Abstract
Uncultivated Nitrospira-like bacteria in different biofilm and activated-sludge samples were investigated by cultivation-independent molecular approaches. Initially, the phylogenetic affiliation of Nitrospira-like bacteria in a nitrifying biofilm was determined by 16S rRNA gene sequence analysis. Subsequently, a phylogenetic consensus tree of the Nitrospira phylum including all publicly available sequences was constructed. This analysis revealed that the genus Nitrospira consists of at least four distinct sublineages. Based on these data, two 16S rRNA-directed oligonucleotide probes specific for the phylum and genus Nitrospira, respectively, were developed and evaluated for suitability for fluorescence in situ hybridization (FISH). The probes were used to investigate the in situ architecture of cell aggregates of Nitrospira-like nitrite oxidizers in wastewater treatment plants by FISH, confocal laser scanning microscopy, and computer-aided three-dimensional visualization. Cavities and a network of cell-free channels inside the Nitrospira microcolonies were detected that were water permeable, as demonstrated by fluorescein staining. The uptake of different carbon sources by Nitrospira-like bacteria within their natural habitat under different incubation conditions was studied by combined FISH and microautoradiography. Under aerobic conditions, the Nitrospira-like bacteria in bioreactor samples took up inorganic carbon (as HCO3− or as CO2) and pyruvate but not acetate, butyrate, and propionate, suggesting that these bacteria can grow mixotrophically in the presence of pyruvate. In contrast, no uptake by the Nitrospira-like bacteria of any of the carbon sources tested was observed under anoxic or anaerobic conditions.
Nitrification, the oxidation of ammonia to nitrate catalyzed by bacteria, is a key part of global nitrogen cycling (37). In the first step of nitrification, chemolithoautotrophic ammonia oxidizers transform ammonia to nitrite, which is subsequently oxidized to nitrate by the nitrite-oxidizing bacteria (8). All isolated chemolithoautotrophic, nitrite-oxidizing bacteria belong to one of four different genera (7) Nitrobacter (alpha subclass of Proteobacteria), Nitrococcus (gamma subclass of Proteobacteria), Nitrospina (delta subclass of Proteobacteria), and Nitrospira (phylum Nitrospira). While species of the genus Nitrobacter have been isolated from a variety of environments, including soil and fresh water, it was long assumed that the other three genera were confined to marine environments (7). In recent studies, however, bacteria related to the genus Nitrospira were also found to occur in different nonmarine habitats. While the first described species of this genus, Nitrospira marina, was isolated from ocean water (49), the second isolated species, N. moscoviensis, was cultured from an iron pipe of a heating system in Moscow, Russia (16). These two species are the only cultivated representatives of the genus Nitrospira, but numerous related bacteria have recently been detected by comparative analysis of 16S rRNA sequences obtained from nitrifying bioreactors (11, 22), rhizosphere (30), a freshwater aquarium filter (21), groundwater contaminated with livestock wastewater (12), deltaic sediment (45), and deep-sea sediments (25). These sequences indicate considerable phylogenetic diversity within the genus Nitrospira; however, a thorough phylogenetic analysis of the phylum Nitrospira considering all of these sequences has not been performed. Furthermore, the retrieval of Nitrospira-related sequences from the above-mentioned environments demonstrates that these bacteria are widely distributed in nature and probably contribute significantly to global nitrite oxidation. However, the lack of pure cultures of Nitrospira-related bacteria from soil, water, sediment, and wastewater treatment plants restricts our knowledge of their physiology and genetics.
In contrast to textbook knowledge (5, 19), Nitrospira-like bacteria, not Nitrobacter spp., are the dominant nitrite oxidizers both in most full-scale wastewater treatment plants and in laboratory scale reactors (22, 36, 43, 48). Based on fluorescence in situ hybridization (FISH) combined with microelectrode measurements, it has been suggested that Nitrospira-like nitrite oxidizers represent K strategists adapted to low nitrite and oxygen concentrations, while Nitrobacter sp., as an r strategist, thrives if nitrite and oxygen are present in higher concentrations (42). Since many wastewater treatment plants suffer from repeated breakdowns of nitrification performance, more insight into the physiology of Nitrospira-like bacteria is required to find measures by which to stabilize this important step of nutrient removal in modern biological sewage treatment.
In this study, we investigated structural and functional features of Nitrospira-like bacteria in nitrifying biofilms and activated sludges from different wastewater treatment plants. Based on an in-depth analysis of the phylogeny of the phylum Nitrospira, 16S rRNA-directed oligonucleotide probes for the phylum and genus Nitrospira were developed. These probes were used to detect Nitrospira-like bacteria in situ in wastewater treatment plants and to study the morphology of their microcolonies by using a confocal laser scanning microscope (CLSM) and digital image analysis. Furthermore, the uptake of different carbon sources by Nitrospira-like bacteria in the bioreactor samples was examined under aerobic and anaerobic conditions by combining FISH and microautoradiography (MAR) (24).
(A preliminary report of this study was presented at the IAWQ Conference on Biofilm Systems [New York, N.Y., 17 to 20 October 1999] and at the IWA 2nd International Symposium on Sequencing Batch Reactor Technology [Narbonne, France, 10 to 11 July 2000].)
MATERIALS AND METHODS
Cultivation of reference organisms.
Bacillus stearothermophilus (DSM 22) and Leptospirillum ferrooxidans (DSM 2705) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, and cultivated in accordance with the supplier's instructions. Paraformaldehyde-fixed N. moscoviensis cells were kindly provided by Eberhard Bock and Gabriele Timmermann (University of Hamburg, Hamburg, Germany).
Bioreactor data, biofilm and activated-sludge sampling, and cell fixation.
Samples of nitrifying biofilm were retrieved from an aerated sequencing batch biofilm reactor (SBBR 1) at a pilot wastewater treatment plant near Ingolstadt, Germany. Additional samples were obtained from a second nitrifying, continuously operated biofilm reactor (Biofor 2) at the same plant. The biofilm grew in both reactors on Biolite expanded clay beads (grain size, 4 to 8 mm) forming a fixed bed with an average volume of 10 m3. Reactor SBBR 1 received reject water from sludge dewatering by the municipal wastewater treatment plant at Ingolstadt with NH4-N concentrations of 300 to 500 mg · liter−1, an average total chemical oxygen demand of 300 mg · liter−1, and an average conductivity of 5,000 to 6,000 μS · cm−1. At the end of each cycle, NH4-N at 40 to 50 mg · liter−1 and NO2-N at up to 70 mg · liter−1 were detected at the outlet of the reactor. The cycle time was 4 to 8 h, and the fixed bed was backwashed every 10 to 48 h. Reactor Biofor 2 received municipal wastewater with an average NH4-N concentration of 13 mg · liter−1, an average chemical oxygen demand of 191 mg · liter−1, and an average conductivity of 500 to 1,000 μS · cm−1. The outlet of the reactor contained NH4-N at 0.3 to 0.8 mg · liter−1 and NO2-N at less than 1 mg · liter−1. The fixed bed was back washed every 10 to 48 h. Activated-sludge samples were taken from the nitrification stage of the Aalborg West (AAV) wastewater treatment plant (Aalborg, Denmark; 275,000 population equivalents). In addition, samples were retrieved from the nitrification stage of the Munich II wastewater treatment plant (Munich, Germany; 106 population equivalents). The average NH4-N concentration in the inlet of the nitrification stage of the AAV plant was 25 mg · liter−1, while the outlet contained NH4-N and nitrite at less than 1 mg · liter−1. The sludge age was 20 days. The average NH4-N concentration was 12 mg · liter−1 at the inlet of the nitrification stage of the Munich II plant and 0.08 mg · liter−1 at the outlet. Nitrite concentrations at the outlet of the nitrification stage are not available for this plant. The sludge age was 7 to 10 days.
