FIG. 2.
GFP fluorescence in cells of Synechocystis strain MpIGisi after 9 days of cultivation in media containing different concentrations of Fe(NH4) citrate. The fluorescence level of precultivated cells [0.7 μM Fe(NH4) citrate] at the beginning of the experiment corresponds to 100%. Error bars denote standard deviations of triplicate samples.
Identification of the isiA gene in various cyanobacterial strains.
In order to use iron-dependent expression of isiA as a molecular marker to monitor iron supply in different cyanobacterial species, further investigations into whether the isiA gene is widespread among cyanobacteria are necessary. The search for this gene was carried out via a PCR approach with degenerate primers. For the generation of degenerate PCR primers, the amino acid sequences deduced from the four known isiA genes were compared to those of the structurally and functionally similar proteins PsbC and Pcb. Several regions of high similarity were detected. A sequence region at the N terminus contains the peptide sequence WWAGNAR, which is completely conserved in all known IsiA sequences and occurs similarily in PsbC. It was chosen to generate the primer isiAfw. After optimization experiments, a degenerate primer that includes the WWA motif and a few bases upstream was derived. In order to create a degenerate primer for isiA-specific amplifications, the second primer (isiArev) was deduced from the peptide sequence (PYFADT) inside the shortened lumenal loop E′ of IsiA, which represents the major difference between IsiA and PsbC (26). The nucleotide sequences encoding these peptides in the different cyanobacterial strains were aligned and used for the primer design. These isiA-specific degenerate primers were tested with DNAs from various cyanobacterial strains of all the basic taxonomic sections (Table 1).
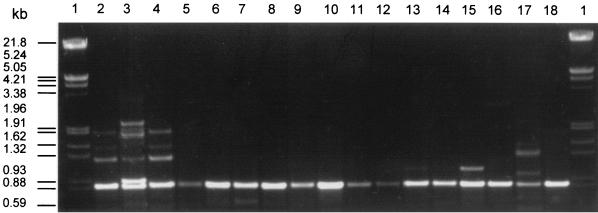
From many of the investigated strains, we amplified an expected 840-bp fragment (Fig. 3) which showed strong hybridization signals with isiA-specific probes in Southern blot experiments (data not shown). Cloning and sequencing revealed that the 840-bp-sized fragments which appeared indeed represented isiA genes. The high specificities of the degenerate isiA primers were further supported by control experiments using the Prochlorococcus marinus strain MED4. Its pcb gene, which possesses very high sequence similarity to isiA, was not amplified, whereas it could be amplified in a PCR using a further degenerate primer, HLWHA-1, which is homologous to a highly conserved region at the N termini of the genes isiA, psbC, and pcb (data not shown).
FIG. 3.
Separation of PCR fragments obtained with DNAs from various organisms (Table 1) using degenerate isiA-specific primers. In all cases, the 840-bp fragment could be verified as an isiA gene fragment by sequencing. Lanes: 1, marker (λ DNA EcoRI/HindIII digested); 2, Synechococcus sp. strain SAG 14.02-1; 3, Synechocystis sp. strain PCC 6803; 4, Synechococcus sp. strain PCC 7942; 5, Synechococcus sp. strain PCC 7002; 6, Anabaena sp. strain PCC 7120; 7, Fischerella muscicola PCC 73103; 8, Anabaena torulosa SAG 26.79; 9, Lyngbya lagerheimii SAG 24.99; 10, Microcystis aeruginosa BM Mi/5; 11, Oscillatoriales sp. strain 99-2/6.2.2 (isolate from Baltic Sea shoreline, Zingst); 12 to 15, BB 1, BB 2, BB 3, and BB 7 (enrichment cultures of estuarine field samples); 16, Oscillatoriales sp. strain 99-3/5.3.1 (isolate from Lake Kummerow); 17, Chlorogloeopsis fritschii SAG 1411-1; 18, Anabaenopsis elenkinii SAG 252.80.
