Abstract
Ammonia oxidation is a rate-limiting step in the biological removal of nitrogen from wastewater. Analysis of microbial communities possessing the amoA gene, which is a small subunit of the gene encoding ammonia monooxygenase, is important for controlling nitrogen removal. In this study, the amoA gene present in Nitrosomonas europaea cells in a pure culture and biofilms in a nitrifying reactor was amplified by in situ PCR. In this procedure, fixed cells were permeabilized with lysozyme and subjected to seminested PCR with a digoxigenin-labeled primer. Then, the amplicon was detected with an alkaline phosphatase-labeled antidigoxigenin antibody and HNPP (2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate), which was combined with Fast Red TR, and with an Alexa Fluor 488-labeled antidigoxigenin antibody. The amoA gene in the biofilms was detected with an unavoidable nonspecific signal when the former method was used for detection. On the other hand, the amoA gene in the biofilms was detected without a nonspecific signal, and the cells possessing the amoA gene were clearly observed near the surface of the biofilm when Alexa Fluor 488-labeled antidigoxigenin antibody was used for detection. Although functional gene expression was not detected in this study, detection of cells in a biofilm based on their function was demonstrated.
The biological removal of nitrogen compounds is an integral part of most modern wastewater treatment facilities to preserve environmental water resources. In this process, ammonia oxidation is a rate-limiting step, where autotrophic ammonia conversion into hydroxylamine is catalyzed by ammonia monooxygenase. Thus, analysis of microbial communities possessing the amoA gene, which encodes ammonia monooxygenase, is important for controlling nitrogen removal.
On the other hand, fluorescent in situ hybridization (FISH) (4, 9) and denaturing gradient gel electrophoresis (10, 17) based on 16S ribosomal DNA and rRNA for molecular analysis have been used in various fields to determine the genetic diversity of a microbial community and to identify individual members. In particular, in situ hybridization with fluorescence-labeled oligonucleotide probes has been widely used for in situ analysis of microbial communities, such as a biofilm in a wastewater treatment process (5, 18, 22, 29). This method relies on the presence of many target sequences within an individual cell. Therefore, bacterial cells containing insufficient rRNA cannot be detected by this approach. Moreover, this taxonomic identification approach cannot be used to detect the presence of single-copy functional genes or their expression at the single-cell level. Hence, in situ hybridization cannot estimate a specific metabolic activity such as ammonia oxidation.
Recently, in situ PCR was developed to amplify and detect functional genes and their expression inside a single cell, thus making it possible to detect a single copy of a functional gene. This method was first developed to amplify and detect a DNA virus inside a cell (11), and Nuovo et al. (21) and Bagasra et al. (6) developed this method for molecular pathology. In environmental microbiology, in situ PCR and in situ reverse transcription-PCR protocols have been used to detect the presence and expression of nahA (12) and todC1 (8) in Pseudomonas cells, lac in Salmonella enterica serovar Typhimurium (28), and slt-I and slt-II in Escherichia coli O157 (27). However, the in situ PCR protocol has been applied only to a dispersed sample of a model microbial community in seawater and river water and has never been used for the analysis of a biofilm. For this reason, little is known about the distribution of a functional gene in biofilms. To determine the distribution of functional genes and their expression in a biofilm is a primary objective in the fields of microbial ecology and wastewater treatment. The purposes of this study were to establish an original in situ PCR protocol for the detection of the amoA gene in a biofilm and to examine the distribution of a microbial community possessing the amoA gene in a biofilm for nitrogen removal.
MATERIALS AND METHODS
Samples and cell fixation.
Nitrosomonas europaea (IFO 14298) as a representative ammonia-oxidizing bacterium was cultured in a nitrification medium according to the method of Watson and Mandel (30) and Bock et al. (7) with minor modifications. The culture was incubated at 28 to 30°C in the dark, and then cells were collected by centrifugation and washed with PBS solution (137 mM NaCl, 8.10 mM Na2HPO4 · 12H2O, 2.68 mM KCl, 1.47 mM KH2PO4; pH 7.4). Biofilms were collected from a laboratory-scale aerobic up-flow nitrifying reactor that treated inorganic-ammonia-rich wastewater when the ammonia load was about 1.5 kg of N/m3/day. The granule-like biofilms were collected through a sampling port and were settled for a few minutes to separate them from the bioreactor liquid phase. The samples were suspended in 4% paraformaldehyde in PBS solution for 16 h (for the pure culture) and 20 h (for the biofilm) at 4°C for fixation and were then washed with PBS solution. The fixed biofilm samples were embedded in Tissu Mount (Chiba Medical, Saitama, Japan) and rapidly frozen at −25°C. Then, the biofilms were cut with a cryostat (CM1850; Leica Microsystems, Nussloch, Germany) into 10-μm-thick sections.
