Abstract
Sterol regulatory element binding protein-1 (SREBP-1), a transcription factor with a basic helix–loop–helix leucine zipper, has two isoforms, SREBP-1a and SREBP-1c, derived from the same gene for regulating the genes of lipogenesis, including acetyl-CoA carboxylase, fatty acid synthase, and stearoyl-CoA desaturase. Importantly, SREBP-1 participates in metabolic reprogramming of various cancers and has been a biomarker for the prognosis or drug efficacy for the patients with cancer. In this review, we first introduced the structure, activation, and key upstream signaling pathway of SREBP-1. Then, the potential targets and molecular mechanisms of SREBP-1-regulated lipogenesis in various types of cancer, such as colorectal, prostate, breast, and hepatocellular cancer, were summarized. We also discussed potential therapies targeting the SREBP-1-regulated pathway by small molecules, natural products, or the extracts of herbs against tumor progression. This review could provide new insights in understanding advanced findings about SREBP-1-mediated lipogenesis in cancer and its potential as a target for cancer therapeutics.
Keywords: SREBP-1, fatty acid synthase, fatty acids, lipogenesis, cancer therapy
Introduction
Sterol regulatory element-binding proteins (SREBPs) are identified as a family of transcription factors with the domain of a basic helix–loop–helix leucine zipper (bHLH-LZ), which regulate genes involved in the pathways of lipid synthesis and uptake (1–3). In mammalian cells, three SREBP isoforms, SREBP-1a, SREBP-1c, and SREBP-2, from two genes, SREBF1 and SREBF2, have been identified, which have overlapping transcriptional programs for the synthesis of fatty acids and cholesterol (4, 5). SREBP-1a and SREBP-1c are derived from the same gene through alternative splicing at transcription start sites (6), but they have different regulations for downstream target genes (4, 7). A study conducted for about 30 years has confirmed that SREBP-1c is involved in regulating fatty acid synthesis and lipogenesis and SREBP-1a can be implicated in two pathways of SREBP-1c and SREBP-2 (specific to cholesterol metabolism) (8). Under pathological conditions, SREBP-1 activation can cause lipid dysfunction to contribute to various metabolic diseases, such as obesity, diabetes mellitus, non-alcoholic fatty liver disease, and cancer (9–12). Recently, more and more evidence has demonstrated that SREBP-1 participates in metabolic reprogramming in different cancer, such as prostate cancer (13), breast cancer (14), and glioblastoma (GBM) (15), which has been a potential target for cancer therapy. Moreover, several essential pathways, such as epidermal growth factor receptor (EGFR), phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB, Akt)/mammalian target of rapamycin (mTOR), and Ras, can regulate SREBP-1 activation to mediate tumor growth and metastasis (15, 16). Importantly, multiple treatment strategies targeting the SREBP-1 signaling pathway, including small molecules or genetic inhibition, have been extensively studied and developed (17, 18). The preparation of the publications in this review was conducted as follows: 1) the articles in English were electronically searched from October 1993 to May 2022 in the databases of PubMed and the Web of Science. 2) “SREBP-1” or “sterol regulatory-element binding protein-1” and “cancer” were used as the search terms. 3) A secondary search was performed by checking the title and the abstract to collect published papers with the inclusion criteria. In this review, we summarized the key upstream signaling pathway and the function of SREBP-1-regulated lipogenesis in different cancer. We also discussed potential therapies targeting the SREBP-1-regulated pathway against tumor progression. This review could provide new insights in understanding advanced findings about SREBP-1-mediated lipid dysfunction in cancer and its potential as a target for cancer therapeutics.
SREBP-1 activation and its downstream targets in cancer
Human gene SREBF1 (26 kb, 22 exons, and 20 introns) at a chromosomal location of 17p11.2 was cloned and characterized in 1995, as a result of alternative splicing at both the 5′ and 3′ ends (19, 20). SREBP-1a and SREBP-1c differ in the extreme N-terminal acidic amino acids, which share a similar structure containing an NH2-terminal transcription factor domain (480 amino acids), a middle hydrophobic region (80 amino acids), and a COOH-terminal regulatory domain (590 amino acids) (3). SREBP-1a and SREBP-1c include 42 amino acids (12 acidic acids) and 24 amino acids (six acidic acids) in the acidic NH2-terminal domain, respectively. SREBP-1c has a much weaker effect in transcription activation than that of SREBP-1a, due to its shortened acidic domain (21). SREBP-1 is conserved from fission yeast to humans (22), and its two isoforms are found in the liver, adipose, and skeletal muscle tissues (20, 23, 24). SREBP-1c is more abundant than SREBP-1a in the liver (21, 25), and SREBP-1a is abundantly expressed in some tissues and cells, such as heart, macrophages, and dendritic cells from bone marrow (26). Recently, it has been revealed that SREBP-1 is obviously activated in different cancers, which is higher than that of non-tumor tissues (13, 27) and has been a biomarker for the prognosis or drug efficacy for patients with cancer (14, 28).
