Abstract
The liver is a vital organ that controls glucose and lipid metabolism, hormone regulation, and bile secretion. Liver injury can occur from various insults such as viruses, metabolic diseases, and alcohol, which lead to acute and chronic liver diseases. Recent studies have demonstrated the implications of long non-coding RNAs (lncRNAs) in the pathogenesis of liver diseases. These newly discovered lncRNAs have various functions attributing to many cellular biological processes via distinct and diverse mechanisms. LncRNA H19, one of the first lncRNAs being identified, is highly expressed in fetal liver but not in adult normal liver. Its expression, however, is increased in liver diseases with various etiologies. In this review, we focused on the roles of H19 in the pathogenesis of liver diseases. This comprehensive review is aimed to provide useful perspectives and translational applications of H19 as a potential therapeutic target of liver diseases.
Keywords: non-coding RNA, LncRNA-H19, liver diseases
1. Introduction
The new landscape of human transcriptome along with the identification of non-coding RNAs (ncRNAs) has uncovered their importance in the pathophysiology of human diseases[1]. These RNA transcripts do not code or transcribe into proteins, however, they have biological functions and regulate intracellular physiological processes. It is estimated that more than 80% of the human genome is generally transcribed of which less than 2% are protein-coding genes[2].
Non-coding RNA transcripts (ncRNA) are broadly categorized based on the number of nucleotides. Short ncRNAs (<200 bp) consist of highly abundant and functionally important RNAs such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and small regulatory RNAs such as microRNAs (miRNAs), and small interfering RNAs (siRNAs). Long non-coding RNAs (lncRNAs) are newly discovered and defined as those with transcripts of more than 200 bp[3]. Similar to mRNAs, lncRNAs have both exons and introns and are transcribed by RNA polymerase II; 5’ capped, 3’ poly-adenylated and spliced[4]. They are tissue and cell-type specific and can also be epigenetically modified and transcriptionally regulated[5].
Several lines of evidence indicated the importance of lncRNA in various biological functions and diseases [6–9]. Several lncRNAs have been identified in hepatocellular carcinoma (HCC) and are closely associated with disease prognosis, such as lncRNA-SNHG7, LINC01278, and MALAT1[10–12]. Differential expression of specific lncRNAs was noted during liver regeneration after partial hepatectomy in rodents[13–15]. LncRNAs also play an important role in cholestatic liver diseases, non-alcoholic steatohepatitis, and alcohol-associated liver disease[8, 16–21].
Among many lncRNAs, H19 was the first to be identified and studied in the liver[6]. Significant progress has been made over the past decade to advance our knowledge of the mechanism of H19 associated liver pathobiology. In this current review, we summarized the most up-to-date literature regarding the regulation and function of H19 in the pathogenesis of liver diseases.
2. LncRNA H19 overview
The H19 gene was originally isolated as a target for the control of embryonic transcriptional regulators regulation of α-fetoprotein (Raf) and regulation of induction of α-fetoprotein (Rif) [22]. As a non-coding RNA, it expresses an mRNA that is incapable of being translated into a protein[23]. The H19 locus belongs to a conserved gene cluster on chromosome 11p15.5 in human, and on chromosome 7 in the mouse. It plays an essential role in growth control and embryonic development[24]. This cluster also contains the insulin-like growth factor 2 (IGF2) gene, located in near 90 kb upstream of the H19 gene. Both H19 and IGF2 genes are coordinately controlled by an intergenic differentially methylated region (DMR), imprinting control region (ICR), and by enhancers located downstream of H19 as shown in Figure 1A[24, 25]. H19 and IGF2 are reciprocally imprinted genes, while IGF2 is expressed from the paternal allele; H19 is only expressed from the maternal allele [24, 25].
Figure 1. Imprinting control region (ICR) on the IGF2-H19 locus determines the expression lncRNA H19.
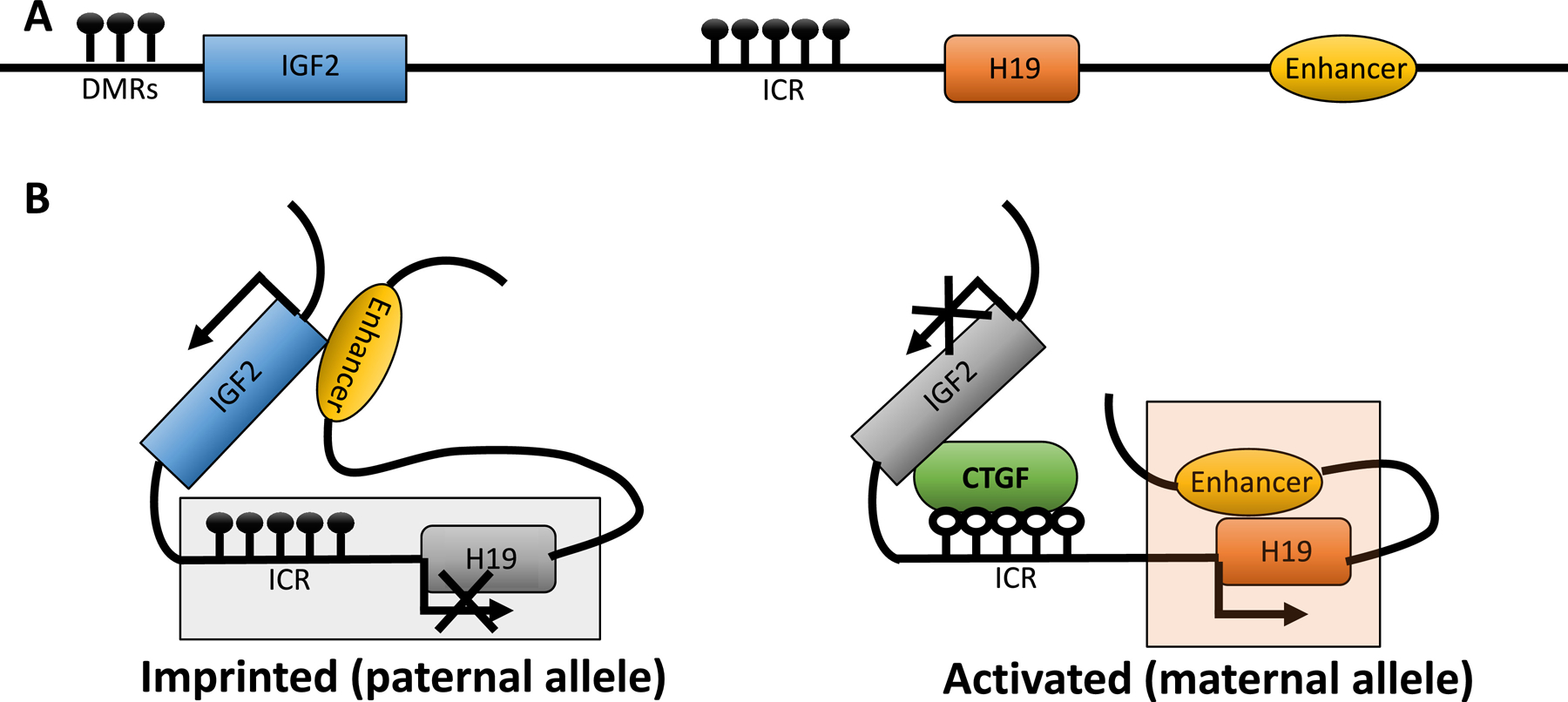
The relative location of DMRs (differentially methylated regions), IGF2 (insulin-like growth factor 2), ICR, H19, and enhancer were presented in Fig. 1A. On the paternal allele (Fig. 1B left), ICR is hypermethylated and prevents the expression of H19. In contrast, on maternal allele (Fig. 1B right), ICR is unmethylated and CCCTC binding factor (CTCF) is recruited. The enhancer binds the H19 promoter and activates its expression.
2.1. Regulation of H19 expression
2.1.1. Regulation by an epigenetic mechanism
In normal conditions, the ICR is hypermethylated at the paternal allele and hypomethylated at the maternal allele. This differential methylation is essential in regulating the normal expression of IGF2 and H19. The ICR carries 67 CpGs that are each methylated in sperm but remain unmethylated in oocytes[26, 27]. Subsequently, the zygote carries one methylated (paternal) and one unmethylated (maternal) copy of the ICR. This parent-of-origin difference in ICR methylation is maintained throughout development in all cell types, except for the primordial germ cells[26]. DNA methylation is not the primary genomic imprint and that the H19 ICR insertion is sufficient to transmit parent-of-origin-dependent DNA methylation patterns independent of its methylation status in sperm[27]. This paternal chromosome-specific ICR methylation is critical for maintaining the parent-of-origin-specific epigenomes and expression patterns at this locus[28].
