Abstract
In the present study, a novel lytic Vibrio parahaemolyticus phage, vB_VpaP_DE10, was isolated from sewage samples collected in Guangzhou city, China. Transmission electron microscopy revealed that phage vB_VpaP_DE10 has an icosahedral head (52.4 ± 2.5 nm) and a short non-contracted tail (21.9 ± 1.0 nm). Phage vB_VpaP_DE10 lysed approximately 31% (8/26) of the antibiotic-resistant V. parahaemolyticus strains tested. A one-step growth curve showed that phage vB_VpaP_DE10 has a relatively long latency time of 25 min and a burst size of ~19 PFU per cell. The genome of phage vB_VpaP_DE10 is a 42,871-bp-long dsDNA molecule with a G + C content of 49.19% and is predicted to contain 46 open reading frames, 26 of which are predicted to be related to functions such as phage structure, packaging, host lysis, and DNA metabolism. Sequence comparisons suggested that vB_VpaP_DE10 is a member of the genus Maculvirus within the family Autographiviridae. Morphological and genomic analysis indicated that vB_VpaP_DE10 is a novel V. parahaemolyticus phage.
Keywords: Vibrio parahaemolyticus, phage, Autographiviridae, genome analysis
1. Introduction
Vibrio parahaemolyticus is a Gram-negative bacterium that exists in marine, riverine, and aquaculture environments [1]. Infection by this bacterium can lead to gastroenteritis, fever, and sepsis [2]. V. parahaemolyticus was first discovered in Osaka, Japan, and has been prevalent ever since [3]. According to the U.S. Centers for Disease Control and Prevention (CDC), approximately 6680 cases of V. parahaemolyticus infection are reported each year with an estimated annual health cost of more than 30 million dollars [4]. In China, V. parahaemolyticus has been the main cause of foodborne disease outbreaks and infectious diarrhea cases in coastal areas [5]. From 2013 to 2017, a total of 1220 strains of V. parahaemolyticus were isolated from 16,504 fecal specimens of patients with acute diarrhea in southeastern China; the annual isolation rate fluctuated between 6.1 and 8.7% [6]. The pathogenic factors of V. parahaemolyticus are mainly related to two virulence factors, namely thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) [7]. Both toxins have hemolytic and enterotoxic activities and are cardiotoxic and cytotoxic [8].
The use of antibiotics is a common practice in treating the disease caused by infection with this bacterium; however, over time, an increasing number of reports have shown that V. parahaemolyticus isolated from various sources is highly resistant to single or multiple antibiotics, particularly ampicillin [9]. Disinfectants are also traditional countermeasures to control V. parahaemolyticus, but they are non-specific, easily destroy the natural ecology of aquaculture water, and the resulting residues pose a threat to the health of consumers [10]. Therefore, environmentally friendly methods are needed to replace traditional measures to control V. parahaemolyticus.
In the past decade, studies have suggested that phage therapy may be a promising strategy to control V. parahaemolyticus [11]. However, at present, there are very limited phage resources. As of 22 April 2022, there were 24,646 phage strains recorded in NCBI, including 24,426 culturable strains, and only 78 V. parahaemolyticus phage strains in total. Therefore, obtaining sufficient phage resources is a top priority. Moreover, it is necessary to clear the genetic information of phages before application. Considering the high genetic diversity and mutation rate of phages, traditional or single microbial analysis methods are no longer applicable. Therefore, it is very important to continuously update research methods and use multiple methods to verify each other.
In this study, we isolated a V. parahaemolyticus phage, vB_VpaP_DE10, from the sewage of the Huangsha Aquatic Product Market in Guangzhou, China. We assessed the basic biological properties of its lytic activity, namely host range and one-step growth curve, and performed a multi-angle comprehensive analysis of its whole genome.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Twenty-six V. parahaemolyticus strains provided by the Institute of Microbiology, Guangdong Academy of Sciences, were used in this study. The serotype, resistance, and origin of each strain were confirmed by previous studies [12] (Table 1). V. parahaemolyticus O4-12 was used as a host for phage isolation and propagation. Strains were stored at −80 °C in 30% (v/v) glycerol. All V. parahaemolyticus strains were cultured in 5 mL of tryptic soy broth (TSB) and incubated at 37 °C for 12 h with shaking.
Table 1.
Antibiotic resistance profile of V. parahaemolyticus strains and host range of phage vB_VpaP_DE10.
| No. | V. parahaemolyticus Strains | Serotype | Antibiotic Resistance | Spot Test | EOP Value | EOP Rank |
|---|---|---|---|---|---|---|
| 1 | O1-1 | O1 | CN-K-CIP-S-AMP | - | NT | Inefficient |
| 2 | O1-3 | O1 | K-CIP-S-AMP-C | + | 0.32 | Medium |
| 3 | O2-5 | O2 | S-AMP | - | NT | Inefficient |
| 4 | O2-7 | O2 | CIP-S-AMP | - | NT | Inefficient |
| 5 | O3-8 | O2 | K-CIP-S-AMP | - | NT | Inefficient |
| 6 | O3-10 | O3 | AMP | - | NT | Inefficient |
| 7 | O3-11 | O3 | CN-S-AMP-C | - | 2.80 | High |
| 8 | O4-12 | O4 | CIP-S-AMP | + | 1 | High |
| 9 | O4-13 | O4 | S-AMP | + | 0.99 | High |
| 10 | O4-14 | O4 | AMP | + | 0.40 | Medium |
| 11 | O5-15 | O5 | CN-K-CIP-S-AMP | + | NT | Inefficient |
| 12 | O6-18 | O6 | K-CIP-S-AMP | + | 0 | Inefficient |
| 13 | O6-20 | O6 | CIP-S-AMP | + | 0.68 | High |
| 14 | O8-21 | O8 | S-AMP | + | 0 | Inefficient |
| 15 | O8-126 | O8 | AMP | - | NT | Inefficient |
| 16 | O9-24 | O9 | CN-CIP-S-AMP | - | NT | Inefficient |
| 17 | O10-25 | O10 | S-AMP | - | NT | Inefficient |
| 18 | O10-26 | O10 | CN-CIP-S-AMP-C | - | NT | Inefficient |
| 19 | O10-28 | O10 | AMP | - | NT | Inefficient |
| 20 | O11-29 | O11 | CN-K-CIP-AMP | - | NT | Inefficient |
| 21 | O11-30 | O11 | S-AMP | - | NT | Inefficient |
| 22 | O11-31 | O11 | CIP-S-AMP | + | 8.91 | High |
| 23 | O12-32 | O12 | S-AMP | - | NT | Inefficient |
| 24 | O12-33 | O12 | CIP-S-AMP | + | 0.065 | Low |
| 25 | O12-34 | O12 | SXT-CIP-AMP-TE-K | - | NT | Inefficient |
| 26 | O12-35 | O12 | CN-CIP-S-AMP | - | NT | Inefficient |
CN: gentamicin, K: kanamycin, CIP: ciprofloxacin, S: streptomycin, AMP: ampicillin, C: chloramphenicol, SXT: sulfamethoxazole-trimethoprim, TE: tetracycline. Clear plaque (+); no plaque (-).
