Abstract
A series of novel pyrazolinone chalcones 3–9 have been synthesized through the condensation of azo pyrazolinone derivatives with various aromatic aldehydes. Spectroscopic techniques and elemental analysis have both corroborated this. Furthermore, all compounds were screened in silico for their ability to inhibit cancer proliferation and metastasis by targeting the PI3K/Akt signaling pathway. This inhibitory pathway might be an efficient approach for the death of cancer cells, angiogenesis, and metastasis prevention. Our results indicated that only compound 6b was the top-ranked. It demonstrated the highest binding energies of −11.1 and −10.7 kcal/mol against the target proteins PI3K and Akt, respectively; thus, it was chosen for in vitro studies. Compound 6b exhibited the most effective cytotoxic impact against the Caco cell line with IC50 of 23.34 ± 0.14 μM. Furthermore, it showed significant inhibition of PI3K/Akt proteins and oxidative stress, leading to elevated Bax and p53 expression, reduced Bcl–2 expression, and triggered cell cycle arrest at the sub-G0/G1 phase. Additionally, it showed significant downregulation of the Raf-1 gene, leading to ERK1/2 protein inhibition. These findings demonstrate that compound 6b obeyed Lipinski’s rule of five and might be used as a favored scaffold for cancer treatment by inhibiting proliferation and metastasis via inhibition of the PI3K/Akt and Raf-1/ERK1/2 signaling pathways.
Introduction
Cancer is a potentially fatal disease that affects people worldwide. The environmental variables account for 90–95% of all cancer cases, with the remaining cases 5–10% attributed to inherited genetic factors. Moreover, surgery, radiation, and chemotherapy were the mainstays of cancer treatment in the past. Even though chemotherapy is the most often used treatment, several chemotherapeutic drugs have harmful side effects, including the indiscriminate death of normal cells and chemotherapeutic resistance.1 As a result, a constant quest for alternative, targeted, novel, efficacious, and less toxic anticancer agents that inhibit metabolic target proteins are urgently needed. Therefore, designing the bioenergetic drugs that disrupt the metabolic route essential for cancer cell survival, metastasis, and proliferation is of critical importance in this context.2
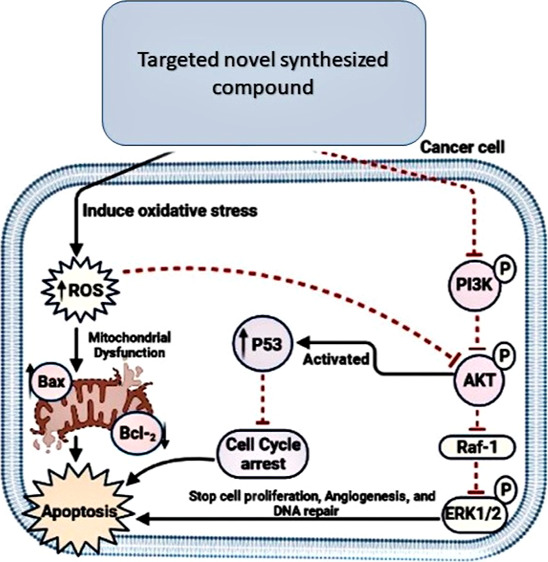
Akt is a proto-oncogenic serine–threonine kinase that plays a role in cell proliferation, glucose metabolism, cell migration, and apoptosis. Akt is activated in tumor cells via phosphorylation, which arises when a PI3K-phosphorylated phosphoinositide (PI) termed PIP3 attaches to the homology Akt domain, followed by translocation to the plasma membrane and phosphorylation by PDK1 and PDK2 at two phosphorylation Ser473 and Thr308 sites. As a result, direct PI3K/Akt degradation was discovered to inhibit Akt activity and induce apoptosis.3 For several years, inhibition of Akt has been regarded as a promising therapeutic approach in oncology as it leads to the inhibition of Raf-1, which inhibits the proliferating and angiogenesis proteins, MEK and ERK1/2. Moreover, oxidative stress inhibits the PI3K/Akt signaling pathway and promotes reactive oxygen species (ROS) generation, resulting in Bax and p53 activation, cell cycle arrest, and cancer cell apoptosis.4 Many efforts have been undertaken to design new anticancer agents that are both selective and effective to inhibit PI3K/Akt and Raf-1/ERK1/2 as they can worsen the proliferation of cancer cells, leading to global programming of cancer cell death and inducing mitochondrial dysfunction because of oxidative stress5,6 (Figure 1).
Figure 1.
Schematic diagram of the design strategy signaling pathways.
Chalcones possess many biological activities, including antibacterial, antifungal, antimalarial, anticonvulsant, and anticancer agents.7 Furthermore, these compounds are of high interest because of their use in synthesizing many biologically active heterocycles such as azepines, pyrazolines, and flavones.8−11
Pyrazolinones have a wide range of biological effects, including anti-inflammatory, antibacterial, antifungal, analgesic, antidiabetic, antioxidant, and anticancer properties. One of the most effective drug-bearing pyrazoline moieties is Axitinib, which is used as an anticancer drug. To create effective anticancer medicines, pyrazoline is also hybridized with other nitrogen-, sulfur-, and oxygen-containing heterocyclic scaffolds such as quinoline, indole, oxazole, and thiazole.12−14
This project aimed to design and synthesize novel anticancer scaffolds of pyrazolinone chalcones due to the importance of pyrazolinones and chalcones in the medical area. Computer-based docking experiments were performed on the produced compounds to study the binding mechanism with the active sites of the target enzymes. In addition, in silico physicochemical and pharmacokinetic properties were carried out to estimate the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of the compounds as well as investigate the structure–activity relationship (SAR). Ultimately, the chosen compound was investigated in vitro to confirm its inhibitory efficacy toward cancer cell proliferation and metastasis by targeting PI3K/Akt and Raf-1/ERK1/2 signaling pathway inhibition.
Materials and Methods
Chemicals and Drugs
Ethyl acetoacetate (EAA), phenylhydrazine, 2,4-dinitrophenylhydrazine (DNP), hydrazine hydrate, p-aminoacetophenone, sodium nitrite, benzaldehyde, 3,5-dimethoxybenzaldehyde, 4-(N,N-dimethylamino)benzaldehyde, 4-hydroxybenzaldehyde, 2-nitrobenzaldehyde, cinnamaldehyde, 4-(N,N-dimethylamino)cinnamaldehyde, sodium hydroxide, ethyl alcohol, glacial acetic acid, trichloroacetic acid (TCA), sodium pyrophosphate, reduced glutathione (GSH), and thiobarbituric acid (TBA) were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Doxorubicin HCl injection, USP, was purchased from Pfizer injectables.
General Information
Thin-layer chromatography monitored the reactions performed on precoated Merck Kieselgel 60 F254 plates (EMD Millipore Corporation, Billerica, MA, USA). A PerkinElmer 1420 spectrophotometer (Waltham, MA, USA) was used to record infrared spectra at the Central Laboratory of Tanta University. The KBr disc technique was used to obtain the spectra. The samples were mounted on a sample holder with a big cavity after drying. The open capillary method was used to estimate melting points, which were calculated using the Gallenkamp melting point and reported uncorrected. Electron impact mass spectrometry (EIMS) was used to measure mass spectra at 70 eV at Al-Azhar University’s Regional Center for Mycology and Biotechnology. A PerkinElmer 240 CHN Elemental Analyzer was used to undertake elemental analysis of substances at Cairo University’s Microanalytical Center. The 1H NMR spectra were recorded on a Bruker AC spectrometer (400 MHz) and 13C NMR (100 MHz at 25 °C in DMSO-d6 with tetramethylsilane as an internal standard. The chemical shifts for 1H NMR are reported in ppm from tetramethylsilane (0 ppm) or referenced to the solvent (DMSO-d6, δ2.50). Chemical shifts (δ) for 13C NMR spectra are referenced to the signals for residual deuterated solvents (DMSO-d6, 37.5). Multiplicities are reported by the following abbreviations: s (singlet), d (doublet), m (multiplet).
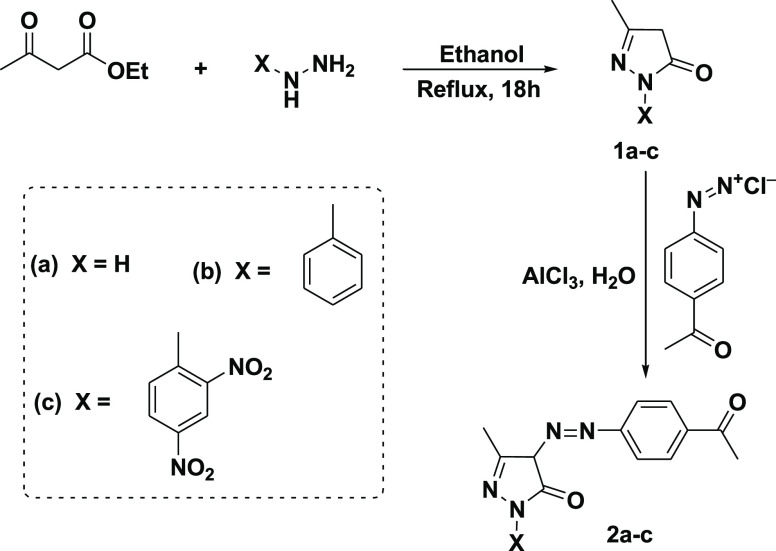
Synthesis of Pyrazolinone Derivatives (1a–c)
A mixture of EAA (10 mmol, 1.28 mL) and hydrazine derivatives (10 mmol) in 30 mL of ethanol was refluxed for 18 h, then cooled, filtered, and recrystallized from ethanol to give compounds 1a–c as described by Alharthy.15
Synthesis of Azo Pyrazolinone Derivatives (2a–c)
Azo pyrazolinone derivatives (2a–c) were prepared by coupling the pyrazolinone (1a–c) with a freshly prepared solution of p-acetyl phenyl diazoniumchloride in the presence of aluminum chloride (AlCl3) as described by Khalil.16
4-((4-Acetylphenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (2a)
The structure of 2a was confirmed as described earlier.16
4-((4-Acetylphenyl) diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (2b)
Reddish-brown powder; yield 91%; mp 129 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.00–7.90 (m, 9H, Ar-H), 2.85 (s, 3H, CH3–CO), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 198.00, 168.30, 155.9, 155.60, 140.70, 134.00, 129.60, 129.30, 124.70, 122.80, 121.90, 62.20, 29.60, 19.80; IR (KBr) ν: 1500–1560 (N=N), 1670 (CO); EIMS m/z: 320.15 [M]+ (C18H16N4O2, calcd 320.35); Anal. Calcd for C18H16N4O2 (320.35): C, 67.49%; H, 5.03%; N, 17.49%. Found: C, 67.17%; H, 4.91%; N, 17.11%.