The regulation of H19 and IGF2 imprinting involves the binding of either CCCTC-binding factor (CTCF) or CpG binding protein 2 (MeCP2) based on the methylation status of the ICR[29]. Hypermethylation of ICR on the paternal chromosome leads to the repression of H19, activation of IGF2, and prohibition of its binding to CTCF[30]. Histone deacetylases (HDACs) interact with MeCP2 binding to methylated CpG dinucleotides and suppress H19 expression [29, 31]. CTCF binds DMR located within IGF2, as a consequence of ICR methylation. This binding promotes the interaction of the proximal IGF2 promoter and the distal enhancers and drives IGF2 expression [29, 30]. On the other hand, hypomethylation of ICR on maternal chromosome results in the opposite effect leading to the expression of H19 and the binding between ICR and CTCF preventing the expression of IGF2[29]. Taken together, the expression of H19 and IGF2 is closely related and their expressions depend on the methylation of the ICR on the either paternal or maternal allele. On the paternal allele, the ICR is methylated, and the enhancer region can interact with IGF2 to allow expression. On the maternal allele, the ICR is not methylated, allowing same enhancer region to, instead, drive H19 expression (Fig. 1B) [29].
2.1.2. Regulation by transcriptional factors
H19 was reported to be regulated by transcriptional factors, including forkhead box A1 (FOXA1) [32, 33], forkhead box F2 (FOXF2)[34], hypoxia-inducible factor 1 subunit alpha (HIF1α) [35], Paxillin (PXN) [36], E2F transcription factor 1 (E2F1) [37], SRY-sex determining region Y-box 2 (SOX2) [38] and paternally expressed gene 3 (PEG3) [39]. FOXA1, a member in the forkhead class of DNA-binding proteins family, is a hepatocyte nuclear factor that transcriptionally activates liver-specific genes [32]. Chromatin immunoprecipitation indicated that FOXA1 binds the H19 enhancer at its two FOXA binding sites [33]. FOXF2, primarily expressed in the lung, can directly bind to the H19 promoter region and accelerate its transcriptional activity [34, 40]. HIF1α transcriptionally activates H19 by directly binding to the H19 promoter or indirectly through the induction of specific protein 1 (SP1), which in turn interacts with the H19 promoter and promotes its expression [35]. Bioinformatics analysis of H19 upstream DNA fragment (−596 to −146bp) using the PROMO algorithm (a program to predict transcription factor binding sites in DNA sequences) identified four SOX2 binding sites which can control H19 expression[38]. Except for those transcriptional activators, PEG3 which is a DNA-binding protein with 12 C2H2 zinc finger motifs can bind to CTCF binding sites in the H19-ICR and act as a transcriptional repressor for H19[39].
2.2. H19 cellular and tissue distribution
H19 is highly expressed in the embryonic tissues and the placenta. Its expression is significantly diminished after birth in most tissues except for the skeletal muscle, cartilage, and cardiac muscle[41, 42]. Under pathological processes such as cancer, connective tissue disorders, and kidney disease, it can re-express[43–45]. In the liver, H19 is also abundantly expressed during the fetal stage and significantly diminished in an adult healthy liver. However, H19 expression is reactivated in chronic liver diseases [46].
To determine the cellular localization of H19 in the liver, we utilized the bile duct ligation model (BDL) in Pat−/− (as wild-type mice) and Mat−/− (as H19 knockout mice or negative controls) and visualized the H19 expression and distribution with RNAScope. No H19 was detected in Mat−/−-BDL and Mat−/−-sham liver. In contrast, H19RNA-positive cells were detected in Pat−/−-BDL liver in the fibrosis areas close to the portal vein adjacent to the bile duct. H19 was found predominantly expressed in the cytosol.[46] Hepatocytes are the major functional cells and contributed roughly 80% of the liver mass. On double staining for H19 and hepatocyte nuclear factor 4-α (HNF4α, a well-established hepatocyte marker), H19 was identified to co-stained with HNF4α, which indicated that H19 could be expressed in hepatocytes. [46]. Both Kupffer cells and infiltrating macrophages have positive F4/80 markers, H19 was found positively expressed in F4/80 positive cells in the liver suggesting the possibility of its expression in both cell populations. Hepatic stellate cells are collagen-producing cells which are responsible for fibrogenesis in responses to toxic stimuli in the liver. H19 was not positively stained in stellate cells. However, H19 was found in the cholangiocytes and could activate stellate cells through exosome delivery, especially in cholestatic liver injury [47]. Of importance, H19 was also identified in SOX9 positive progenitor cells in the liver[46]. Due to the ability of progenitor cells to differentiate into hepatocytes or cholangiocytes, it is also plausible that the presence of H19 in the cholangiocytes and hepatocytes is derived from these progenitor cells, although the hypothesis needs to be tested in future studies.
A microarray analysis of sexually dimorphic gene expression in somatic mouse tissues indicated that H19 was the top female-biased gene in the mouse eye, but not in other tissues (lung, liver, kidney, etc.). However, the upregulation of H19 was still from the maternal allele instead of escaped from its silencing genomic imprint on the paternal allele. IGF2 was also upregulated in the female mouse eye which indicated that it was not due to loss of imprinting of ICR[48]. An additional study found the upregulation of H19 and IGF2 in the female eye was caused by the DNA methylation status of IGF2 DMRs, instead of ICR[49]. Mouse stain differentially expression of H19 was also observed among BALB/cJ and other mouse strains. BALB/cJ mice have higher adult liver H19 and alpha-fetoprotein levels due to the loss of zinc fingers and homeoboxes 2 (ZHX2) gene expression[50].
2.3. The regulatory functions of H19
The regulatory functions of lncRNAs are diverse due to their various binding abilities. They can bind to DNA or RNA through the base pair matching, as well as bind to proteins through their docking structures. Through those interactions, lncRNAs modulated various molecular and cellular processes, including chromatin modification, epigenetic regulation, transcription complex recruitment, translation, and post-translation regulation[51]. LncRNA H19 regulates these processes through the interaction with miRNAs and proteins as illustrated in Figure 2. MiR-675 is embedded in H19’s first exon and had been shown to promote cancer cell growth and inhibit apoptosis[52–54]. H19 can bind to HuR, an RNA-binding protein, and suppress the processing of miR-675. In turn, the miR-675’s target genes were upregulated due to this suppression [52]. H19 can also interact with other miRNAs, such as miR-148a, to serve as the “sponge” and block these microRNA’s ability to inhibit their targets [55]. H19 is involved in multiple molecular processes through its ability to interact with different proteins[56]. For example, H19 interacts with methyl-CpG-binding domain protein 1 (MBD1) and recruits histone lysine methyltransferases to methylate target DNA region and silence the target genes [56]. H19 can interact with RNA-binding proteins, like polypyrimidine tract-binding protein 1 (PTBP1), to stabilize the target mRNA and prevent its degradation [57].
Figure 2. The regulatory function of H19.