2.2. Isolation of Phage
Phage vB_VpaP_DE10 was isolated from sewage samples collected in the Huangsha aquatic product market in Guangzhou city, China, by a previously described method [13]. Sewage samples were subjected to centrifugation to remove large particulates (5000× g for 10 min), and the supernatant was filtered using a 0.45 μm filter (HuanKai Microbial, Guangzhou, China). Thereafter, MgSO4 was added to the filtrate to a final concentration of 50 mM and allowed to stand for 10 min before filtering using a 0.22-μm filter (HuanKai Microbial, Guangzhou, China). The filter membranes were cut and eluted with a broth containing 3% (w/v) Bacto beef extract, 3% (v/v) Tween 80, and 50 mM NaCl. Subsequently, 2 mL of eluent was mixed with 2 mL of sterile double-strength TSB (4 mM CaCl2) and an 80-µL V. parahaemolyticus O4-12 culture (OD600 = 0.2). The mixtures were incubated overnight at 37 °C with shaking. After incubation, the mixed cultures were centrifuged at 8000× g for 5 min, and the supernatant was filtered through a 0.45 μm syringe filter. The purified phages were incubated with V. parahaemolyticus again at least twice to enrich the phage particles. A spot test assay was performed to test for the presence of lytic phages. The double-layer agar method was used to obtain single plaques and a plaque was purified four times to ensure the isolated phage was pure [14].
2.3. Phage Concentration and Morphological Observation
Phage concentration was performed as previously described, with modifications [15]. First, 0.1 mL phage lysate sediment was incubated in 50 mL TSB at 37 °C for 4 h, then centrifuged at 5000× g for 10 min and filtered through a 0.45-μm-pore membrane. Thereafter, polyethylene glycol (15% w/v PEG8000, 0.5 M NaCl) was added to it and placed at 4 °C overnight.
The overnight phage concentrate was centrifuged at 12,000× g for 20 min at 4 °C. The sediment was resuspended in SM buffer. The CsCl density gradient centrifugation method was used to purify and concentrate phages [16]. Phage particles located near the 1.5-g/mL density band were collected and their morphological features were observed under a transmission electron microscope (Hitachi H-7650; Hitachi, Tokyo, Japan).
2.4. Spot Assay
First, 100 μL of bacterial culture in the mid-log phase was mixed with 5 mL TSB (0.4% agar and 2 mM CaCl2), poured onto tryptic soy agar plates, and placed at room temperature for 5 min or until the agar solidified. Then, 2 µL of tenfold diluted phage stock were pipetted on each plate. Thereafter, the plates were incubated at 37 °C for 6 h. Based on the clarity of plaques, the results were divided into two categories: clear (+) and no plaques (-) in the spotting area.
2.5. Efficiency of Plating (EOP)
All the V. parahaemolyticus isolates which were sensitive to the phage vB_VpaP_DE10 in the spot test were selected for the determination of EOP by the double-agar layer method [17]. Briefly, tenfold serial dilutions of phage suspensions (100 μL) were added to 5 mL TSB-soft agar with 100 μL V. parahaemolyticus culture (108 CFU/mL). The mixture was poured onto TSA plates and incubated at 37 °C for 6 h. The EOP values were calculated as the ratio of lysis plaques produced in each susceptible strain divided by the number of the plaques produced in V. parahaemolyticus O4-12 and then were ranked as “high efficiency” (EOP ≥ 0.5), “medium efficiency” (0.1 ≤ EOP < 0.5), “low efficiency” (0.001 < EOP < 0.1), or “inefficient” (EOP ≤ 0.001).
2.6. Phage Nucleotide Extraction
Phage whole-genome extraction was performed as described by Xing S et al. [18]. The phage concentrate was centrifuged at 12,000× g for 20 min at 4 °C, and the supernatant was discarded. Subsequently, 1 mL SM buffer was added to fully wash the tube wall and precipitate DNA, following which DNase I and RNase A were added to final concentrations of 0.1 units/µL and 3 µg/mL, respectively. The mixture was incubated at 37 °C for 1 h. Next, purified phages were treated with 10% SDS, EDTA, and proteinase K for 30 min at 65 °C. Then, an equal volume of Tris-saturated phenol solution was added and the mixture was centrifuged at 12,000× g for 5 min. The upper aqueous phase was transferred to another centrifuge tube, an equal volume of premixed phenol-chloroform-isoamyl alcohol (25:24:1) was added to it, and the mixture was centrifuged at 12,000× g for 5 min. The upper aqueous phase was again transferred to a fresh centrifuge tube, and an equal volume of chloroform was added (this step was repeated thrice). Thereafter, the upper aqueous phase was pipetted into a fresh centrifuge tube, an equal volume of isopropanol was added to it, and the tube was then incubated at −20 °C for 30 min. Next, the DNA sediment was collected by centrifugation at 12,000× g for 5 min at 4 °C, washed with 70% ethanol, and centrifuged at 12,000× g for 5 min again (this step was repeated twice). Subsequently, the DNA sediment was dried at room temperature and 50 μL deionized water (preheated to 65 °C) was added to it. The extracted DNA was stored at −20 °C.
2.7. Genome Sequencing and Bioinformatics Analysis
The phage DNA was sequenced using Ion Torrent S5 platform (Thermo Fisher Scientific, Waltham, MA, USA). High-quality reads were assembled using SPAdes v. 3.12.0 [19]. The whole-genome sequence was aligned with phage sequences in GenBank using BLASTN and then analyzed by average nucleotide identity (ANI). Genome annotation was conducted using Prokka [20]. Putative tRNA-encoding genes were predicted using tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/ accessed on 18 November 2021). The virulence factors and antibiotic genes were predicted using the virulence factor database [21] (VFDB, http://www.mgc.ac.cn/VFs/ accessed on 18 November 2021) and antibiotic resistance gene database [22] (ARDB, http://ardb.cbcb.umd.edu/ accessed on 18 November 2021), respectively. A circular map of the phage vB_VpaP_DE10 genome was generated using CGView [23]. For phylogenetic analysis, the amino acid sequences of DNA polymerase and RNA polymerase were selected for multiple alignments, firstly using the Clustal W algorithm [24]. Based on the alignments, a neighbor-joining (NJ) phylogenetic tree was constructed in MEGA-X using the Jones–Taylor–Thornton model [25,26]. Bootstrap values were calculated from 1000 replicates [27]. The Newick format was used to create the phylogenetic tree by iTOL (https://itol.embl.de accessed on 18 June 2022). Finally, the whole phage genome was visualized using Easyfig2.2.3 [28].