DNA extraction.
DNA extraction was carried out according to the methods of Smalla (26) with minor modifications. DNA was extracted from a 0.5-g (wet weight) biofilm pellet. The biofilms were harvested by centrifugation at 10,000 × g for 10 min. The harvested cells were sonicated with an ultrasonic disrupter (type UR-20P; probe diameter, 2 mm; TOMY Seiko Co., Ltd., Tokyo, Japan) at 20 W and 28 kHz for 30 s in 5 ml of sucrose lysis buffer (0.3 M sucrose, 0.7 M NaCl, 40 mM EDTA, 50 mM Tris-HCl) and then centrifuged at 2,000 × g for 10 min. The supernatant was incubated at 55°C in the presence of sodium dodecyl sulfate (0.5%), proteinase K (0.1 mg/ml), and hexadecylmethylammonium bromide (1%). DNA was purified by applying phenol, chloroform, and isoamyl alcohol and precipitated by the addition of ethanol and sodium acetate.
Oligonucleotides.
The following three primers were used: amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′), amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′ [K = G or T; S = G or C]) (25), and amoA-1FF (5′-CAATGGTGGCCGGTTGT-3′). These primers target stretches corresponding to positions 332 to 349, 802 to 822, and 187 to 203 of the open reading frame published previously for the amoA gene sequence of N. europaea, respectively. The amoA-1FF primer was designed specifically for ammonia-oxidizing bacteria belonging to the β subclass of Proteobacteria. Specificity was examined using FASTA (24) and BLAST (1) programs to compare the primers with the complete sequence data registered in GenBank. As for FISH analysis, the oligonucleotide probe Nso190 (16) labeled with CY3 was used for in situ detection of ammonia-oxidizing bacteria. Nso190 is a probe specific for a region in the 16S rRNA of ammonia-oxidizing bacteria.
Cell wall permeabilization.
Cell wall permeabilization and in situ PCR were carried out according to the method of Hodson et al. (12) with minor modifications. The washed samples were resuspended in PBS solution. A 30-μl aliquot was spotted onto amino alkylsilane-coated slides (Applied Biosystems, Foster City, Calif.) and dried in an oven at 50°C for 5 min. Prior to cell wall permeabilization, the fixed samples were dehydrated sequentially in 50, 80, and 100% ethanol. The dehydrated samples were treated with the lysozyme solution (0.5 mg [for the pure culture] and 1.0 mg [for the biofilm] of lysozyme per ml, 100 mM Tris-HCl [pH 8.2], and 50 mM EDTA) for 30 min at room temperature. Lysozyme was removed by consecutive washes with PBS solution. Further permeabilization was carried out by treatment with proteinase K at a final concentration of 0.1 μg/ml for 10 min at room temperature, followed by heat inactivation of proteinase K at 94°C for 3 min. Then, the samples were washed with the PBS solution and dehydrated sequentially in 50, 80, and 100% ethanol. The samples on the slides were ready for the addition of PCR mixture.
In situ PCR procedures.
A GeneAmp in situ PCR core kit (Applied Biosystems) was used for amplification of the amoA gene in cells. Then, the following seminested PCR protocol was used. First, a 50-μl aliquot of PCR buffer (10 mM Tris-HCl, 50 mM KCl; pH 8.3) containing 0.6 μM (each) amoA-1FF and amoA-2R, 2.5 mM MgCl2, 0.6 mM (each) deoxynucleoside triphosphates, and 10 U of AmpliTaq DNA polymerase, IS (Applied Biosystems), was added to each sample spot on the slide and covered with an AmpliCover disk and AmpliCover clip (Applied Biosystems). The concentrations of polymerase and MgCl2 were higher than those used in the conventional PCR because polymerase and MgCl2 tend to adhere to a glass slide or a coverslip (20). PCR was performed with the following temperature profile using a GeneAmp in situ PCR system 1000 (Applied Biosystems): initial denaturation at 94°C for 90 s and then 20 cycles of denaturation at 94°C for 40 s, annealing at 53°C for 30 s, and extension at 72°C for 40 s. The slide was then washed with 0.5× SSC (750 mM NaCl, 75 mM trisodium citrate; pH 7.0) at 45°C for 10 min. In the second PCR, the reaction mixture was the same as that for the first amplification except that amoA-1FF was replaced by digoxigenin-labeled amoA-1F.