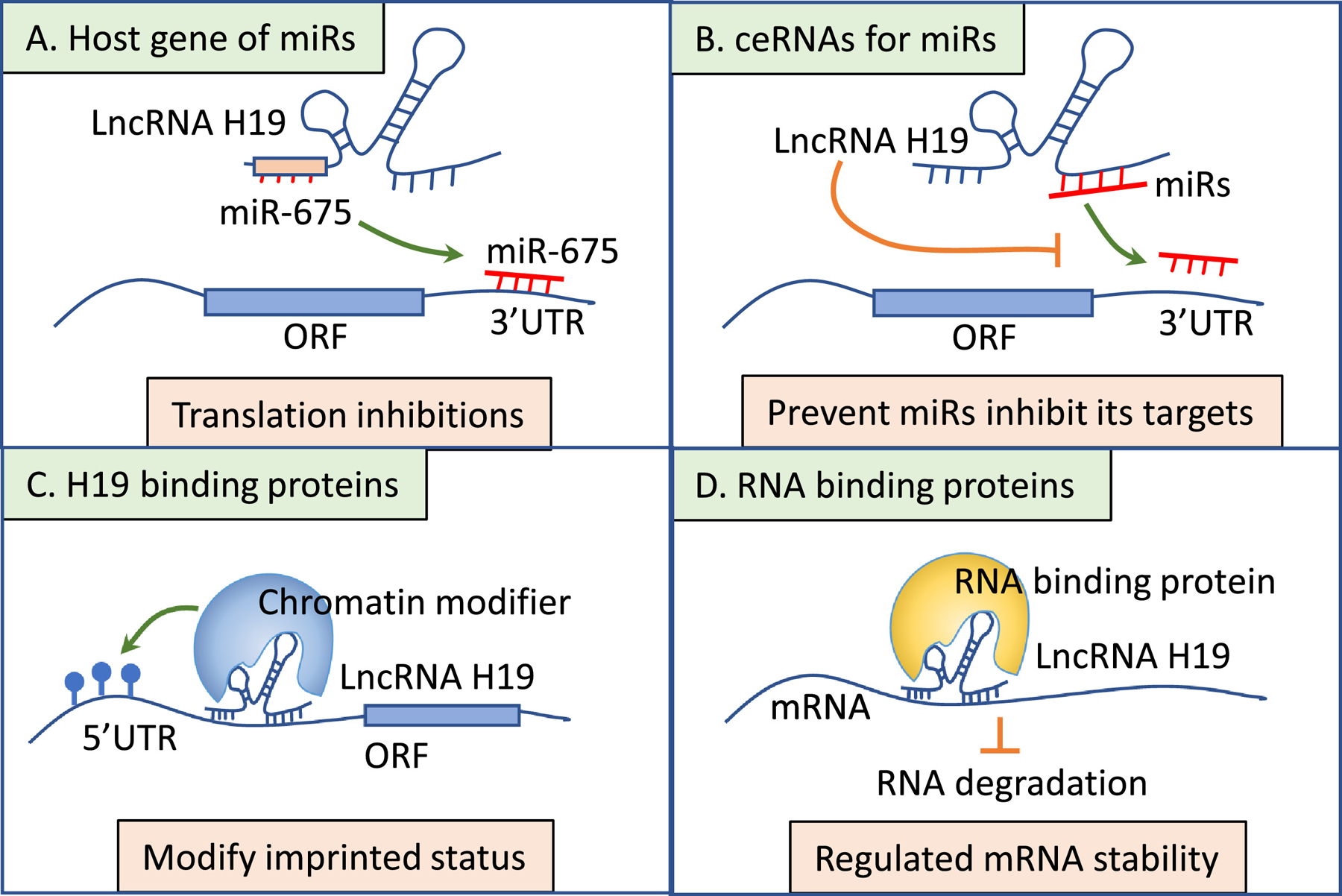
A. H19 regulates miR-675’s targets through the host expression of this microRNA. B. H19 serves as a “sponge” to block the interaction of miRNAs and their targets. C. H19 interacts with chromatin modifiers resulting in gene silencing. D. H19 binds to RNA-binding protein to regulate targeted mRNA stability.
3. Roles of LncRNA H19 in the pathogenesis of liver diseases
3.1. Roles of H19 in non-alcoholic fatty liver disease (NAFLD)
NAFLD, one of the most common causes of chronic liver disease, represents a spectrum of histopathological disorders ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), which can progress to cirrhosis and liver cancer[58]. The mechanism of NAFLD is complex[59, 60] and recent studies suggested the implications of H19 in its pathogenesis. In a mouse model of NAFLD using high-fat feeding, H19 as well as genes that are involved in hepatic lipogenesis are upregulated[61, 62]. In addition, free fatty acids (FFAs) promote H19 expression and facilitates lipogenesis in the hepatocytes, while knockdown of H19 inhibits FFA-induced lipid accumulation and triglyceride secretion[61, 62]. Mechanistically, H19 directly downregulates the expression of miR-130a which in turn inhibits the expression of peroxisome proliferator-activated receptor γ (PPARγ) level in the liver[61]. H19 also promotes hepatic steatosis by upregulating transcription factor MLX-interacting protein-like (MLXIPL) or carbohydrate-responsive element-binding protein (ChREBP)[62]. H19-induced lipid accumulation is effectively inhibited by PF-04691502, an PI3K/mTOR inhibitor, in a dose-dependent manner suggesting that H19 regulates hepatic lipid metabolism through the activation of mTOR signaling[62]. H19 also serves as a lipid sensor by synergizing with RNA-binding protein, PTBP1, to modulate hepatic metabolic homeostasis[57]. PTBP1 regulates mRNA stability and pre-mRNA splicing and is implicated in directing cell reprogramming[63, 64]. H19 can interact with PTBP1 to increase SREBP1 protein cleavage, stability, and transcriptional activity to activate lipogenic genes [57]. In summary, H19 is one of the key regulators in the pathogenesis of hepatic steatosis through its regulation of other non-coding RNAs (such as miR-130a) or transcription factors involved in lipogenesis.
H19-induced hepatic steatosis may also result from the indirect effect secondary to its role in diabetes. RNA sequencing of db/db mice liver reveals H19 as a key regulator of gluconeogenesis and hepatic glucose output via regulation of forkhead box O1 (FOXO1) nuclear levels[65]. Hepatic H19 expression is chronically increased in diabetic mice and liver-specific H19 overexpression promotes hepatic glucose production, hyperglycemia, and insulin resistance[66]. Patients with type 2 diabetes also have a higher level of circulating plasma H19[67].
3.2. Roles of H19 in cholestasis
Cholestasis is defined as a condition where bile flow is decreased, either due to impaired secretion by hepatocytes or by obstruction to the bile flow[68]. Bile acids (BA), synthesized from cholesterol in the liver, play an essential role in eliminating cholesterol and facilitating intestinal digestion and absorption of dietary fat, steroids, and lipophilic vitamins. BA metabolism is tightly regulated by nuclear receptor signaling to coordinate BA synthetic enzymes and transporters[69]. BA also serves as important signaling molecules in the regulation of glucose and lipid metabolism. Excessive accumulation of cytotoxic BAs in the hepatocytes can cause cell damage leading to inflammation and fibrosis[69]. The nuclear receptor small heterodimer partner (SHP, NR0B2) is an essential component in the negative feedback regulation of bile acid synthesis. In response to the intrahepatic accumulation of BA, nuclear receptor farnesoid X receptor (FXR) activates SHP to repress the expression of two regulatory enzymes sterol 12α -hydroxylase (CYP8B1) and cholesterol 7α -hydroxylase (CYP7A1) for de novo synthesis of BA[70, 71]. The activation of SHP also represses the transactivation of basolateral BA transporter of the hepatocytes such as sodium-taurocholate cotransporting polypeptide (NTCP) to block uptake of BA[72]. Emerging evidence demonstrated the role of H19 in cholestasis through various mechanisms (Fig. 3).
Figure 3. LncRNA H19 regulation network in liver cells under pathological conditions.