2.8. Phage One-Step Growth Curve Assay
The one-step growth curve of phage vB_VpaP_DE10 was tested according to a previous method with minor modifications [15]. Briefly, 1 mL of the diluted bacterial culture (1 × 108 CFU/mL) was centrifuged at 8000× g for 5 min, the sediment was resuspended in 1 mL SM buffer, and 100 µL phage at multiplicity of infection (MOI) = 0.1 was added to it. This mixture was incubated at 37 °C for 10 min and the mixture was then centrifuged and resuspended in 1 mL TSB. Next, 0.2 mL of this mixture was added to 19.8 mL of TSB and incubated at 37 °C and 200 r/min for 50 min. Samples were taken every 5 min to determine the phage titer from 0 min. The experiment was repeated thrice. The burst size was calculated as follows:
2.9. Accession Number
The whole-genome sequence of phage vB_VpaP_DE10 was deposited in GenBank under the accession number MZ516827.
2.10. Statistical Analysis
The data were expressed as mean ± standard deviation (SD) and the differences were analyzed with two-way ANOVA using GraphPad Prism 8.4.2. Significance was considered at p < 0.05.
3. Results
3.1. Isolation, Identification, and General Characterization of Phages
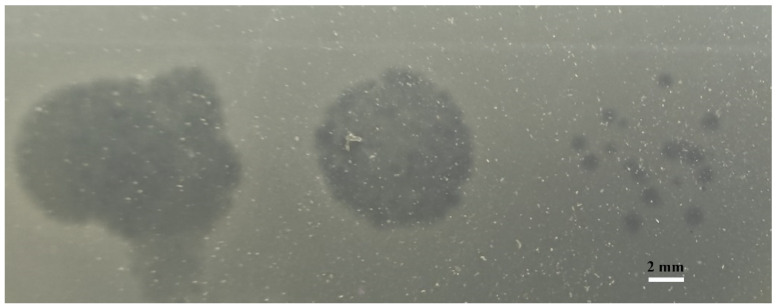
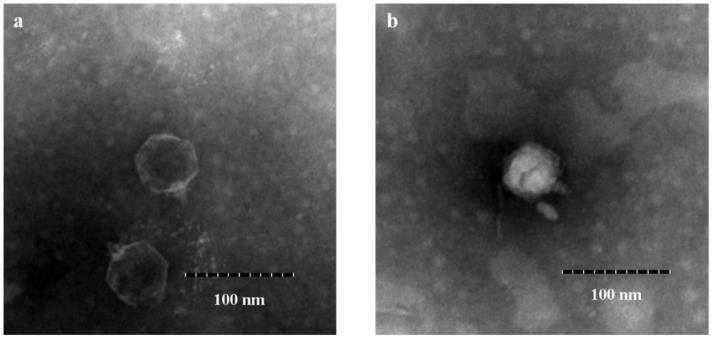
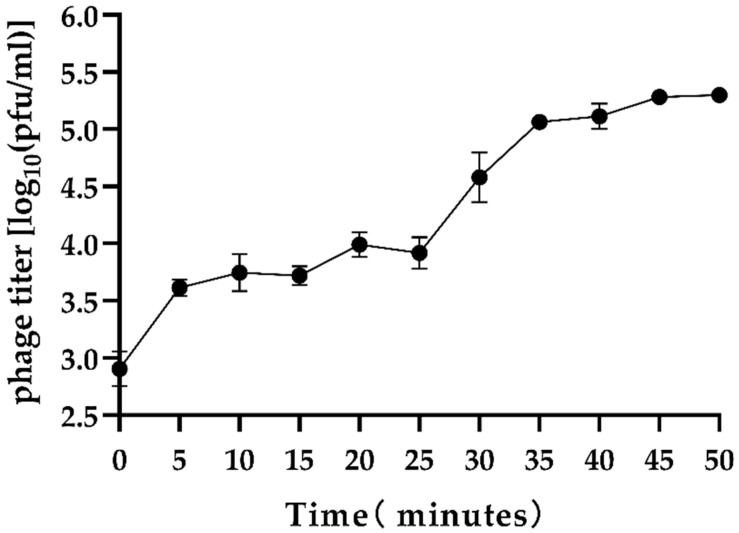
The phage vB_VpaP_DE10 was isolated from sewage using antibiotic-resistant V. parahaemolyticus O4-12 as the host cell. It was able to form clear plaques approximately 1–2 mm in diameter (Figure 1). Transmission electron microscopy (Figure 2) revealed that phage vB_VpaP_DE10 has an icosahedral head of 52.4 ± 2.5 nm and a short non-contracted tail of 21.9 ± 1.0 nm. Based on the one-step growth curves shown in Figure 3, phage vB_VpaP_DE10 was characterized by a latency period of 0–25 min, a lysis period of 25–35 min, and a burst size of approximately 19 PFU per cell.
Figure 1.
The serial dilution spot test of phage vB_VpaP_DE10 against V. parahaemolyticus O4-12.
Figure 2.
Morphology of phage vB_VpaP_DE10 observed using transmission electron microscopy (a) after DNA injection and (b) without DNA injection.
Figure 3.
One-step growth curve of phage vB_VpaP_DE10.
3.2. Host Range
A total of 26 strains of antibiotic-resistant V. parahaemolyticus, which belonged to 11 different kinds of O antigens, were used to evaluate the host range of phage vB_VpaP_DE10. In spot tests, phage vB_VpaP_DE10 formed clear spots on the lawns of 10 out of 26 V. parahaemolyticus strains (Table 1), indicating that these strains were sensitive to the phage suspensions. Thus, these 10 V. parahaemolyticus strains were used for the EOP determination. Results showed that vB_VpaP_DE10 only formed plaques on 8 V. parahaemolyticus strains by the double-layer agar plate method but did not form plaques on V. parahaemolyticus O6-20 and O8-21, indicating that though it inhibited the growth of V. parahaemolyticus O6-20 and O8-21, it did not lyse them.