4-((4-Acetylphenyl)diazenyl)-1-(2,4-dinitrophenyl)-3-methyl-1H-pyrazol-5(4H)-one (2c)
Reddish-brown powder; yield 89%; mp 89 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.40–9.11 (m, 7H, Ar-H), 2.85 (s, 3H, CH3–CO), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 198.00, 168.30, 155.9, 155.60, 150.20, 142.30, 141.50, 134.00, 129.60, 127.7, 123.70, 122.80, 111.20, 62.20, 29.60, 19.80; IR (KBr) ν: 1509–1600 (N=N), 1673 (CO); EIMS m/z: 410.23 [M]+ (C18H14N6O6, calcd 410.34); Anal. Calcd for C18H14N6O6 (410.34): C, 52.69%; H, 3.44%; N, 20.48%. Found: C, 52.27%; H, 3.22%; N, 20.18%.
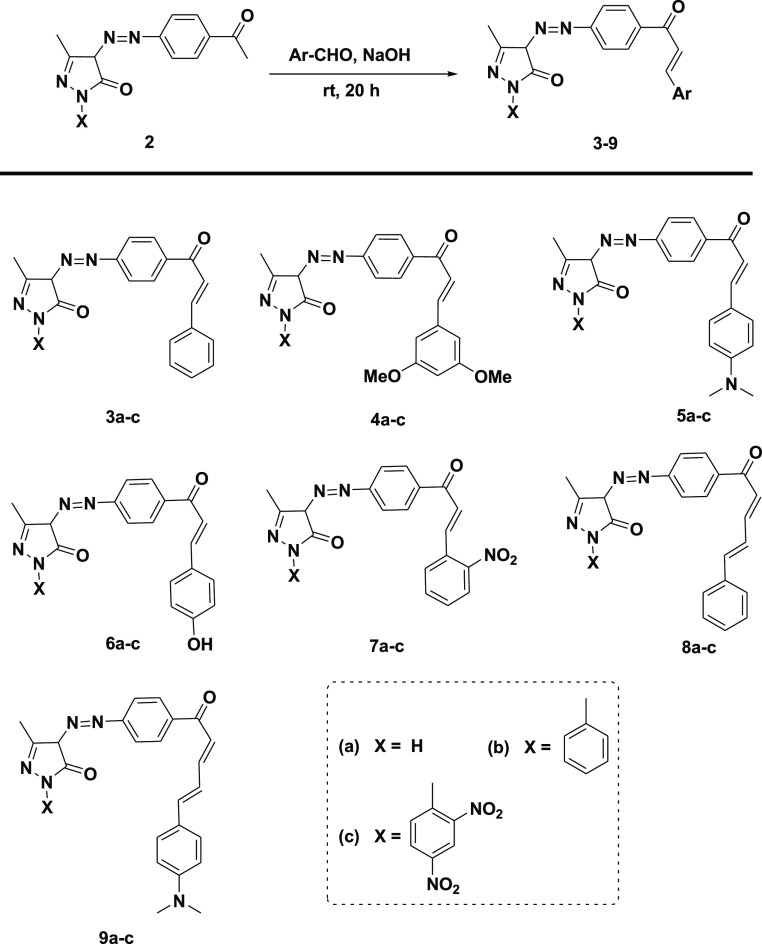
General Procedure for the Synthesis of Pyrazolinone Chalcones (3–9)
In a 50 mL conical flask, a mixture of aromatic aldehyde (1 mmol), azo pyrazolinone derivatives (2a–c, 1 mmol), sodium hydroxide (0.08 g, 2 mmol), and ethanol (10.0 mL) was stirred at room temperature for 20 h (TLC control, petroleum ether/ethyl acetate: 8:2). Then the reaction mixture was poured into ice water, filtered off, and dried.
(E)-4-((4-Cinnamoylphenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (3a)
Reddish-brown powder; yield 91%; mp 174 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.00 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 6.90 (s, 1H, NH), 7.10–7.80 (m, 9H, Ar-H), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 157.00, 155.90, 145.50, 135.50, 135.20, 130.70, 129.00, 128.00, 126.70, 123.40, 121.70, 64.80, 19.50; IR (KBr) ν: 1560 (N=N), 1677 (CO); EIMS m/z: 332.16 [M]+ (C19H16N4O2, calcd 332.36); Anal. Calcd for C19H16N4O2 (332.36): C, 68.66%; H, 4.85%; N, 16.86%. Found: C, 68.16%; H, 4.65%; N, 16.46%.
(E)-4-((4-Cinnamoylphenyl)diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (3b)
Reddish-brown powder; yield 92%; mp 112 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.10 (d, 1H, CH-Ph), 7.65 (d, 1H, CH–CO), 7.00–7.80 (m, 14H, Ar-H), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 145.50, 140.70, 135.50, 135.20, 132.40, 130.70, 129.30, 129.00, 128.00, 126.70, 124.70, 123.40, 121.90, 121.70, 62.20, 19.80; IR (KBr) ν: 1552 (N=N), 1657 (CO); EIMS m/z: 408.28 [M]+ (C25H20N4O2, calcd 408.45); Anal. Calcd for C25H20N4O2 (408.45): C, 73.51%; H, 4.94%; N, 13.72%. Found: C, 73.11%; H, 4.84%; N, 13.44%.
(E)-4-((4-Cinnamoylphenyl)diazenyl)-1-(2,4-dinitrophenyl)-3-methyl-1H-pyrazol-5(4H)-one (3c)
Brown powder; yield 91%; mp 108 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.10–9.05 (m, 12H, Ar-H), 7.90 (d, 1H, CH-Ph), 7.50 (d, 1H, CH–CO), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 145.50, 145.20, 142.30, 141.50, 135.50, 135.20, 130.70, 129.00, 128.00, 126.70, 123.70, 123.40, 121.70, 111.20, 62.20, 19.80; IR (KBr) ν: 1570 (N=N), 1666 (CO); EIMS m/z: 498.11 [M]+ (C25H18N6O6, calcd 498.45); Anal. Calcd for C25H18N6O6 (498.45): C, 60.24%; H, 3.64%; N, 16.86%. Found: C, 59.98%; H, 3.48%; N, 16.56%.
(E)-4-((4-(3-(3,5-Dimethoxyphenyl)acryloyl)phenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (4a)
Reddish-brown powder; yield 90%; mp 155 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 7.00 (s, 1H, NH), 6.10–7.80 (m, 7H, Ar-H), 3.70 (s, 6H, 2CH3–O), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 161.90, 157.00, 155.90, 145.50, 137.50, 135.20, 130.70, 123.40, 121.70, 103.20, 99.90, 70.70, 56.20, 19.50; IR (KBr) ν: 1451 (N=N), 1598 (CO), 2931 (OMe); EIMS m/z: 392.19 [M]+ (C21H20N4O4, calcd 392.41); Anal. Calcd for C21H20N4O4 (392.41): C, 64.28%; H, 5.14%; N, 14.28%. Found: C, 63.98%; H, 4.98%; N, 14.08%.
(E)-4-((4-(3-(3,5-Dimethoxyphenyl)acryloyl)phenyl)diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (4b)
Reddish-brown powder; yield 89%; mp 172 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 6.30–7.80 (m, 12H, Ar-H), 3.70 (s, 6H, 2CH3–O), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 161.90, 157.00, 155.90, 145.50, 140.60, 137.50, 135.20, 130.70, 129.30, 124.70, 123.40, 121.90, 121.70, 103.20, 99.90, 62.20, 56.20, 19.80; IR (KBr) ν: 1595 (N=N), 1655 (CO), 2837 (OMe); EIMS m/z: 468.37 [M]+ (C27H24N4O4, calcd 468.50); Anal. Calcd for C27H24N4O4 (468.50): C, 69.22%; H, 5.16%; N, 11.96%. Found: C, 68.98%; H, 5.02%; N, 11.86%.
(E)-4-((4-(3-(3,5-Dimethoxyphenyl)acryloyl)phenyl)diazenyl)-1-(2,4-dinitrophenyl)-3-methyl-1H-pyrazol-5(4H)-one (4c)
Brown powder; yield 90%; mp 154 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.95 (d, 1H, CH-Ph), 7.65 (d, 1H, CH–CO), 6.30–9.10 (m, 10H, Ar-H), 3.70 (s, 6H, 2CH3–O), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 161.90, 155.90, 145.50, 145.20,142.30, 137.50, 135.20, 127.70, 123.70, 121.70, 103.20, 99.90, 62.20, 56.20, 19.80; IR (KBr) ν: 1580 (N=N), 1670 (CO), 2857 (OMe); EIMS m/z: 558.38 [M]+ (C27H22N6O4, calcd 558.50); Anal. Calcd for C27H22N6O4 (558.50): C, 58.06%; H, 3.97%; N, 15.05%. Found: C, 57.86%; H, 3.77%; N, 14.91%.