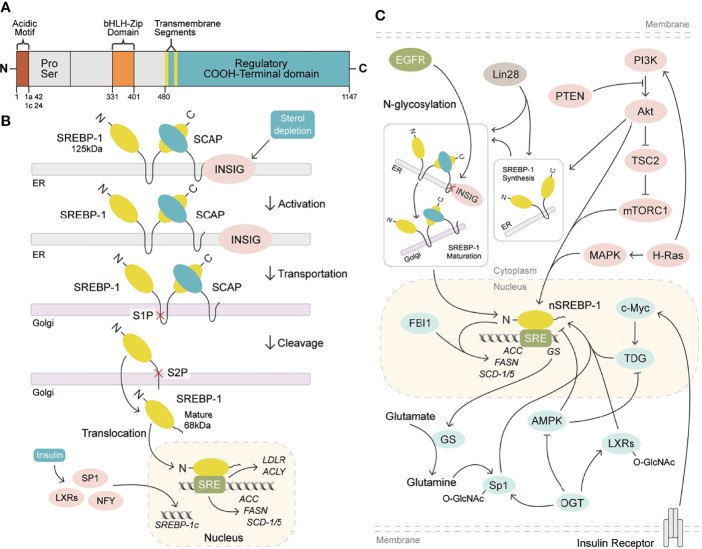
SREBP-1a and SREBP-1c are synthesized as 125-kDa precursors in endoplasmic reticulum membrane and cleaved into an NH2-terminal fragment (68 kDa) by site 1 and site 2 proteases to translocate into the nucleus for lipogenesis gene transcription, when kept in the environment from sterol depletion (29–31). SREBP-1 can form a complex with SREBP cleavage-activating protein (SCAP) to mediate its cleavage in sterol depletion or in the nutritional environment (10, 25). Insulin increase by carbohydrate ingestion can cause the decrease of Insig-2a, which leads to the release of the SREBP-1c/SCAP complex to stimulate SREBP-1c cleavage in the Golgi for the activation of lipogenic/glycolytic genes (25, 32). Moreover, insulin can increase SREBP-1c expression by activating the promoters containing the binding sites for the liver X receptor, Sp1, nuclear factor-Y, and SREBP-1c (33). In addition, the cleavage of SREBP-1c is regulated by polyunsaturated fatty acids, not as SREBP-2 by SCAP or Insig-1 (34). After the cleavage of SREBP-1, the nuclear form of SREBP-1 binds to sterol regulatory elements (SREs) to stimulate the transcriptions of lipogenic genes (25, 29). As reported, SREBP-1 can activate a series of genes for fatty acid synthesis, including ATP citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), and stearoyl-CoA desaturase 1 and 2 (rodent)/5 (human) (SCD-1 and -2/5), and for lipid uptake, such as low-density lipoprotein receptor (LDLR) (4, 30–34). Furthermore, it has been reported that SREBP-1 activation can be involved in various cellular functions, such as cell proliferation, cell cycle, muscle cell differentiation, foam cell formation, and somatic cell reprogramming (35–39). Critically, the activation of SREBP-1 obviously stimulates de novo lipogenesis to regulate cell growth, cell cycle progression, and metastatic characteristics for cancer progression (12, 13, 40–42). Meanwhile, these genes regulated by SREBP-1, including ACLY, ACC, FASN, and SCD, are highly expressed in cancer tissues compared to adjacent non-tumor tissues, which play essential roles in the growth, metastasis, and survival of various cancers (43–46). Taken together, SREBP-1 and its target genes are essential in cancer progression, which have been potential targets for pharmacological or genetic therapy ( Figures 1A, B ).
Figure 1.
SREBP-1 structure, activation, and signaling pathways. (A) SREBP-1 structure and activation. SREBP-1 contains the NH2-terminal domain (the bHLH-Zip motif and an acidic motif), a middle hydrophilic region, and the COOH-terminal domain. (B) After INSIG dissociation from SCAP, SREBP-1 translocates to the Golgi apparatus and is cleaved by site 1 protease (S1P) and site 2 protease (S2P) to form a nuclear form (nSREBP-1) for activating the transcription of its downstream targets, such as FASN, ACC, SCD-1/5, ACLY, and LDLR. (C) Multiple signaling pathways regulate SREBP-1 expression, translocation, and maturation, including EGFR, PI3K/Akt/mTORC1, and others. OGT: O-GlcNAc transferase, GS: glutamate synthetase, TDG: thymine DNA glycosylase.
SREBP-1-regulated lipogenesis in cancers
Currently, multiple signaling pathways control SREBP-1 activation to regulate the downstream genes for lipogenesis, including the EGFR, PI3K/Akt/mTOR, and p53 mutation pathways. EGFR signaling promotes N-glycosylation of SCAP to reduce the association with Insig-1, which consequently induces the proteolytic activation of SREBP-1 (47). Importantly, the hyperactive mutation of PI3K/Akt/mTOR signaling can inhibit oxidative stress and ferroptotic death by regulating SREBP-1/SCD-1-mediated lipogenesis (48). Akt activation can induce the synthesis of full-length SREBP-1 and cause the accumulation of nuclear SREBP-1 to increase the accumulation of intracellular lipids, such as fatty acids and phosphoglycerides, which is sterol sensitive for activating the FASN promoter (16). The mammalian/mechanistic target of rapamycin complex 1 (mTORC1) regulates the transcription and translation of SREBP-1 and SREBP-2 to promote de novo lipid biosynthesis in response to nutrients or insulin, which regulate cell proliferation, mitochondrial metabolism, or glycolytic capacity in cancer (49, 50). Similarly, insulin/glutamine deprivation activates SREBP-1 to bind to the promoter of glutamine synthetase (GS) for its transcription. GS results in the O-linked N-acetylglucosaminylation (O-GlcNAcylation) of specificity protein 1 (Sp1) for the induction of the SREBP-1/ACC1 pathway, which increases lipogenesis and lipid droplet accumulation in liver and breast cancer (51). Based on the key role of O-GlcNAcylation, it has been found that O-GlcNAc transferase (OGT) regulates SREBP-1 expression and its targets in a proteasomal and AMP-activated protein kinase (AMPK)-dependent manner (52). Meanwhile, OGT increases the O-GlcNAcylation of liver X receptors to induce the expression of secretory clusterin (sCLU), which is closely associated with SREBP-1c and its mediated activation of the CLU promoter for the contribution of cisplatin resistance (53). In addition, insulin can activate c-Myc to promote the transactivation of thymine DNA glycosylase (TDG), which decreases the abundance of 5-carboxylcytosine in the SREBP-1 promoter for the upregulation of SREBP-1 and ACC1. On the other hand, AMPK can inhibit DNA demethylation of the SREBP-1 promoter mediated by TDG, suggesting that TDG regulated by insulin or metformin is a potential therapeutic target for T2DM-related cancers (54). In the tumor immune microenvironment, Treg cells maintain the metabolic fitness and mitochondrial integrity of M2-like tumor-associated macrophages indirectly by repressing IFNγ production derived from CD8+ T cells, which is dependent on SREBP-1 and its mediated fatty acid synthesis (55). Under the conditions of serum starvation and hypoxia, cholesterol or O2 deficiency mediates the expression of the FVII gene encoding coagulation factor VII via the SREBP-1/glucocorticoid-induced leucine zipper, which leads to the production and shedding of procoagulant extracellular vesicles (56). A tumor gene, H-ras can upregulate mitogen-activated protein (MAP) kinase and PI3K signaling, which increases SREBP-1 and FASN levels to drive mammary epithelial cell malignant transformation and tumor virulence (57). A proto-oncogenic transcription factor, FBI-1 can directly interact with SREBP-1 via DNA-binding domains to synergistically activate FASN transcription for maintaining rapid cancer cell proliferation (58). An RNA-binding protein, Lin28 enhances SREBP-1 translation and maturation to promote cancer progression by directly binding to the mRNAs of both SREBP-1 and SCAP (59). Taken together, EGFR, PI3K/Akt, and mTOR signal pathways can control SREBP-1 expression and activation to regulate its downstream pathways in cancers ( Figure 1C , Table 1 ).