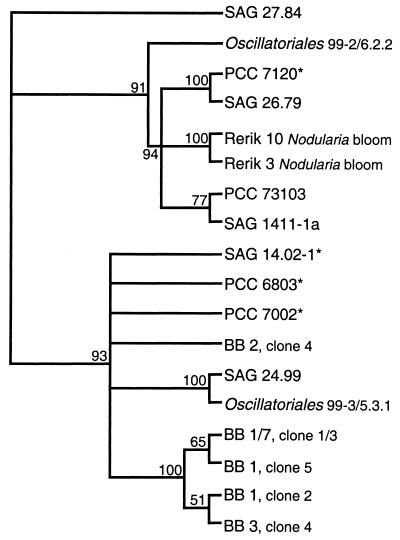
Fragments of isiA genes from representatives of all five taxonomic sections of cyanobacteria (33) could be amplified, indicating a widespread distribution of this gene. In further experiments, these degenerate primers were applied to DNAs from new isolates and mixed phytoplankton cultures of different waters. In isolates from brackish water (shoreline, Zingst, Baltic Sea; salinity, 10‰) and a freshwater lake (Lake Kummerow) as well as in mixed samples from the brackish Bodstedter Bodden (Fig. 3, lanes 12 to 15), isiA fragments could be amplified. After being cloned and sequenced, three different isiA sequences appeared from the mixed phytoplankton cultures (Bodstedter Bodden). The newly obtained sequences of isiA fragments from additional strains and the field samples fit very well into a joint phylogram with already known isiA sequences (Fig. 4). The clustering of isiA sequences from field samples basically reflects expectations with respect to their taxonomic relationships. The partial isiA sequences obtained from a Nodularia bloom fit into the cluster of filamentous cyanobacteria, whereas the isiA sequences obtained from enriched phytoplankton from a eutrophic estuary in which unicellular and colony-forming cyanobacteria dominate (38) fit in the coccoid strains (Fig. 4).
FIG. 4.
Rooted cladogram (TreeView version 1.5; R. D. M. Page, 1998) after phylogenetic analysis of translated IsiA protein sequences (PAUP software package, neighbor-joining method). The isiA sequences (830-bp internal fragment) come from databases (marked by  ) and from the present study of different cyanobacterial strains, clones from field samples, and enrichment cultures. Bootstrap values were calculated after 1,000 replications; only branchings with values above 50% are shown.
) and from the present study of different cyanobacterial strains, clones from field samples, and enrichment cultures. Bootstrap values were calculated after 1,000 replications; only branchings with values above 50% are shown.
Expression of isiA mRNA in enrichment cultures of field samples.
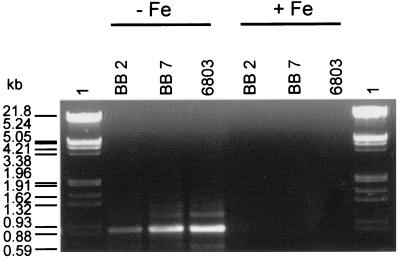
Although the presence of isiA in field samples is an important precondition for its use as a marker gene, its proper function cannot be assumed in advance. The expression of isiA in enriched cultures originating from brackish phytoplankton was tested first in experiments by means of RT-PCR. The four cultures BB 1, BB 2, BB 3, and BB 7 containing three different isiA genes (Fig. 4) were cultivated in iron-depleted medium. In order to ensure that isiA-bearing organisms were still present after the iron starvation period in spite of potential competition with other algae and/or bacteria, DNAs were extracted separately at the sampling times and used in PCRs with isiA-specific primers. At the beginning of and after 12 days of iron-deficient cultivation, RNA was extracted from harvested cells. RT-PCR led to the 840-bp internal isiA fragments from the iron-starved enrichment cultures BB 2 and BB 7 (Fig. 5). A control PCR excluded DNA contamination and confirmed isiA expression. For comparison, we analyzed a sample from an iron-starved laboratory culture of the cyanobacterial model strain Synechocystis sp. strain PCC 6803 that indicated synthesis of a corresponding target mRNA (Fig. 5). These RT-PCR experiments resulted in the first proof of iron-dependent isiA expression in noncharacterized phytoplankton species from eutrophic coastal waters. This indicates similar responses to reduced iron availability in at least parts of the coastal population of cyanoplankton and in frequently investigated cyanobacterial model strains.