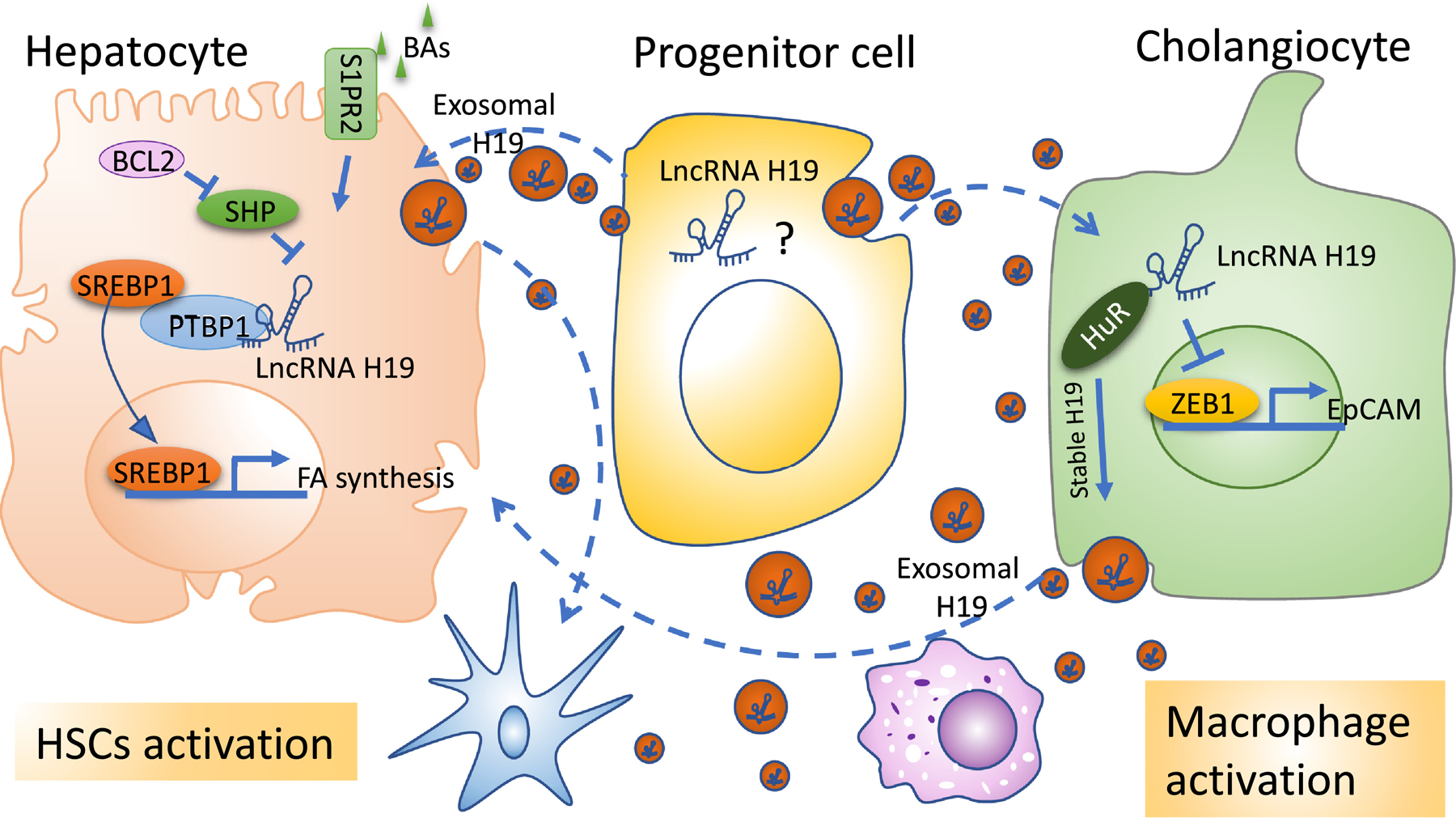
Red sphere: exosomal H19; BAs: bile acids; S1PR2: sphingosine-1-phosphate receptor 2; BCL2: B-cell lymphoma 2; SREBP1: sterol regulatory element-binding transcription factor 1; PTBP1: polypyrimidine tract binding protein 1; FA: fatty acid; HuR: human antigen R; ZEB1: zinc finger E-box binding homeobox 1; EpCAM: epithelial cellular adhesion molecule.
Apoptosis is important in regulating tissue homeostasis. B-cell lymphoma protein 2 (BCL2), an antiapoptotic family member containing four conserved α -helical motifs known as BCL-2 homology (BH1–4) domains and a transmembrane domain (TM), is known to inhibit apoptosis by binding to the pro-apoptotic proteins BCL2-associated X protein (BAX) and BCL2-antagonist/killer 1 (BAK)[72, 73]. BCL2 expression is low in normal hepatocytes; however, its expression is highly induced in cholestasis[74, 75]. Overexpression of hepatic BCL2 disrupts BA homeostasis as manifested by the accumulation of serum BA and bilirubin levels and leads to the induction of hepatic H19 expression[73]. Furthermore, knockdown H19 or re-expressed SHP largely rescued BCL2-induced cholestatic liver injury[73].
Using the BDL-induced cholestasis model, we found that the hepatic let-7 family, a family of miRNAs required for development timing, tumor suppression, and metabolism regulation[76], is markedly induced by H19[17]. H19 promotes the biogenesis and expression of a cluster of let-7 miRNAs, including let-7a-5p, let-7d-5p, and let-7f-5p, in hepatic cholestasis and functionally suppresses their bioavailability. H19 antagonizes the expression of its binding partner, PTBP1, which associates with pre-let-7d and pre-let-7a-1 and inhibits let-7 biogenesis but promotes their bioavailability[17]. H19 also facilitates the development of BDL-induced hepatic cholestasis by preventing Zinc finger E-box binding homeobox 1 (ZEB-1) mediated inhibition of epithelial cell adhesion molecule (EpCAM). ZEB1 is a transcriptional repressor that plays a role in epithelial-to-mesenchymal transition (EMT) during fibrogenesis [77]. Its expression is modulated by SHP; which in turn inhibits H19 gene transcription as described previously[73]. EpCAM is a cell to cell adhesion molecule that is expressed in many human epithelial tissues and is associated with enhanced cellular proliferation[78]. H19 induces EpCAM but inhibits ZEB1 expression in the BDL model and overexpression of ZEB1 or knockdown of EpCAM ameliorates H19-induced cholestasis[78].
H19 is also implicated in biliary atresia, a severe liver disease commonly seen in neonates. Biliary atresia is caused by the obliteration of the intra-and the extrahepatic biliary duct leading to cholestasis and fibrosis[79]. Hepatic H19 expression and the level of serum H19 are associated with the severity of fibrosis in patients with biliary atresia[79]. Additionally, its expression is correlated with the up-regulation of hepatic sphingosine 1-phosphate receptor 2 (S1PR2) and sphingosine kinase 2 (SPHK2) which are known to attribute to cholestatic liver injury by enhancing cholangiocyte proliferation and inflammatory responses[79–81]. H19 deficiency abrogates BDL-induced liver fibrosis and cholangiocyte hyperplasia and attenuates the S1PR2/SPHK2 signaling pathway in the cholestatic liver[79].
Mdr2−/− mouse is another reproducible model to study chronic biliary liver disease[82]. In this model, the expression of hepatic H19 in 100-days-old WT mice was very low with a slight but significant increase in male Mdr2−/− mice. However, there was a marked increase in its expression by 200-fold in female Mdr2−/− mice [83]. Gender difference in the susceptibility for cholestatic liver disease in this mouse model has been observed and that female Mdr2−/− mice develop more severe cholestasis-induced liver injury especially with a more hydrophobic bile acid composition and taurocholate (TCA) when compared to their male counterparts[84]. Further mechanistic experiments revealed that TCA and 17β-estradiol up-regulate H19 expression and that knocking down H19 ameliorates cholestatic liver injury in female Mdr2−/− mice[83]. The mechanism of H19 regulation by 17β-estradiol is still elusive and deserved further studies. The induction of H19 in the cholestatic mouse model appears to be cell-type specific in the liver. H19 expression level in cholangiocytes is significantly up-regulated in both BDL and Mdr2−/− mice and closely related to fibrotic liver injury[47]. Cholangiocyte-derived H19-enriched exosomes promote hepatic fibrosis through hepatic stellate cell (HSC) activation, proliferation, and inflammation through macrophage activation[18, 47]. In summary, aberrant expression of H19 not only contributes to the bile duct ligation (BDL) and Mdr2−/−-induced fibrotic liver injury but also is associated with disease progression in patients with cholestatic liver disease and biliary atresia patients [79].
3.3. Role of H19 in hepatic fibrosis
Hepatic fibrosis is a consequence of the dysregulation of the extracellular matrix (ECM) synthesis secondary to HSC activation. HSCs activated by various toxic stimuli undergo proliferation and myofibroblastic transformation, lose their retinols and produce a considerable amount of ECM, such as alpha-smooth muscle actin (α-SMA) and type I collagen[85, 86]. Hepatocyte damage can also drive HSCs to transform from the quiescent state to activated type by releasing transforming growth factor-β (TGF-β)[86]. As previously mentioned, the regulation of H19 imprinting involves the binding of CpG binding protein MeCP2 based on the methylation status of the ICR[29]. MeCP2 has been shown to orchestrate liver fibrosis and HSC proliferation[87]. In the carbon tetrachloride (CCl4)-induced liver fibrosis model, H19 expression is markedly reduced while the expression of MeCP2 is increased in the activated HSCs; suggesting the negative regulation of H19 by MeCP2[88]. In addition to MeCP2, DNA methyltransferases1 (DNMT1) silencing of H19 also occurs in activated HSCs via the ERK1/2 signaling pathway[89]. More emerging evidence supported that H19 could promote the HSC activation. Recently, H19 was reported to promote the activation of HSC in vitro. The upregulation of H19 aggravates EMT in the liver of CCl4-induced fibrosis through sponged miR-148a[55]. Activation of H19 also enhances retinoic acid signals to induce HSC activation and is associated with retinol metabolism during HSC activation through alcohol dehydrogenase III (ADH3)[90].