3.3. Genomic Signature of Phage vB_VpaP_DE10
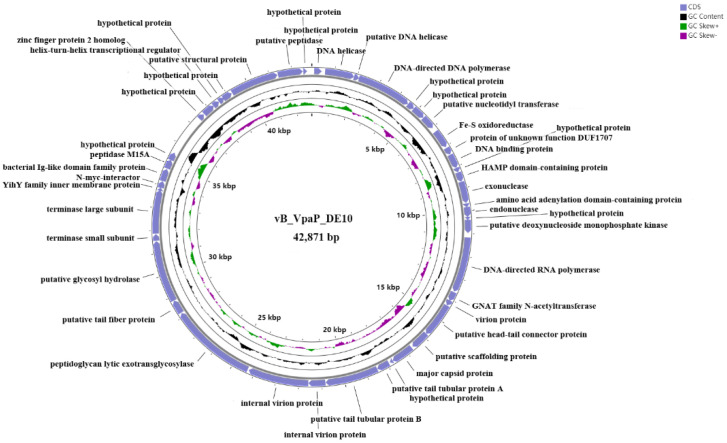
To better understand the phage vB_VpaP_DE10, its genomic DNA was extracted and sequenced. As shown in Figure 4, the phage vB_VpaP_DE10 genome consisted of 42,871 bp of double-stranded DNA with an average G + C content of 49.19% and 46 open reading frames (ORFs) with an average length of 861 bp and varying in size from 93 to 3855 bp. A total of 39,599 nucleotides (accounting for 92.37% of the total genome) formed the coding sequence. As shown in Figure 5 and Table 2, among the annotated 46 ORFs, 26 were predicted as functional proteins, including ten phage DNA metabolism-related proteins (ORF2, ORF3, ORF4, ORF7, ORF10, ORF13, ORF15, ORF17, ORF18, and ORF19), ten morphogenesis-related proteins (ORF20, ORF21, ORF22, ORF23, ORF25, ORF26, ORF27, ORF28, ORF30, and ORF44), four lysis-related functional proteins (ORF29, ORF31, ORF37, and ORF45), and two packaging-related proteins (ORF32 and ORF33).The presence of the RNA polymerase gene suggested that vB_VpaP_DE10 may belong to the described family Autographiviridae. Autographivirinae was once considered a subfamily of the Podoviridae family whose defining feature was the presence of virally encoded RNA polymerases [29]. Further common characteristics of these phages include conservation of gene arrangement and apparently genus-specific lysis cassettes and RNAP specificity loops. In 2019, ICTV removed the Autographivirinae and Autographivirinae-like viruses from the family Podoviridae and assigned a family rank, “Autographiviridae” [30]. In addition, no tRNA-encoding, antibiotic resistance, lysogenic, or virulence genes were observed upon genomic analysis.
Figure 4.
Vibrio parahaemolyticus phage vB_VpaP_DE10 genome. Circles display (from the outside) the following: (1) Open reading frames transcribed in the clockwise or the counterclockwise direction. (2) G + C % content. Values > 49.19 % (average) are shown as outward peaks, and smaller values are shown as inward peaks. (3) GC skew. (4) Physical map scaled in kbp.
Figure 5.
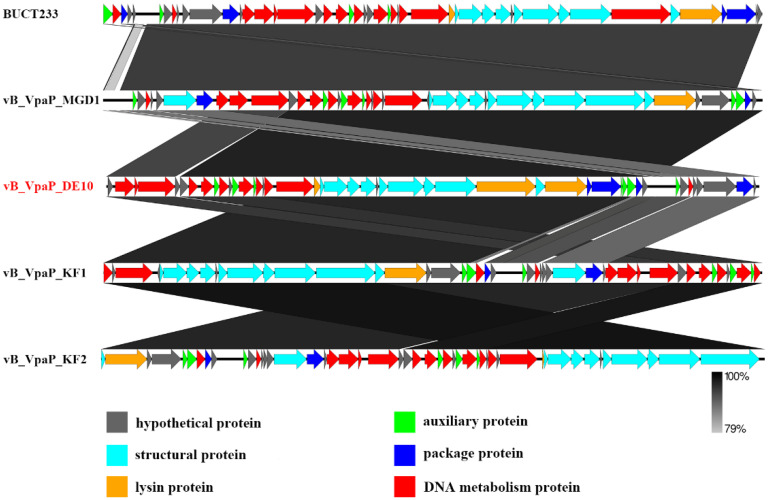
Genome comparison of vB_VpaP_DE10, BUCT233, vB_VpaP_MGD1, and vB_VpaP_KF1–2 phages using the Easyfig tool. Different colored arrows represent 46 predicted open reading frames with different functions.
Table 2.
Predicted ORFs of V. parahaemolyticus phage vB_VpaP_DE10.