FIG. 5.
Separation of internal isiA gene fragments obtained by RT-PCR using RNAs isolated from different cyanobacterial samples cultivated for 12 days under restricted iron availability (−Fe) in contrast to conditions for control cells (+Fe). Lanes BB, enrichment cultures of estuarine field samples (numbers signify the different cultures); lanes 6803, Synechocystis sp. strain PCC 6803; lanes 1, marker (λ DNA EcoRI and HindIII digested).
DISCUSSION
Reliability of isiA expression as a molecular marker of cellular iron supply.
The coincidence of isiA expression and iron-dependent growth limitation in semicontinuous cultures of the model strain Synechocystis sp. strain PCC 6803 suggests a close relationship of the two processes. However, the almost complete activation of transcription as well as the high level of isiA transcription after the onset of iron depletion, when the growth rate was reduced by only a few percentage points, characterizes the system as a very sensitive one with regard to reduced iron availability in the model strain Synechocystis. In addition, the fast disappearance of isiA mRNA under iron-replete growth conditions is a clear indication of a high rate of turnover of isiA mRNA and cannot be explained by mRNA dilution due to cell division. Under optimum growth conditions for Synechocystis, expression of isiA is a fast and sensitive means to detect even early stages of iron deficiency. This efficacy is similar to that of the molecular marker flavodoxin in diatoms (31), which is applied to oceanic phytoplankton.
In the cyanobacterial model strain, the changing isiA expression in response to fluctuating extracellular concentrations of Fe(NH4) citrate further indicates a well-balanced system of iron supply. With regard to its intracellular iron demand, Synechocystis possesses a sensitive regulating system. Iron concentrations above and below the critical limit for the Synechocystis strain MpIGisi caused gradually changing steady-state levels of GFP fluorescence. The regulation of intracellular iron metabolism seems to be adapted (without “on-off” limits) to the extracellular iron supply continuously. Since a specific ferric-citrate receptor is known in Synechocystis (19), the potential variability of iron uptake had been reduced in our experiments in advance. Thus, the estimated threshold for concentration-dependent isiA expression depends on ferric citrate as the iron source. However, our demonstration of the basic regulatory scheme opens perspectives to the probing of the sophisticated relations of iron availability and iron sequestration in the future, when the extracellular iron speciation is more under the control of the model organism or competing organisms themselves. The independence of isiA transcript decline and organism growth rate after the cessation of iron depletion is an important feature in distinguishing critical nutrients in the environment, since the cessation of iron limitation does not necessarily result in growth promotion when a second nutrient has an immediate limiting effect (colimitation).
Distribution of isiA in extant cyanobacteria.
Provisional objections to isiA being able to serve as a molecular marker of iron limitation (23) with regard to the distribution of the gene among cyanobacteria can now be rejected. The screening of different cyanobacterial strains for isiA served mainly to give us the initial impression that we could amplify gene fragments with one set of degenerate PCR primers. Using optimized isiA-specific degenerate primers, internal isiA fragments of cyanobacterial strains could be obtained without amplification of any other gene coding for a chlorophyll-binding protein like PsbC or Pcb. The isiA-bearing strains belong to all five cyanobacterial sections (33), which indicates a widespread distribution of the gene. The available sequences cover at least one taxon in each of the five sections of cyanobacterial taxonomy as well as heterocystous strains, indicating that isiA is common in extant cyanobacteria.
Though the strains can also be separated into three different groups of 16S rRNA clustering (27), most of the strains cannot be designated correspondingly, since 16S rRNA sequences are not available and cannot be easily obtained from nonaxenic strains. Although the obtained fragments show a sequence homology of 65 to 73% and reveal different clusters in the phylogram (Fig. 4), they could be amplified easily from miscellaneous and distantly related cyanobacterial species by a simple PCR protocol. However, in some cyanobacteria no isiA fragments could be amplified. These strains do not necessarily lack the isiA gene, as no efforts have so far been made to vary the DNA extraction method. Although the successful DNA extraction method was tested with 16S rRNA-specific primers, in cases of nonaxenic cultures the results can reflect the presence of contaminating DNAs of heterotrophic bacteria. Furthermore, the degenerate primers might have restrictions for a simultaneous amplification of isiA genes from the different strains. In two cases, isiA fragments were found only after we applied the universal primers for the genes psbC, pcb, and isiA to DNAs (Stanieria sp., Baltic Nodularia bloom), but the reproducible amplification of isiA fragments from field samples verifies that the utility of our degenerate primers is not restricted to the few culture strains which served as sources for the primers. The degenerate primers might already be very useful for monitoring isiA expression in cyanobacterial blooms dominated by one or a few species by means of semiquantitative RT-PCR. Clear proof of the existence of a cyanobacterial strain lacking an isiA gene has not yet been found, but the ongoing complete-genome sequencing projects of several cyanobacterial strains will provide definitive information in the near future.