(E)-4-((4-(3-(4-(Dimethylamino)phenyl)acryloyl)phenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (5a)
Reddish-brown powder; yield 91%; mp 149 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 7.00 (s, 1H, NH), 6.50–7.80 (m, 8H, Ar-H), 2.85 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 157.00, 155.90, 149.10, 145.50, 135.20, 130.70, 127.70, 125.00, 123.40, 121.70, 114.50, 64.80, 40.60, 19.50; IR (KBr) ν: 1580 (N=N), 1670 (CO), 1179 (C–N); EIMS m/z: 375.24 [M]+ (C21H21N5O2, calcd 375.42); Anal. Calcd for C21H21N5O2 (375.42): C, 67.18%; H, 5.64%; N, 18.65%. Found: C, 67.08%; H, 5.48%; N, 18.53%.
(E)-4-((4-(3-(4-(Dimethylamino)phenyl)acryloyl)phenyl)diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (5b)
Reddish-brown powder; yield 91%; mp 214 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 6.50–7.80 (m, 13H, Ar-H), 2.90 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 149.10, 145.50, 140.70, 135.20, 130.70, 129.30, 127.60, 125.00, 124.20, 123.40, 121.90, 121.70, 114.50, 62.20, 40.60, 19.80; IR (KBr) ν: 1562 (N=N), 1653 (CO), 1159 (C–N); EIMS m/z: 451.32 [M]+ (C27H25N5O2, calcd 451.52); Anal. Calcd for C27H25N5O2 (451.52): C, 71.82%; H, 5.58%; N, 15.51%. Found: C, 71.62%; H, 5.46%; N, 15.31%.
(E)-4-((4-(3-(4-(Dimethylamino)phenyl)acryloyl)phenyl)diazenyl)-1-(2,4-dinitrophenyl)-3-methyl-1H-pyrazol-5(4H)-one (5c)
Brown powder; yield 92%; mp 134 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.95 (d, 1H, CH-Ph), 7.65 (d, 1H, CH–CO), 6.50–9.10 (m, 11H, Ar-H), 2.85 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 149.10, 145.20, 145.50, 142.30, 141.50, 135.20, 130.70, 127.70, 127.60, 125.00, 123.70, 123.40, 121.70, 119.20, 114.50, 62.20, 40.60, 19.80; IR (KBr) ν: 1570 (N=N), 1655 (CO), 1165 (C–N); EIMS m/z: 541.07 [M]+ (C27H23N7O6, calcd 541.17); Anal. Calcd for C27H23N7O6 (541.17): C, 59.89%; H, 4.28%; N, 18.11%. Found: C, 59.49%; H, 4.16%; N, 17.95%.
(E)-4-((4-(3-(4-Hydroxyphenyl)acryloyl)phenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (6a)
Reddish-brown powder; yield 92%; mp 202 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 7.00 (s, 1H, NH), 6.50–7.80 (m, 8H, Ar-H), 5.00 (s, 1H, OH), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 158.00, 157.00, 155.90, 145.50, 135.20, 130.70, 128.10, 123.40, 121.70, 116.10, 64.80, 19.50; IR (KBr) ν: 1385 (N=N), 1641 (CO), 3457 (OH); EIMS m/z: 348.23 [M]+ (C19H16N4O3, calcd 348.36); Anal. Calcd for C19H16N4O3 (348.36): C, 65.51%; H, 4.63%; N, 16.08%. Found: C, 65.33%; H, 4.43%; N, 15.94%.
(E)-4-((4-(3-(4-Hydroxyphenyl)acryloyl)phenyl)diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (6b)
Reddish-brown powder; yield 93%; mp 193 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 6.50–7.80 (m, 13H, Ar-H), 5.00 (s, 1H, OH), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 158.00, 157.00, 155.90, 145.50, 140.70, 135.20, 130.70, 129.30, 124.70, 121.90, 116.10, 62.20, 19.80; IR (KBr) ν: 1559 (N=N), 1664 (CO), 3166 (OH); EIMS m/z: 424.32 [M]+ (C25H20N4O3, calcd 424.45); Anal. Calcd for C25H20N4O3 (424.45): C, 70.74%; H, 4.75%; N, 13.20%. Found: C, 70.44%; H, 4.55%; N, 13.08%.
(E)-1-(2,4-Dinitrophenyl)-4-((4-(3-(4-hydroxyphenyl)acryloyl)phenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (6c)
Brown powder; yield 92%; mp 124 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.95 (d, 1H, CH-Ph), 7.65 (d, 1H, CH–CO), 6.50–9.10 (m, 11H, Ar-H), 5.00 (s, 1H, OH), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 158.00, 157.00, 155.90, 145.50, 145.20, 142.30, 141.50, 135.20, 130.70, 128.10, 127.70, 123.70, 123.40, 121.70, 119.20, 116.10, 62.20, 19.80; IR (KBr) ν: 1560 (N=N), 1671 (CO), 3240 (OH); EIMS m/z: 514.25 [M]+ (C25H18N6O7, calcd 514.45); Anal. Calcd for C25H18N6O7 (514.45): C, 58.37%; H, 3.53%; N, 16.34%. Found: C, 58.17%; H, 3.23%; N, 16.22%.
(E)-3-Methyl-4-((4-(3-(2-nitrophenyl)acryloyl)phenyl)diazenyl)-1H-pyrazol-5(4H)-one (7a)
Reddish-brown powder; yield 93%; mp 175 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.50 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 7.00 (s, 1H, NH), 7.40–8.00 (m, 8H, Ar-H), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 157.00, 155.90, 146.40, 135.20, 135.10, 130.70, 130.30, 129.20, 127.50, 123.40, 121.70, 121.30, 64.80, 19.50; IR (KBr) ν: 1580 (N=N), 1690 (CO), 1490–1520 (N–O); EIMS m/z: 377.25 [M]+ (C19H15N5O4, calcd 377.35); Anal. Calcd for C19H15N5O4 (377.35): C, 60.47%; H, 4.01%; N, 18.56%. Found: C, 60.27%; H, 3.93%; N, 18.44%.
(E)-3-Methyl-4-((4-(3-(2-nitrophenyl)acryloyl)phenyl)diazenyl)-1-phenyl-1H-pyrazol-5(4H)-one (7b)
Reddish-brown powder; yield 91%; mp 135 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.50 (d, 1H, CH-Ph), 7.60 (d, 1H, CH–CO), 7.40–8.00 (m, 13H, Ar-H), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 146.40, 145.50, 140.70, 135.20, 135.10, 130.70, 130.30, 129.30, 129.20, 127.50, 124.70, 123.40, 121.90, 121.70, 121.30, 62.20, 19.80; IR (KBr) ν: 1570 (N=N), 1680 (CO), 1480–1515 (N–O); EIMS m/z: 453.22 [M]+ (C25H19N5O4, calcd 453.45); Anal. Calcd for C25H19N5O4 (453.45): C, 66.22%; H, 4.22%; N, 15.44%. Found: C, 66.10%; H, 4.08%; N, 15.26%.
(E)-1-(2,4-Dinitrophenyl)-3-methyl-4-((4-(3-(2-nitrophenyl)acryloyl)phenyl)diazenyl)-1H-pyrazol-5(4H)-one (7c)
Brown powder; yield 90%; mp 145 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.50 (d, 1H, CH-Ph), 7.65 (d, 1H, CH–CO), 7.40–9.10 (m, 11H, Ar-H), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 146.40, 145.50, 145.20, 142.30, 141.50, 135.20, 135.10, 130.70, 130.30, 129.20, 127.70, 127.60, 123.70, 123.40, 121.70, 121.30, 119.20, 62.20, 19.80; IR (KBr) ν: 1560 (N=N), 1670 (CO), 1470–1530 (N–O); EIMS m/z: 543.28 [M]+ (C25H17N7O8, calcd 543.44); Anal. Calcd for C25H17N7O8 (543.44): C, 55.25%; H, 3.15%; N, 18.04%. Found: C, 55.13%; H, 3.05%; N, 17.94%.
3-Methyl-4-((4-((2E,4E)-5-phenylpenta-2,4-dienoyl)phenyl)diazenyl)-1H-pyrazol-5(4H)-One (8a)
Reddish-brown powder; yield 92%; mp 182 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.90 (t, 1H, CH=CH–CO), 7.30 (d, 1H, CH=CH–CO), 7.00 (s, 1H, NH), 7.10–7.70 (m, 9H, Ar-H), 6.85 (t, 1H, CH=CH–Ph), 6.70 (d, 1H, CH=CH–Ph), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 157.00, 155.90, 144.70, 135.50, 135.20, 131.50, 130.70, 129.30, 129.00, 128.30, 126.70, 125.60, 123.40, 70.70, 19.50; IR (KBr) ν: 1552 (N=N), 1668 (CO), 2930 (=C–H); EIMS m/z: 358.29 [M]+ (C21H18N4O2, calcd 358.39); Anal. Calcd for C21H18N4O2 (358.39): C, 70.38%; H, 5.06%; N, 15.63%. Found: C, 70.22%; H, 4.96%; N, 15.45%.