Detection of the amplified gene with Alexa Fluor 488-labeled antidigoxigenin antibody.
A digoxigenin-labeled amplicon was detected with an Alexa Fluor 488 (Molecular Probes Inc., Eugene, Oreg.)-labeled antidigoxigenin antibody. Antidigoxigenin Fab fragments (Roche Diagnostics, Mannheim, Germany) were labeled with an Alexa Fluor 488 protein labeling kit (Molecular Probes) according to the manufacturer's instructions. First, 50 μl of a blocking buffer (30 μg of bovine serum albumin per ml in PBS solution) was spotted onto the samples and incubated for 30 min at room temperature. Then, 25 μl of the buffer was replaced by 25 μl of alkaline phosphatase-conjugated antidigoxigenin Fab fragments (Roche Diagnostics) diluted to 68 nM (for biofilm) or 0.27 μM (for pure culture) in a dilution buffer (100 mM Tris-HCl [pH 7.5], 150 mM NaCl), and incubated at 37°C for 1 h. After that, the slides were washed with buffer I (100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) three times and then washed with buffer II (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10 mM MgCl2) twice. Then, the samples were counterstained with TO-PRO-3 (Molecular Probes) for 5 min. For preparation of a working solution, TO-PRO-3 was diluted with double-distilled water (ddH2O) 1,000-fold. After washing, the samples were mounted in FluoroGuard antifade reagent (Bio-Rad, Richmond, Calif.) for observation under a confocal laser scanning microscope (TCS4D; Leica Lasertechnik, Heidelberg, Germany) equipped with an Ar-Kr ion laser (488, 568, and 647 nm). Confocal images were obtained with a PL FLUOTAR ×10/0.30 objective (total magnification, ×100) and PL APO ×63/1.40 oil objective (total magnification, ×630). Figures were composed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, Calif.) on a Macintosh PowerPC (Apple Computer Co., Cupertino, Calif.).
Detection of the amplified gene with alkaline phosphatase-labeled antidigoxigenin antibody.
The detection of the amplicon with an alkaline phosphatase-labeled antidigoxigenin antibody and 2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate (HNPP), which was combined with Fast Red TR according to the methods of Yamaguchi et al. (31) with minor modifications. HNPP is converted to HNP by alkaline phosphatase. Then, HNP combines with Fast Red TR and produces a bright red fluorescent material, HNP-TR (13). The procedure was almost the same as that for detection using the Alexa Fluor 488-labeled antidigoxigenin antibody. After blocking, 25 μl of the buffer was replaced by 25 μl of alkaline phosphatase-labeled antidigoxigenin antibodies diluted to 60 in the dilution buffer and incubated at 37°C for 1 h. After the slides had been washed, the samples were counterstained with TO-PRO-3 for 5 min. After being washed with buffer II, the samples were incubated in HNPP-Fast Red TR (Roche Diagnostics) solution (100 μg of HNPP per ml and 250 μg of Fast Red TR per ml in buffer II) for 15 min at room temperature, and the slides were washed with ddH2O. The samples were mounted in Macllavaine buffer (53.2 mM citric acid and 93.6 mM Na2HPO4 [pH 4.5]) for observation under a confocal laser scanning microscope.
FISH targeting 16S ribosomal DNA.