Table 1.
Main key pathways regulate SREBP-1 activation for lipogenesis.
| Key upstream signal pathways | Molecular mechanism | Refs |
|---|---|---|
| EGFR | Promotes SCAP N-glycosylation to reduce the interaction of Insig-1 for SREBP-1 activation | (47) |
| Mutation of PI3K/Akt/mTOR | Inhibits oxidative stress and ferroptotic death by regulating the SREBP-1/SCD-1 pathway | (48) |
| Akt | Induces full-length SREBP-1 synthesis and causes nuclear SREBP-1 accumulation for increased intracellular lipid | (16) |
| mTORC1 | Activates SREBP-1 transcription to regulate lipid synthesis in vitro | (49) |
| mTORC1 | Regulates the translation and transcription of PGC-1α, SREBP-1/2, and HIF-1α and the translation of nucleus-encoded mitochondrial mRNAs | (50) |
| Glutamine synthetase | Causes the O-linked N-acetylglucosaminylation of the specificity protein 1 to induce the SREBP-1/ACC1 pathway for increased lipogenesis under the conditions of insulin/glutamine deprivation-induced SREBP-1 activation | (51) |
| O-GlcNAc transferase | Regulates SREBP-1 expression and its transcriptional targets, FASN, ACLY, ACC, and lipoprotein lipase via AMPK | (52) |
| O-GlcNAc transferase | Enhances secretory clusterin expression via liver X receptors and SREBP-1 for regulating cell cycle, apoptosis, and cisplatin resistance | (53) |
| Insulin | Upregulates c-Myc, SREBP-1, and ACC1 expression to transactivate thymine DNA glycosylase for the decrease of 5-carboxylcytosine abundance in the SREBP-1 promoter | (54) |
| Treg cells | Promotes SREBP-1-mediated lipid metabolism of M2-like tumor-associated macrophages by suppressing interferon-γ secretion from CD8+ T cells | (55) |
| Cholesterol starvation and hypoxia | Mediates coagulation factor VII expression by regulating the SREBP-1/glucocorticoid-induced leucine zipper pathway | (56) |
| H-ras | Upregulates MAPK and PI3K signaling to increase the levels of SREBP-1 and FASN for driving malignant transformation and tumor virulence | (57) |
| FBI-1 | Directly interacts with the DNA-binding domains of SREBP-1 to activate FASN transcription for rapid cancer cell proliferation | (58) |
| Lin28 | Enhances the translation and maturation of SREBP-1 by directly binding the mRNAs of both SREBP-1 and SCAP for cancer progression | (59) |
Colorectal cancer
The SREBP-1 expression in colorectal cancer (CRC) tissues is significantly higher than that in non-cancerous or normal tissues (60, 61). The colocalization of SREBP-1 and FASN is found in CRC tissues, and the transcriptional regulation of SREBP-1 on the FASN promoter can respond to the deficiency in endogenous fatty acid synthesis in colorectal neoplasia (62). Moreover, the combination of SREBP-1 and ACLY can significantly promote CRC cell proliferation, migration, and invasion, which is mediated by lipid synthesis modulation (63). In addition, the overexpression of SREBP-1 promotes the angiogenesis and invasion of CRC cells by promoting MMP-7 expression related to NF-κB p65 phosphorylation (61). SREBP-1 silencing decreases the levels of fatty acid and SREBP-1 targets genes to inhibit the initiation and tumor progression of CRC, which might be associated with the alternation of cellular metabolism, including mitochondrial respiration, glycolysis, and fatty acid oxidation (64). SREBP-1 also can interact with c-MYC to enhance its binding ability to the snail family transcriptional repressor 1 (SNAIL) promoter to regulate its expression and promote epithelial–mesenchymal transition in CRC (65). For the resistance of chemotherapy in CRC, SREBP-1 is overexpressed in chemoresistant CRC samples and is correlated with poor survival. SREBP-1 can increase the chemosensitivity to gemcitabine through downregulation of caspase-7, which might be a new prognostic biomarker of CRC (66). Meanwhile, the overexpression of SREBP-1 enhances the resistance of 5-FU through regulation of caspase-7-dependent PARP1 cleavage in CRC (60). On the other hand, radiation stimuli can rapidly increase SREBP-1/FASN signaling to cause cholesterol accumulation, cell proliferation, and cell death, suggesting that targeting the SREBP-1/FASN/cholesterol axis is a potential strategy for CRC patients undergoing radiotherapy (67). Recently, results from colon cancer cells and xenograft tumor models have demonstrated that RAS protein activator-like 1 inhibits colon cancer cell proliferation by regulating LXRα/SREBP1c-mediated SCD1 activity (68). An lncRNA ZFAS1 can bind polyadenylate-binding protein 2 to stabilize and increase the levels of SREBP-1 and its targets, FASN and SCD1, for the promotion of lipid accumulation in CRC (69). In rapamycin-resistant colon cancer cells, diacylglycerol kinase zeta can promote mTORC1 activation and cell-cycle progression, which are essential for SREBP-1 expression (70). Together, these findings demonstrate that SREBP-1 participates in colon cancer growth, invasion, and the resistance of chemotherapy or radiation, which have been potential therapeutic targets for CRC treatment.