The successful demonstration of isiA mRNA accumulation under iron-restricted conditions in enriched cultures from the estuary indicates a relevant supplemental tool for gaining greater knowledge of estuarine ecosystems with respect to the interaction of iron level and cyanobacterial development. Though the total iron supply in ecosystems can be very high, this possibility does not prevent cyanobacteria from showing significant sensitivity to the micronutrient (15), and corresponding investigations would be supported by noninvasive methodologies.
Any conclusions regarding isiA expression in the field would require balanced interpretations. Scanlan and Wilson (37) have given an overview of the categorizeation of indicators of nutrient deficiency in phytoplankton as sensitive or nonsensitive. In addition, the coherence of signal intensity and the extent of iron limitation have yet to be characterized. According to our study, isiA can be placed into the category of indicators which are sensitive to the onset of limitation. Thus, signals can potentially be obtained before field measurements of growth and primary production indicate statistically significant cyanoplankton responses to iron deficiency. However, such situations can also be expected when flavodoxin is used as a corresponding marker in eukaryotic algae (31). Our study is one contribution to overcoming the provisional reservations concerning isiA's suitability as a molecular indicator of iron stress, owing to methodological problems (23), since specific isiA signals can now be obtained in spite of their high similarity to those of other genes coding for chlorophyll-binding proteins. As the flavodoxin approach (7) has been further characterized to perfect its practice (8, 31), further investigations with respect to isiA expression in cyanobacteria will likewise be directed to the detection of signals in experimental phytoplankton systems.
ACKNOWLEDGMENTS
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG).
The excellent technical support of B. Brzezinka and K. Sommerey is acknowledged. We are grateful for the provision of strains and isolates by B. Kolpers, S. Mundt, (Pharmaceutical Institute, University of Griefswald) and K. Ribbeck (FB Biowissenschaften, University of Rostock). Cells of Prochlorococcus strain MED4 were kindly provided by W. Hess (Department of Genetics, Humboldt University, Berlin, Germany), and Microcystis aeruginosa was kindly provided by B. Meyer (Max-Planck-Institute for Limnology, Plön, Germany). We thank M. Blank (FB Biowissenschaften, University of Rostock) for introducing the PAUP software package. Samples of the Nodularia bloom were kindly provided by H. Schubert (Plant Ecology, University of Greifswald).