3.4. Role of H19 in acute liver failure
Acute liver failure is a life-threatening medical condition manifested by the sudden loss of hepatic function especially in those with underlying liver diseases[91]. The study on the role of H19 in acute liver failure is limited. One study demonstrated that human adipose-derived stem cells secreted extracellular vesicles (EVs) improve survival in the liver failure model (by injecting D-aminogalactose plus lipopolysaccharides intraperitoneally) in rats by releasing H19[92]. H19 reduces hepatocytes’ apoptosis and the apoptotic rate of the H19-EVs group is lower than that of the EVs group without H19, indicating that H19 can enhance the protective effect of EVs on hepatocytes[92].
3.5. Role of H19 in hepatitis B viral (HBV) infection
HBV infection is a major public health problem worldwide, approximately 257 million of the world’s population show serological evidence of chronic past infection. HBV is a double-stranded DNA virus with several serological markers: HBsAg and anti-HBs, HBeAg and anti-HBe, and anti-HBc IgM and IgG. It is by itself not cytopathic and liver damage results from the complex interaction between virus replication and host immune response[93]. H19 is associated with HBV-associated liver injury through its interaction with miRNAs[53, 94]. H19 and miR-675 are up-regulated in chronic HBV patients. Inhibition of H19 or miR-675 in L02 cells increases cell viability, suppresses hepatitis B X protein (HBx)-induced cell apoptosis, oxidative stress, inflammatory cytokine production, and oxidative stress through miR-675’s target gene, peroxisome proliferator-activated receptor-α (PPARα) [53]. The expression of PPARα is decreased in patients with chronic HBV, and there was a negative association between the expression of H19 and PPARα, or between miR-675 and PPARα. Knocking down PPARα leads to the reversal ofthe effects of H19 or miR-675 inhibition in HBx-induced L02 cells through Akt/mTOR signaling[53]. H19 is also highly expressed in HBV-related hepatocellular carcinoma (see more detail on the roles of H19 and hepatocellular carcinoma below)[94]. Its expression is associated with lymph node invasion and distant metastasis and inversely correlated with the overall survival[94]. H19 knockdown represses cellular proliferation and invasion but upregulates apoptosis of HBV-related HCC cells. The function of H19 is mediated through its target, miR-22 with the downstream effects on the expression of genes related to the EMT pathway[94].
3.6. Roles of H19 in hepatocellular carcinoma (HCC)
HCC is one of the leading causes of cancer-related mortality worldwide. H19 expression is increased in several cancers including HCC and that its involvement in cancer initiation, progression, and development suggests the potential of H19 as a diagnostic and prognostic biomarker. [95, 96]. Hypomethylated and hypermethylated profiles of H19 DMR are associated with the aberrant imprinting of IGF2 and H19 in human HCC[97]. H19 induces oxidative stress and MAPK/ERK signaling pathway[98]. It also activates cell division control protein 42 homolog (CDC42)/serine/threonine-protein kinase PAK 1 (P21 Activated Kinase 1) pathway to promote cell proliferation, migration and invasion by targeting miR-15b[99]. H19 can also serve as a molecular sponge for tumor suppressor miRNAs such as miR-326 for HCC pathogenesis[100]. Cancer stem-cell-like (CSC) liver cancer cells displayed aggressive and metastatic phenotype with CD90 positive expression. LncRNA profiling indicated that H19 was enriched in CD90+ cells and released through exosomes. Gain and loss of function experiments revealed H19 played an important role in the exosome-mediated tumor microenvironment and angiogenesis[101]. A recent meta-analysis showed a significant association between H19 polymorphism and cancer susceptibility[102]. The association of single nucleotide polymorphisms (SNPs) of H19 with the risk and prognosis of HCC was determined in 944 samples, 472 with HCC and 472 matched controls. Three SNPs on H19 gene, rs2735971 (G→A), rs2839698 (C→T), rs3024270 (G→C), were studied. The rs2839698(T;T) and rs2839698(C;T) genotypes were found to be associated with a 1.32-fold increased HCC risk comparing to rs2839698(C;C) homozygotes (T;T and C;C representing homozygotes, while C;T representing heterozygote). In the stratified analysis, rs2839698 and rs3024270 were found to be a significant increase in HCC risk especially in those less than 60 years old. Those with rs2839698 had significantly poor prognosis among smokers with HCC[103]. A comprehensive review of H19, tumorigenesis, and HCC has been described elsewhere[96, 104].
4. The roles of IGF2 in liver diseases.
As previously stated, H19 and IGF2 are closely linked and regulated [105]. IGF2, a gene in the H19/IGF2 cluster, shows highly similar patterns of gene expression with H19. IGF2 is the predominant IGF with serum concentration approximately 3 times higher than that of IGF1 [106]. Similar to H19, IGF2 also plays a critical role in the pathogenesis of liver diseases, notably NAFLD and HCC [106]. In the NASH-HCC model, liver IGF2 level started to rise at 3 months at the development of steatosis and significantly increased when HCC developed [107]. Overexpression of IGF2 increases lipid accumulation and promotes hepatic steatosis [108]. The expression of hepatic IGF2 was increased by 40–100 folds in the human HCC liver compared to normal controls [109].
5. Translational applications
Detection of circulating H19 raises the possibility for its use as the biomarkers for diagnosis, risk prediction, and prognosis in liver diseases. Serum H19 was associated with poor disease-free survival in patients with HCC [110]. Modulation of H19 has a profound impact on the liver disease phenotypes. BC819 is a plasmid carrying H19 regulatory sequences which contain 814 bp 5’-flanking region. It was constructed and linked with a fragment of diphtheria toxin (DT-A). Intravesical administration of BC-819 resulted in a 50% reduction of the marker lesion in a phase I/II human study[111] Since the presence of H19 transcription factors only found in tumor cells, the designed plasmid was tested in early phase clinical trials for several types of cancers and HCC [112, 113]. Intra-arterial treatment with the DTA-H19 plasmid significantly delayed the tumor growth and resulted in tumor regression in animals with liver metastases of colon cancer[114]. Furthermore, a double promoter contained both H19 and IGF2-P4 (IGF2-promoter-4) regulatory sequences were constructed to drive DT-A (H19-DTA-IGF2-P4-DTA). These double promoter constructs exhibited superior tumor growth inhibition activity compared to the single promoter vectors [115]. These ongoing studies suggest clinical applicability in the use of H19 as the potential target for liver diseases.
6. Perspectives and conclusions
Since its discovery, we now have a much better understanding of the roles of H19 in the pathogenesis of liver diseases. H19 is implicated in various types of liver diseases through different mechanisms such as epigenetic modification, its effect on target gene promoters and mRNA stability, its role as the sponge for other miRNAs, and its implication in the cross-talk between hepatic and non-hepatic parenchymal cells in the liver (Fig. 4). These regulations trigger downstream, signaling pathways to control lipid and bile acid metabolism, fibrogenesis, cellular proliferation, and tumorigenesis. An improved understanding from these studies will certainly provide us a useful perspective that will eventually lead to therapeutic intervention by targeting H19 in liver diseases.
Figure 4. Summary of the regulation of H19 in liver diseases.