| Label | Length (nt|aa) | Product | Organism | Identity |
|---|---|---|---|---|
| ORF2 | 1281|426 | DNA helicase | Vibrio phage vB_VpP_DE18 | 100% |
| ORF3 | 234|77 | putative DNA helicase | Vibrio phage vB_VpP_DE18 | 100% |
| ORF4 | 2427|808 | DNA-directed DNA polymerase | Vibrio phage vB_VpP_DE18 | 99% |
| ORF7 | 597|198 | putative nucleotidyl transferase | Vibrio phage vB_VpaP_KF1 | 98% |
| ORF10 | 630|209 | DNA binding protein | Vibrio phage BUCT233 | 98% |
| ORF13 | 951|316 | exonuclease | Vibrio phage vB_VpP_DE18 | 99% |
| ORF15 | 441|146 | endonuclease | Vibrio phage BUCT233 | 100% |
| ORF17 | 585|194 | putative deoxynucleoside monophosphate kinase | Vibrio phage vB_VpaP_KF1 | 80% |
| ORF18 | 2451|816 | DNA-directed RNA polymerase | Vibrio phage BUCT233 | 99% |
| ORF19 | 420|139 | GNAT family N-acetyltransferase | Vibrio phage vB_VpP_DE18 | 100% |
| ORF20 | 246|81 | virion protein | Vibrio phage vB_VpP_DE18 | 100% |
| ORF21 | 1533|510 | putative head–tail connector protein | Vibrio phage vB_VpP_DE17 | 99% |
| ORF22 | 816|271 | putative scaffolding protein | Vibrio phage vB_VpaP_MGD1 | 99% |
| ORF23 | 999|332 | major capsid protein | Vibrio phage vB_VpaP_MGD1 | 99% |
| ORF25 | 561|186 | putative tail tubular protein A | Vibrio phage vB_VpaP_MGD1 | 100% |
| ORF26 | 2343|780 | putative tail tubular protein B | Vibrio phage vB_VpaP_MGD1 | 99% |
| ORF27 | 741|246 | internal virion protein | Vibrio phage vB_VpP_FE11 | 86% |
| ORF28 | 2679|892 | internal virion protein | Vibrio phage vB_VpaP_KF1 | 95% |
| ORF29 | 3855|1284 | peptidoglycan lytic exotransglycosylase | Vibrio phage vB_VpP_DE18 | 99% |
| ORF30 | 612|203 | putative tail fiber protein | Vibrio phage VP93 | 99% |
| ORF31 | 2733|910 | putative glycosyl hydrolase | Vibrio phage vB_VpaP_KF1 | 99% |
| ORF32 | 300|99 | terminase small subunit | Vibrio phage vB_VpP_DE18 | 98% |
| ORF33 | 1920|639 | terminase large subunit | Vibrio phage vB_Vc_SrVc2 | 100% |
| ORF37 | 414|137 | peptidase M15A | Vibrio phage vB_VpP_DE18 | 100% |
| ORF44 | 2136|711 | putative structural protein | Vibrio phage vB_VpP_DE18 | 98% |
| ORF45 | 1092|363 | putative peptidase | Vibrio phage vB_VpP_DE18 | 100% |
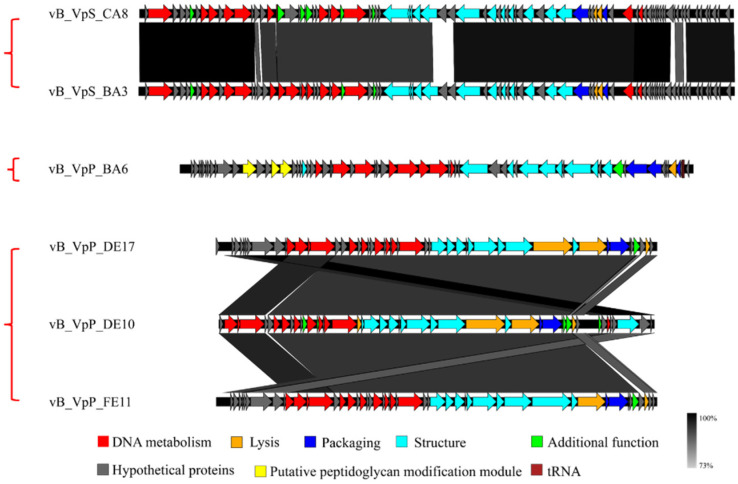
In order to investigate the genetic diversities of Vibrio phages isolated by our group, the whole genome alignment between phage vB_VpaP_DE10 and phages vB_VpP_BA6 [13], vB_VpP_DE17 [15], vB_VpP_FE11 [16], vB_VpS_BA3, and vB_VpS_CA8 [12] was performed. As showed in Figure 6, sequence comparison allowed for clear discrimination of three kinds of genomes. Genomes of Siphoviridae phages vB_VpS_BA3 and vB_VpS_CA8 represented type 1, the genome of Podoviridae phage vB_VpS_BA6 represented type 2, and genomes of Autographiviridae phage vB_VpaP_DE10, vB_VpP_DE17, and vB_VpP_FE11 all represented type 3.
Figure 6.
Genome comparison of vB_VpaP_DE10, vB_VpP_BA6, vB_VpS_BA3, vB_VpS_CA8, and vB_VpP_DE17, using Easyfig.
3.4. Phage Taxonomy and Phylogeny Analysis
In order to know the genome profile of phage vB_VpaP_DE10, the whole-genome BLASTn analysis was performed first. The genome of vB_VpaP_DE10 showed a high homology (>95%) with genomes of eight distinct species of the genus Maculvirus (Table 3). Among them, the highest sequence similarity was given by the phage vB_VpaP_MGD1 (97.02% identity, 86% query coverage). When comparing the 46 ORFs of vB_VpaP_DE10 with vB_VpaP_MGD1, 32 showed ≥95% identity, 4 showed 85% to 95% identity, 3 showed <85% identity; and 7 showed no homology at all. In the 7 ORFs with no homology, 4 were annotated as hypothetical proteins (ORF 1, 5, 42, 46); the others were annotated as DNA helicase (ORF 2), YihY family inner membrane protein (ORF 19), and GNAT family N-acetyltransferase (ORF 34), respectively. They were all present in vB_VpaP_DE10 and absent in vB_VpaP_MGD1.
Table 3.
Phages with high genome homology (>95%) to phage vB_VpaP_DE10.
| Accession Number | Virus Name | Identity (%) |
|---|---|---|
| MT501516.1 | vB_VpaP_MGD1 | 96.41 |
| NC_048035.1 | vB_VpaP_KF1 | 95.97 |
| MZ592921.1 | vB_VpP_NS8 | 95.87 |
| MZ020222.1 | BUCT233 | 95.64 |
| MW331544.1 | vB_Vc_SrVc2 | 95.55 |
| LR794124.1 | vB_Vc_SrVc9 | 95.55 |
| NC_048036.1 | vB_VpaP_KF2 | 95.21 |
| MZ182247.1 | vB_VpP_DE18 | 95.06 |
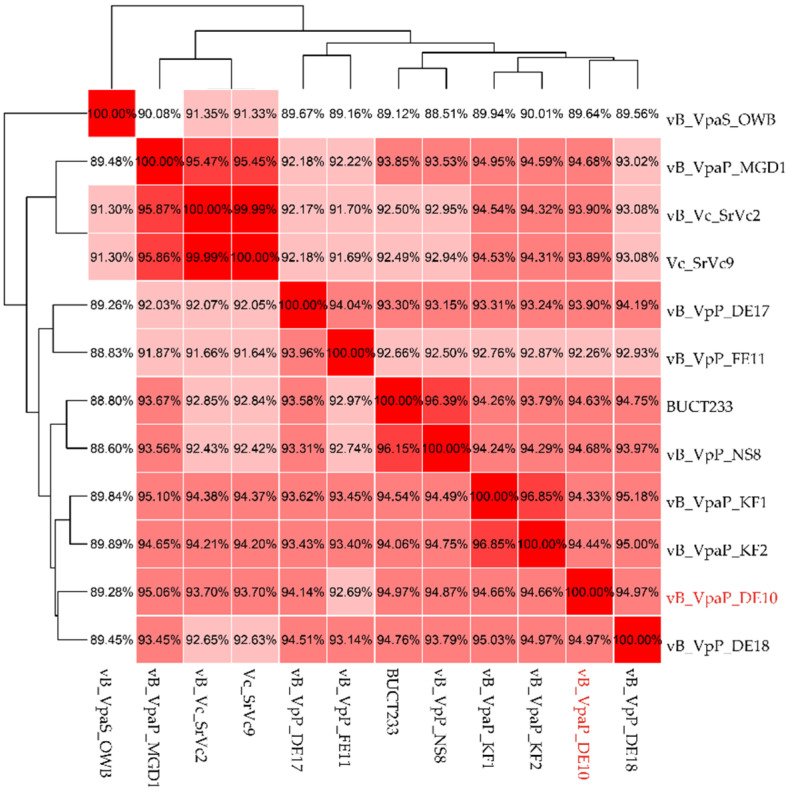
Due to the high genetic diversity of viruses, ANI analysis was suggested for their taxonomy in recent years. Thus, in order to accurately classify the phage vB_VpaP_DE10, ANI analysis between Vibrio phages of the genus Maculvirus was performed. As shown in Figure 7, the ANI values of vB_VpaP_DE10 with the 11 phages which had the highest BLASTN similarities were between 89.46% and 94.97% (Figure 7). As the threshold of ANI values to distinguish the viral species was below 95%, phage vB_VpaP_DE10 was considered as a new phage species [30,31].