Prostate cancer
During the castrated progression of a prostate cancer xenograft model, SREBP-1a and SREBP-1c are significantly greater than precastrated levels. The staining for the clinical specimens shows a high level of SREBP-1 compared with non-cancerous prostate tissue and SREBP-1 is decreased after hormone withdrawal therapy for 6 weeks (71). SREBP-1 and its downstream effectors, FASN and ACC, are upregulated in a TCGA cohort of prostate cancer and late-stage transgenic adenocarcinoma of mouse prostate tumor (72). Furthermore, SREBP-1 expression is positively associated with the clinical Gleason grades and promotes prostate cancer cell growth, migration, and castration-resistant progression, which may be mediated by the alterations of metabolic signaling networks, including androgen receptor (AR), lipogenesis, and NAPDH oxidase 5-induced oxidative stress (13). More importantly, the results from overexpression or RNA interference of SREBP-1c demonstrate that SREBP-1c specifically inhibits AR transactivation by binding to the endogenous AR target promoter, which may be associated with the recruitment of histone deacetylase 1, suggesting that SREBP-1 plays an important role in regulating AR-dependent prostate cancer growth (73). On the other hand, AR and mTOR bind to the regulatory region of SREBP-1 to promote its cleavage and translocation to the nucleus. The synthetic androgen R1881 can induce SREBP-1 recruitment to regulatory regions of its targets, which is abrogated by the inhibition of the mTOR signaling pathway. These data suggest that the AR/mTOR complex can promote SREBP-1 expression and activity to activate its downstreams for reprogramming lipid metabolism (74). Additionally, phospholipase Cϵ, Kruppel-like factor 5 (KLF5), and protein kinase D3 can directly or indirectly interact with SREBP-1 to control lipid metabolism in the occurrence and progression of prostate cancer (75–77). Considering the key roles of miRNAs or lncRNAs, miRNA-21 transcriptionally regulates insulin receptor substrate 1/SREBP-1 signaling pathway to promote prostate cancer cell proliferation and tumorigenesis (18). Two other miRNAs, miRNA-185 and miRNA-342, can block SREBP-1/2 and its downregulated targets, FASN and hydroxy-3-methylglutaryl CoA reductase (HMGCR), to inhibit prostate cancer tumorigenicity (78). A long non-coding RNA molecule, PCA3 is inversely correlated with miRNA-132-3p to regulate the antimony-induced lipid metabolic imbalance, which is closely related to the SREBP-1 protein level, not influencing the SREBP-2 expression in mRNA and protein levels (79). Collectively, these data from cell and animal models or clinical specimens demonstrate that SREBP-1 plays a very important role in prostate cancer progression and that multiple signaling pathways, such as AR and miRNAs, can regulate SREBP-1 activation to mediate the dysfunction of lipid metabolism in prostate cancer.
Breast cancer
As the pattern in other cancers, the mRNA and protein levels of SREBP-1 are higher in tissues of breast cancer, compared with paracancerous tissues. A higher expression of SREBP-1 is positively correlated with tumor differentiation, metastatic stage, and metastasis in lymph nodes. Moreover, in vitro studies reveal that SREBP-1 inhibition can inhibit the cell migration and invasion of breast cancer cells, such as MDA-MB-231 and MCF-7 (14). In aromatase inhibitor-resistant breast cancer cells, SREBP-1 can drive Keratin-80 upregulation to directly promote cytoskeletal rearrangements, cellular stiffening, and cell invasion (80). In MCF-10a breast epithelial cells with H-ras transformation and inhibition of MAP kinase, SREBP-1c and FASN are upregulated and SREBP-1a shows less change. In a panel of samples with primary human breast cancer, SREBP-1c and FASN are increased and correlated with each other (81). miRNA expression profiling shows that miRNA-18a-5p is most evidently decreased in breast cancer cells, which can target SREBP-1 to repress E-cadherin expression. This suggests that the miR-18a-5p/SREBP-1 axis plays a crucial role in the epithelial–mesenchymal transition and metastasis of advanced breast cancer (82). A hormone derived from adipose tissue, leptin can induce ATP production from fatty acid oxidation and intracellular lipid accumulation in MCF-7 cells and tumor xenograft models, which is mediated by SREBP-1 induction and autophagy in breast cancer (83). Additionally, two key molecules, glucose-regulated protein 94 (GRP94) and nuclear protein p54(nrb)/Nono, can regulate the SREBP-1 signaling pathway in in vitro and in vivo models, which have been potential targets for breast cancer treatment (84, 85). GRP94 can upregulate genes for fatty acid synthesis and degradation, including SREBP-1, LXRα, and acyl-CoA thioesterase 7, to favor the progression of breast cancer metastasis (84). As shown in immunohistological staining for human breast cancer tissues, the level of p54(nrb) is positively correlated with SREBP-1a, and its conserved Y267 residue is required for the interaction with nuclear SREBP-1a. It suggests that p54(nrb) is a key regulator for the nuclear form of SREBP-1a, which might be a potential target for treating breast cancer (85). Taken together, SREBP-1 can favor breast cancer progression, which can be regulated by multiple signal molecules, such as miRNA-18a-5p, GRP94, and p54(nrb).
Glioblastoma
In clinic, the therapeutic efficacy of drugs on GBM might be closely related to EGFR amplification, its mutations, or PI3K hyperactivation (86). EGFR induces the SREBP-1 cleavage and nuclear translocation, which is mediated by Akt but is independent on mTORC1 (40). EGFR-activating mutation (EGFRvIII) can promote SREBP-1 cleavage and cause a higher expression of nuclear SREBP-1 to elevate LDLR expression in GBM patient samples, tumors cells, and mouse models, which is dependent on the PI3K/Akt pathway (15, 87). Moreover, EGFR signaling binds to specific sites of the enhanced miRNA-29 promoter to increase its expression in GBM cells by upregulating SCAP/SREBP-1 whereas miRNA-29 can suppress SCAP and SREBP-1 expression to inhibit lipid synthesis in GBM (88, 89). Additionally, it is found that SREBP-1 depletion can induce apoptosis and endoplasmic reticulum stress in U87 GBM cells and tumor xenograft models, which is the result of Akt/mTORC1 signaling-mediated cancer growth and survival (90). Except EGFR and Akt/mTORC1 signaling, sterol O-acyltransferase highly expressed in GBM can control the cholesterol esterification and storage as a key player via upregulation of SREBP-1 (91). Collectively, the oncogenic EGFR/PI3K/Akt pathway upregulates SREBP-1 activation to control lipid metabolism in GBM.