3-Methyl-1-phenyl-4-((4-((2E,4E)-5-phenylpenta-2,4-dienoyl)phenyl)diazenyl)-1H-pyrazol-5(4H)-one (8b)
Reddish-brown powder; yield 92%; mp 136 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.90 (t, 1H, CH=CH–CO), 7.20 (d, 1H, CH=CH–CO), 7.00–7.75 (m, 14H, Ar-H), 6.85 (t, 1H, CH=CH–Ph), 6.70 (d, 1H, CH=CH–Ph), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 144.70, 140.70, 135.50, 135.20, 131.50, 130.70, 129.30, 129.00, 128.30, 126.70, 125.60, 124.70, 123.40, 121.90, 68.20, 19.80; IR (KBr) ν: 1572 (N=N), 1688 (CO), 2920 (=C–H); EIMS m/z: 434.29 [M]+ (C27H22N4O2, calcd 434.49); Anal. Calcd for C27H22N4O2 (434.49): C, 74.64%; H, 5.10%; N, 12.89%. Found: C, 74.44%; H, 4.98%; N, 12.77%.
1-(2,4-Dinitrophenyl)-3-methyl-4-((4-((2E,4E)-5-phenylpenta-2,4-dienoyl)phenyl)diazenyl)-1H-pyrazol-5(4H)-one (8c)
Reddish-brown powder; yield 94%; mp 117 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.90 (t, 1H, CH=CH–CO), 7.20 (d, 1H, CH=CH–CO), 7.10–9.10 (m, 12H, Ar-H), 6.85 (t, 1H, CH=CH–Ph), 6.67 (d, 1H, CH=CH–Ph), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 145.20, 144.70, 142.30, 141.50, 135.50, 135.20, 131.50, 130.70, 129.30, 129.00, 128.30, 127.70, 126.70, 125.60, 123.70, 123.40, 119.20, 68.20, 19.80; IR (KBr) ν: 1562 (N=N), 1678 (CO), 2918 (=C–H); EIMS m/z: 524.32 [M]+ (C27H20N6O6, calcd 524.48); Anal. Calcd for C27H20N6O6 (524.48): C, 61.83%; H, 3.84%; N, 16.02%. Found: C, 61.67%; H, 3.72%; N, 15.92%.
4-((4-((2E,4E)-5-(4-(Dimethylamino)phenyl)penta-2,4-dienoyl)phenyl)diazenyl)-3-methyl-1H-pyrazol-5(4H)-one (9a)
Reddish-brown powder; yield 89%; mp 165 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.05 (t, 1H, CH=CH–CO), 7.65 (d, 1H, CH=CH–CO), 7.00 (s, 1H, NH), 6.35–7.85 (m, 8H, Ar-H), 6.80 (t, 1H, CH=CH–Ph), 6.60 (d, 1H, CH=CH–Ph), 2.85 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 173.30, 157.00, 155.90, 149.20, 144.70, 135.20, 131.50, 130.70, 127.60, 125.60, 125.00, 123.40, 114.50, 70.80, 40.60, 19.50; IR (KBr) ν: 1570 (N=N), 1660 (CO), 1189 (C–N), 2915 (=C–H); EIMS m/z: 401.26 [M]+ (C23H23N5O2, calcd 401.46); Anal. Calcd for C23H23N5O2 (401.46): C, 68.81%; H, 5.77%; N, 17.44%. Found: C, 68.65%; H, 5.55%; N, 17.24%.
4-((4-((2E,4E)-5-(4-(Dimethylamino)phenyl)penta-2,4-dienoyl)phenyl)diazenyl)-3-methyl-1-phenyl-1H-pyrazol-5(4H)-one (9b)
Reddish-brown powder; yield 88%; mp 209 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.00 (t, 1H, CH=CH–CO), 7.60 (d, 1H, CH=CH–CO), 6.50–7.80 (m, 13H, Ar-H), 6.80 (t, 1H, CH=CH–Ph), 6.60 (d, 1H, CH=CH–Ph), 2.85 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 149.10, 144.70, 140.70, 135.20, 131.50, 130.70, 129.30, 127.70, 125.60, 125.00, 124.70, 123.40, 121.90, 114.50, 68.20, 40.60, 19.80; IR (KBr) ν: 1560 (N=N), 1665 (CO), 1182 (C–N), 2925 (=C–H); EIMS m/z: 477.38 [M]+ (C29H27N5O2, calcd 477.56); Anal. Calcd for C29H27N5O2 (477.56): C, 72.94%; H, 5.70%; N, 14.66%. Found: C, 72.78%; H, 5.52%; N, 14.48%.
4-((4-((2E,4E)-5-(4-(Dimethylamino)phenyl)penta-2,4-dienoyl)phenyl)diazenyl)-1-(2,4-dinitrophenyl)-3-methyl-1H-pyrazol-5(4H)-one (9c)
Reddish-brown powder; yield 88%; mp over 300 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.95 (t, 1H, CH=CH–CO), 7.65 (d, 1H, CH=CH–CO), 6.50–9.10 (m, 11H, Ar-H), 6.90 (t, 1H, CH=CH–Ph), 6.70 (d, 1H, CH=CH–Ph), 2.90 (s, 6H, (CH3)2–N), 2.35 (s, 1H, CH), 1.20 (s, 3H, CH3); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 190.00, 168.30, 157.00, 155.90, 149.10, 145.20, 144.70, 142.30, 141.50, 135.20, 131.50, 130.70, 129.30, 127.70, 127.60, 125.60, 125.00, 123.70, 123.40, 119.20, 114.50, 68.20, 40.60, 19.80; IR (KBr) ν: 1565 (N=N), 1680 (CO), 1179 (C–N), 2935 (=C–H); EIMS m/z: 567.39 [M]+ (C29H25N7O6, calcd 567.55); Anal. Calcd for C29H25N7O6 (567.55): C, 61.37%; H, 4.44%; N, 17.28%. Found: C, 61.19%; H, 4.26%; N, 17.12%.
In Silico Study
In the current research, molecular docking studies17−24 were performed to explore the binding modes of the ligand molecules toward the target proteins PI3K and Akt. The crystal structures of the targets were retrieved from the RCSB protein data bank.25 The target files were optimized by removing the co-crystalized ligands, heteroatoms, and water molecules. In addition, their energies were minimized using CHARM Force Field26 in Discovery Studio 3.5 Visualizer. Further, the 2D structures of the prepared analogues were generated in cdx format (2D structures) using ChemDraw Ultra 8.0 and then converted to sdf files (3D structures) using the Open Babel GUI 2.4.1 tool.27 Furthermore, the UFF force field28 in the PyRx tool was used to minimize their energies. An in-house library of 12 ligand molecules was generated for the docking. The in silico docking technique was performed using PyRx—a virtual screening tool.29 Grid maps of 25 × 25 × 25 Å3 were generated around the active site region of the target proteins, resulting in nine conformers for each docked molecule, and the minimum binding energy was selected for further study. The 2D and 3D representations of docking results were visualized using Discovery Studio 3.5. Finally, the drug-like properties of the newly prepared molecules were calculated using mol inspiration, Swiss ADME, and Admet SAR web tools.
Anticancer Investigations on the Expected Compound (In Vitro)
The anticancer impact was studied using the tetrazolium 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay on the most effective compound resulting from the in silico studies and then submitted to further analyses.
Cell Line Maintenance and Treatment
The lung cancer cell line (A549), triple-negative breast cancer cell line (MDA-231), pancreatic cancer cell line (PCL), estrogen receptor-positive breast cancer cell line (MCF-7), colon cancer cell line (Caco), and WISH normal cell line were seeded with (1 × 104 cells/well) separately using complete media containing (Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 1% penicillin/streptomycin) in a 5% CO2 incubator and a 95% humidified environment at 37 °C. All cell lines were provided by the Center of Excellence for Research in Regenerative Medicine and Its Applications, Alexandria University, Egypt. The cell lines were incubated with the selected compound at several concentrations (0–200 μM) and doxorubicin (DOX) as a standard chemotherapeutic drug (0–100 μM) for 48 h, and then, the viability of the cells was determined using the MTT assay (Gibco-BRL, New York, NY, USA).30,31
Cell Morphology Study
Briefly, 1 × 105 of the Caco cell line was seeded in a six-well plate, incubated for 24 h, and then treated dose-dependently with 1/4IC50, 1/2IC50, and IC50 of the selected compound. After 48 h incubation, morphological alterations of treated and untreated cells were evaluated and captured using an inverted light microscope (Olympus, USA).
Cell Cycle Examination
Flow cytometry was used to analyze cell cycle phases using an Accuri C6 flow cytometer (Becton Dickinson BD, USA) on Caco cells 1 × 105 that were trypsinized, centrifuged at 5000 rpm, then washed with 1× cold phosphate-buffered saline (PBS), and fixed with cold absolute ethanol as described by Noser et al. and Darzynkiewicz et al.31,32
qPCR Assessment
The Caco 1 × 105 control and treated cells were trypsinized, centrifuged at 4500 rpm, and washed with 1× PBS. Then the pelleted Caco cells were subjected to RNA extraction and transcription to cDNA as described by Kvastad et al.33 The expression of Raf-1, p53, Bax, and Bcl–2 mRNA was measured using Applied qPCR Biosystems (Foster City, USA) on treated and control Caco cells according to Livak and Schmittgen.34 The primer sequences were designed using primer 3plus as in Table 1.