Figure 7.
Heatmap of the ANI values for 12 whole genomes of the family Autographiviridae. Values range from 0 (0% ANI) to 1 (100% ANI): clusters of highly similar phages are highlighted in pink and red.
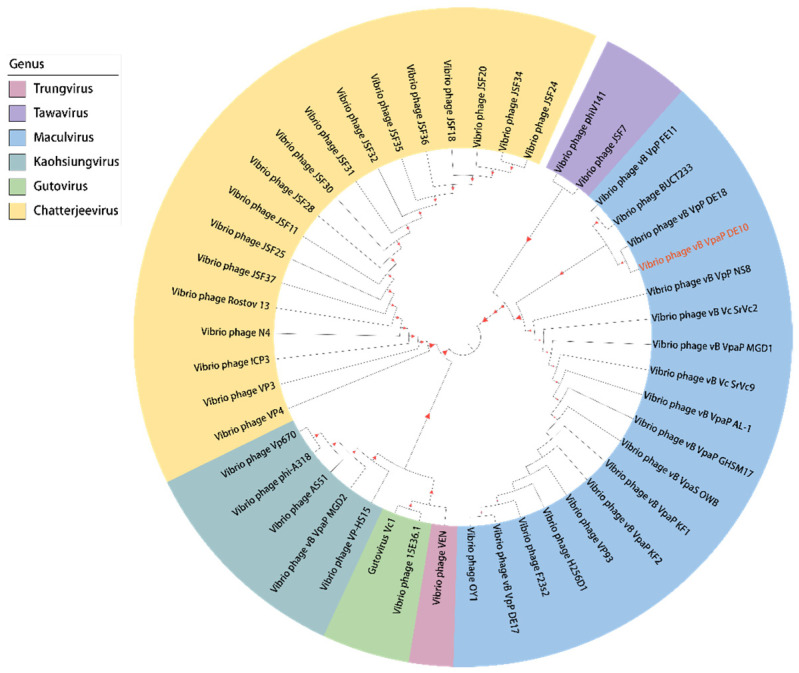
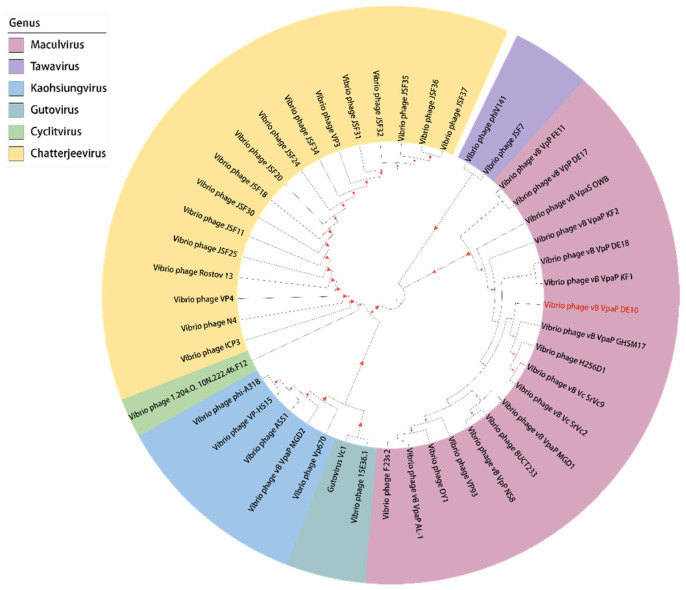
For phylogenetic analysis of phage vB_VpaP_DE10, the DNA polymerase and RNA polymerase were analyzed with all the Vibrio phages of the family Autographiviridae family. In the DNA polymerase-based phylogenetic tree, phage vB_VpaP_DE10 was most closely related to the phage vB_VpP_DE18 of the genus Maculvirus (Figure 8). When in the RNA polymerase-based tree, vB_VpaP_DE10 was also clustered with four Maculvirus phages (vB_VpaP_GHSM17, H256D1, vB_Vc_SrVc2, and vB_Vc_SrVc9 (Figure 9). Therefore, phage vB_VpaP_DE10 was considered as a member of the genus Maculvirus, which belongs to the family Autographiviridae.
Figure 8.
Phylogenetic tree based on the DNA polymerase.
Figure 9.
Phylogenetic tree based on the RNA polymerase.
4. Discussion
In this study, phage vB_VpaP_DE10 was obtained from sewage in the aquatic market, from where several Vibrio phages were isolated by our group at a different time. Although originating from the same place, genomes of six Vibrio phages showed high genetic diversities, and taxonomy varied. For example, even within the group of short-tail phages, the genome of vB_VpP_BA6 was completely different from vB_VpP_DE17, vB_VpP_FE11, and vB_VpaP_DE10.
Based on the virionic morphology and genome analysis, phage vB_VpaP_DE10 was a member of the Autographiviridae family. Several Autographiviridae phages specific for V. parahaemolyticus have been studied, such as phage vB_VpP_DE17 [15], phage vB_VpP_FE11 [16], phage vB_VpaP_MGD2 [14], phage vB_VpaP_AL-1 [32], phage VP93 [33], phage OWB [34], phage vB_VpaP_KF1, and phage vB_VpaP_KF2 [35]. Each phage has a different growth profile that can be used to evaluate the potential for application. Previous studies on V. parahaemolyticus phages of Autographiviridae family showed a latency period of 5–30 min and burst size of 37–244 PFU per cell [14,15,16,32]. Compared with these reports, vB_VpaP_DE10 had a relatively long latency period and a small burst size.