Biofilm and activated sludge were fixed for 5 h at 4°C in 3% paraformaldehyde as described by Amann (2) immediately after sampling (the biofilm was first detached from the expanded clay beads by gentle swirling). Unfixed sample aliquots used for DNA extraction were centrifuged (10 min, 4,550 × g), the supernatant was discarded, and the biomass was stored at −20°C. Pure bacterial cultures were harvested by centrifugation (10 min, 10,000 × g), resuspended in phosphate-buffered saline (PBS), and centrifuged again (10 min, 10,000 × g). The supernatant was removed prior to fixation of the cells in 3% paraformaldehyde. B. stearothermophilus cells were fixed with ethanol (40). Fixed samples and fixed pure cultures were stored in PBS-ethanol (1:1) at −20°C.
PCR amplification, cloning, sequencing, and phylogenetic analysis of 16S rDNAs.
DNA was extracted from frozen biofilm samples in accordance with the protocol described by Zhou et al. (51). Almost complete (1,497 to 1,533 nucleotides) bacterial 16S rDNAs were amplified by PCR, cloned, and sequenced as detailed by Juretschko et al. (22). The 16S rDNA sequences obtained were added to the ARB 16S rRNA sequence database (http://www.arb-home.de). The sequences were aligned by the ARB_EDIT module of the program, and the alignments were refined by visual inspection. Nucleic acid similarities were calculated by using the respective tool of the ARB program. Phylogenetic trees were computed by application of the ARB neighbor-joining and maximum-parsimony tools and by maximum-likelihood analysis of different sets of data. Treeing analyses were performed with and without application of a 50% conservation filter for the Nitrospira phylum. This filter was based on all sequences affiliated with the phylum Nitrospira that were longer than 1,400 nucleotides and was used to exclude highly variable alignment columns that are not conserved in at least 50% of the Nitrospira phylum sequences. For calculation of consensus trees, all sequences longer than 1,300 nucleotides were first processed by the maximum-likelihood method to determine the basic topology of the tree. The shorter sequences were added subsequently by use of the ARB_PARSIMONY function of the ARB program without changing the tree topology (26). Bootstrap values were determined by 100 iterations of maximum-parsimony bootstrapping analysis without application of a conservation filter. Neighbor-joining trees were calculated based on different sequence sets to verify the results obtained by the other treeing methods. Different sets of outgroup sequences that represented members of several other phyla of the domain Bacteria and were longer than 1,500 nucleotides were used in all treeing calculations. Checks for chimeric sequences were performed by independent phylogenetic analyses of the first 513 5′ base positions, the middle 513 base positions, and the last 513 3′ base positions of the sequences.
Incubation of activated sludge and biofilm with radioactive substrates.
Substrate uptake experiments were performed with living activated-sludge and biofilm samples. The radioactively labeled substrates used were [3H]acetate, [14C]pyruvate (Amersham, Little Chalfont, United Kingdom), [14C]butyrate, [14C]propionate, and [14C]bicarbonate (NEN Life Science, Boston, Mass.). Fresh activated-sludge and biofilm samples were harvested the day before the experiments were performed, kept at 4°C, and brought to the laboratory. The dry-matter content (suspended solids) of activated sludge and biofilm was adjusted to a concentration of 1 g of suspended solids · liter−1 with sterile-filtered supernatant from the respective bioreactors as described by Lee et al. (24). For incubation with all of the substrates but bicarbonate, 3 ml of diluted activated sludge or biofilm was transferred to 9-ml glass serum vials. For incubation with bicarbonate, 5 ml of a diluted sample was transferred to 25-ml serum vials. The incubation conditions applied were (i) aerobic, (ii) anoxic (i.e., anaerobic in the presence of 1 mM nitrate), and (iii) anaerobic. The nitrate concentrations in the samples were estimated by use of a nitrate test kit (Merckoquant Nitrate-test; Merck, Darmstadt, Germany) prior to the anaerobic incubations to ensure that these experiments were not influenced by nitrate present in the sludge or biofilm liquid. No nitrate was detected in the samples from the AAV plant and from reactor Biofor 2, but biofilm from reactor SBBR1 contained nitrate at approximately 500 mg · liter−1. This biofilm was centrifuged (10 min, 5,000 × g), the supernatant containing nitrate was discarded, and the biofilm was resuspended in sterile-filtered, nitrate-free biofilm liquid. This biofilm liquid had previously been obtained by anaerobic overnight incubation of an aliquot of the same biofilm sample. Following this pretreatment, no nitrate was detected in the biofilm liquid by the nitrate test kit. All serum vials for anoxic and anaerobic incubations were closed with thick, gas-tight butyl rubber stoppers and flushed with pure nitrogen gas prior to incubation, and all following steps were performed by using strict anaerobic techniques. Unlabeled organic substrates were added to a final concentration of 1 mM. This concentration is slightly higher than the concentrations of fatty acids in most wastewater treatment plants, but it was used to ensure maximal substrate uptake rates and to avoid starvation of the bacteria in the incubated samples due to substrate depletion. The respective labeled substrates were then added to a final activity of 10 μCi (the specific activities of the purchased radioactive tracers were 30, 60, 58, 10, and 5 mCi · mmol−1 for [14C]pyruvate, [14C]propionate, [3H]acetate, [14C]butyrate, and [14C]bicarbonate, respectively). Aerobically incubated preparations were vigorously shaken for 3 h, while anoxically and anaerobically incubated preparations were incubated for 4 h without agitation. All samples containing [14C]bicarbonate, however, were incubated for 5 h to compensate for the possibly lower fixation rates of inorganic carbon and because preliminary experiments (data not shown) demonstrated that 5 h of incubation was more suitable than 3 h for monitoring of the uptake of inorganic carbon by Nitrospira-like bacteria. Moreover, 1 mM NH4Cl was added to these samples as a substrate for the indigenous ammonia oxidizers. Nitrite resulting from their activity could serve as an energy source for the nitrite oxidizers during incubation with bicarbonate. The incubated samples were centrifuged (10 min, 5,000 × g) and fixed in 3% (wt/vol) paraformaldehyde as described above. The samples were then resuspended in a 1:1 mixture of PBS and 96% (vol/vol) ethanol and stored at −20°C.
The amounts of organic substrates taken up by the biomass were determined to confirm that the substrates were not depleted during incubation. Aliquots (0.5 ml) of the incubated samples were taken before and after the experiments. The aliquots were immediately cooled on ice, centrifuged (5 min, 10,000 × g), and filtered (0.2-μm-pore-size sterile filters; Millipore), and the supernatant was frozen for later analysis. Amounts of pyruvate, acetate, propionate, and butyrate in the supernatants of the respective incubations were measured by high-performance liquid chromatography on a Dionex ion chromatograph with a suppressed conductivity detector, 1 mM NaOH as the mobile phase, and an IonPac AS11-HC column. The uptake of [14C]bicarbonate (as HCO3− or CO2) was estimated by measuring the amount of [14C]bicarbonate bound by the biomass after incubation. The radioactive bicarbonate in the biomass was quantified by liquid scintillation counting (Tri-Carb Analyzer 1600TR; Packard Instrument) of an aliquot. Unbound CO2 had been removed from these aliquots prior to measurement by lowering of the pH to less than 1 with 1 N HCl and thorough flushing with N2 for 30 min. Aliquots of pasteurized biofilm and sludge were incubated with the respective radioactive substrates to test for adsorption and precipitation phenomena in all experiments as described by Lee et al. (24).
16S rRNA-directed oligonucleotide probes.