Table 1. Primer Sequences Used in qRT-PCR.
| gene | forward primer (/5---/3) | reverse primer (/5---/3) |
|---|---|---|
| Raf-1 | GCAGGATAACAACCCATTC | GGTCAGCGTGCAAGCATT |
| P53 | TAACAGTTCCTGCATGGGCGGC | AGGACAGGCACAAACACGCACC |
| Bax | GGCTGGACACTGGACTTCCT | GGTGAGGACTCCAGCCACAA |
| Bcl–2 | TTCGCAGAGATGTCCAGTCA | TTCAGAGACAGCCAGGAGAA |
| GAPDH | TGTGTCCGTCGTGGATCTGA | CCTGCTTCACCACCTTCTTGA |
Biomarker Estimation of Antioxidant/Oxidative Stress
The 1 × 105 Caco cells were treated for 48 h. Subsequently, the cells were scraped, and pelleted cells were washed twice with 1× cold PBS. The scraped pelleted cells were incubated in the lysis buffer as described by Noser et al.31 to measure the levels of malondialdehyde (MDA) and the activity of reduced glutathione (GSH).35 The protein content was determined using Bradford.36
Western Blot Analysis
The method of Mruk and Cheng37 was used for immunoblotting. Proteins are removed from Caco control and treated cells using cold RIPA lysis buffer and quantified using Bradford.36 Equal amounts of proteins (20 μg) were separated and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking the membrane, the primary antibodies as phospho-PI3K (ab182651), phospho-Akt (ab81283), and phospho-ERK1/2 (ab214362) were added and incubated with it. Then, the primary antibodies were removed, carefully washed several times, and incubated with the secondary antibody horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H + L) (ab205718). The bands were visualized and normalized with β-actin as described by Noser et al.31
Statistical Analysis
The experimental results are presented as mean ± SE. GraphPad Prism 6 software was used to determine the significance of differences between the control and treated groups using one and two-way ANOVA.
Results and Discussion
Chemistry of the Synthesized Compounds
The reaction of EAA with hydrazine derivatives led to the formation of pyrazolinone derivatives (1a–c) with high yields as described by Alharthy.15p-Acetyl phenyl diazonium chloride was coupled with the pyrazolinone derivatives (1a–c) to give the azo pyrazolinone derivatives (2a–c) with 92, 91, and 89% yields, respectively, as described in Scheme 1.
Scheme 1. Synthesis Pathway of Compounds 1 and 2.
The chalcones (3–9) were synthesized from the reaction of azo pyrazolinone derivatives (2a–c) with different substituted aromatic aldehydes in a basic medium as described in Scheme 2.
Scheme 2. Synthesis Pathway of Compounds 3–9.
All synthesized compounds 2–9 were characterized using different spectroscopic techniques and elemental analysis.
In Silico Docking Study
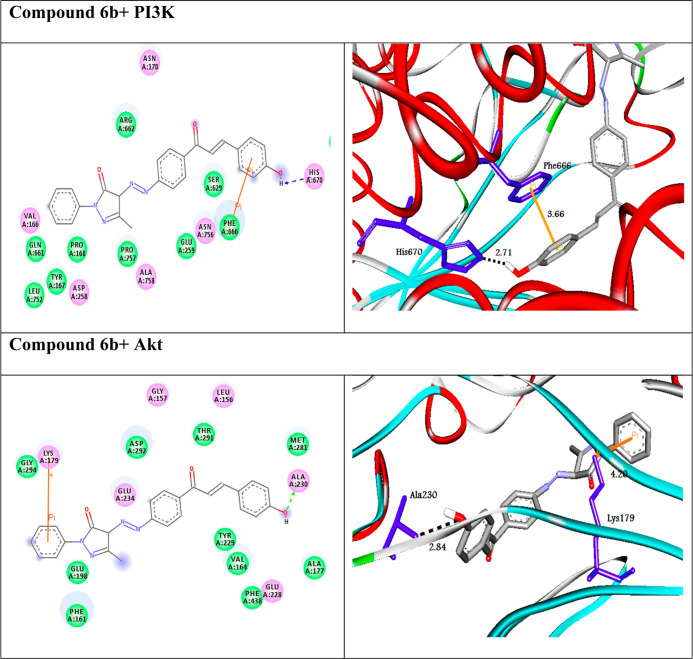
PI3K/Akt is one of the major signaling pathways associated with tumor proliferation in human cancer.38 Therefore, these proteins are selected as pivotal therapeutic targets for identifying cancer agents. In the present study, the molecular docking approach was achieved to explore new PI3K/Akt drug candidates. The screened compounds exhibited docking scores between −8.3 and −11.1 kcal/mol and between −8.6 and −10.7 kcal/mol against PI3K and Akt targets, respectively. The molecular docking studies resulted in compound 6b having the highest binding energy against the proteins. Figure 2 shows the 2D and 3D representations of the best-docked compound 6b intermolecular interactions with both targets. Compound 6b docked to the protein PI3K through five H-bonds and two π-cation interactions with the amino acid residues Asn114, Val125, Cys126, Glu78, Ser690, Arg140, and Arg693 at distances of 2.99, 2.98, 2.95, 2.44, 2.71, 5.86, and 4.17, respectively (Table 2). In addition, compound 6b interacted with the protein Akt through five H-bonds and one π-cation. The SAR study showed that heterocyclic rings as pyrazolinone and phenolic moieties in compound 6b play a significant role for enhancing its activity.
Figure 2.
(Left side) 2D and (right side) 3D representations of interactions of the best-docked compound 6b with amino acid residues of the targets PI3K and Akt.
Table 2. Calculated Docking Scores (in kcal/mol) of the Best-Docked Compound with the Targets.
| PI3K |
Akt |
|||||
|---|---|---|---|---|---|---|
| compounds | docking score (ΔGbind) | docked complex (amino acid–ligand) interactions | distance (Å) | docking score (ΔGbind) | docked complex (amino acid–ligand) interactions | distance (Å) |
| 6b | –11.1 | H-bond | –10.7 | H-bond | ||
| Asn114---compound 6b | 2.99 | Thr195:OG1---compound 6b | 2.12 | |||
| Val125---compound 6b | 2.98 | Thr195:OG1---compound 6b | 2.98 | |||
| Cys126---compound 6b | 2.95 | Thr195:OG1---compound 6b | 2.84 | |||
| Glu78:OE1---compound 6b | 2.44 | Ala230:N---compound 6b | 2.79 | |||
| Ser690:OG---compound 6b | 2.71 | Glu228:O---compound 6b | 2.35 | |||
| π-cation | π-cation | |||||
| Arg140:NH1---compound 6b | 5.86 | Lys179:NZ---compound 6b | 4.20 | |||
| Arg693:NH2---compound 6b | 4.17 | |||||
Moreover, we performed further analysis for the compounds such as ADMET and drug-like properties, as represented in Table 3. The results suggested that the intestinal barrier may well absorb all compounds but not at the blood–brain barrier level. In addition, all compounds except 4c, 5c, 6c, 7c, 8c, and 9c obeyed the Lipinski rule of five (Ro5) by not having over one violation.
Table 3. ADMET and Drug-likeness Profiles of the Compounds.
| molecular weight (g/mol) | blood–brain barrier (BBB+) | Caco-2 permeability (Caco2+) | % human intestinal absorption (HIA+) | TPSA A2 | logp | HBA | HBD | N rotatable | N violations | GI absorption | carcinogenicity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| acceptable ranges | 130–500 | –3 to 1.2 | <25 poor, 500 great | <80% high, >25% low | ≤140 | <5 | 2.0–20.0 | 0.0–6.0 | ≤10 | ≤1 | noncarcinogenic | |
| 3a | 332.36 | 0.97 | 55.68 | 99.53 | 83.25 | 1.78 | 5 | 1 | 5 | 0 | high | noncarcinogenic |
| 3b | 408.45 | 0.98 | 61.78 | 100.00 | 74.46 | 3.15 | 5 | 0 | 6 | 0 | high | noncarcinogenic |
| 3c | 498.45 | 0.90 | 50.86 | 99.39 | 166.12 | 1.47 | 9 | 0 | 8 | 1 | low | noncarcinogenic |
| 4a | 392.41 | 0.83 | 51.72 | 98.24 | 101.71 | 1.18 | 7 | 1 | 7 | 0 | high | noncarcinogenic |
| 4b | 468.50 | 0.87 | 53.48 | 99.66 | 92.92 | 2.50 | 7 | 0 | 8 | 0 | high | noncarcinogenic |
| 4c | 558.50 | 0.80 | 50.00 | 99.26 | 184.66 | 0.92 | 11 | 0 | 10 | 2 | low | noncarcinogenic |
| 5a | 375.42 | 0.88 | 57.66 | 99.13 | 86.49 | 1.70 | 5 | 1 | 6 | 0 | high | noncarcinogenic |
| 5b | 451.52 | 0.94 | 62.83 | 100.00 | 77.70 | 3.02 | 5 | 0 | 7 | 0 | high | noncarcinogenic |
| 5c | 541.51 | 0.74 | 51.45 | 99.29 | 169.36 | 1.39 | 9 | 0 | 9 | 2 | low | noncarcinogenic |
| 6a | 348.36 | 0.79 | 55.60 | 98.95 | 103.48 | 1.25 | 6 | 2 | 5 | 0 | high | noncarcinogenic |
| 6b | 424.45 | 0.89 | 53.85 | 100.00 | 94.69 | 2.61 | 6 | 1 | 6 | 0 | high | noncarcinogenic |
| 6c | 514.45 | 0.73 | 52.45 | 99.13 | 186.35 | 0.99 | 10 | 1 | 8 | 2 | high | noncarcinogenic |
| 7a | 377.35 | 0.85 | 50.75 | 95.12 | 129.07 | 0.90 | 7 | 1 | 6 | 0 | high | noncarcinogenic |
| 7b | 453.45 | 0.90 | 50.86 | 99.39 | 120.28 | 2.27 | 7 | 0 | 7 | 0 | high | noncarcinogenic |
| 7c | 543.44 | 0.90 | 50.86 | 99.39 | 211.94 | 0.72 | 11 | 0 | 9 | 2 | low | noncarcinogenic |
| 8a | 358.39 | 0.97 | 55.68 | 99.53 | 83.25 | 2.16 | 5 | 1 | 6 | 0 | high | noncarcinogenic |
| 8b | 434.49 | 0.98 | 61.78 | 100.00 | 74.46 | 3.48 | 5 | 0 | 7 | 0 | high | noncarcinogenic |
| 8c | 524.48 | 0.90 | 50.86 | 99.31 | 166.12 | 1.81 | 9 | 0 | 9 | 2 | low | noncarcinogenic |
| 9a | 401.46 | 0.88 | 57.66 | 99.13 | 86.49 | 2.07 | 5 | 1 | 7 | 0 | high | noncarcinogenic |
| 9b | 477.56 | 0.94 | 62.83 | 100.00 | 77.70 | 3.35 | 5 | 0 | 8 | 0 | high | noncarcinogenic |
| 9c | 567.55 | 0.74 | 51.45 | 99.29 | 169.36 | 1.72 | 9 | 0 | 10 | 2 | low | noncarcinogenic |
Conversely, the bioavailability radar (Figure 3) of the best-docked molecule 6b showed that the pink-colored zone is the perfect space for each property. The plot suggested that compound 6b could be a potential new anticancer drug candidate.