REFERENCES
- 1.Abraham E R, Law C S, Boyd P W, Lavender S J, Maldonado M T, Bowie A R. Importance of stirring in the development of an iron-fertilized phytoplankton bloom. Nature. 2000;407:727–730. doi: 10.1038/35037555. [DOI] [PubMed] [Google Scholar]
- 2.Allen M B, Arnon D I. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 1955;30:366–372. doi: 10.1104/pp.30.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand L E. Minimum iron requirements of marine phytoplankton and the implications for the biogeochemical control of new production. Limnol Oceanogr. 1991;36:1756–1771. [Google Scholar]
- 5.Burnap R L, Troyan T, Sherman L A. The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC 7942 (CP43′) is encoded by the isiA gene. Plant Physiol. 1993;103:893–902. doi: 10.1104/pp.103.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler A. Acquisition and utilization of transition metal ions by marine organisms. Science. 1998;281:207–210. doi: 10.1126/science.281.5374.207. [DOI] [PubMed] [Google Scholar]
- 7.Doucette G J, Erdner D L, Peleato M L, Hartmann J J, Anderson D M. Quantitative analysis of iron-stress related proteins in Thalassiosira weissflogii: measurement of flavodoxin and ferredoxin using HPLC. Mar Ecol Prog Ser. 1996;130:269–276. [Google Scholar]
- 8.Erdner D L, Price N M, Doucette G J, Peleato M L, Anderson D M. Characterization of ferredoxin and flavodoxin as markers of iron limitation in marine phytoplankton. Mar Ecol Prog Ser. 1999;184:43–53. [Google Scholar]
- 9.Falk S, Samson G, Bruce D, Huner N P, Laudenbach D E. Functional analysis of the iron-stress induced cp 43′ polypeptide of PS II in the cyanobacterium Synechococcus sp. PCC 7942. Photosynth Res. 1995;45:51–60. doi: 10.1007/BF00032235. [DOI] [PubMed] [Google Scholar]
- 10.Geiß U, Vinnemeier J, Schoor A, Hagemann M. The iron-regulated isiA gene of Fischerella muscicola strain PCC 73103 is linked to a likewise regulated gene encoding a Pcb-like chlorophyll-binding protein. FEMS Microbiol Lett. 2001;197:123–129. doi: 10.1111/j.1574-6968.2001.tb10593.x. [DOI] [PubMed] [Google Scholar]
- 11.Guikema J A, Sherman L A. Organization and function of chlorophyll in membranes of cyanobacteria during iron starvation. Plant Physiol. 1983;73:250–256. doi: 10.1104/pp.73.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchins D A, Bruland K W. Grazer-mediated regeneration and assimilation of Fe, Zn and Mn from planktonic prey. Mar Ecol Prog Ser. 1994;110:259–269. [Google Scholar]
- 13.Hutchins D A, Ditullio G R, Zhang Y, Bruland K W. An iron limitation mosaic in the California upwelling regime. Limnol Oceanogr. 1998;43:1037–1054. [Google Scholar]
- 14.Hutchins D A, Witter A E, Butler A, Luther G W., III Competition among marine phytoplankton for different chelated iron species. Nature. 1999;400:858–861. [Google Scholar]
- 15.Kawaguchi T, Lewitus A J, Aelion C M, McKellar H N. Can urbanization limit iron availability to estuarine algae? J Exp Mar Biol Ecol. 1997;213:53–69. [Google Scholar]
- 16.Kolber Z S, Barber R T, Coale K H, Fitzwater S E, Greene R M, Johnson K S, Lindley S, Falkowski P G. Iron limitation of phytoplankton photosynthesis in the Equatorial Pacific Ocean. Nature. 1994;371:145–149. [Google Scholar]
- 17.Kratz W A, Myers J. Nutrition and growth of several blue-green algae. Am J Bot. 1955;42:282–287. [Google Scholar]
- 18.Kunert A, Hagemann M, Erdmann N. Construction of promoter probe vectors for Synechocystis sp. PCC 6803 applying the light-emitting reporter systems Gfp and LuxAB. J Microbiol Methods. 2000;41:184–194. doi: 10.1016/s0167-7012(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 19.Labouré A M, Briat J F. Uptake of iron from ferric-citrate in the cyanobacteria Synechocystis PCC 6803. C R Acad Sci Ser III. 1993;316:661–666. [PubMed] [Google Scholar]
- 20.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 21.La Roche J, van der Staay G W M, Partensky F, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W D, Green B R. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc Natl Acad Sci USA. 1996;93:15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Roche J, Boyd P W, McKay R M L, Geider R J. Flavodoxin as an in-situ marker for iron stress in phytoplankton. Nature. 1996;382:802–805. [Google Scholar]