The program ARB and the current version of the ARB 16S rRNA sequence database (approximately 15,000 entries) were used to develop new 16S rRNA-directed oligonucleotide probes (Table 1). The probes used for in situ hybridization were 5′ labeled with the dye FLUOS [5(6)-carboxyfluorescein-N-hydroxysuccinimide ester] or with the sulfoindocyanine dye Cy3 or Cy5. Labeled probes and unlabeled competitor oligonucleotides were obtained from Thermo Hybaid (Interactiva Division, Ulm, Germany) or MWG (Ebersberg, Germany). The following oligonucleotide probes were used in addition to the probes developed in this study: (i) NEU, which is specific for halophilic and halotolerant Nitrosomonas spp. and Nitrosococcus mobilis (47); (ii) Nso1225, which is specific for ammonia oxidizers in the beta subclass of Proteobacteria (33), except for N. mobilis (38); (iii) NIT3, which is complementary to a sequence region of all Nitrobacter species (48); (iv) the EUB338 probe mixture, which consists of probes EUB338 (3), EUB338-II, and EUB338-III (14) covering the domain Bacteria; and (v) NON338, which is complementary to probe EUB338 (29) and was used as a negative control in all FISH experiments. All of the probes developed in this study were named in conformance with the standard introduced by Alm et al. (1), while the names of previously published probes were left unchanged to avoid confusion.
TABLE 1.
16S rRNA-targeted oligonucleotide probes developed in this studya
| Probe | Sequence (5′–3′) | Target site (16S rRNA position)b | % formamide/NaCl concn (mM) |
|---|---|---|---|
| S-G-Ntspa-0662-a-A-18 | GGAATTCCGCGCTCCTCT | 662–679 | 35/80 |
| Comp-Ntspa-0662 | GGAATTCCGCTCTCCTCT | 662–679 | —c |
| S-∗-Ntspa-0712-a-A-21 | CGCCTTCGCCACCGGCCTTCC | 712–732 | 50/28 |
| Comp-Ntspa-0712 | CGCCTTCGCCACCGGTGTTCC | 712–732 | —d |
The names, sequences, and target sites of the probes, the formamide concentrations in the hybridization buffer, and the salt concentrations in the wash buffers required for specific in situ hybridization are specified.
E. coli numbering (10).
Used as unlabeled competitor together with probe S-G-Ntspa-0662-a-A-18.
Used as unlabeled competitor together with probe S-∗-Ntspa-0712-a-A-21.
FISH and MAR.
To determine oligonucleotide probe dissociation profiles, 5-μl samples of fixed reference cells from pure cultures were spotted onto microscope slides (Paul Marienfeld, Bad Mergentheim, Germany) and dried for 10 min at 46°C. Thereupon, in situ hybridization was performed as detailed by Manz et al. (29). Different concentrations of formamide in the hybridization buffers and of sodium chloride in the washing buffers were used to measure probe binding at increasing hybridization stringencies.
Initially, all biofilm and activated-sludge samples were hybridized with probe NON338 derivatives labeled with FLUOS, Cy3, and Cy5 to exclude nonspecific probe binding. In none of the samples was nonspecific labeling of cells observed.
A modified FISH protocol was used to preserve the three-dimensional structure of bacterial aggregates in biofilm and activated-sludge flocs. Silicone tube segments with a diameter of 5 mm and a length of 5 to 8 mm were glued onto microscope coverslips by using bicomponent glue. These hybridization chambers were filled with 20 μl of the biofilm or activated-sludge samples. After sedimentation (by gravity) of the biomass, approximately 10 μl of the supernatant was removed and replaced with 20 μl of 1% (wt/vol) molten agarose (Gibco BRL ultraPure agarose; Life Technologies, Paisley, Scotland) at a temperature of approximately 37°C. After solidification of the agarose, the embedded samples were dehydrated by dipping the slips successively into 50, 80, and 96% (vol/vol) ethanol for 3 min each. Subsequently, FISH was performed as described previously (29) but 20 μl of hybridization buffer and increased amounts of probes (60 ng of probes labeled with Cy3 or Cy5 and 100 ng of probes labeled with FLUOS) were applied. Following the hybridization and washing steps, the slips were immersed for 10 s in ice-cold deionized water to remove buffer salts from the slip surface.
Fixed biofilm or activated-sludge samples that had been incubated with radioactive substrates were embedded in cryoembedding compound (catalog no. 350100; Microm, Walldorf, Germany) and sliced with a microtome (model HM 505E; Microm). Sections with thicknesses of 5 to 10 μm were applied to microscope coverslips, and FISH was performed with suitable oligonucleotide probes. MAR was performed as described by Lee et al. (24). Different exposure times (2, 5, and 7 days) before the development of the radiographic film emulsion were tested for all samples incubated with the different substrates (shorter exposure resulted in weaker signals, while prolonged exposure caused increased background noise). Coverslips with developed film were stored at 4°C until microscopic analysis.
Microscopy and digital image analysis.
All samples hybridized with oligonucleotide probes were embedded in Citifluor (Citifluor, Canterbury, United Kingdom) prior to microscopic observation. Alternatively, biofilm was embedded in a 1:1 mixture of Citifluor and a 0.01% (wt/vol) fluorescein solution for negative staining. Fluorescence signals were recorded with an LSM 510 CLSM (Zeiss, Oberkochen, Germany) equipped with two HeNe lasers (543 and 633 nm, respectively) for detection of Cy3 and Cy5 and one Ar ion laser (450 to 514 nm) for detection of FLUOS. Probe dissociation profiles were obtained as described by Daims et al. (14). Diameters of cell aggregates were determined by using the measurement tools of the software delivered with the CLSM (LSM 510, version 2.01). For this purpose, optical sections were acquired in those planes where the colonies to be measured were largest and the diameters of the colonies in these images were determined. The average diameters of the Nitrospira colonies in the different samples were obtained by processing of 50 to 100 colonies per sample. Three-dimensional reconstructions of Nitrospira cell aggregates were generated from stacked optical sections through biofilm samples. The optical sections with thicknesses of 0.5 to 0.7 μm were acquired with the CLSM and saved as monochrome 8-bit tagged image file format files. The single images of these image stacks were imported in consecutive order by a three-dimensional image analysis and visualization program (H. Daims, unpublished data). During this step, the intensities of the image pixels were stored in a three-dimensional array in computer memory. Thereupon, background noise was reduced by three-dimensional median filtering and the pixel intensity data were analyzed to distinguish cell material from voids within cell clusters. Isosurfaces (i.e., surfaces connecting points of the same intensity) were extracted from the pixel intensity array by a modified implementation of the marching-tetrahedron algorithm (18) to display the surfaces of cell clusters. The isosurface calculation requires that the intensity level of the isosurface be supplied by the user, who must find appropriate intensity levels for visualization of the real surfaces of an object as precisely as possible. These intensity levels were determined with the aid of a second visualization method that displayed the real interfaces between the cell aggregates, the surrounding medium, and the internal voids as semitransparent layers in one image. This method is based on a modification of a volume-rendering algorithm published elsewhere (13) that draws surfaces automatically and independently of user-supplied intensity levels. The results of the two visualization techniques were evaluated by generating isosurfaces with different intensity levels and by comparing these isosurfaces visually with the surfaces drawn by the volume-rendering algorithm. During this evaluation, the most suitable intensity levels were selected for drawing of the cell clusters as isosurfaces in the final images. All voids within cell clusters were drawn colored and semitransparent to visualize the cavities and channels in the cell aggregates.
RESULTS
Phylogenetic affiliation of Nitrospira-like bacteria in the sequencing batch biofilm reactor.