Hybridization was performed according to the standard protocol described by Amann (2) at 46°C for 2.5 h in a hybridization buffer containing NaCl (0.9 M), formamide (40%) (5, 16), Tris-HCl (20 mM, pH 7.4), and sodium dodecyl sulfate (0.01%). The probe concentration was 0.5 ng/μl. Hybridization was followed by a stringent washing step at 48°C for 20 min in a washing buffer containing Tris-HCl (20 mM, pH 7.4), NaCl (35 mM) (5, 16), and sodium dodecyl sulfate. The washing buffer was removed by rinsing the slides with ddH2O. Then, the samples were counterstained with TO-PRO-3, mounted in FluoroGuard antifade reagent (Bio-Rad), and observed under the confocal laser scanning microscope.
PCR and agarose gel electrophoresis.
To confirm specificity, extracted DNA was amplified by PCR with amoA-1F, amoA-2R, and amoA-1FF. Extracted DNA was added to 50 μl of a PCR buffer (5 mM Tris-HCl, 5 mM KCl, 10 μM EDTA, 0.1 mM dithiothreitol, 0.0001% Tween 20, 0.0001% Nonidet P-40) containing 0.5 μM (each) primers, 200 μM deoxynucleotide triphosphate, 1 mM MgSO4, and 1 U of KOD DNA polymerase (Toyobo Co., Ltd., Osaka, Japan). PCR was carried out in a GeneAmp PCR system 9700 (Applied Biosystems) using the same cycle program as the in situ PCR cycle program. PCR products were electrophoresed in a 2% agarose gel and visualized by ethidium bromide staining.
RESULTS
Amplification of amoA from extracted DNA.
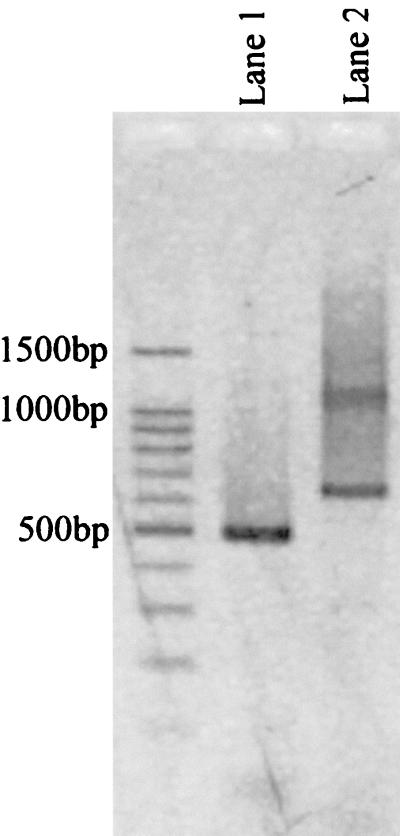
Amplification of the amoA gene from extracted DNA of the biofilms using the oligonucleotide primer set amoA-1FF and amoA-2R resulted in a 636-bp DNA fragment with a slight smear (Fig. 1, lane 2). The 636-bp amplicon was reamplified using the seminested primer set amoA-1F and amoA-2R, and as a result, only a 491-bp amplicon was generated (Fig. 1, lane 1).
FIG. 1.
Gel electrophoresis of amoA gene products amplified from extracted DNA of the biofilms. Lane 1, 491-bp amoA gene fragment amplified by primers amoA-1F and amoA-2R; lane 2, 636-bp amoA gene fragment amplified by primers amoA-1FF and amoA-2R
amoA gene detection with in situ PCR.
To establish the in situ PCR protocol, the amoA gene in N. europaea was amplified by in situ PCR. At first, detection of the amplicon generated by amoA-1F and amoA-2R was attempted by in situ hybridization with the fluorescein isothiocyanate-labeled probe. However, no signal was observed under the confocal laser scanning microscope. An insufficient amount of amplicon or problems related to hybridization were thought to be the cause of this failure. To solve these problems, the seminested PCR protocol has been used to increase the specificity and sensitivity of in situ PCR (12, 17, 28). The amoA-1F and amoA-2R primers were used in the first PCR, and amoA-1F and a fluorescein isothiocyanate-labeled primer that was complementary to a region internal to amoA-1F and amoA-2R were used in the second PCR. As a result, although the level of sensitivity was certainly increased, the level of signal intensity was still very low. To increase the sensitivity further, a digoxigenin-labeled primer was used in the second PCR, and the amplicon was detected with the alkaline phosphatase-labeled antidigoxigenin antibody and HNPP combined with Fast Red TR. The level of sensitivity was increased significantly, and thus, this method was applied to the subsequent in situ PCR procedure.