Figure 3.
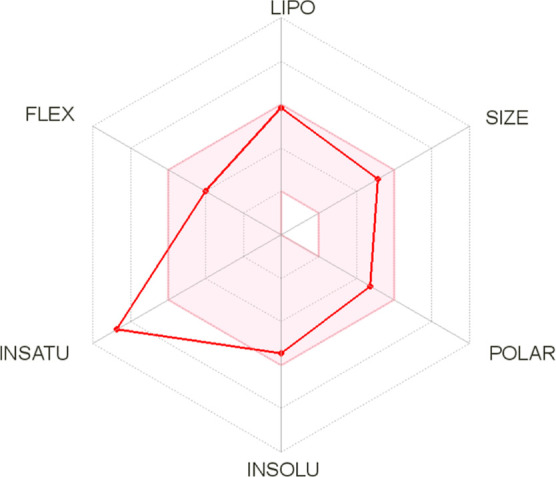
Oral bioavailability radar of the best compound 6b.
Antitumor/Cytotoxic (In Vitro) Studies
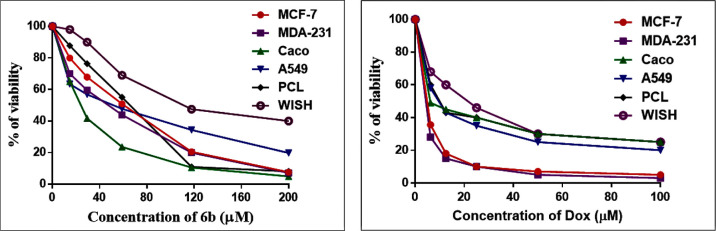
Compound 6b was chosen from the molecular docking studies to investigate its potential as a new anticancer drug via inhibiting the PI3K/Akt and Raf-1/ERK1/2 signaling pathways. Compound 6b showed significant antitumor effects on A549, MDA-231, Caco, PCL, and MCF-7 cancer cell lines with IC50 values equal to 40.91 ± 0.35, 38.45 ± 0.29, 23.34 ± 0.14, 56.33 ± 0.22, and 50.15 ± 0.14 μM, respectively. Our results showed that compound 6b had more significant inhibitory effects on the Caco colon cancer cell line compared with DOX as a reference drug (IC50 = 6.713 ± 0.27 μM). Moreover, compound 6b showed lower cytotoxic effects on WISH normal cells (IC50 = 124.4 ± 1.7 μM). This means that it is more powerful against cancer cell proliferation while having no damaging side effects on healthy cells; this is in line with the in silico results. DOX, on the other hand, has an extremely cytotoxic effect on normal cells (IC50 = 19.27 ± 0.31 μM) (Figure 4). Thus, the Caco cell line was selected for further analysis.
Figure 4.
Antitumor/cytotoxic effect of compound 6b and doxorubicin on various cell lines.
Alternation in Morphological Features
Cytotoxic agents infrequently alter the cell morphology, resulting in an abnormal cell morphology, increased cell debris, and decreased cell numbers. In the present study, detectable morphological features of apoptosis were observed in Caco cells treated with compound 6b in a dose-dependent manner (1/4IC50, 1/2IC50, IC50), including cellular shrinkage with the cell number reduced and detachment, cell rounding, and cytoplasmic condensation. However, the morphology of the untreated cells appeared normal and confluent, as shown in Figure 5. These results further elaborated the ability of compound 6b in inducing apoptosis.
Figure 5.
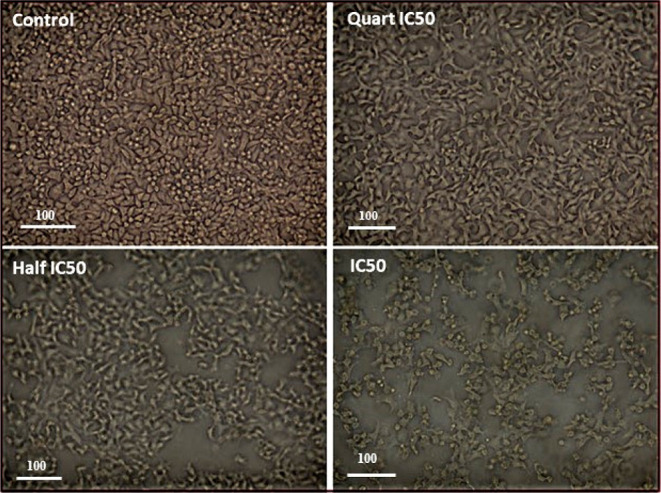
Morphological features of apoptosis in the Caco cells treated with compound 6b in a dose-dependent manner (1/4IC50, 1/2IC50, IC50) after 48 h treatment.
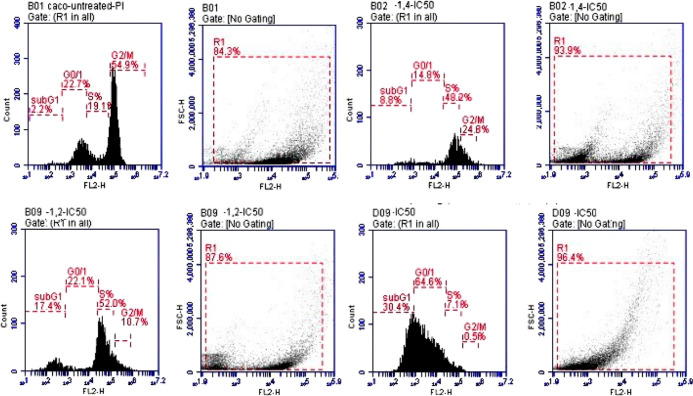
Cell Cycle Arrest Detection
The inhibition of the PI3K/Akt protein kinases causes cell cycle arrest. Moreover, this arrest confirmed the reduction in proliferation and metastasis of the Caco cancer cell line via inhibiting the Raf-1/ERK1/2 signaling pathway. Compound 6b enhanced the percentage of cells in the sub-G0/G1 phase in Caco cells at all doses when compared with the untreated cells (this is the phase in which cells wait to enter the cell cycle to duplicate; when the number of cells in this phase rises, the cell cycle has been stopped, and division and replication are impossible). The 1/4IC50, 1/2IC50, and IC50 of compound 6b showed cell cycle arrest at rates of 8.8, 17.4, and 30.4%, respectively, in the sub-G0/G1 phase compared to untreated Caco cells (2.2%). This means that compound 6b inhibits cell growth, arrest in cell cycle progression, and the increase of cells in G1 reflecting its apoptotic effect, and the G2/M checkpoint blocks the entry into mitosis when DNA is damaged, as illustrated in Figure 6. Collectively, these results demonstrated that compound 6b can induce inhibition to PI3K/Akt and Raf-1/ERK1/2 signaling pathways and trigger apoptosis via arresting the cell cycle in the sub-G0/G1 phase in a dose-dependent manner.38,39
Figure 6.
Cell cycle phases of compound 6b in the Caco cell line in a dose-dependent manner (1/4IC50, 1/2IC50, IC50) after 48 h treatment.