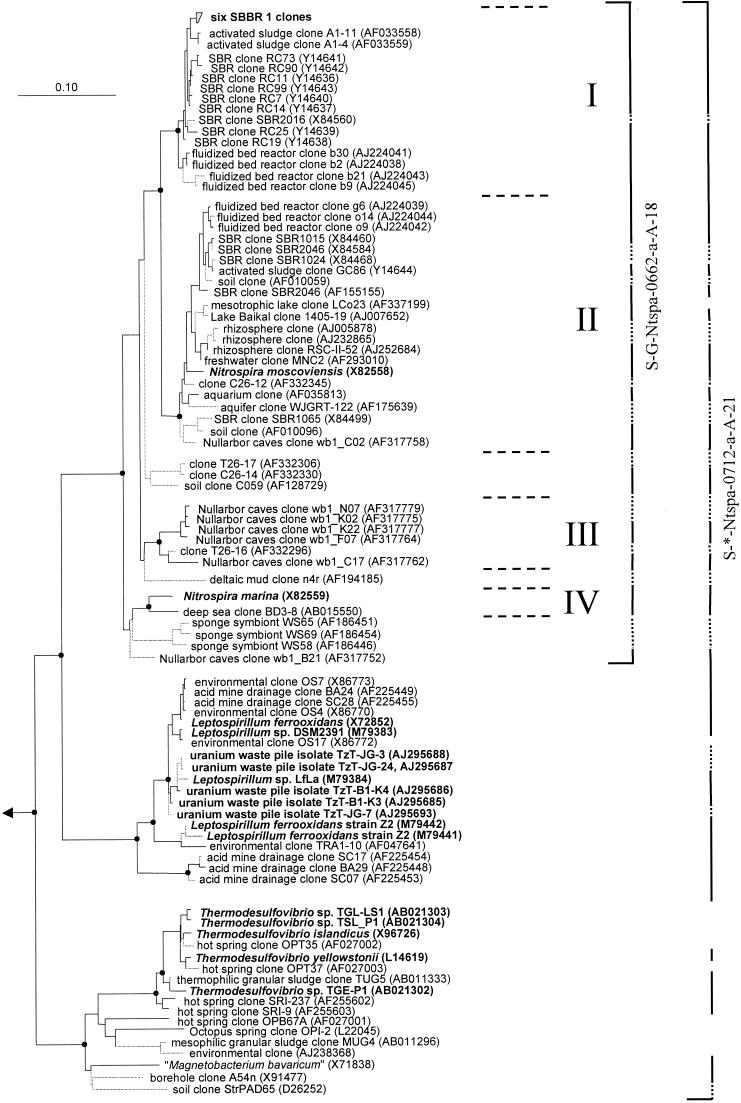
Biofilm was taken from nitrifying sequencing batch biofilm reactor SBBR 1, total DNA was extracted, and a 16S rDNA clone library was established. Among the 129 cloned and phylogenetically analyzed 16S rDNA sequences, 6 almost identical sequences shared high overall similarities (greater than 97%) with 16S rRNA sequences of Nitrospira-like bacteria obtained from nitrifying bioreactors in other studies (Fig. 1). None of the 129 16S rRNA sequences analyzed that were retrieved from reactor SBBR 1 grouped with the genus Leptospirillum and the Thermodesulfovibrio-“Magnetobacterium ” lineage.
FIG. 1.
Phylogenetic tree of the phylum Nitrospira based on comparative analysis of 16S rRNA sequences. The basic tree topology was determined by maximum-likelihood analysis of all sequences longer than 1,300 nucleotides. Shorter sequences were successively added by use of the ARB_PARSIMONY module of the ARB program without changing the overall tree topology. Branches leading to sequences shorter than 1,000 nucleotides are dotted to point out that the exact affiliation of these sequences cannot be determined. Black spots on tree nodes symbolize high-parsimony bootstrap support above 90% based on 100 iterations. The scale bar indicates 0.1 estimated change per nucleotide. Sequences of Nitrospira-like bacteria retrieved in this study from reactor SBBR 1 and sequences that belong to isolated strains are in boldface. The four sublineages of the genus Nitrospira are delimited by horizontal dashed lines and numbered I to IV. The brackets illustrate the coverage of the 16S rRNA-targeted oligonucleotide probes developed in this study. Dotted bracket segments indicate that the corresponding partial sequences do not include the probe target site. Brackets are interrupted where sequences are not targeted by the respective probe.
16S rRNA-directed oligonucleotide probes for in situ detection of the genus and the phylum Nitrospira.
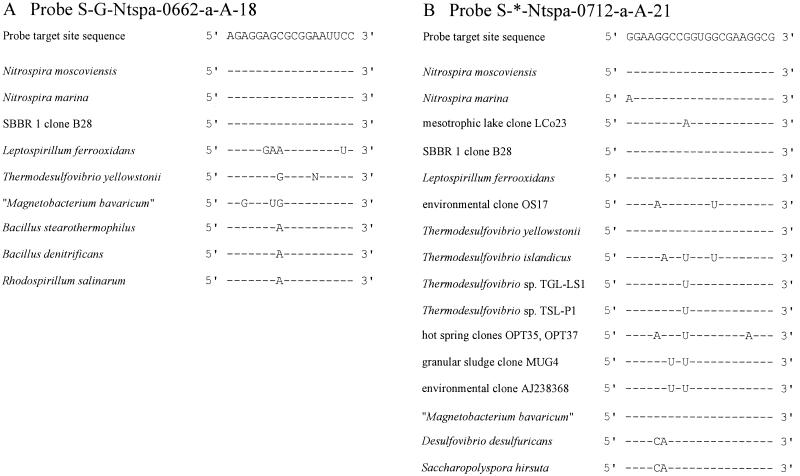
In this study, two 16S rRNA-directed oligonucleotide probes were developed for in situ identification of members of the genus and phylum Nitrospira, respectively (for details, see Table 1). The organisms target by each probe are indicated by brackets in Fig. 1.
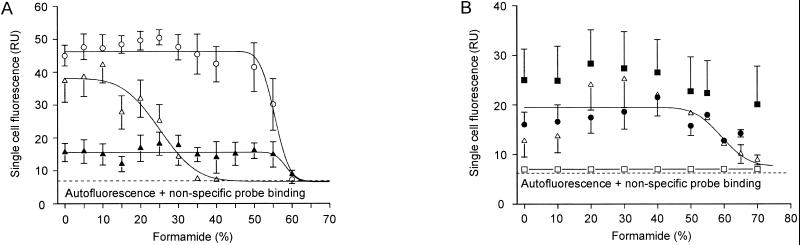
Probe S-G-Ntspa-0662-a-A-18 targets the described species N. moscoviensis and N. marina, as well as all environmental 16S rRNA sequences clustering with the genus Nitrospira line of descent for which the probe target site has been sequenced (Fig. 1). The 16S rRNA sequences of these organisms are fully complementary to probe S-G-Ntspa-0662-a-A-18 at the probe binding site, while all members of the other main lineages in the phylum Nitrospira possess at least one central mismatch with this probe (Fig. 2A). Several additional nontarget bacteria, for example, B. stearothermophilus, have a single C→A transversion at Escherichia coli position 669 (Fig. 2A). The probe dissociation profile of probe S-G-Ntspa-0662-a-A-18 indicates that this single mismatch does not prevent binding of the probe to the 16S rRNA (Fig. 3A). Fluorescence emitted by probe-stained cells of target species N. moscoviensis was clearly visible after FISH with up to 55% formamide in the hybridization buffer. However, cells of nontarget organism B. stearothermophilus were also stained by the probe and appeared even brighter. The probe-conferred fluorescence of the Bacillus cells was probably stronger, because these heterotrophic cells contained more ribosomes than the slow-growing, autotrophic cells of N. moscoviensis. Therefore, a competitor oligonucleotide (Comp-Ntspa-0662) was designed that is fully complementary to the 16S rRNAs of all nontarget organisms with the same sequence as B. stearothermophilus between E. coli positions 662 and 679. This competitor completely suppressed the hybridization of probe S-G-Ntspa-0662-a-A-18 with B. stearothermophilus if applied in equimolar amounts together with the probe in a hybridization buffer containing at least 35% formamide (Fig. 3A). Under these conditions, probe S-G-Ntspa-0662-a-A-18 also did not bind to Leptospirillum ferrooxidans and Thermodesulfovibrio yellowstonii (data not shown). Bacterial probe EUB338 stained N. moscoviensis and B. stearothermophilus at formamide concentrations between 0 and 60% without any visible decrease in the fluorescence signal intensity (data not shown).