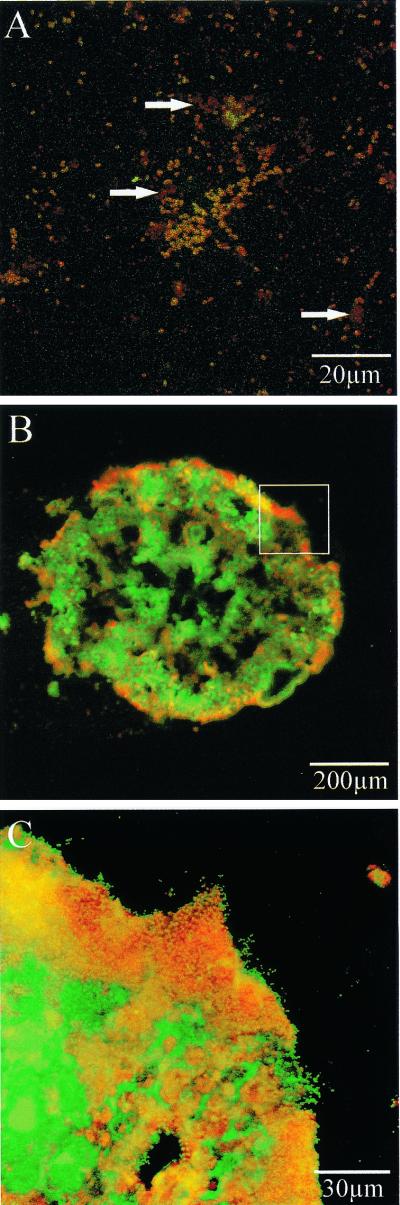
First, the amoA gene in an N. europaea cell was amplified and detected. In situ PCR requires that the polymerase be able to penetrate into the cells and act on the nucleic acid in cells without the PCR amplicon diffusing away from the cells. Therefore, to determine the optimal pretreatment conditions, the time of fixation with 4% paraformaldehyde was varied from 4 to 20 h and the lysozyme concentration was varied from 0.05 to 5.0 mg/ml. As a result, the amoA gene was successfully detected inside intact N. europaea cells that had been fixed for 16 h and permeabilized with 0.5-mg/ml lysozyme buffer (Fig. 2A). Moreover, the protocol was applied to the biofilm that was collected from the up-flow aerobic nitrifying reactor. The fixation time was varied from 4 to 32 h, and the lysozyme concentration was varied from 0.05 to 5.0 mg/ml. The amoA gene was detectable in the sample fixed for 20 h and permeabilized with 1.0 mg of lysozyme per ml. However, an unavoidable nonspecific signal was particularly observed in the case of the biofilm (Fig. 2B and C).
FIG. 2.
Detection of ammonia-oxidizing bacterial cells possessing amoA gene in pure culture (A) and in the biofilm (B and C), with alkaline phosphatase-labeled antidigoxigenin antibody. Panels B and C show the same biofilm. The yellow color indicates positive cells, while the green shows negative cells (arrows indicate the nonspecific signal). The portion of panel B that was magnified to produce panel C is indicated with a white box.