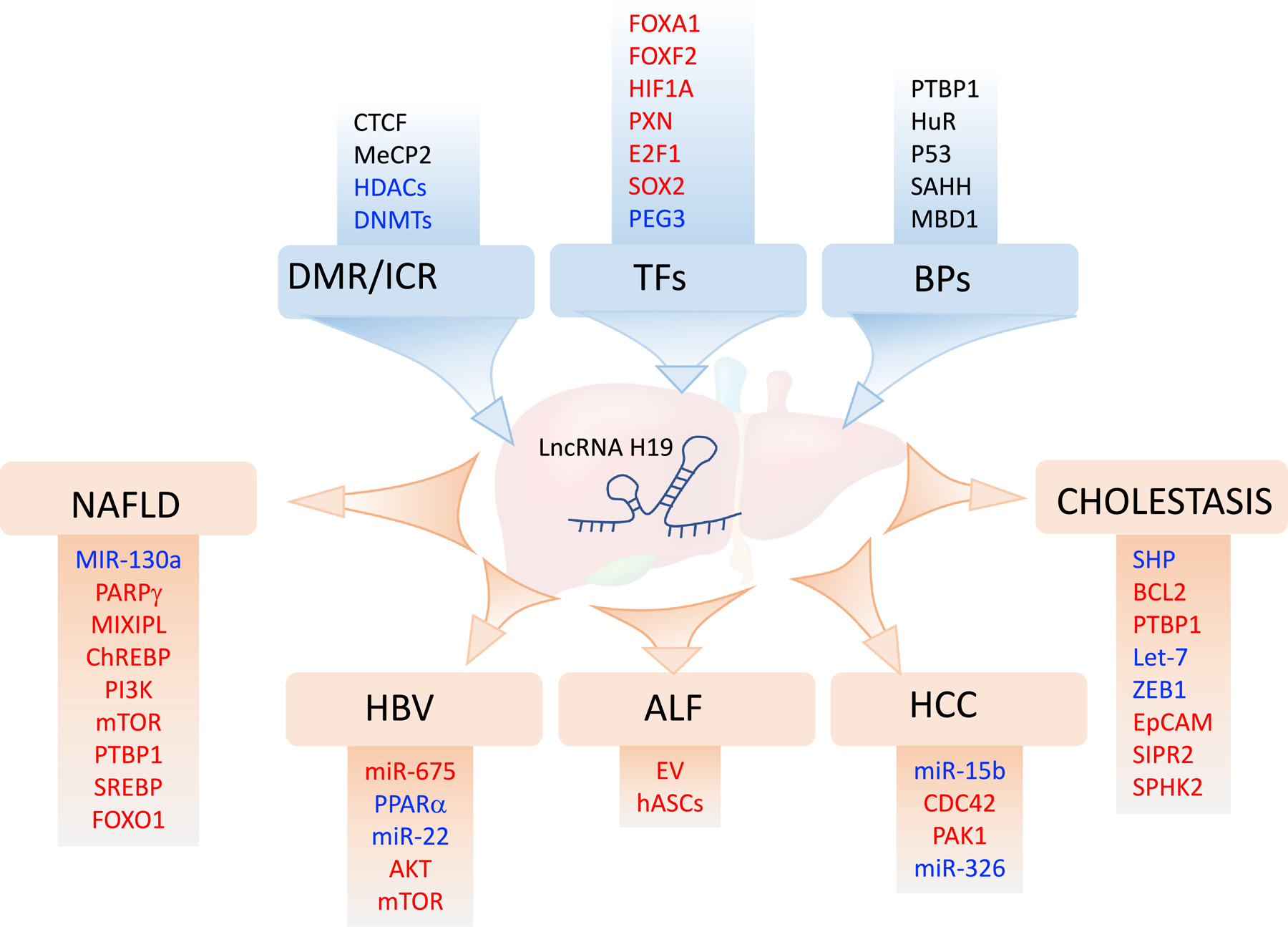
The blue boxes are mechanisms regulating H19 expression. DMR: differentially methylated region; ICR: imprinting control region; TFs: transcriptional factors; BPs: binding partners. The red boxes are the mechanisms of H19 in liver diseases. NAFLD: non-alcoholic fatty liver disease; HBV: hepatitis B virus; ALF: acute liver failure; HCC: hepatocellular carcinoma. The factors in blue represented negative regulators while the factors in red represented positive regulators. The factors in black indicated unknown changes in expression levels.
Acknowledgments
ZY is supported by NIH K01AA26385, Indiana University Research Support Fund Grant (IU RSFG), and the Ralph W. and Grace M. Showalter Research Trust, and Indiana Institute for Medical Research; SL is supported in part by R01 DK107682, R01 AA025208, U01 AA026917, UH2/UH3 AA026903, VA Merit Award 1I01CX000361 and Indiana Clinical and Translational Sciences Institute, UL1TR002529, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award and Showalter Scholar Indiana University School of Medicine and IUSM Dean’s Scholar in Medical Research; PK is supported by the grant from Indiana Institute for Medical Research (IIMR). All the authors have read the journal’s authorship agreement and declare that the manuscript has been reviewed and approved.
Abbreviation
- α-SMA
alpha smooth muscle actin
- ADH3
alcohol dehydrogenase 3
- AFP
alpha fetoprotein
- BA
bile acids
- BAK
BCL2-antagonist/killer 1
- BAX
BCL2-associated X protein
- BCL2
B-cell lymphoma protein 2
- BDL
bile duct ligation
- CCl4
carbon tetrachloride
- CDC42
cell division control protein 42 homolog
- CK19
cytokeratin 19
- ChREBP
carbohydrate-responsive element-binding protein
- CTCF
CCCTC-binding factor
- CYP8B1
cytochrome P450 family 8 subfamily B member 1
- CYP7A1
cytochrome P450 family 7 subfamily A member 1
- DMR
differentially methylated region
- DNMT1
DNA methyltransferases1
- E2F1
E2F transcription factor 1
- EMT
epithelial–mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- EVs
extracellular vesicles
- FFAs
free fatty acids
- FOXA
forkhead box A1
- FOXF2
forkhead box F2
- FOXO1
forkhead box O1
- FXR
farnesoid X receptor
- HBV
hepatitis B viral
- HBx
hepatitis B X protein
- HCC
hepatocellular carcinoma
- HDACs
histone deacetylases
- HIF1α
hypoxia inducible factor 1 subunit alpha
- HNF4α
hepatocyte nuclear factor 4-α
- HSC
hepatic stellate cell
- ICR
imprinting control region
- IGF2
insulin-like growth factor 2
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- Mat
maternal
- MDR2
multidrug resistance 2
- MeCP2
CpG binding protein 2
- MLXIPL
MLX-interacting protein-like
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- ncRNAs
non-coding RNAs
- NTCP
sodium-taurocholate cotransporting polypeptide
- Pat
paternal
- PAK1
P21 activated kinase 1
- PEG3
paternally expressed gene 3
- PPARα
peroxisome proliferator-activated receptor-α
- PPARγ
peroxisome proliferator-activated receptor γ
- PTBP1
polypyrimidine tract-binding protein 1
- PSC
primary sclerosing cholangitis
- PXN
Paxillin
- RAF
regulation of α-fetoprotein
- RIF
regulation of induction of α-fetoprotein
- rRNAs
ribosomal RNAs
- siRNAs
small interfering RNAs
- SOX2
SRY-sex determining region Y- box 2
- SP1
specific protein 1
- SREBP1
sterol regulatory element binding protein1
- SHP
small heterodimer partner
- S1PR2
sphingosine 1-phosphate receptor 2
- SPHK2
sphingosine kinase 2
- SNPs
single nucleotide polymorphisms
- TCA
taurocholate
- TGF-β
transforming growth factor-β
- TM
transmembrane domain
- tRNAs
transfer RNAs
- ZEB-1
zinc finger E-box binding homeobox 1
- ZHX2
zinc fingers and homeoboxes 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All the authors have read the journal’s policy on disclosure of potential conflicts of interest and declare that there are no conflicts of interest to report.
Conflict of interest: None of the authors have any conflicts of interest with this work
Part of the work in this manuscript was presented as an oral presentation at the Annual Meeting of the Central Society for Clinical and Translational Research and Midwest American Federation for Medical Research (CSCTR/AFMR) meeting, Chicago, IL in 2019.