FIG. 2.
Target site sequences and corresponding 16S rRNA sequence regions of target and nontarget organisms for probe S-G-Ntspa-0662-a-A-18 (A) and probe S-*-Ntspa-0712-a-A-21 (B). Hyphens represent identical nucleotides. Mismatches between the rRNA sequences of organisms and the probe target site sequence are indicated by capital letters.
FIG. 3.
Probe dissociation profiles of the oligonucleotide probes developed in this study with reference organisms under increasingly stringent hybridization and washing conditions. For each data point, the mean fluorescence intensity of at least 100 cells was determined. Regression curves were calculated by the plotting software based on a sigmoidal curve fit model. Error bars indicate 1 standard deviation. Error bars that are smaller than the marker symbols are not shown. (A) Hybridization of target organism N. moscoviensis with probe S-G-Ntspa-0662-a-A-18 in the presence of Comp-Ntspa-0662 (▴). Hybridization of nontarget bacterium B. stearothermophilus with probe S-G-Ntspa-0662-a-A-18 without (○) and with (Δ) competitor Comp-Ntspa-0662. (B) Hybridization of target organisms N. moscoviensis (●) and L. ferrooxidans (■) with probe S-*-Ntspa-0712-a-A-21 in the presence of competitor Comp-Ntspa-0712. Hybridization of nontarget bacterium D. desulfuricans with probe S-*-Ntspa-0712-a-A-21 without (▵) and with (□) competitor Comp-Ntspa-0712. The regression curve refers to the data points obtained for D. desulfuricans without addition of the competitor. RU, relative units.
Probe S-*-Ntspa-0712-a-A-21 targets all members of the phylum Nitrospira except T. islandicus; Thermodesulfovibrio sp. strains TGL-LS1 and TSL-P1; hot-spring clone OPB67A; environmental sequences OPT35, OPT37, MUG4, AJ238368, and OS17; mesotrophic-lake clone LCo23 (Fig. 2B); and Octopus Spring clone OPI-2. N. marina has one G→A transition at position 712 (Fig. 2B). We assume that this single mismatch does not prevent the binding of probe S-*-Ntspa-0712-a-A-21 to N. marina because it is located marginally at the 5′ end of the probe target region. Target species N. moscoviensis and L. ferrooxidans were stained by probe S-*-Ntspa-0712-a-A-21 with up to 70% formamide in the hybridization buffer (Fig. 3B). Probably due to the length (21 nucleotides) and high G+C content of this probe (76.2%), no clear dissociation of the probe from the target organisms was observed, even at the highest formamide concentration tested. In addition, the competitor Comp-Ntspa-0712 (see below), which was applied together with probe S-*-Ntspa-0712-a-A-21 in these experiments, did not prevent hybridization of the probe to the target organisms. Figure 3B also contains the dissociation profile of probe S-*-Ntspa-0712-a-A-21 with Desulfovibrio desulfuricans. This species represents a group of nontarget organisms with two mismatches in the binding region of the probe (Fig. 2B). These mismatches did not prevent hybridization of the probe, but instead, this species was stained as efficiently as N. moscoviensis with up to 60% formamide in the hybridization buffer and detectable signals were still observed when more formamide was applied (Fig. 3B). Therefore, a competitor oligonucleotide (Comp-Ntspa-0712) complementary to the 16S rRNAs of all organisms with the same sequence as D. desulfuricans between E. coli positions 712 and 732 was designed to improve the specificity of probe S-*-Ntspa-0712-a-A-21. When Comp-Ntspa-0712 was added in equimolar concentrations to the hybridization buffer, probe S-*-Ntspa-0712-a-A-21 did not stain D. desulfuricans, even under conditions of low stringency (Fig. 3B).
In situ detection and morphology of Nitrospira microcolonies in biofilm and activated sludge.
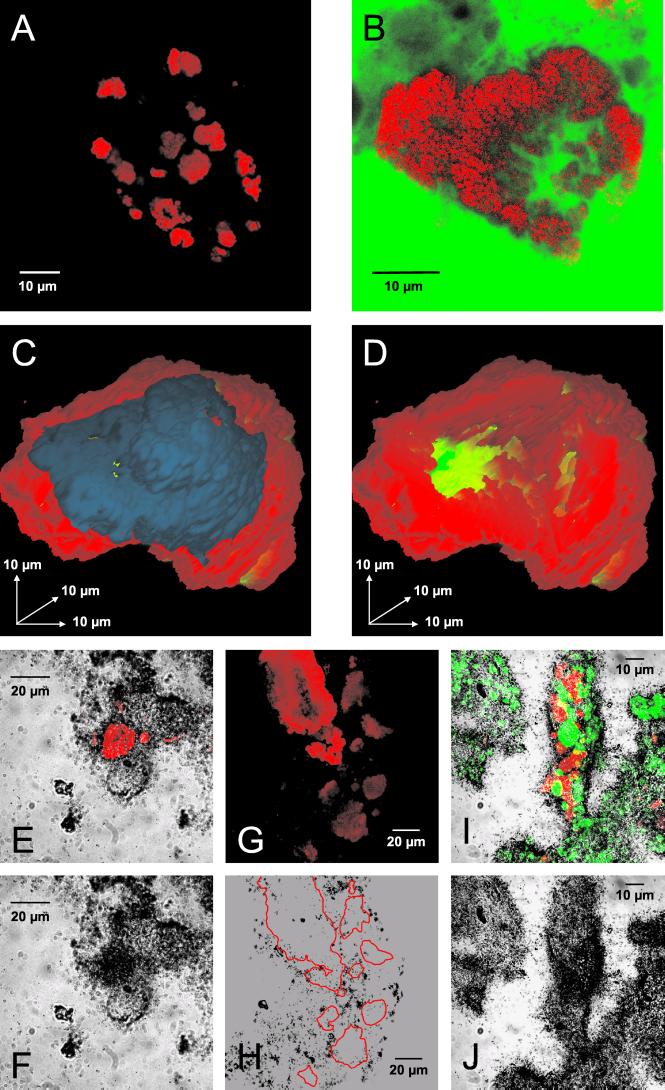
Activated sludge and biofilm from four different nitrifying bioreactors were screened for Nitrospira-related bacteria by FISH with the newly developed oligonucleotide probes. The presence of Nitrospira-like organisms in reactor SBBR 1 was confirmed by FISH with probes S-G-Ntspa-0662-a-A-18 and S-*-Ntspa-0712-a-A-21. Both probes exclusively stained the same cell aggregates with average diameters of 12.4 ± 7.0 μm (the mean diameters with standard errors are specified) (Fig. 4B). Similar results were obtained for an additional biofilm reactor (Biofor 2), where the cell aggregates had an average diameter of 11.4 ± 7.6 μm. The smallest and largest measured diameters of the Nitrospira cell clusters were 4.9 and 38.1 μm, respectively, in reactor SBBR 1 and 1.6 and 31.7 μm, respectively, in reactor Biofor 2. Some of the Nitrospira microcolonies in these biofilms had a spherical appearance, but most of them were shaped more irregularly. Their most remarkable features were cavities and channels within the cell colonies. Negative staining of biofilm from reactor SBBR 1 with fluorescein after FISH with probe S-G-Ntspa-0662-a-A-18 revealed that the fluorescein, which did not cross the boundaries of the Nitrospira cells, could penetrate these voids (Fig. 4B). The absence of bacterial cells from the cavities was confirmed by additional hybridization with the EUB338 probe mixture (data not shown). The impression that the Nitrospira microcolonies could be interlaced with a network of microscopic channels was verified by three-dimensional reconstruction of a probe-stained Nitrospira microcolony (Fig. 4C and D). Nitrospira microcolonies with this intricate morphology occurred frequently in the biofilms but were rare in both activated-sludge samples. Nitrospira aggregates detectable with both probes (S-G-Ntspa-0662-a-A-18 and S-*-Ntspa-0712-a-A-21) were abundant in these sludges, but compared to the biofilm samples, the cells were packed more tightly and the aggregates were significantly smaller and had a more spherical shape (Fig. 4A). The average diameter of a Nitrospira microcolony in the AAV plant was 2.8 ± 1.6 μm, with a minimum of 0.9 μm and a maximum of 7.5 μm. In the activated-sludge sample from the Munich II plant, their average diameter was 3.9 ± 2.3 μm, with a minimum of 1.0 μm and a maximum of 13.6 μm.