Hepatocellular carcinoma
Lipogenesis participates in the initiation and progression of HCC, one of the leading causes of cancer-related deaths worldwide. Multiple pathways can control SREBP-1 to regulate lipogenesis in HCC, such as hepatoma-derived growth factor (HDGF), apoptosis-antagonizing transcription factors (AATF), or acyl-CoA synthetases. HDGF is found to be closely associated with HCC prognosis, which is coexpressed with SREBP-1. The changes of the first amino acid or the type of PWWP domain are crucial for HDGF-mediated SREBP-1 activation for the lipogenesis promotion in HCC (92). In human non-alcoholic steatohepatitis, HCC tissues, and various human HCC cell lines, AATF expression is higher, which can be induced by TNF-α. Promoter analysis reveals that AATF can bind SREBP-1c to regulate HCC cell proliferation, migration, colony formation, and xenograft tumor growth and metastasis (93). As an HCC-associated tumor suppressor, zinc fingers and homeoboxes 2 (ZHX2) is negatively associated with SREBP-1c in HCC cell lines and human specimens. ZHX2 can increase miRNA-24-3p at the transcription level to induce SREBP-1c degradation for the suppression of HCC progression (94). A member of the acyl-CoA synthetase (ACS) family, acyl CoA synthetase 4 (ACSL4) has been identified as an oncogene and a novel marker of alpha-fetoprotein-high subtype HCC. ACSL4 upregulates SREBP-1 and its downstream lipogenic enzymes via c-Myc to modulate de novo lipogenesis during the progression of HCC (95). A tissue microarray analysis shows that 60 of 80 HCC tissues have a positive staining and high expression of spindlin 1 (SPIN1), which are positively associated with malignancy of HCC. SPIN1 coactivates and interacts with SREBP-1c to increase the levels of intracellular triglycerides, cholesterols, and lipid droplets, which can enhance HCC growth (96). Hepatitis B virus (HBV)-encoded X protein (HBx) significantly promotes HepG2 cell proliferation and lipid accumulation through increased protein levels of C/EBPα, SREBP-1, FASN, and ACC1 (97). Histone deacetylase 8 (HDAC8) is directly upregulated by SREBP-1 where it is coexpressed in dietary obesity models of HCC. HDAC8 can inhibit cell death mediated by p53/p21 and cell-cycle arrest at the G2/M phase and promote β-catenin-dependent cell proliferation by the interaction with chromatin modifier EZH2 to repress the Wnt antagonist in HCC (98). The deficiency of signal transducers and activators of transcription 5 (STAT5) causes the upregulation of SREBP-1 and peroxisome proliferator-activated receptor γ signaling. The combined loss of STAT5/glucocorticoid receptor can cause approximately 60% HCC at 12 months, which is associated with enhanced TNF-α and ROS and the activation of c-Jun N-terminal kinase 1 and STAT3 (99). Additionally, neddylation by UBC12 can prolong SREBP-1 stability with decreased ubiquitination. In human specimens of HCC and the Gene Expression Omnibus (GEO) database, the SREBP-1 level is positively correlated with UBC12 and contributes to HCC aggressiveness, which can be blocked by an inhibitor of NEDD8-activating enzyme E1, MLN4924 (100). Importantly, both mTOR signaling and SREBP-1 can increase fatty acid desaturase 2 expression to activate sapienate metabolism in the HCC cells and xenograft models (101). Certainly, SREBP-1 expression in HCC tissues is significantly higher than that in adjacent tissues, especially in large tumor sizes, high histological grade, and stage of tumor node metastasis (TNM). The level of SREBP-1 is correlated with a worse 3-year overall and disease-free survival of HCC patients, which is not dependent on the prediction for the prognosis. Moreover, SREBP-1 overexpression or knockdown can regulate cell proliferation, invasion, and migration of HCC cells (42). Especially, SREBP-1c, rather than SREBP-1a, is elevated and activated in HCC (102). Collectively, multiple upstream molecules, such as HDGF, AATF, ZHX2, ACSL4, SPIN, HBx, HDAC8, and STAT5, can regulate SREBP-1 expression, activation, and stability to promote HCC proliferation, invasion, and migration for tumor growth and metastasis.
Lung cancer
The phosphorylation of Akt-mediated phosphoenolpyruvate carboxykinase 1 (PCK1) at S90 can reduce its gluconeogenic activity and use GTP as a phosphate donor to phosphorylate Insig1/2 in the endoplasmic reticulum, which activates SREBP signaling for lipid synthesis in cancer (103). In non-small cell lung cancer (NSCLC), PCK1 can promote nuclear SREBP-1 activation by phosphorylating Insig1/2, which is associated with TNM stage and progression of NSCLC (104). The mature form of SREBP-1 is overexpressed and correlated with Akt phosphorylation at Ser473 for lipid synthesis, which may be reversed by protein arginine methyltransferase 5 (PTMI5) knockdown in lung adenocarcinoma (105). B7-H3, a glycoprotein, results in aberrant lipid metabolism in lung cancer, which is mediated by the SREBP-1/FASN signaling pathway (106). In mutant KRAS-expressing NSCLC, KRAS increases SREBP1 expression in part by MEK1/2 signaling and SREBP-1 knockdown significantly inhibits cell proliferation through regulating mitochondrial metabolism (107). In EGFR-mutant NSCLC cells, SREBP-1 is a key feature of the resistance to gefitinib, which might be a promising target for the treatment of EGFR mutation in lung cancer (108). As that in other cancers, miRNA-29 reduces lung cancer proliferation and migration through inhibition of SREBP-1 expression by interacting with the 3′-UTR of SREBP-1 (109). Taken together, Akt activation and its mediated PCK1, B7-H3, and KRAS can regulate SREBP-1 expression and activation in lung cancer progression.
Skin cancer
In melanomas, SREBP-1 predominantly binds to the transcription start sites of genes of de novo fatty acid biosynthesis (DNFA). The inhibition of DNFA or SREBP-1 by enzyme inhibitors or antisense oligonucleotides exerts obvious anticancer effects on both BRAFi-sensitive and -resistant melanoma cells (27). BRAF inhibition can induce SREBP-1 downregulation to inhibit lipogenesis, and SREBP-1 inhibition can overcome the acquired resistance to BRAF-targeted therapy in both in vitro and in vivo models (110). In addition, the PI3K/Akt/mTORC1 pathway, not by BRAF mutation, can regulate SREBP-1 activity and subsequent cholesterogenesis in melanoma (111). In another type of skin cancer, squamous cell carcinomas (SCCs), SREBP-1 can link tumor protein 63 (TP63)/KLF5 to regulate fatty acid metabolism, including fatty acid, sphingolipid, and glycerophospholipid biosynthesis, by binding to hundreds of cis-regulatory elements, which is associated with SCC viability, migration, and poor survival of patients (112). Thus, multiple signal pathways, such as BRAF, PI3K, and TP63/KLF5, can control SREBP-1 for the regulation of skin cancer progression.