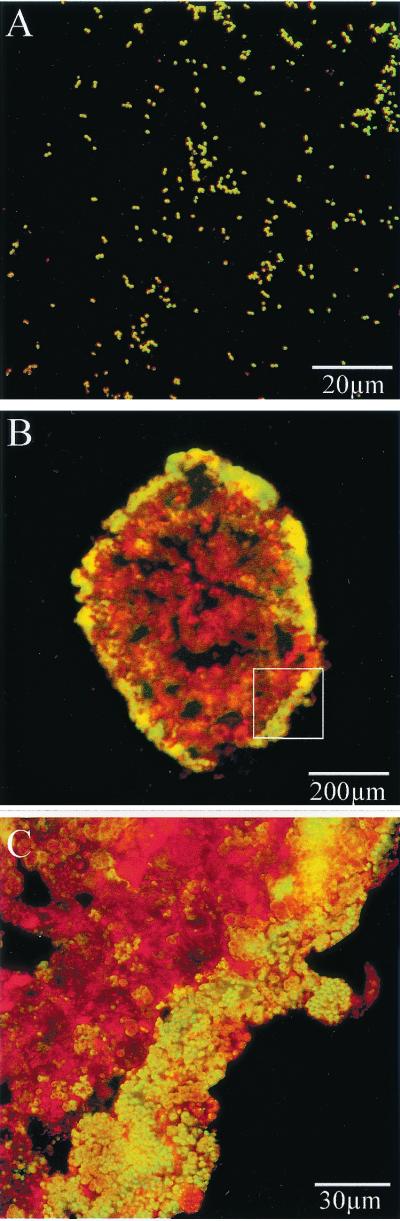
To reduce the intensity of the nonspecific signal, the digoxigenin-labeled amplicon in the cell was detected with antidigoxigenin antibodies labeled with Alexa Fluor 488. As a result, the amoA gene was detected inside N. europaea cells without the nonspecific signal (Fig. 3A). Moreover, the amoA gene in the biofilm was also detected with a weak nonspecific signal, as shown in Fig. 3B and C. The green fluorescence in Fig. 3C (upper right) is the supposed nonspecific signal caused by adsorption of the antibodies. However, it was clear that cells possessing the amoA gene were found near the surface of the biofilm.
FIG. 3.
Detection of the cells possessing amoA in pure culture (A) and in the biofilm (B and C), with Alexa Fluor 488-labeled antidigoxigenin antibody. Panels B and C show the same biofilm. The yellow color indicates positive cells, while the red shows negative cells. The portion of panel B that was magnified to produce panel C is indicated with a white box.
FISH.
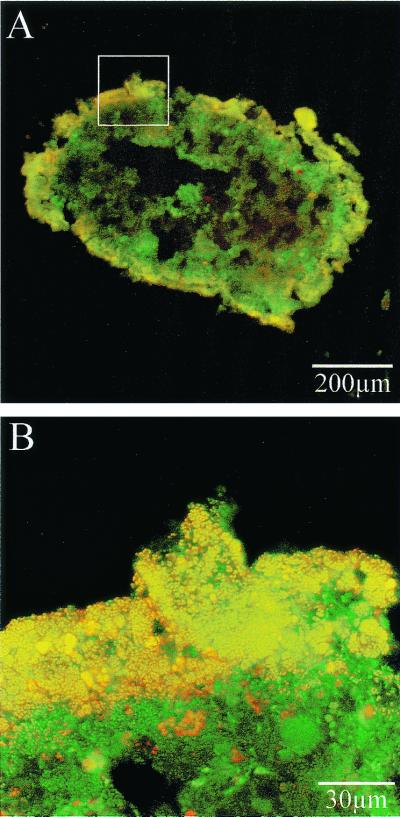
To confirm the validity of microbial communities detected by in situ PCR, ammonia-oxidizing bacteria in the biofilm were visualized by FISH analysis (Fig. 4). The yellow shows ammonia-oxidizing bacteria, and the green shows other bacteria. It was observed that ammonia-oxidizing bacteria were the dominant population on the surface of the biofilm.
FIG. 4.
Detection of ammonia-oxidizing bacteria with a CY3-labeled Nso190 probe. Panels A and B show the same biofilm. The yellow color indicates ammonia oxidizers, while the green shows other bacteria. The portion of panel A that was magnified to produce panel B is indicated with a white box.
DISCUSSION
At first, the amoA gene in the N. europaea cells (pure culture) and the biofilm (mixed culture) was detected with an alkaline phosphatase-labeled antidigoxigenin antibody; however, an unavoidable nonspecific signal was observed, which has also been reported by Tani et al. (27). This was probably due to the fluorescent HNP-TR diffusing out of the cells.
To reduce the intensity of the nonspecific signal, a new strategy of detecting the amplicon in the cells with an Alexa Fluor 488-labeled antidigoxigenin antibody was proposed. First, detection of the amoA gene in the cell was conducted completely under the same conditions as for the alkaline phosphatase-labeled antidigoxigenin antibody. However, a sufficient signal intensity was not obtained, which might be due to the lower penetration efficiency and/or the lower sensitivity of the antibody compared with the alkaline phosphatase-labeled antidigoxigenin antibody. Therefore, several concentrations (0.035, 0.07, and 0.14 μM) were attempted to increase signal intensity. In consequence, when the concentration of the antibody was increased to 0.14 μM, for the N. europaea cells, the amoA gene was detected without the nonspecific signal. As for the biofilm, the amoA gene was successfully detected with a low background signal, although the concentration of the antibody was increased to 35 nM. These results indicate that the use of Alexa Fluor 488-labeled antibody enables the construction of a promising detection system that can be applied to in situ PCR.