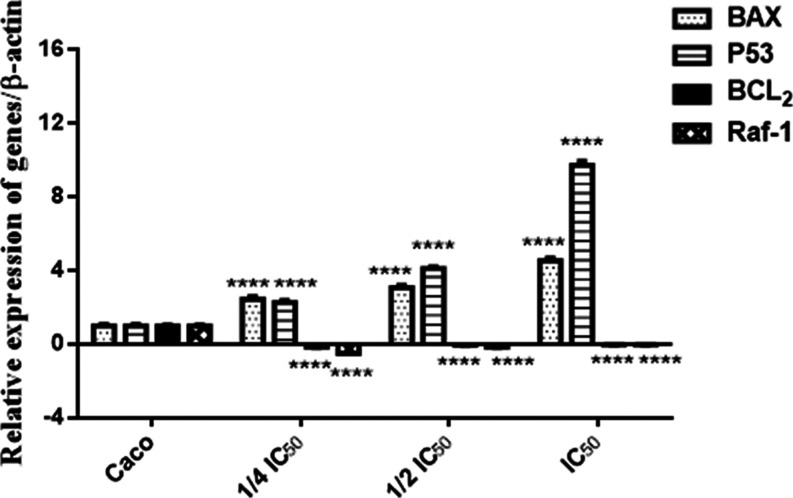
qRT-PCR Study
qRT-PCR was used to measure the mRNA expression of the Bax, p53 (apoptotic markers), Bcl–2 (antiapoptotic marker), and Raf-1 (proliferative marker) genes in the Caco cell line. In cells treated with compound 6b, the expression of Bax and p53 was considerably (p < 0.0001) increased in a dose-dependent manner, with the highest expression in the IC50 of cells treated with compound 6b compared to untreated cells. Bcl–2 and Raf-1 gene expressions were dramatically (p < 0.0001) downregulated in compound 6b-treated cells in a dose-dependent manner, with a little expression in the IC50 of compound 6b-treated cells compared to untreated cells, as shown in Figure 7. As a result, the upregulation of Bax expression and downregulation of Bcl–2 expression indicate that compound 6b causes mitochondrial membrane dysfunction that causes release of cytochrome C and activation of the caspase cascade that finally led to apoptosis. In contrast, the upregulation of p53 expression and downregulation of the expression of the proliferative gene Raf-1 result from the inhibition of PI3K/Akt and lead to inhibit ERK1/2 signaling pathways. This clarifies the ability of compound 6b to inhibit cell proliferation, angiogenesis, and metastasis, leading to cell cycle arrest and apoptosis induction.39,40
Figure 7.
Relative expression of Raf-1, P53, Bax, and Bcl–2 in the Caco cell line (compound 6b) in a dose-dependent manner (1/4IC50, 1/2IC50, IC50) after 48 h treatment.
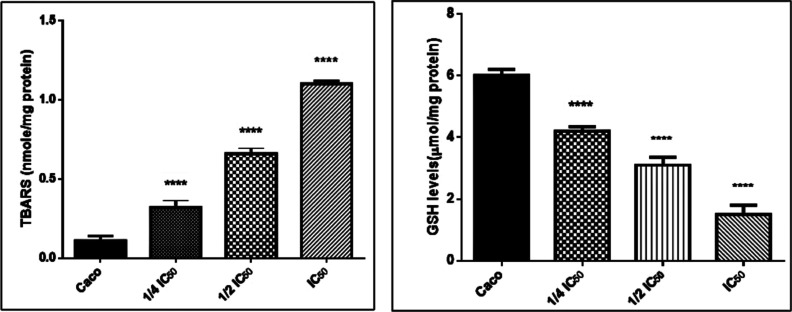
Antioxidant/Oxidative Stress Biomarkers
Our results revealed that the level of MDA that results from the lipid peroxidation process occurring in Caco cells treated with compound 6b in a dose-dependent manner (1/4IC50, 1/2IC50, IC50) was increased significantly, while the specific activity of reduced glutathione (GSH) was remarkably diminished as compared with the untreated Caco cells, as shown in Figure 8. This implies that compound 6b could promote programming cell death in cancer cells by generating intracellular ROS and blocking the antioxidant endogenous enzymes. According to this theory, the high ROS production in cancer cells causes malfunction of the mitochondrial membrane, inhibits the PI3K/Akt protein kinase, and leads to the Raf-1/ERK1/2 signaling pathway inhibition. Finally, apoptosis is induced, and cell survival, proliferation, and metastasis are arrested.38,41−43
Figure 8.
MDA and GSH levels in the Caco cell line (compound 6b) in a dose-dependent manner (1/4IC50, 1/2IC50, IC50) after 48 h treatment.
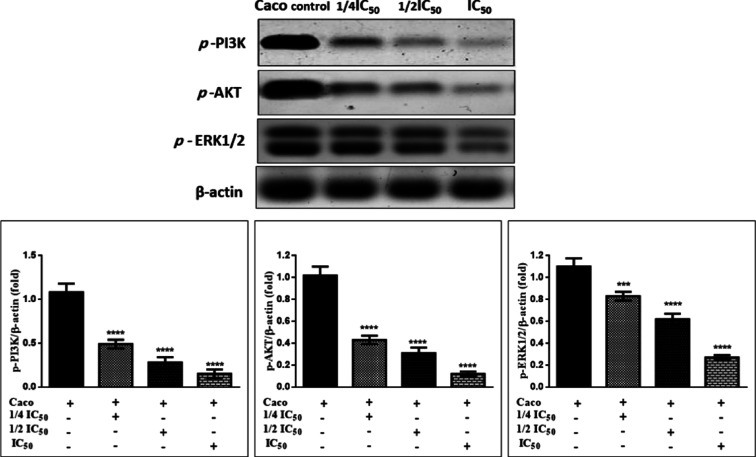
Immunoblotting Confirms the Inhibition of the PI3K/Akt/ERK1/2 Signaling Pathway
Our results elucidated that compound 6b induces a remarkable drop in PI3K/Akt protein kinase with a significant decrease in ERK1/2 in the Caco colon cancer cell line dose-dependently as compared to untreated cells, Figure 9. These findings confirmed compound 6b’s mechanical routes of inhibition of PI3K, which inhibits PIP3 and promotes ROS intracellular production, resulting in Akt inhibition via dephosphorylation. When Akt is dephosphorylated, p53 is activated due to blocking MDM2 protein and the cell cycle is stopped. Additionally, it inhibits Raf-1, which leads to inhibition of MEK protein and finally causes ERK1/2 inhibition, arresting cell proliferation, angiogenesis, and metastasis.44−48
Figure 9.
Effects of compound 6b on the phosphorylation of PI3K/Akt/ERK1/2 in Caco cells. The cells were treated with the 1/4IC50, 1/2IC50, and IC50 for 48 h, and the protein phosphorylation levels were relative to β-actin protein (internal control) using western blot analysis.
Conclusions
The condensation of azo pyrazolinone derivatives with various aromatic aldehydes yielded a series of novel pyrazolinone chalcones 3–9. This has been confirmed by spectroscopic techniques as well as elemental analyses. The newly synthesized pyrazolinone chalcone (6b) was selected according to its in silico molecular binding energy toward PI3K/Akt protein kinases. Consequently, in vitro studies approved that the chosen compound 6b causes apoptosis and cell death by inducing ROS generation-mediated inhibition of PI3K/Akt. The cellular mechanism of interdependence between PI3K/Akt inhibition and Raf-1/ERK1/2 proliferative inhibition, p53 activation, and cell cycle arrest involved in the Caco colon cancer cell line has been elucidated. According to our results, compound 6b could be used as a promising anticancer agent.
Acknowledgments
This in vitro study was conducted in the Center of Excellence for Research in Regenerative Medicine and Its Applications (CERRMA), Faculty of Medicine, Alexandria University, Egypt.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02181.
Spectral data for all compounds and docking and biological studies (PDF)
Author Contributions
A.A.N., I.A.S., A.H.A., and M.M.S contributed in this work
This research received no external funding.
The authors declare no competing financial interest.