Other cancers
In esophageal tumors, SREBP1 is overexpressed and associated with worse overall survival rate of patients. Overexpressing SREBP1 in OE33 cells leads to increased biological phenotypes, including colony formation, migration, and invasion, which is mediated by reduced mesenchymal markers (vimentin, zinc finger E-box binding homeobox 1 (ZEB1), N-cadherin) and increased epithelial markers (E-cadherin). Mechanistically, SREBP-1 can regulate T-cell factor 1/lymphoid enhancer factor 1 and their target proteins, such as CD44 and cyclin D1, and increase the levels of SCD-1, phosphorylated GSK-3β, and nuclear β-catenin for the proliferation and metastasis of esophageal carcinoma (113, 114). SREBP-1 and ZEB1 are potential targets of miRNA-142-5p, a tumor suppressor, and they are associated with tumor progression and poor prognosis (114). Moreover, the levels of PCK1 pS90, Insig1 pS207/Insig2 pS151, and SREBP-1 are associated with the tumor, metastasis stage, and progression of esophageal squamous cell carcinoma, suggesting PCK1 activity-regulated SREBP-1 as a potential target for the diagnosis and treatment of esophageal cancer (115). In gastric cancer tissues and cells, SREBP-1c is activated and promotes the expressions of FASN and SCD-1 to mediate malignant phenotypes, which has been identified as a potential target for the treatment of gastric cancer (116). In the poorest cancer of the digestive system, pancreatic cancer, the lipogenic liver X receptor (LXR)–SREBP-1 axis controls polynucleotide kinase/phosphatase (PNKP) transcription for regulating cancer cell DNA repair and apoptosis. Compared with the adjacent tissues, the levels of LXRs and SREBP-1 are significantly reduced in the tumor tissues, which are inhibited by a small molecule, triptonide, by activating p53 and DNA strand break for cell death (117). Insulin induces transgelin-2 via SREBP-1-mediated transcription, which has been a novel therapeutic target for diabetes-associated pancreatic ductal adenocarcinoma (118). High glucose can enhance the SREBP-1 level to promote tumor proliferation and suppress apoptosis and autophagy (119). A high expression of TNF-α is associated with significant reductions of SREBP-1, FASN, and ACC at the mRNA level in pancreatic ductal adenocarcinoma (120). Importantly, SREBP-1 is overexpressed in both pancreatic cancer tissues and cell lines, which is critical for cancer proliferation, apoptosis, tumor growth, and overall survival (41). Therefore, the SREBP-1 signaling axis has been a promising target for the prevention and treatment of pancreatic cancer.
Compared with controls, SREBP-1 is significantly increased and positively correlated with serum triglyceride in endometrial cancer (121). Ten single-nucleotide polymorphisms (SNPs) of SREBP-1 are associated with endometrial cancer, and rs2297508 SNP with the C allele accounts for 14%, which may serve as a genetic marker for early detection (122). In a progestin-resistant Ishikawa cell line, sirtuin 1 (SIRT1) and SREBP-1 at the mRNA and protein levels are upregulated, while progesterone receptor and forkhead transcription factor 1 (FoxO1) are downregulated, suggesting that the SIRT1/FoxO1/SREBP-1 pathway is involved in the progestin resistance of endometrial cancer (123). It is also found that FoxO1 can inhibit proliferative and invasive capacities by directly inhibiting SREBP-1 in this cancer (124). Salt-inducible kinase 2 can upregulate SREBP-1c to enhance fatty acid synthesis and SREBP-2 to promote cholesterol synthesis in in vitro and in vivo ovarian cancer models (125). In the obesity host, increased SREBP-1 is linked to ovarian cancer progression and metastasis (126) and meditates malignant characteristics, such as cell proliferation, migration, invasion, and tumor growth (127). In clear cell renal cell carcinoma (ccRCC) patients, high levels of E2F transcription factor 1 (E2F1) and SREBP-1 are associated with poor prognosis. E2F1 can increase ccRCC cell proliferation and epithelial–mesenchymal transition (EMT) and promote tumor progression of a mouse xenograft model by inhibiting SREBP-1-mediated aberrant lipid metabolism (128). In bladder cancer, PKM2 interacts with SREBP-1c through the regulation of the Akt/mTOR signaling pathway, which in turn activates FASN transcription for tumor growth (129). Collectively, SREBP-1-regulated lipid metabolism is essential for the cancer progression of the urinogenital system, including endometrial, ovarian, and bladder cancer and renal cell carcinomas. The results from cell lines and clinical tissues demonstrate that long intergenic non-protein-coding RNA 02570 can adsorb miRNA-4649-3p to upregulate SREBP-1 and FASN, which promotes the progression of nasopharyngeal carcinoma (NPC) (130). Epstein–Barr virus-encoded latent membrane protein 1 is expressed in NPC and increases the maturation and activation of SREBP-1 to induce de novo lipid synthesis in the tumor growth and metastasis, which can be inhibited by two inhibitors, luteolin and fatostatin (131). In oral squamous cell carcinoma, glutathione peroxidase 4 (GPX4) can reduce the ferroptosis-mediated cell number via downregulation of SREBP-1 (132). Compared with normal thyroid tissue or benign thyroid nodules, SREBP1 expression is significantly higher in invasive thyroid cancer, which is associated with extrathyroidal extension, advanced stage, and shorter survival of patients (28, 133). The in vitro results demonstrate that SREBP-1 can obviously increase the rate of oxygen consumption and the capacities of invasion and migration in thyroid cancer cells, which is mediated by the upregulation of the Hippo-YAP/CYR61/CTGF pathway (133). In addition, the nuclear form of SREBP-1 (nSREBP-1) can bind to the promoter of protein kinase RNA-like endoplasmic reticulum kinase (PERK) to augment its expression and phosphorylation, which might be the main reason that nSREBP-1 regulates cell proliferation, cell cycle arrest, apoptosis, and autophagy under endoplasmic reticulum (ER) stress in sarcomas (chondrosarcoma and osteosarcoma) (134). Together, SREBP-1 is upregulated and participates in the progression of NPC, oral squamous cell carcinoma, thyroid cancer, and sarcomas. Collectively, these findings indicate that the various key pathways that control SREBP-1 activation and SREBP-1-regulated lipogenesis significantly participate in tumor growth and progression and may be a promising target in multiple malignancies ( Table 2 , Figure 2 ).