FIG. 4.
In situ analyses of Nitrospira-like bacteria within activated sludge and biofilms. (A) Nitrospira cell aggregates detected in activated sludge by FISH with probe S-G-Ntspa-0662-a-A-18 (red). (B) Nitrospira cell aggregate detected in biofilm from reactor SBBR 1 by FISH with probe S-G-Ntspa-0662-a-A-18 (red). All of the Nitrospira colonies detected in SBBR 1 with probe S-G-Ntspa-0662-a-A-18 also hybridized with probe S-*-Ntspa-0712-a-A-21 (data not shown). The biofilm was also stained with fluorescein (green). (C and D) Three-dimensional reconstruction of a Nitrospira cell aggregate from reactor SBBR 1 stained by FISH with probe S-G-Ntspa-0662-a-A-18. Nitrospira cells are red; the blue part of the microcolony in panel C was digitally removed to allow insight into the aggregate (D). Voids within the aggregate are green. (E and F) Uptake of bicarbonate by Nitrospira-like bacteria in biofilm from reactor SBBR 1 under aerobic incubation conditions. (E) Nitrospira cells stained by probe S-G-Ntspa-0662-a-A-18 (red) combined with the micrograph of the radiographic film at the same position. Panel F shows only the film to visualize the MAR signal at the position of the Nitrospira cells. Other MAR signals were caused by CO2-fixing bacteria, which were not detected by the Nitrospira-specific probe. (G and H) No uptake of acetate by Nitrospira-like bacteria in biofilm from reactor SBBR 1 under aerobic incubation conditions. (G) Nitrospira cells stained by probe S-G-Ntspa-0662-a-A-18 (red). Panel H is a micrograph of the radiographic film at the same position. The localization of the Nitrospira microcolonies in panel G is indicated by red borderlines in panel H. (I and J) Uptake of pyruvate by Nitrospira-like bacteria (stained by probe S-G-Ntspa-0662-a-A-18; red) and by ammonia oxidizers (stained by probes NEU and Nso1225; green) in biofilm from reactor SBBR 1 under aerobic incubation conditions. The fluorescence recorded in a stack of images by the CLSM is combined by orthographic projection in panel I. Stacked cells of Nitrospira-like bacteria and ammonia oxidizers appear therefore yellow. The image stack was acquired to ensure that all of the nitrifiers that contributed to the MAR signal would be visible in the final image.
All samples were also screened by FISH with a set of probes targeting nitrifying bacteria other than Nitrospira spp. Nitrobacter spp. were detected in neither the biofilm from reactor Biofor 2 nor the two activated-sludge samples when probe NIT3 was used. In reactor SBBR 1, however, probe-stained Nitrobacter cells occurred frequently but were less abundant than Nitrospira-like bacteria (data not shown). Furthermore, ammonia oxidizers from the beta subclass of Proteobacteria were detected frequently in all samples when probes NEU and Nso1225 were used (Fig. 4I).
Uptake of carbon sources by Nitrospira-like bacteria in wastewater treatment plants.
The uptake of substrates by Nitrospira-like bacteria in wastewater treatment plants was studied by incubation of living nitrifying biofilm and activated sludge with radioactive carbon sources, followed by FISH and MAR. Biofilm samples from reactors SBBR 1 and Biofor 2 and activated sludge from the AAV plant were incubated with different carbon sources. The liquid scintillation counts and high-performance liquid chromatography measurements performed before and after all incubations confirmed that the substrates were not depleted during the incubation time (data not shown). The incubated samples were hybridized with the Nitrospira-specific probes developed in this study and the EUB338 probe mixture. After completion of the FISH-MAR procedure, the samples were screened for probe-stained microcolonies of Nitrospira-like bacteria. Silver grain formation above these aggregates indicated that the cells took up the respective substrates during the incubation period. All substrate uptake experiments were qualitative, except for one experiment with bicarbonate and activated sludge from the AAV plant, which was evaluated quantitatively by counting MAR-positive and -negative Nitrospira microcolonies (see below). Under aerobic incubation conditions, most of the Nitrospira colonies detected in the biofilm sample from reactor SBBR 1 took up inorganic carbon (Fig. 4E and F). No uptake of acetate, propionate, or butyrate by Nitrospira-like bacteria was observed after aerobic incubation (Fig. 4G and H). Most of the Nitrospira colonies in the biofilm, however, were clearly MAR positive after aerobic incubation with 14C-labeled pyruvate (Fig. 4I and J). Uptake of pyruvate by the remaining Nitrospira colonies was uncertain because only weak silver grain formation was observed above these aggregates. Results similar to those obtained with reactor SBBR 1 were obtained with the biofilm from reactor Biofor 2 (data not shown). Nitrospira-like bacteria did not take up any substrate tested in all three samples under anoxic or anaerobic conditions. Other bacteria in the samples, which were stained by the EUB338 probe mixture, took up the different substrates under the anoxic incubation conditions and, to a much lesser extent, under the anaerobic incubation conditions (data not shown). These bacteria were not identified by FISH with more specific probes but served as a positive control for the incubation experiments and the FISH-MAR procedure. In contrast, no MAR-positive cells were detected in the control experiments performed with pasteurized biofilm (data not shown). Therefore, adsorption of labeled substrates to cells or other organic components did not account for the MAR signals observed with living biofilm and activated sludge.
An additional experiment was performed to determine the fraction of detectable Nitrospira microcolonies taking up bicarbonate in activated sludge from the AAV wastewater treatment plant. The sludge was incubated with [14C]bicarbonate under aerobic conditions, and the subsequent steps of the FISH-MAR protocol were performed. Different microscope slips covered with the radiographic film emulsion were exposed for 5, 6, or 10 days. Following film development, 200 Nitrospira microcolonies were investigated for fixation of inorganic carbon at each exposure time in several microscope fields per slip to obtain the fraction of colonies with inorganic carbon uptake activity. After 5 days of exposure, 56% of the Nitrospira colonies counted were MAR positive. These colonies usually had diameters greater than 3.2 μm, while most of the smaller colonies were MAR negative. Similar results were obtained after 6 days of exposure, when 51% of the colonies counted were MAR positive, with the same correlation of colony size and MAR signals. After 10 days of exposure, however, the percentage of clearly MAR-positive colonies had increased to 77%. The remaining cell aggregates were very small (diameters of less than 2.0 μm), and silver grain formation above these colonies possibly indicating bicarbonate uptake could not be distinguished from side effects due to background radiation or radiation emitted by larger adjacent colonies.