- 23.La Roche J, McKay R M L, Boyd P. Immunological and molecular probes to detect phytoplankton responses to environmental stress in nature. Hydrobiologia. 1999;401:117–198. [Google Scholar]
- 24.Laudenbach D E, Straus N A. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J Bacteriol. 1988;170:5018–5026. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonhardt K, Straus N A. An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. J Gen Microbiol. 1992;138:1613–1621. doi: 10.1099/00221287-138-8-1613. [DOI] [PubMed] [Google Scholar]
- 26.Leonhardt K, Straus N A. Photosystem II genes isiA, psbDI and psbC in Anabaena sp. PCC 7120: cloning, sequencing and the transcriptional regulation in iron-stressed and iron-repleted cells. Plant Mol Biol. 1994;24:63–73. doi: 10.1007/BF00040574. [DOI] [PubMed] [Google Scholar]
- 27.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Lie B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin J H, Fitzwater S E. Iron deficiency limits phytoplankton growth in the northeast Pacific subarctic. Nature. 1988;331:341–343. [Google Scholar]
- 29.Martin J H, Gordon R M, Fitzwater S E. Iron in Antarctic waters. Nature. 1990;345:156–158. [Google Scholar]
- 30.Martin J H, Coale K H, Johnson K S, Fitzwater S E, Gordon R M, Tanner S J, Hunter C N, Elrod V A, Nowicki J L, Coley T L, Barber R T, Lindley S, Watson A J, Van Scoy K, Law C S, Liddicoat M I, Ling R, Stanton T, Stockel J, Collins C, Anderson A, Bidigare R, Ondrusek M, Latasa M, Millero F J, Lee K, Yao W, Zhang J Z, Fredrich G, Sakamoto C, Chavez F, Buck K, Kolber Z, Green R, Falkowski P G, Chisholm S W, Hoge F, Swift R, Yungle J, Turner S, Nightingale P I, Hatton A, Liss P, Tindale N W. Testing the iron hypothesis in ecosystems of the equatorial Pacific. Nature. 1994;371:123–129. [Google Scholar]
- 31.McKay R M, Geider R J, La Roche J. Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to Fe stress. Plant Physiol. 1997;114:615–622. doi: 10.1104/pp.114.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park Y I, Sandström S, Gustafsson P, Öquist G. Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol Microbiol. 1999;32:123–129. doi: 10.1046/j.1365-2958.1999.01332.x. [DOI] [PubMed] [Google Scholar]
- 33.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–16. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sandmann G, Peleato M L, Fillat M F, Lazaro M C, Gomez-Moreno C. Consequences of the iron-dependent formation of ferredoxin and flavodoxin on photosynthesis and nitrogen fixation on Anabaena strains. Photosynth Res. 1990;26:119–125. doi: 10.1007/BF00047083. [DOI] [PubMed] [Google Scholar]
- 36.Scanlan D J, Silman N J, Donald K M, Wilson W H, Carr N G, Joint I, Mann N H. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–2420. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scanlan D J, Wilson W H. Application of molecular techniques to addressing the role of P as a effector in marine ecosystems. Hydrobiologia. 1999;401:149–175. [Google Scholar]
- 38.Schiewer U. 30 Years' eutrophication in shallow brackish waters—lessons to be learned. Hydrobiologia. 1998;363:73–79. [Google Scholar]
- 39.Turner D R. Problems in trace metal speciation modeling. In: Tessier A, Turner D R, editors. Metal speciation and bioavailability in aquatic systems. Chichester, United Kingdom: John Wiley & Sons; 1995. pp. 149–203. [Google Scholar]
- 40.Vinnemeier J, Kunert A, Hagemann M. Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol Lett. 1998;169:323–330. doi: 10.1111/j.1574-6968.1998.tb13336.x. [DOI] [PubMed] [Google Scholar]
- 41.Vinnemeier J, Hagemann M. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch Microbiol. 1999;172:377–386. doi: 10.1007/s002030050774. [DOI] [PubMed] [Google Scholar]
- 42.Wells M L, Mayer L M, Guillard R R L. A chemical method for estimation the availability of iron to phytoplankton in seawater. Mar Chem. 1991;33:23–40. [Google Scholar]
- 43.Wells M L, Price N M, Bruland K W. Iron limitation and the cyanobacterium Synechococcus in equatorial Pacific waters. Limnol Oceanogr. 1994;39:1481–1486. [Google Scholar]
- 44.Wilhelm S W. Ecology of iron-limited cyanobacteria: a review of physiological responses and implications for aquatic systems. Aquat Microb Ecol. 1995;9:295–303. [Google Scholar]