Table 2.
SREBP-1-regulated lipogenesis in different cancers.
| Cancer type | Targets | Molecular mechanism | Refs |
|---|---|---|---|
| Colorectal cancer | SREBP-1/FASN | Is higher than that in non-cancerous tissues and regulates the binding on FASN promoters for cancer proliferation, invasion, and migration | (60, 62) |
| Colorectal cancer | SREBP-1/MMP-7/NF-κB p65 phosphorylation | Promotes angiogenesis, invasion, and metastasis | (61) |
| Colorectal cancer | SREBP-1/ACLY | Enhances biological behaviors, including cell proliferation, DNA reproductions, apoptosis, invasion, and migration and regulates lipid production and related metabolic pathways | (63) |
| Colorectal cancer | SREBP-1-mediated the alternation of cellular metabolism | Decreases fatty acid levels to inhibit the initiation and tumor progression | (64) |
| Colorectal cancer | SREBP-1/c-MYC/SNAIL | Enhances the binding to the SNAIL promoter to increase its expression and promote epithelial–mesenchymal transition | (65) |
| Colorectal cancer | SREBP-1/caspase-7 | Regulates the chemosensitivity to gemcitabine or the resistance of 5-FU | (60, 66) |
| Colorectal cancer | SREBP-1/FASN | Is increased by radiation stimuli and causes cholesterol accumulation, cell proliferation, and apoptosis | (67) |
| Colorectal cancer | RAS protein activator like 1/SREBP-1c/LXRα/SCD-1 | Inhibits cell proliferation and tumor growth by directly binding SREBP-1c, LXRα, and SCD-1 promoter | (68) |
| Colorectal cancer | LncRNA ZFAS1/PABP-2/SREBP-1 | Binds polyadenylate-binding protein 2 (PABP-2) to stabilize SREBP-1 at the mRNA level to regulate lipid accumulation for malignant phenotype | (69) |
| Colorectal cancer | Diacylglycerol kinase zeta/mTORC1/SREBP-1 | Promotes mTORC1 activation to regulate diacylglycerol and phosphatidic acid metabolism for maintaining tumor cell growth and survival | (70) |
| Prostate cancer | SREBP-1a/SREBP-1c | Are significantly greater than non-cancerous tissues and precastration and is decreased after hormone withdrawal therapy | (71) |
| Prostate cancer | SREBP-1/FASN/ACC | Are upregulated and associated with clinical Gleason grades, and castration-resistant progression | (13, 72) |
| Prostate cancer | SREBP-1/AR | Promotes cell growth, migration, and progression by regulating metabolic and signaling networks, including AR, lipogenesis, and oxidative stress | (13) |
| Prostate cancer | SREBP-1c/AR/HDAC1 | Inhibits AR transactivation and is associated with the recruitment of histone deacetylase 1 for AR-dependent cancer growth | (73) |
| Prostate cancer | AR/mTOR/SREBP-1 | Binds the regulatory region of SREBP-1 to promote the cleavage and nuclear translocation for lipid metabolism reprogramming | (74) |
| Prostate cancer | PLCϵ/AMPKα/SREBP-1 | Is elevated and induce SREBP-1 nuclear translocation through the inhibition of AMPKα phosphorylation for cancer lipid metabolism and malignant progression | (75) |
| Prostate cancer | KLF5/SREBP-1/FASN | Binds to SREBP-1 to enhance the promoter activity of FASN for androgen-dependent induction | (76) |
| Prostate cancer | Protein kinase D3/SREBP-1/FASN/ACLY | Interacts with SREBP-1 to regulate the levels of lipogenic genes, such as FASN and ACLY for de novo lipogenesis and cancer progression | (77) |
| Prostate cancer | miRNA-21/IRS1/SREBP-1/FASN/ACC | Downregulates insulin receptor substrate 1 (IRS1)-medicated transcription to decrease the levels of SREBP-1, FASN, and ACC for tumorigenesis | (18) |
| Prostate cancer | miRNA-185/-342/SREBP-1/2 | Blocks SREBP-1/2 and its downstream targets to inhibit tumorigenicity | (78) |
| Prostate cancer | lncRNA PCA3/miRNA-132-3p/SREBP-1 | Is inversely correlated with the microRNA-132-3p level to interact SREBP-1 for antimony-induced lipid metabolic disorder | (79) |
| Breast cancer | SREBP-1 | Is higher than paracancerous tissues and positively correlated with tumor differentiation, metastatic stage, and lymph node metastasis | (14) |
| Breast cancer | SREBP-1/keration-80 | Drives keration-80 upregulation to promote cytoskeletal arrangement and invasive characteristic for aromatase inhibitor resistance | (80) |
| Breast cancer | MAPK/PI3K/SREBP-1c/FASN | Are upregulated and correlated with each other, and participate in the H-ras transformation and MAP kinase inhibition | (81) |
| Breast cancer | miRNA-18a-5p/SREBP-1/Snail/HDAC1/2 | Modulates epithelial–mesenchymal transition and metastasis by regulating a complex formation of Snail and HDAC1/2 | (82) |
| Breast cancer | SREBP-1/autophagy | Contributes to leptin-mediated fatty acid metabolic reprogramming | (83) |
| Breast cancer | GRP94/SREBP-1/LXRα/ACOT7 | Regulates the protein levels of genes encoding fatty acid synthesis and degradation, such as SREBP-1, LXRα and ACOT7 for brain metastasis | (84) |
| Breast cancer | Nuclear protein p54(nrb)/Nono/SREBP-1 | Is positively corelated with SREBP-1, and conserved Y267 residue is required for nuclear SREBP-1a binding and protein stability for increased lipid demand of cancer growth | (85) |
| Glioblastoma | EGFR/SREBP-1/Akt | Induces SREBP-1 cleavage and nuclear translocation by Akt activation, which is independent in mTORC1 for fatty acid synthesis and rapamycin’s poor efficacy | (40) |