DISCUSSION
Phylogenetic trees containing all of the publicly available 16S rRNA sequences related to the phylum Nitrospira were calculated by using the maximum-parsimony and maximum-likelihood treeing methods on different data sets. The resultant consensus tree of the phylum shown in Fig. 1 is almost identical in topology to the respective neighbor-joining tree (data not shown). Consistent with the original definition of the phylum Nitrospira proposed by Ehrich et al. (16), the phylum currently consists of three main monophyletic lineages that are supported by all treeing methods and high bootstrap values. All of the sequences in the first main lineage are affiliated with N. moscoviensis and N. marina, the sequences in the second main lineage are related to L. ferrooxidans, and the organisms in the third main lineage are relatives of T. yellowstonii and “M. bavaricum”. Our analyses demonstrate that the line of descent containing both Nitrospira species can be further subdivided into at least four monophyletic sublineages, which are supported by all treeing methods and high bootstrap values above 90% (I to IV; Fig. 1). The sequences grouping together in each sublineage share 16S rRNA similarities of at least 94.9%. In contrast, the similarities of sequences that belong to different sublineages are always below 94.0%. Therefore, it can be postulated, based on the suggestion by Stackebrandt and Goebel (44), that the genus Nitrospira contains, in addition to N. marina and N. moscoviensis, at least two new candidate species represented by sublineages I and III, respectively. However, valid description of these species must await the isolation and phenotypic characterization of these organisms. Comparison of sublineage affiliation and sampled environments suggests that sublineages I, III, and IV each encompass a specialized group of nitrite oxidizers adapted to a certain habitat while sublineage II contains nitrite oxidizers that can thrive in different systems. One might speculate that the apparently unequal distribution of the sublineages in nature is a direct consequence of evolutionary events or of limited searches that have been done for these organisms.
Specifically, sublineage I contains only uncultivated organisms found in nitrifying bioreactors. These bacteria were detected in a laboratory scale sequencing batch reactor (11), a laboratory scale fluidized bed reactor (43), and an industrial full-scale activated-sludge basin (22). In addition, all six of the Nitrospira-related 16S rRNA sequences that were obtained from reactor SBBR 1 in this study group with this sublineage (Fig. 1). Sublineage II contains the cultivated species N. moscoviensis, as well as 16S rRNA sequences of uncultivated bacteria retrieved from diverse habitats, including bioreactors (9, 11, 43), freshwater aquaria (21), soil, rhizosphere (30), and lake water. The topology of the tree implies that these two sublineages emerged from the same ancestor after the divergence of the other two sublineages (Fig. 1). Sequences retrieved from different nitrifying bioreactors occur in sublineages I and II, but no obvious correlation between the types or operational modes of the source reactors and the affiliation of the corresponding sequences to one of the sublineages exists. For example, each sublineage contains organisms found in sequencing batch reactors. Furthermore, the sequences retrieved from the same fluidized bed reactor by Schramm et al. (43) are distributed over both sublineages (clones b30, b2, b21, b9, g6, o14, and o9; Fig. 1). Sublineage III consists mainly of sequences found in aquatic samples from Nullarbor caves in Australia (20) (Fig. 1). Sublineage IV hosts the cultivated species N. marina and a related organism detected in the deep sea. It should be noted that eight environmentally retrieved 16S rDNA sequences affiliated with the Nitrospira lineage are possibly not members of the above-described four sublineages. These sequences, however, are incomplete, and their exact phylogenetic affiliation with the other sublineages of the genus, thus, cannot be determined.
Based on the available 16S rRNA sequences, both a Nitrospira phylum-specific oligonucleotide probe and a Nitrospira genus-specific oligonucleotide probe suitable for FISH were developed. Probe S-*-Ntspa-0712-a-A-21 is specific for most known members of the phylum Nitrospira. A probe targeting the phylum Nitrospira has, to our knowledge, not been published before, and thus, probe S-*-Ntspa-0712-a-A-21 extends the current set of bacterial group-specific probes (28, 29, 32, 35, 40), which have found widespread application in microbial ecology (4, 46). Probe S-G-Ntspa-0662-a-A-18 covers the whole genus Nitrospira, including the four sublineages mentioned above. Nontarget organisms without nucleotide mismatches at the probe binding site, or elsewhere on the rRNA, are not known. A similar probe that targets positions 664 to 685 on the 16S rRNA of Nitrospira-like bacteria was described in a previous report (21). This probe is also suitable for in situ hybridization (36), but it does not cover the Nitrospira-like bacteria detected in an industrial activated-sludge plant by Juretschko et al. (22). Several other 16S rRNA-targeted oligonucleotide probes for the in situ detection of Nitrospira-like bacteria were published previously (22, 43). These probes, however, were designed to target only certain Nitrospira-related sequences retrieved from wastewater treatment plants and do not cover the whole genus Nitrospira.
Application of the newly developed Nitrospira-specific probes revealed that the microcolony architecture of the target organisms differed significantly between the activated-sludge and biofilm samples. Constant turbulence, shearing, and the limited sludge age most likely prevent large, morphologically complex cell aggregates of slowly growing nitrite oxidizers in activated-sludge flocs. Accordingly, the Nitrospira microcolonies in the activated-sludge samples were relatively small, compact, and almost spherical. In contrast, the Nitrospira aggregates in the biofilm samples were significantly larger and had a more complex and irregular morphology with internal cavities and a network of cell-free channels. Structurally similar networks formed by channels and cavities between cell aggregates were previously detected by confocal laser scanning microscopy in biofilms of different origins and compositions (23, 31). Channels were also found in microbial granules and have been interpreted as facilitating the exchange of nutrients and gases between the surface and deeper regions (27, 39, 50). This idea was strongly supported by de Beer et al. (15), who showed that O2 concentrations inside a biofilm are significantly higher in voids than in cell clusters. The network of channels and voids we observed within cell clusters of Nitrospira-like bacteria may have similar effects. Staining with fluorescein demonstrated that the channels are permeable for low-molecular-weight, water-soluble substances and thus could facilitate the diffusion of nitrite, gases, and metabolic waste compounds throughout the aggregates. Furthermore, the voids and channels in Nitrospira cell clusters indicate that the modern concept of biofilm architecture as a complex assembly of cell aggregates, organic polymers, and cavities may also apply on a smaller scale to single bacterial microcolonies.
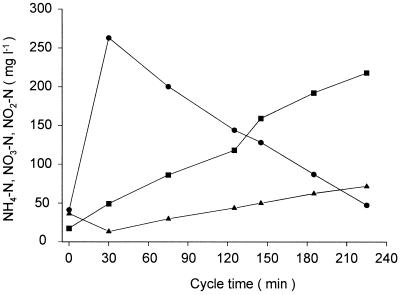
The Nitrospira-specific probes were used together with a previously published probe for in situ detection of Nitrobacter bacteria (48) to investigate the community structure of nitrite-oxidizing bacteria in nitrifying activated-sludge and biofilm samples. While Nitrospira was present in significant amounts in all of the samples analyzed, Nitrobacter cells were detected only in sequencing batch biofilm reactor SBBR 1. Reactor SBBR 1 receives reject water from sludge dewatering, which is particularly rich in ammonia and dissolved salts. Due to the batch performance of the reactor, ammonia, nitrite, and nitrate concentrations vary significantly during an operating cycle (Fig. 5). The repeated, pronounced temporal nitrite concentration shifts within SBBR 1 create an ecological niche for nitrite oxidizers adapted to high nitrite concentrations that does not occur in continuously operated bioreactors. This niche is obviously filled by Nitrobacter sp., which, according to Schramm et al. (42), is a putative r strategist for nitrite and oxygen. In contrast, the Nitrospira-like bacteria were postulated to be K strategists that can grow with lower nitrite (and oxygen) concentrations and can thus coexist with Nitrobacter bacteria in SBBR 1.
FIG. 5.
Concentrations of NH4-N (●), NO3-N (▪), and NO2-N (▴) in reactor SBBR 1 during a representative reactor operating cycle.
The activity and in situ physiology of Nitrospira-like organisms in wastewater treatment plants were studied by a recently developed combination of FISH and MAR (24). The FISH-MAR experiments with radioactive bicarbonate demonstrated that Nitrospira-like bacteria growing in aerated bioreactors fix inorganic carbon. This finding was not unexpected, since pure cultures of N. moscoviensis and N. marina grow autotrophically under laboratory conditions (16, 49), but inorganic carbon fixation of the recently discovered Nitrospira-like bacteria has not been demonstrated previously. Furthermore, uptake of inorganic carbon can be exploited as a method by which to demonstrate the in situ activity of nitrifying bacteria. Such a method is urgently required, since positive FISH signals were also observed for starved or metabolically inhibited nitrifying bacteria (34, 47). In the municipal activated sludge analyzed, more than 75% of the Nitrospira microcolonies detected in situ were unambiguously active, as visualized by the incorporation of inorganic carbon. The high proportion of active Nitrospira cell aggregates in the activated sludge may result from the permanent removal of inactive, nonproliferating cells due to continuous reactor operation.