In addition, the phage was also tested for the ability to lyse other bacteria for assessment of host range [17,36]. Both spot assay and EOP test of phage vB_VpaP_DE10 were performed. Results showed that the EOP values varied widely among the 10 antibiotic-resistant strains. Values of V. parahaemolyticus O3-11 and O11-31 were higher than the indicated host V. parahaemolyticus O4-12, whereas others were lower. Changes of surface molecules that serve as phage receptors would significantly affect EOP and reduce phage adsorption [37,38]. Therefore, EOP differences are likely the result of differences in bacterial receptors.
For the host range difference, phages vB_VpaP_DE10 and vB_VpaP_FE11 both belong to the Maculvirus genus of the family Autographiviridae, and have short tails and high sequence similarity; however, the phage vB_VpaP_FE11 could infect V. parahaemolyticus O6-20, O8-21, and O11–30 strains whereas vB_VpaP_DE10 could not. The host range of phages has been reported to be associated with tail fiber or receptor-binding proteins [39]. Mutations of tail-related proteins could alter the host range of phages [40]. In the structure module of the vB_VpaP_DE10 genome, three tail-related proteins were predicted, including the tail tubular protein A/B (TTPA/TTPB, Gp25/26) and tail fiber protein (Gp30). These three proteins were also predicted in the genome of phage FE11, and their amino acid sequences have high similarity to that of phage of vB_VpaP_DE10 (96.77%, 95.00%, and 96.55%, respectively). The reason why the host ranges of the two phages differ may be due to the differences in their amino acid residues of 3–5%. Further studies are required to confirm this hypothesis. In conclusion, we isolated and characterized a novel lytic V. parahaemolyticus phage, vB_VpaP_DE10, having a short tail, and belonging to the genus Maculvirus within the family Autographiviridae.
Acknowledgments
Thanks to the Institute of Microbiology, Guangdong Academy of Sciences for providing Vibrio parahaemolyticus.
Author Contributions
W.H. and M.Y. conceived the study. Y.Y. and H.C. designed and conducted the experiments and wrote the paper. Q.H., S.H. and. J.H. performed the genome and comparative genome analysis. J.Z., Q.W. and X.L. contributed to the data analysis. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The findings of this study are available within this paper. The complete genome sequence of phage vB_VpaP_DE10 was submitted to the GenBank database under accession number MZ516827.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Funding Statement
The authors are grateful to the National Natural Science Foundation of China (NSFC) (31730070), Guangdong Major Project of Basic and Applied Basic Research (2020B0301030005), Key Laboratory of Guangdong Province (2020B121201009), and the Guangdong Province Academy of Sciences Special Project for Capacity Building of Innovation Driven Development (2020GDASYL-20200401002) for financial support.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bacian C., Verdugo C., García K., Perez-Larruscain J., de Blas I., Cachicas V., Lopez-Joven C. Longitudinal Study of Total and Pathogenic Vibrio parahaemolyticus (tdh+ and/or trh+) in Two Natural Extraction Areas of Mytilus chilensis in Southern Chile. Front. Microbiol. 2021;12:621737. doi: 10.3389/fmicb.2021.621737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda S., Hiyoshi H., Tandhavanant S., Kodama T. Advances on Vibrio parahaemolyticus research in the postgenomic era. Microbiol. Immunol. 2020;64:167–181. doi: 10.1111/1348-0421.12767. [DOI] [PubMed] [Google Scholar]
- 3.Han D., Yu F., Chen X., Zhang R., Li J. Challenges in Vibrio parahaemolyticus infections caused by the pandemic clone. Future Microbiol. 2019;14:437–450. doi: 10.2217/fmb-2018-0308. [DOI] [PubMed] [Google Scholar]
- 4.Matamp N., Bhat S.G. Phage Endolysins as Potential Antimicrobials against Multidrug Resistant Vibrio alginolyticus and Vibrio parahaemolyticus: Current Status of Research and Challenges Ahead. Microorganisms. 2019;7:84. doi: 10.3390/microorganisms7030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paudyal N., Pan H., Liao X., Zhang X., Li X., Fang W., Yue M. A Meta-Analysis of Major Foodborne Pathogens in Chinese Food Commodities between 2006 and 2016. Foodborne Pathog. Dis. 2018;15:187–197. doi: 10.1089/fpd.2017.2417. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Zhu Q., Liu Y., Wang R., Xie H., Chen J., Cheng Y., Zhang H., Cao L., Chen Y. Pathogenic Characteristics of and Variation in Vibrio parahaemolyticus Isolated from Acute Diarrhoeal Patients in Southeastern China from 2013 to 2017. Infect. Drug Resist. 2020;13:1307–1318. doi: 10.2147/IDR.S234186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paria P., Chakraborty H.J., Behera B.K., Mohapatra P.K.D., Das B.K. Computational characterization and molecular dynamics simulation of the thermostable direct hemolysin-related hemolysin (TRH) amplified from Vibrio parahaemolyticus. Microb. Pathog. 2019;127:172–182. doi: 10.1016/j.micpath.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Meng H., Gu D., Li Y., Jia M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 2019;222:43–51. doi: 10.1016/j.micres.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Lei T., Jiang F., He M., Zhang J., Zeng H., Chen M., Pang R., Wu S., Wei L., Wang J., et al. Prevalence, virulence, antimicrobial resistance, and molecular characterization of fluoroquinolone resistance of Vibrio parahaemolyticus from different types of food samples in China. Int. J. Food Microbiol. 2020;317:108461. doi: 10.1016/j.ijfoodmicro.2019.108461. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Li M., Li Y. Intervention Strategies for Reducing Vibrio parahaemolyticus in Seafood: A Review. J. Food Sci. 2015;80:R10–R19. doi: 10.1111/1750-3841.12727. [DOI] [PubMed] [Google Scholar]
- 11.Liu R., Han G., Li Z., Cun S., Hao B., Zhang J., Liu X. Bacteriophage therapy in aquaculture: Current status and future challenges. Folia Microbiol. 2022;67:573–590. doi: 10.1007/s12223-022-00965-6. [DOI] [PubMed] [Google Scholar]