Supplementary Material
References
- Abu-Khudir R.; Ismail G. A.; Diab T. Antimicrobial, antioxidant, and anti-tumor activities of Sargassum linearifolium and Cystoseira crinita from Egyptian Mediterranean Coast. Nutr. Cancer 2021, 73, 829–844. 10.1080/01635581.2020.1764069. [DOI] [PubMed] [Google Scholar]
- von Meyenfeldt M. Cancer-associated malnutrition: An introduction. Eur. J. Oncol. Nurs. 2005, 9, S35–S38. 10.1016/j.ejon.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Noser A. A.; El-Naggar M.; Donia T.; Abdelmonsef A. H. Synthesis, In Silico and In Vitro Assessment of New Quinazolinones as Anticancer Agents via Potential AKT Inhibition. Molecules 2020, 25, 4780. 10.3390/molecules25204780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Li R.; Zhao X.; Yu X.; Sun Q. Metformin Promotes HaCaT Cell Apoptosis through Generation of Reactive Oxygen Species via Raf-1-ERK1/2-Nrf2 Inactivation. Inflammation 2018, 41, 948–958. 10.1007/s10753-018-0749-z. [DOI] [PubMed] [Google Scholar]
- An W.; et al. Apoptotic pathway as the therapeutic target for anticancer traditional Chinese medicines. Front. Pharmacol. 2019, 10, 758. 10.3389/fphar.2019.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braicu C.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. 10.3390/cancers11101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim M. J.; Alam O.; Nawaz F.; Alam M. J.; Alam P. Current status of pyrazole and its biological activities. J. Pharm. BioAllied Sci. 2016, 8, 2–17. 10.4103/0975-7406.171694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y.; et al. Chalcone derivatives: Role in anticancer therapy. Biomolecules 2021, 11, 894. 10.3390/biom11060894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthee C.; Terre’Blanche G.; Legoabe L. J.; Janse van Rensburg H. D. Exploration of chalcones and related heterocycle compounds as ligands of adenosine receptors: therapeutics development. Mol. Diversity 2021, 10.1007/s11030-021-10257-9. [DOI] [PubMed] [Google Scholar]
- Elkhalifa D.; Al-Hashimi I.; Al Moustafa A.-E.; Khalil A. A comprehensive review on the antiviral activities of chalcones. J. Drug Target. 2021, 29, 403–419. 10.1080/1061186x.2020.1853759. [DOI] [PubMed] [Google Scholar]
- Tong Y.; Lyu Y.; Xu S.; Zhang L.; Zhou J. Optimum chalcone synthase for flavonoid biosynthesis in microorganisms. Crit. Rev. Biotechnol. 2021, 41, 1194–1208. 10.1080/07388551.2021.1922350. [DOI] [PubMed] [Google Scholar]
- Farooq S.; Ngaini Z. One-pot and two-pot synthesis of chalcone based mono and bis-pyrazolines. Tetrahedron Lett. 2020, 61, 151416. 10.1016/j.tetlet.2019.151416. [DOI] [Google Scholar]
- Ozmen Ozgun D.; Gul H. I.; Yamali C.; Sakagami H.; Gulcin I.; Sukuroglu M.; Supuran C. T. Synthesis and bioactivities of pyrazoline benzensulfonamides as carbonic anhydrase and acetylcholinesterase inhibitors with low cytotoxicity. Bioorg. Chem. 2019, 84, 511–517. 10.1016/j.bioorg.2018.12.028. [DOI] [PubMed] [Google Scholar]
- Haider K.; Shafeeque M.; Yahya S.; Yar M. S. A comprehensive review on pyrazoline based heterocyclic hybrids as potent anticancer agents. Eur. J. Med. Chem. Rep. 2022, 5, 100042. 10.1016/j.ejmcr.2022.100042. [DOI] [Google Scholar]
- Alharthy R. D. Design and Synthesis of Novel Pyrazolo[3,4-d]Pyrimidines: In Vitro Cytotoxic Evaluation and Free Radical Scavenging Activity Studies. Pharm. Chem. J. 2020, 54, 273–278. 10.1007/s11094-020-02190-2. [DOI] [Google Scholar]
- Khalil A.; Hassan M.; Mohamed M.; Elsayed A. Metal salt-catalyzed diazocoupling of 3-substituted-1-pyrazol-2-in-5-ones in aqueous medium. Dyes Pigm. 2005, 66, 241–245. 10.1016/j.dyepig.2004.10.005. [DOI] [Google Scholar]
- Rashdan H. R. M.; Shehadi I. A.; Abdelmonsef A. H. Synthesis, Anticancer Evaluation, Computer-Aided Docking Studies, and ADMET Prediction of 1,2,3-Triazolyl-Pyridine Hybrids as Human Aurora B Kinase Inhibitors. ACS Omega 2021, 6, 1445–1455. 10.1021/acsomega.0c05116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashdan H. R. M.; Abdelmonsef A. H.; Abou-Krisha M. M.; Yousef T. A. Synthesis, Identification, Computer-Aided Docking Studies, and ADMET Prediction of Novel Benzimidazo-1,2,3-triazole Based Molecules as Potential Antimicrobial Agents. Molecules 2021, 26, 7119. 10.3390/molecules26237119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmonsef A. H.; Mosallam A. M. Synthesis, in vitro biological evaluation and in silico docking studies of new quinazolin-2,4-dione analogues as possible anticarcinoma agents. J. Heterocycl. Chem. 2020, 57, 1637–1654. 10.1002/jhet.3889. [DOI] [Google Scholar]
- Abo-Bakr A. M.; Alsoghier H. M.; Abdelmonsef A. H. Molecular Docking, Modeling, Semiempirical Calculations Studies and In Vitro Evaluation of New Synthesized Pyrimidin-imide Derivatives. J. Mol. Struct. 2022, 1249, 131548. 10.1016/j.molstruc.2021.131548. [DOI] [Google Scholar]
- Abdelmonsef A. H.; et al. A search for antiinflammatory therapies: Synthesis, in silico investigation of the mode of action, and in vitro analyses of new quinazolin-2,4-dione derivatives targeting phosphodiesterase-4 enzyme. J. Heterocycl. Chem. 2022, 59, 474–492. 10.1002/jhet.4395. [DOI] [Google Scholar]
- Haredi Abdelmonsef A.; Eldeeb Mohamed M.; El-Naggar M.; Temairk H.; Mohamed Mosallam A. Novel Quinazolin-2,4-Dione Hybrid Molecules as Possible Inhibitors Against Malaria: Synthesis and in silico Molecular Docking Studies. Front. Mol. Biosci. 2020, 7, 105. 10.3389/fmolb.2020.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehadi I. A.; Rashdan H. R. M.; Abdelmonsef A. H. Homology Modeling and Virtual Screening Studies of Antigen MLAA-42 Protein: Identification of Novel Drug Candidates against Leukemia-An In Silico Approach. Comput. Math. Methods Med. 2020, 2020, 8196147. 10.1155/2020/8196147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haredi Abdelmonsef A. Computer-aided identification of lung cancer inhibitors through homology modeling and virtual screening. Egypt. J. Med. Hum. Genet. 2019, 20, 6. 10.1186/s43042-019-0008-3. [DOI] [Google Scholar]
- Berman H. M.; et al. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; et al. CHARMM: Molecular dynamics simulation package. J. Comput. Chem. 2009, 30, 1545–1614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; et al. Open Babel: An Open chemical toolbox. J. Cheminf. 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé A. K.; Casewit C. J.; Colwell K. S.; Goddard W. A.; Skiff W. M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Molecular Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. 10.1021/ja00051a040. [DOI] [Google Scholar]
- Dallakyan S.; Olson A. J.. Small-Molecule Library Screening by Docking with PyRx. Chemical Biology; Springer, 2015; Vol. 1263, pp 243–250. [DOI] [PubMed] [Google Scholar]
- Dash S. K.; Ghosh T.; Roy S.; Chattopadhyay S.; Das D. Zinc sulfide nanoparticles selectively induce cytotoxic and genotoxic effects on leukemic cells: Involvement of reactive oxygen species and tumor necrosis factor alpha. J. Appl. Toxicol. 2014, 34, 1130–1144. 10.1002/jat.2976. [DOI] [PubMed] [Google Scholar]
- Noser A. A.; Abdelmonsef A. H.; El-naggar M.; Salem M. M. New Amino Acid Schiff Bases as Anticancer Agents via Potential Mitochondrial Complex I-Associated Hexokinase Inhibition and Targeting AMP-Protein Kinases/mTOR Signaling Pathway. Molecules 2021, 26, 5332. 10.3390/molecules26175332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z.; Halicka H. D.; Zhao H.. Analysis of Cellular DNA Content by Flow and Laser Scanning Cytometry. Polyploidization and Cancer. Advances in Experimental Medicine and Biology; Poon R. Y. C., Ed.; Springer: New York, 2010; pp 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvastad L.; et al. Single cell analysis of cancer cells using an improved RT-MLPA method has potential for cancer diagnosis and monitoring. Sci. Rep. 2015, 5, 16519. 10.1038/srep16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Salem M. M.; et al. Propolis Potentiates Methotrexate Anticancer Mechanism and Reduces its Toxic Effects. Nutr. Cancer 2020, 72, 460–480. 10.1080/01635581.2019.1640884. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Mruk D. D.; Cheng C. Y. Enhanced chemiluminescence (ECL) for routine immunoblotting. Spermatogenesis 2011, 1, 121–122. 10.4161/spmg.1.2.16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; et al. Inhibition of PI3K/AKT signaling via ROS regulation is involved in rhein-induced apoptosis and enhancement of oxaliplatin sensitivity in pancreatic cancer cells. Int. J. Biol. Sci. 2021, 17, 589–602. 10.7150/ijbs.49514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W.; Song G. Inhibitory effects of delphinidin on the proliferation of ovarian cancer cells via PI3K/AKT and ERK 1/2 MAPK signal transduction. Oncol. Lett. 2017, 14, 810–818. 10.3892/ol.2017.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey J. A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta, Mol. Cell Res. 2007, 1773, 1263–1284. 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.; et al. ROS-mediated inactivation of the PI3K/AKT pathway is involved in the antigastric cancer effects of thioredoxin reductase-1 inhibitor chaetocin. Cell Death Dis. 2019, 10, 809. 10.1038/s41419-019-2035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.; et al. Phosphoinositide 3-kinase (Pi3k) reactive oxygen species (ros)-activated prodrug in combination with anthracycline impairs pi3k signaling, increases dna damage response and reduces breast cancer cell growth. Int. J. Mol. Sci. 2021, 22, 2088. 10.3390/ijms22042088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X.; et al. Alantolactone induces apoptosis through ROS-mediated AKT pathway and inhibition of PINK1-mediated mitophagy in human HepG2 cells. Artif. Cells, Nanomed., Biotechnol. 2019, 47, 1961–1970. 10.1080/21691401.2019.1593854. [DOI] [PubMed] [Google Scholar]
- Yao W.; et al. Delicaflavone induces ROS-mediated apoptosis and inhibits PI3K/AKT/mTOR and Ras/MEK/Erk signaling pathways in colorectal cancer cells. Biochem. Pharmacol. 2020, 171, 113680. 10.1016/j.bcp.2019.113680. [DOI] [PubMed] [Google Scholar]
- Su X.; et al. Vitamin C kills thyroid cancer cells through ROS-dependent inhibition of MAPK/ERK and PI3K/AKT pathways via distinct mechanisms. Theranostics 2019, 9, 4461–4473. 10.7150/thno.35219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; et al. ROS Promote ox-LDL-induced platelet activation by up-regulating autophagy through the inhibition of the PI3K/AKT/mTOR pathway. Cell. Physiol. Biochem. 2018, 50, 1779–1793. 10.1159/000494795. [DOI] [PubMed] [Google Scholar]
- Deng S.; et al. Dexamethasone induces osteoblast apoptosis through ROS-PI3K/AKT/GSK3β signaling pathway. Biomed. Pharmacother. 2019, 110, 602–608. 10.1016/j.biopha.2018.11.103. [DOI] [PubMed] [Google Scholar]
- Gao X.; et al. Cocoa tea (Camellia ptilophylla) induces mitochondria-dependent apoptosis in HCT116 cells via ROS generation and PI3K/Akt signaling pathway. Food Res. Int. 2020, 129, 108854. 10.1016/j.foodres.2019.108854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.