Qualitative microscopic observation of the samples from reactors SBBR 1 and Biofor 2 after MAR with radioactive pyruvate showed that most of the Nitrospira colonies took up pyruvate. As the parallel experiments with [14C]bicarbonate had shown that most of the Nitrospira colonies in these samples also took up inorganic carbon, this suggests that at least a fraction of these bacteria can grow mixotrophically (defined in this context as the ability to simultaneously incorporate inorganic and organic carbon sources). This ability might contribute to the competitiveness of Nitrospira-like bacteria in wastewater treatment plants. Growth with pyruvate as a carbon source has also been reported for pure cultures of Nitrobacter (6) and N. marina bacteria, which reached the highest cell densities in nitrite media containing pyruvate (49). However, since the FISH-MAR technique cannot be used to monitor the simultaneous uptake of two or more different substrates, the possibility cannot be excluded that different populations of Nitrospira-like bacteria with different abilities to take up inorganic carbon or pyruvate existed in the samples investigated. In contrast to the aerobic incubation experiments, no uptake of organic or inorganic carbon sources by Nitrospira-like bacteria was observed in the absence of oxygen. However, combined FISH and microsensor measurements revealed that high numbers of Nitrospira-like bacterial cells can persist in biofilm zones with low oxygen pressure (17, 36, 41). The ability to switch from aerobic respiration to an anaerobic metabolism should be advantageous in such habitats. According to our experiments, Nitrospira-like bacteria seemingly do not use acetate, propionate, butyrate, or pyruvate in the presence of nitrate and the absence of oxygen. Thus, these results provide no indication that the Nitrospira-like bacteria respired anaerobically with nitrate, but they might employ other strategies to survive periods of limited oxygen availability. Pure-culture experiments indicated that N. marina is obligately aerobic (49) while N. moscoviensis can oxidize H2 with nitrate as the electron acceptor and CO2 as the sole carbon source (16). Hydrogenase activity was not examined in this study, but we do not exclude the possibility that the Nitrospira-like bacteria living in engineered systems can take advantage of this or other alternatives to aerobic nitrite oxidation.
ACKNOWLEDGMENTS
The present research was supported by projects A1 and A2 of Sonderforschungsbereich 411 from the Deutsche Forschungsgemeinschaft (Research Center of Fundamental Studies of Aerobic Biological Wastewater Treatment), the D76 project WA1558/1-1, and by the Danish Technical Research Council (Framework Program, Activity and Diversity in Complex Microbial Systems).
The excellent technical assistance of Beatrix Schlatter, Sibylle Schadhauser, and Jutta Elgner is acknowledged. We thank Eva Arnold for providing concentrations of nitrogen compounds in reactors SBBR 1 and Biofor 2. We also thank Linda Blackall for critically reading the manuscript.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkeman A D C, van Elsas J D, de Bruigin F J, editors. Molecular microbial ecology manual. 3.3.6. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bever J, Stein A, Teichmann H, editors. Weitergehende Abwasserreinigung. Munich, Germany: R. Oldenbourg Verlag; 1995. [Google Scholar]
- 6.Bock E. Growth of Nitrobacter in the presence of organic matter. II. Chemoorganotrophic growth of Nitrobacter agilis. Arch Microbiol. 1976;108:305–312. doi: 10.1007/BF00454857. [DOI] [PubMed] [Google Scholar]
- 7.Bock E, Koops H-P. The genus Nitrobacter and related genera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1992. pp. 2302–2309. [Google Scholar]
- 8.Bock E, Koops H-P, Ahlers B, Harms H. Oxidation of inorganic nitrogen compounds as energy source. In: Balows H, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1992. pp. 414–430. [Google Scholar]
- 9.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 11.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J C, Kim S J. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl Environ Microbiol. 2000;66:956–965. doi: 10.1128/aem.66.3.956-965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csebfalvi B, Gröller E. Interactive volume rendering based on a “bubble” model. Technical report TR-186-2-00-23. Vienna, Austria: Institute of Computer Graphics and Algorithms, Vienna University of Technology; 2000. [Google Scholar]
- 14.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 15.de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transfer. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 16.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium. Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 17.Gieseke A, Purkhold U, Wagner M, Amann R, Schramm A. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl Environ Microbiol. 2001;67:1351–1362. doi: 10.1128/AEM.67.3.1351-1362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guéziec A, Hummel R. Exploiting triangulated surface extraction using tetrahedral decomposition. IEEE Trans Visualization Comput Graphics. 1995;1:328–342. [Google Scholar]
- 19.Henze M, Harremoës P, la Cour Jansen J, Arvin E. Wastewater treatment. 2nd ed. Berlin, Germany: Springer-Verlag; 1997. [Google Scholar]
- 20.Holmes A J, Tujula N A, Holley M, Contos A, James J M, Rogers P, Gillings M R. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ Microbiol. 2001;3:256–264. doi: 10.1046/j.1462-2920.2001.00187.x. [DOI] [PubMed] [Google Scholar]
- 21.Hovanec T A, Taylor L T, Blakis A, Delong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommering-Röser A, Koops H-P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence J R, Korber D R, Hoyle B D, Costerton J W, Caldwell D E. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K-H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 26.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 27.MacLeod F A, Guiot S R, Costerton J W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Environ Microbiol. 1990;56:1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 30.Marilley L, Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. [Google Scholar]
- 31.Massol-Deyá A A, Whallon J, Hickey R F, Tiedje J M. Channel structures in aerobic biofilms of fixed-film reactors treating contaminated groundwater. Appl Environ Microbiol. 1995;61:769–777. doi: 10.1128/aem.61.2.769-777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier H, Amann R, Ludwig W, Schleifer K-H. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 33.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgenroth E, Obermayer A, Arnold E, Brühl A, Wagner M, Wilderer P A. Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci Technol. 2000;41:105–113. [Google Scholar]
- 35.Neef A, Amann R, Schlesner H, Schleifer K-H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 36.Okabe S, Satoh H, Watanabe Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1999;65:3182–3191. doi: 10.1128/aem.65.7.3182-3191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosser J I. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 38.Purkhold U, Pommering-Röser A, Juretschko S, Schmid M C, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson R W, Akin D E, Nordstedt R A, Thomas M V, Aldrich H C. Light and electron microscopic examinations of methane-producing biofilms from anaerobic fixed-bed reactors. Appl Environ Microbiol. 1984;48:127–136. doi: 10.1128/aem.48.1.127-136.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 41.Schramm A, De Beer D, Gieseke A, Amann R. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ Microbiol. 2000;2:680–686. doi: 10.1046/j.1462-2920.2000.00150.x. [DOI] [PubMed] [Google Scholar]
- 42.Schramm A, de Beer D, van den Heuvel J C, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 45.Todorov J R, Chistoserdov A Y, Aller J Y. Molecular analysis of microbial communities in mobile deltaic muds of southeastern Papua New Guinea. FEMS Microbiol Ecol. 2000;33:147–155. doi: 10.1111/j.1574-6941.2000.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 46.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 48.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]
- 49.Watson S W, Bock E, Valois F W, Waterbury J B, Schlosser U. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. [Google Scholar]
- 50.Wu W M, Hickey R F, Zeikus J G. Characterization of metabolic performance of methanogenic granules treating brewery wastewater: role of sulfate-reducing bacteria. Appl Environ Microbiol. 1991;57:3438–3449. doi: 10.1128/aem.57.12.3438-3449.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]