The images obtained both by in situ PCR analysis with the Alexa Fluor 488-labeled antibody and by FISH analysis show that the ammonia-oxidizing bacteria were found near the surface of the biofilms. This result suggests that the bacteria possessing the amoA gene were successfully detected by in situ PCR. Although FISH analysis is a very important tool in microbial ecology, cells with an insufficient ribosomal content cannot be detected by this method. Practically, a fluorescent signal was observed only near the surface of the biofilms by FISH analysis with EUB338 (a probe targeting all bacteria except Archaea), which was probably due to the low activity of the bacteria living in a deep part of the biofilm (data not shown). In spite of extensive attempts to increase the intensity of the fluorescent signal from FISH by sample enrichment (25) and using a single probe with multiple fluorochromes (4) and multiple probes (3), low-activity cells are difficult to detect. In contrast, a few copies of the amoA gene in the cells were successfully detected by in situ PCR. This indicates that cells possessing a functional gene can be detected by in situ PCR regardless of its activity. Actually, when the biofilm cultivated without ammonia for a month was examined by FISH and in situ PCR, ammonia-oxidizing bacteria were not detected by FISH at all, whereas the cells possessing the amoA gene were detected near the surface of the biofilms by in situ PCR (data not shown).
However, the images obtained by in situ PCR analysis are less distinct than those obtained by FISH analysis. This was probably because cells were damaged by permeabilization and/or thermal cycling, as well as (i) diffusion of the labeled amplicon and its adhesion to negative cells, (ii) adhesion of the digoxigenin-labeled primer to negative cells, and (iii) adhesion of the Alexa Fluor 488-labeled antibody to negative cells.
Cases 2 and 3 are practically inconceivable because no signal was detected when the negative control without polymerase was examined. As for case 1, this diffusion artifact has been reported as a problem of in situ PCR (14, 20). On the other hand, Nuovo (19) reported that the movement of DNA segments was restricted not by the pore size but rather by biochemical forces, such as hydrogen bonding and ionic charges. Therefore, under optimal fixation and permeabilization conditions, there is minimal migration of the amplicon from its site of origin, and diffusion artifacts are significantly reduced by reduction of the PCR cycle (≤30), generation of longer PCR products, or incorporation of biotinylated nucleotides to generate bulkier and thus less diffusible PCR products (15, 23). In this study, to increase the level of sensitivity, the seminested PCR protocol was used, and the total number of thermal cycles was 40 cycles. This led to the destruction of the cell morphology, which probably caused the diffusion artifacts. However, taking into account that fewer cycles caused a very low signal, it is important to optimize these parameters very carefully and to simplify the PCR protocol to the furthest extent possible.