- 12.Yang M., Liang Y., Huang S., Zhang J., Wang J., Chen H., Ye Y., Gao X., Wu Q., Tan Z. Isolation and Characterization of the Novel Phages vB_VpS_BA3 and vB_VpS_CA8 for Lysing Vibrio parahaemolyticus. Front. Microbiol. 2020;11:259. doi: 10.3389/fmicb.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang M., Liang Y., Su R., Chen H., Wang J., Zhang J., Ding Y., Kong L., Zeng H., Xue L., et al. Genome characterization of the novel lytic Vibrio parahaemolyticus phage vB_VpP_BA6. Arch. Virol. 2019;164:2627–2630. doi: 10.1007/s00705-019-04351-5. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y., Zhang Y., Lan W., Sun X. Characterization of vB_VpaP_MGD2, a newly isolated bacteriophage with biocontrol potential against multidrug-resistant Vibrio parahaemolyticus. Arch. Virol. 2021;166:413–426. doi: 10.1007/s00705-020-04887-x. [DOI] [PubMed] [Google Scholar]
- 15.Yang M., Chen H., Guo S., Tan S., Xie Z., Zhang J., Wu Q., Tan Z. Characterization and genome analysis of a novel Vibrio parahaemolyticus phage vB_VpP_DE17. Virus Res. 2022;307:198580. doi: 10.1016/j.virusres.2021.198580. [DOI] [PubMed] [Google Scholar]
- 16.Yang M., Chen H., Huang Q., Xie Z., Liu Z., Zhang J., Ding Y., Chen M., Xue L., Wu Q., et al. Characterization of the Novel Phage vB_VpaP_FE11 and Its Potential Role in Controlling Vibrio parahaemolyticus Biofilms. Viruses. 2022;14:264. doi: 10.3390/v14020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciacci N., D’Andrea M.M., Marmo P., Demattè E., Amisano F., Di Pilato V., Fraziano M., Lupetti P., Rossolini G.M., Thaller M.C. Characterization of vB_Kpn_F48, a Newly Discovered Lytic Bacteriophage for Klebsiella pneumoniae of Sequence Type 101. Viruses. 2018;10:482. doi: 10.3390/v10090482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing S., Zhang X., Sun Q., Wang J., Mi Z., Pei G., Huang Y., An X., Fu K., Zhou L., et al. Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 2017;162:3843–3847. doi: 10.1007/s00705-017-3503-1. [DOI] [PubMed] [Google Scholar]
- 19.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B., Pop M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stothard P., Grant J.R., Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 2019;20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Yazdi M., Bouzari M., Ghaemi E.A. Genomic analyses of a novel bacteriophage (VB_PmiS-Isfahan) within Siphoviridae family infecting Proteus mirabilis. Genomics. 2019;111:1283–1291. doi: 10.1016/j.ygeno.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Adriaenssens E.M., Sullivan M.B., Knezevic P., van Zyl L.J., Sarkar B.L., Dutilh B.E., Alfenas-Zerbini P., Łobocka M., Tong Y., Brister J.R., et al. Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2020;165:1253–1260. doi: 10.1007/s00705-020-04577-8. [DOI] [PubMed] [Google Scholar]
- 30.Pan L., Li D., Sun Z., Lin W., Hong B., Qin W., Xu L., Liu W., Zhou Q., Wang F., et al. First Characterization of a Hafnia Phage Reveals Extraordinarily Large Burst Size and Unusual Plaque Polymorphism. Front. Microbiol. 2022;12:754331. doi: 10.3389/fmicb.2021.754331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E.V., Kropinski A.M., Krupovic M., Kuhn J.H., Lavigne R., Brister J.R., Varsani A., et al. Minimum Information about an Uncultivated Virus Genome (MIUViG) Nat. Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Gómez J.P., López-Cuevas O., Campo N.C., González-López I., Martínez-Rodríguez C.I., Gomez-Gil B., Chaidez C. Genomic and biological characterization of the novel phages vB_VpaP_AL-1 and vB_VpaS_AL-2 infecting Vibrio parahaemolyticus associated with acute hepatopancreatic necrosis disease (AHPND) Virus Res. 2022;312:198719. doi: 10.1016/j.virusres.2022.198719. [DOI] [PubMed] [Google Scholar]
- 33.Bastías R., Higuera G., Sierralta W., Espejo R.T. A new group of cosmopolitan bacteriophages induce a carrier state in the pandemic strain of Vibrio parahaemolyticus. Env. Microbiol. 2010;12:990–1000. doi: 10.1111/j.1462-2920.2010.02143.x. [DOI] [PubMed] [Google Scholar]
- 34.Hu M., Zhang H., Gu D., Ma Y., Zhou X. Identification of a novel bacterial receptor that binds tail tubular proteins and mediates phage infection of Vibrio parahaemolyticus. Emerg. Microbes Infect. 2020;9:855–867. doi: 10.1080/22221751.2020.1754134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., Lim J.A., Kwak S.J., Park J.H., Chang H.J. Comparative genomic analysis of novel bacteriophages infecting Vibrio parahaemolyticus isolated from western and southern coastal areas of Korea. Arch. Virol. 2018;163:1337–1343. doi: 10.1007/s00705-018-3756-3. [DOI] [PubMed] [Google Scholar]
- 36.Park D., Park J. Characterization and Food Application of the Novel Lytic Phage BECP10: Specifically Recognizes the O-polysaccharide of Escherichia coli O157:H7. Viruses. 2021;13:1469. doi: 10.3390/v13081469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denes T., den Bakker H.C., Tokman J.I., Guldimann C., Wiedmann M. Selection and Characterization of Phage-Resistant Mutant Strains of Listeria monocytogenes Reveal Host Genes Linked to Phage Adsorption. Appl. Environ. Microbiol. 2015;81:4295–4305. doi: 10.1128/AEM.00087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Melo A.C.C., Da Mata Gomes A., Melo F.L., Ardisson-Araújo D.M.P., de Vargas A.P.C., Ely V.L., Kitajima E.W., Ribeiro B.M., Wolff J.L.C. Characterization of a bacteriophage with broad host range against strains of Pseudomonas aeruginosa isolated from domestic animals. BMC Microbiol. 2019;19:134. doi: 10.1186/s12866-019-1481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim A.N.W., Yen M., Seed K.D., Lazinski D.W., Camilli A. A Tail Fiber Protein and a Receptor-Binding Protein Mediate ICP2 Bacteriophage Interactions with Vibrio cholerae OmpU. J. Bacteriol. 2021;203:e0014121. doi: 10.1128/JB.00141-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobrega F.L., Vlot M., de Jonge P.A., Dreesens L.L., Beaumont H.J.E., Lavigne R., Dutilh B.E., Brouns S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018;16:760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The findings of this study are available within this paper. The complete genome sequence of phage vB_VpaP_DE10 was submitted to the GenBank database under accession number MZ516827.