References
- [1].Szabo G, Bala S. MicroRNAs in liver disease. Nature reviews Gastroenterology & hepatology 2013;10:542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett 2014;344:20–7. [DOI] [PubMed] [Google Scholar]
- [4].Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 2013;50:212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Morlando M, Fatica A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology 2014;60:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020;19:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang Z, Ross RA, Zhao S, Tu W, Liangpunsakul S, Wang L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun 2017;1:513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao Y, Wu J, Liangpunsakul S, Wang L. Long Non-coding RNA in Liver Metabolism and Disease: Current Status. Liver Res 2017;1:163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shen A, Ma J, Hu X, Cui X. High expression of lncRNA-SNHG7 is associated with poor prognosis in hepatocellular carcinoma. Oncol Lett 2020;19:3959–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang WJ, Tian XP, Bi SX, Zhang SR, He TS, Song LY, et al. The beta-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene 2020;39,4538–50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chang HL, Bamodu OA, Ong JR, Lee WH, Yeh CT, Tsai JT. Targeting the Epigenetic Non-Coding RNA MALAT1/Wnt Signaling Axis as a Therapeutic Approach to Suppress Stemness and Metastasis in Hepatocellular Carcinoma. Cells 2020;9(4):1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu Y, Qiu Z, Zhang Y, Li B, Jiang X. Partial hepatectomyinduced upregulation of SNHG12 promotes hepatocyte proliferation and liver regeneration. Mol Med Rep 2020;21:1089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li C, Chang L, Chen Z, Liu Z, Wang Y, Ye Q. The role of lncRNA MALAT1 in the regulation of hepatocyte proliferation during liver regeneration. Int J Mol Med 2017;39:347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang L, Damle SS, Booten S, Singh P, Sabripour M, Hsu J, et al. Partial Hepatectomy Induced Long Noncoding RNA Inhibits Hepatocyte Proliferation during Liver Regeneration. PLoS One 2015;10:e0132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chiang JYL. Linking long noncoding RNA to control bile acid signaling and cholestatic liver fibrosis. Hepatology 2017;66:1032–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang L, Yang Z, Huang W, Wu J. H19 potentiates let-7 family expression through reducing PTBP1 binding to their precursors in cholestasis. Cell Death Dis 2019;10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Liu R, Wang Y, Zhu W, Zhao D, Wang X, et al. Cholangiocyte-Derived Exosomal lncRNA H19 Promotes Macrophage Activation and Hepatic Inflammation under Cholestatic Conditions. Cells 2020;9(1):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Di Mauro S, Scamporrino A, Petta S, Urbano F, Filippello A, Ragusa M, et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int 2019;39:1742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Leti F, Legendre C, Still CD, Chu X, Petrick A, Gerhard GS, et al. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl Res 2017;190:25–39 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Atanasovska B, Rensen SS, van der Sijde MR, Marsman G, Kumar V, Jonkers I, et al. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology 2017;66:794–808. [DOI] [PubMed] [Google Scholar]
- [22].Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A 1984;81:5523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 1988;7:673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A 2013;110:20693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 1998;12:3693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 1995;9:407–13. [DOI] [PubMed] [Google Scholar]
- [27].Gebert C, Kunkel D, Grinberg A, Pfeifer K. H19 imprinting control region methylation requires an imprinted environment only in the male germ line. Mol Cell Biol 2010;30:1108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet 2004;36:883–8. [DOI] [PubMed] [Google Scholar]
- [29].Pope C, Mishra S, Russell J, Zhou Q, Zhong XB. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases 2017;5(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A 2006;103:10684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Drewell RA, Goddard CJ, Thomas JO, Surani MA. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res 2002;30:1139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Azmi AS, Bao GW, Gao J, Mohammad RM, Sarkar FH. Network insights into the genes regulated by hepatocyte nuclear factor 4 in response to drug induced perturbations: a review. Curr Drug Discov Technol 2013;10:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Long L, Spear BT. FoxA proteins regulate H19 endoderm enhancer E1 and exhibit developmental changes in enhancer binding in vivo. Mol Cell Biol 2004;24:9601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xu JL, Hua T, Ding J, Fan Y, Liu ZJ, Lian JW. FOXF2 aggravates the progression of non-small cell lung cancer through targeting lncRNA H19 to downregulate PTEN. Eur Rev Med Pharmacol Sci 2019;23:10796–802. [DOI] [PubMed] [Google Scholar]
- [35].Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep 2017;7:45029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Marasek P, Dzijak R, Studenyak I, Fiserova J, Ulicna L, Novak P, et al. Paxillin-dependent regulation of IGF2 and H19 gene cluster expression. J Cell Sci 2015;128:3106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005;280:29625–36. [DOI] [PubMed] [Google Scholar]
- [38].Zhang J, Han C, Ungerleider N, Chen W, Song K, Wang Y, et al. A Transforming Growth Factor-beta and H19 Signaling Axis in Tumor-Initiating Hepatocytes That Regulates Hepatic Carcinogenesis. Hepatology 2019;69:1549–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ye A, He H, Kim J. PEG3 binds to H19-ICR as a transcriptional repressor. Epigenetics 2016;11:889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Z, Yang X, Kai J, Wang F, Wang Z, Shao J, et al. HIF-1alpha-upregulated lncRNA-H19 regulates lipid droplet metabolism through the AMPKalpha pathway in hepatic stellate cells. Life Sci 2020:117818. [DOI] [PubMed] [Google Scholar]
- [41].Choong OK, Chen CY, Zhang J, Lin JH, Lin PJ, Ruan SC, et al. Hypoxia-induced H19/YB-1 cascade modulates cardiac remodeling after infarction. Theranostics 2019;9:6550–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeira E, Abramovitch R, Meir K, Even Ram S, Gil Y, Bulvik B, et al. The knockdown of H19lncRNA reveals its regulatory role in pluripotency and tumorigenesis of human embryonic carcinoma cells. Oncotarget 2015;6:34691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lynch CA, Tycko B, Bestor TH, Walsh CP. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol Cancer 2002;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gao Y, Li S, Zhang Z, Yu X, Zheng J. The Role of Long Non-coding RNAs in the Pathogenesis of RA, SLE, and SS. Front Med (Lausanne) 2018;5:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ignarski M, Islam R, Muller RU. Long Non-Coding RNAs in Kidney Disease. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jiang Y, Huang Y, Cai S, Song Y, Boyer JL, Zhang K, et al. H19 Is Expressed in Hybrid Hepatocyte Nuclear Factor 4alpha(+) Periportal Hepatocytes but Not Cytokeratin 19(+) Cholangiocytes in Cholestatic Livers. Hepatol Commun 2018;2:1356–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, et al. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 2019;70:1317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Reinius B, Kanduri C. Elevated expression of H19 and Igf2 in the female mouse eye. PLoS One 2013;8:e56611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang D, Wang G, Yang H, Liu H, Li C, Li X, et al. DNA methylation modulates H19 and IGF2 expression in porcine female eye. Genet Mol Biol 2017;40:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Perincheri S, Dingle RW, Peterson ML, Spear BT. Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci U S A 2005;102:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, et al. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018;9:18648–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu Y, Xu L, Lu B, Zhao M, Li L, Sun W, et al. LncRNA H19/microRNA-675/PPARalpha axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol Immunol 2019;116:18–28. [DOI] [PubMed] [Google Scholar]
- [54].Yang X, Lou Y, Wang M, Liu C, Liu Y, Huang W. miR675 promotes colorectal cancer cell growth dependent on tumor suppressor DMTF1. Mol Med Rep 2019;19:1481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang Z, et al. H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-beta signaling in both hepatic stellate cells and hepatocytes. J Cell Physiol 2019;234:9698–710. [DOI] [PubMed] [Google Scholar]
- [56].Zhang L, Zhou Y, Huang T, Cheng AS, Yu J, Kang W, et al. The Interplay of LncRNA-H19 and Its Binding Partners in Physiological Process and Gastric Carcinogenesis. Int J Mol Sci 2017;18(2):450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 2018;67:1768–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Khashab MA, Liangpunsakul S, Chalasani N. Nonalcoholic fatty liver disease as a component of the metabolic syndrome. Curr Gastroenterol Rep 2008;10:73–80. [DOI] [PubMed] [Google Scholar]