In this study, the cells possessing the amoA gene were successfully detected by in situ PCR with the Alexa Fluor 488-labeled antibody. As a result, bacteria could be detected by in situ PCR regardless of its rRNA content and could be detected based on their function. On the other hand, problems that are not observed when in situ PCR is applied to dispersed samples were revealed when in situ PCR was applied to biofilms. However, the fact that the spatial distribution of a functional gene in the biofilm becomes detectable represents significant progress in this field. This in situ PCR is expected to be a valuable method in cases such as those in which the ammonia oxidation rate has dropped although the number of ammonia-oxidizing bacteria is not changed, and it will be essential for explaining such phenomena. It is anticipated that in future studies, functional gene expression (mRNA) will be detected by in situ PCR, and consequently the spatial distribution of the “actually functioning” bacteria in a biofilm from the natural environment or from a wastewater treatment system will be elucidated.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic probes. In: Akkermans A D C, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoi Y, Miyoshi T, Okamoto T, Tsuneda S, Hirata A, Kitayama A, Nagamune T. Microbial ecology of nitrifying bacteria in wastewater treatment process examined by fluorescence in situ hybridization. J Biosci Bioeng. 2000;90:234–240. [PubMed] [Google Scholar]
- 6.Bagasra O, Seshamma T, Hansen J. Application of in situ PCR methods in molecular biology. I. Details of methodology for general use. Cell Vision. 1994;1:324–335. [Google Scholar]
- 7.Bock E, Sundermeyer-Klinger H, Stackebrandt E. New facultative lithoautotrophic nitrite-oxidizing bacteria. Microbiology. 1983;136:281–284. [Google Scholar]
- 8.Chen F, Dustman W A, Hodson R E. Microbial detection of the toluene dioxygenase gene and its expression inside bacterial cells in seawater using prokaryotic in situ PCR. Hydrobiologia. 1999;401:131–138. [Google Scholar]
- 9.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 10.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasse A T, Retzel E F, Staskus K A. Amplifications and detection of lentiviral DNA inside cells. Proc Natl Acad Sci USA. 1990;87:4971–4975. doi: 10.1073/pnas.87.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodson R E, Dustman W A, Garg R P, Moran M A. In situ PCR for visualization distribution of specific genes and gene products in prokaryotic communities. Appl Environ Microbiol. 1995;61:4074–4082. doi: 10.1128/aem.61.11.4074-4082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagiyama N, Yoshida K, Hamabata T, Juni N, Awasaki T, Fujita S, Momiyama M, Yoshida M C, Hori S H. A novel fluorescent method for in situ hybridization. Acta Histochem Cytochem. 1993;26:441–445. [Google Scholar]
- 14.Komminoth P, Long A, Ray R, Wolfe H. In situ polymerase chain reaction detection of viral DNA, single copy genes and gene rearrangements in cell suspension and cytospins. Diagn Mol Pathol. 1992;1:85–97. [PubMed] [Google Scholar]
- 15.Luehrsen K R. In situ PCR as a tool to amplify DNA and RNA target in cells and tissues attached to slides. Cell Vision. 1995;2:230–235. [Google Scholar]
- 16.Mobarry B, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Shleifer K. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuovo G J. The foundations of successful RT in situ PCR. Front Biosci. 1996;1:c4–c15. doi: 10.2741/a110. [DOI] [PubMed] [Google Scholar]
- 20.Nuovo G J. In situ localization of PCR-amplified DNA and cDNA. Mol Biol. 1998;10:49–62. doi: 10.1007/BF02745862. [DOI] [PubMed] [Google Scholar]
- 21.Nuovo G J, Gallery F, MacConnell P. An improved technique for the in situ detection of DNA after polymerase chain reaction amplification. Am J Pathol. 1991;139:1239–1244. [PMC free article] [PubMed] [Google Scholar]
- 22.Okabe S, Itoh T, Satou H, Watanabe Y. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl Environ Microbiol. 1999;65:5107–5116. doi: 10.1128/aem.65.11.5107-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson B, Till M, Otto P, Goolsby C, Furtado M, McBride L, Wolinsky S. Detection of HIV-1 DNA and messenger RNA in individual cells by PCR-driven in situ hybridization and flow cytometry. Science. 1993;260:976–979. doi: 10.1126/science.8493534. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1999;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smalla K. Extraction of microbial DNA from sewage and manure slurries. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. p. 1.1.3. [Google Scholar]
- 27.Tani K, Kurokawa K, Nasu M. Development of a direct in situ PCR method for detection of specific bacteria in natural environments. Appl Environ Microbiol. 1998;64:1536–1540. doi: 10.1128/aem.64.4.1536-1540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolker-Nielsen T, Holmstrøm K, Molin S. Visualization of specific gene expression in individual Salmonella typhimurium cells by in situ PCR. Appl Environ Microbiol. 1997;63:4196–4203. doi: 10.1128/aem.63.11.4196-4203.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson S W, Mandel M. Comparison of the morphology and deoxyribonucleic acid composition of 27 strains of nitrifying bacteria. J Bacteriol. 1971;107:563–569. doi: 10.1128/jb.107.2.563-569.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi N, Inaoka S, Tani K, Kenzaka T, Nasu M. Detection of specific bacterial cells with 2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate and fast red TR in situ hybridization. Appl Environ Microbiol. 1996;62:275–278. doi: 10.1128/aem.62.1.275-278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]