- [59].Liangpunsakul S, Chalasani N. Lipid mediators of liver injury in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol 2019;316:G75–G81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pan X, Zhang Y, Kim HG, Liangpunsakul S, Dong XC. FOXO transcription factors protect against the diet-induced fatty liver disease. Sci Rep 2017;7:44597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu J, Tang T, Wang GD, Liu B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARgamma axis in non-alcoholic fatty liver disease. Biosci Rep 2019;39(7):BSR20181722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang H, Cao Y, Shu L, Zhu Y, Peng Q, Ran L, et al. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J Cell Mol Med 2020;24:1399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol 2012;47:360–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013;152:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Goyal N, Sivadas A, Shamsudheen KV, Jayarajan R, Verma A, Sivasubbu S, et al. RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci Rep 2017;7:8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang N, Geng T, Wang Z, Zhang R, Cao T, Camporez JP, et al. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 2018;3(10):e120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fawzy MS, Abdelghany AA, Toraih EA, Mohamed AM. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn J Basic Med Sci 2020;20(3):365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Goldstein J, Levy C. Novel and emerging therapies for cholestatic liver diseases. Liver Int 2018;38:1520–35. [DOI] [PubMed] [Google Scholar]
- [69].Chiang JY. Recent advances in understanding bile acid homeostasis. F1000Res 2017;6:2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 2000;6:517–26. [DOI] [PubMed] [Google Scholar]
- [71].Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta 2011;1812:893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, et al. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 2001;121:140–7. [DOI] [PubMed] [Google Scholar]
- [73].Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 2016;6:20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kurosawa H, Que FG, Roberts LR, Fesmier PJ, Gores GJ. Hepatocytes in the bile duct-ligated rat express Bcl-2. Am J Physiol 1997;272:G1587–93. [DOI] [PubMed] [Google Scholar]
- [75].Sanchez-Munoz D, Castellano-Megias VM, Romero-Gomez M. Expression of bcl-2 in ductular proliferation is related to periportal hepatic stellate cell activation and fibrosis progression in patients with autoimmune cholestasis. Dig Liver Dis 2007;39:262–6. [DOI] [PubMed] [Google Scholar]
- [76].Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016;7:100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- [78].Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology 2017;66:1183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xiao Y, Liu R, Li X, Gurley EC, Hylemon PB, Lu Y, et al. Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology 2019;70:1658–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nagahashi M, Takabe K, Liu R, Peng K, Wang X, Wang Y, et al. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology 2015;61:1216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liu R, Zhao R, Zhou X, Liang X, Campbell DJ, Zhang X, et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014;60:908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ikenaga N, Liu SB, Sverdlov DY, Yoshida S, Nasser I, Ke Q, et al. A new Mdr2(−/−) mouse model of sclerosing cholangitis with rapid fibrosis progression, early-onset portal hypertension, and liver cancer. Am J Pathol 2015;185:325–34. [DOI] [PubMed] [Google Scholar]
- [83].Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, et al. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology 2017;66:869–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].van Nieuwerk CM, Groen AK, Ottenhoff R, van Wijland M, van den Bergh Weerman MA, Tytgat GN, et al. The role of bile salt composition in liver pathology of mdr2 (−/−) mice: differences between males and females. J Hepatol 1997;26:138–45. [DOI] [PubMed] [Google Scholar]
- [85].Lee YS, Jeong WI. Retinoic acids and hepatic stellate cells in liver disease. J Gastroenterol Hepatol 2012;27 Suppl 2:75–9. [DOI] [PubMed] [Google Scholar]
- [86].Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 2017;14:397–411. [DOI] [PubMed] [Google Scholar]
- [87].Tao H, Huang C, Yang JJ, Ma TT, Bian EB, Zhang L, et al. MeCP2 controls the expression of RASAL1 in the hepatic fibrosis in rats. Toxicology 2011;290:327–33. [DOI] [PubMed] [Google Scholar]
- [88].Yang JJ, Liu LP, Tao H, Hu W, Shi P, Deng ZY, et al. MeCP2 silencing of LncRNA H19 controls hepatic stellate cell proliferation by targeting IGF1R. Toxicology 2016;359–360:39–46. [DOI] [PubMed] [Google Scholar]
- [89].Yang JJ, She Q, Yang Y, Tao H, Li J. DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate cell activation and fibrosis. Toxicol Lett 2018;295:325–34. [DOI] [PubMed] [Google Scholar]
- [90].Wang ZM, Xia SW, Zhang T, Wang ZY, Yang X, Kai J, et al. LncRNA-H19 induces hepatic stellate cell activation via upregulating alcohol dehydrogenase III-mediated retinoic acid signals. Int Immunopharmacol 2020;84:106470. [DOI] [PubMed] [Google Scholar]
- [91].Lee WM, Squires RH, Jr., Nyberg SL, Doo E, Hoofnagle JH. Acute liver failure: Summary of a workshop. Hepatology 2008;47:1401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Jin Y, Wang J, Li H, Gao S, Shi R, Yang D, et al. Extracellular Vesicles Secreted by Human Adipose-derived Stem Cells (hASCs) Improve Survival Rate of Rats with Acute Liver Failure by Releasing lncRNA H19. EBioMedicine 2018;34:231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol 2019;4:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li L, Han T, Liu K, Lei CG, Wang ZC, Shi GJ. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci 2019;23:5392–401. [DOI] [PubMed] [Google Scholar]
- [95].Ariel I, de Groot N, Hochberg A. Imprinted H19 gene expression in embryogenesis and human cancer: the oncofetal connection. Am J Med Genet 2000;91:46–50. [DOI] [PubMed] [Google Scholar]
- [96].Ghafouri-Fard S, Esmaeili M, Taheri M. H19 lncRNA: Roles in tumorigenesis. Biomed Pharmacother 2020;123:109774. [DOI] [PubMed] [Google Scholar]
- [97].Wu J, Qin Y, Li B, He WZ, Sun ZL. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics 2008;91:443–50. [DOI] [PubMed] [Google Scholar]
- [98].Ding K, Liao Y, Gong D, Zhao X, Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun 2018;502:194–201. [DOI] [PubMed] [Google Scholar]
- [99].Zhou Y, Fan RG, Qin CL, Jia J, Wu XD, Zha WZ. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics 2019;111:1862–72. [DOI] [PubMed] [Google Scholar]
- [100].Wei LQ, Li L, Lu C, Liu J, Chen Y, Wu H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol 2019;234:5153–62. [DOI] [PubMed] [Google Scholar]
- [101].Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 2015;14:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li XF, Yin XH, Cai JW, Wang MJ, Zeng YQ, Li M, et al. Significant association between lncRNA H19 polymorphisms and cancer susceptibility: a meta-analysis. Oncotarget 2017;8:45143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yang ML, Huang Z, Wang Q, Chen HH, Ma SN, Wu R, et al. The association of polymorphisms in lncRNA-H19 with hepatocellular cancer risk and prognosis. Biosci Rep 2018;38(5):BSR20171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J 2018;17:900–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kaffer CR, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol 2001;21:8189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Adamek A, Kasprzak A. Insulin-Like Growth Factor (IGF) System in Liver Diseases. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].De Minicis S, Agostinelli L, Rychlicki C, Sorice GP, Saccomanno S, Candelaresi C, et al. HCC development is associated to peripheral insulin resistance in a mouse model of NASH. PLoS One 2014;9:e97136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kessler SM, Laggai S, Van Wonterg E, Gemperlein K, Muller R, Haybaeck J, et al. Transient Hepatic Overexpression of Insulin-Like Growth Factor 2 Induces Free Cholesterol and Lipid Droplet Formation. Front Physiol 2016;7:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Cariani E, Lasserre C, Seurin D, Hamelin B, Kemeny F, Franco D, et al. Differential expression of insulin-like growth factor II mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosis. Cancer Res 1988;48:6844–9. [PubMed] [Google Scholar]
- [110].Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers 2015;2015:191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol 2008;180:2379–83. [DOI] [PubMed] [Google Scholar]
- [112].Amit D, Tamir S, Hochberg A. Development of targeted therapy for a broad spectrum of solid tumors mediated by a double promoter plasmid expressing diphtheria toxin under the control of IGF2-P4 and IGF2-P3 regulatory sequences. Int J Clin Exp Med 2013;6:110–8. [PMC free article] [PubMed] [Google Scholar]
- [113].Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther 2010;12:607–16. [PubMed] [Google Scholar]
- [114].Sorin V, Ohana P, Mizrahi A, Matouk I, Birman T, Hochberg A, et al. Regional therapy with DTA-H19 vector suppresses growth of colon adenocarcinoma metastases in the rat liver. Int J Oncol 2011;39:1407–12. [DOI] [PubMed] [Google Scholar]
- [115].Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med 2010;8:134. [DOI] [PMC free article] [PubMed] [Google Scholar]