| Glioblastoma | EGFR activating mutation/SREBP-1/LDLR | Promotes the cleavage of SREBP-1 to increase the expression of the nuclear form for increased LDLR by activating the PI3K/Akt pathway | (15) |
| Glioblastoma | EGFR/SCAP/SREBP-1/miRNA-29 | Binds specific sites of microRNA-29 for its increased expression by upregulating SCAP/SREBP-1 | (88) |
| Glioblastoma | miRNA-29/SCAP/SREBP-1 | Represses the levels of SCAP and SREBP-1 for the inhibition of de novo lipid metabolism | (89) |
| Glioblastoma | SREBP-1/Akt/mTORC1 | Regulates endoplasmic reticulum stress and apoptosis by Akt/mTORC1 signaling | (90) |
| Glioblastoma | Sterol O-acyltransferase/SREBP-1 | Controls cholesterol esterification and storage by upregulating SREBP-1 | (91) |
| Hepatocellular carcinoma | Hepatoma-derived growth factor/SREBP-1 | Is coexpressed with and activates SREBP-1 by changing first amino acid or the type of PWWP domain for cancer development and prognosis | (92) |
| Hepatocellular carcinoma | Apoptosis-antagonizing transcription factor/SREBP-1 | Interacts with the SREBP-1c binding site to regulate cell proliferation and survival for driving hepatocarcinogenesis | (93) |
| Hepatocellular carcinoma | Zinc fingers and homeoboxes 2/miRNA-24-3p/SREBP-1c | Increases the microRNA-24-3p mRNA level to induce SREBP-1c degradation for suppressing cancer progression | (94) |
| Hepatocellular carcinoma | Acyl CoA synthetase 4/c-Myc/SREBP-1 | Upregulates SREBP-1 and its downstreams by c-Myc for the regulation of de novo lipogenesis | (95) |
| Hepatocellular carcinoma | Spindlin 1/SREBP-1c | Coactivates and interacts SREBP-1c to increase the contents of intracellular triglycerides, cholesterols, and lipid droplets for cancer growth | (96) |
| Hepatocellular carcinoma | Hepatitis B virus-encoded X protein/C/EBPα/SREBP-1/FASN/ACC1 | Promotes cell proliferation and lipid accumulation by increasing C/EBPα, SREBP-1, FASN, and ACC1 | (97) |
| Hepatocellular carcinoma | SREBP-1/HDAC8 | Directly upregulates histone deacetylase 8 and are coexpressed in tumor models induced dietary obesity | (98) |
| Hepatocellular carcinoma | Signal transducers and activators of transcription 5 (STAT5)/PPARγ/JNK1/STAT3 | Regulates SREBP-1 and PPARγ signaling in 60% HCC by enhancing TNF-α, ROS, c-Jun N-terminal kinase 1, and STAT3 | (99) |
| Hepatocellular carcinoma | UBC12 neddylation/SREBP-1 | Prolong SREBP-1 stability with decreased ubiquitination for the contribution of cancer aggressiveness | (100) |
| Hepatocellular carcinoma | mTOR/SREBP-1/FADS2 | Increase fatty acid desaturase 2 expression to upregulate sapienate metabolism in the cell and xenograft models | (101) |
| Hepatocellular carcinoma | SREBP-1c | Is higher in large tumor size, high histological grade, and stage of tumor-node metastasis, correlates with overall and disease-free survival, and regulates cell proliferation, migration, and invasion | (42, 102) |
| Lung cancer | Phosphoenolpyruvate carboxykinase 1/Insig1/2/SREBP-1 | Phosphorylates Insig1/2 to activate SREBP-1 signaling for lipid biosynthesis, which contributes to the stage of tumor-node metastasis and progression | (103, 104) |
| Lung cancer | Protein arginine methyltransferase 5/SREBP-1/Akt | Catalyzes the symmetric dimethyl arginine modification of mature form of SREBP-1 to induce its activation, which is correlated with Akt phosphorylation at Ser473 for de novo lipogenesis and cancer growth | (105) |
| Lung cancer | B7-H3/SREBP-1/FASN | Modulates the SREBP-1/FASN pathway to mediate abnormal lipid metabolism | (106) |
| Lung cancer | KRAS/MEK1/2/SREBP-1 | Increases SREBP-1 expression partly by regulating MEK1/2 signaling for cell proliferation and mitochondrial metabolism | (107) |
| Lung cancer | SREBP-1 | Is an essential characteristic for acquired gefitinib resistance in EGFR-mutant cancer | (108) |
| Lung cancer | miRNA-29 | Interacts with the 3′-UTR of SREBP-1 to reduce SREBP-1 expression for the inhibitions of cell proliferation, migration, and tumor growth | (109) |
| Melanomas | SREBP-1/ACLY/FASN/SCD | Binds to the transcription start sites of genes encoding de novo fatty acid biosynthesis in both BRAFi-sensitive and -resistant cells | (27) |
| Melanomas | SREBP-1 | Mediates the acquired resistance to BRAF-targeted therapy in the in vitro and in vivo models | (110) |
| Melanomas | PI3K/Akt/mTORC1/SREBP-1 | Regulates SREBP-1 activity and subsequent cholesterogenesis for reactive oxygen species-induced damage and lipid peroxidation | (111) |
| Squamous cell carcinomas | SREBP-1/tumor protein 63/KLF5 | Links tumor protein 63/KLF5 to regulate fatty acid metabolism by binding hundreds of cis-regulatory elements for cancer cell viability, migration, and poor survival of patients | (112) |
| Esophageal cancer | SREBP-1/SCD/Wnt/β-catenin | Is overexpressed and correlated with worse overall survival of patients and regulates cell proliferation and metastasis by inducing epithelial–mesenchymal transition via SCD1-induced activation of the Wnt/β-catenin pathway | (113) |
| Esophageal cancer | SREBP-1/miR-142-5p/ZEB1 | Are targeted by miR-142-5p for the migration and sphere formation via the reduction of epithelial–mesenchymal transition markers | (114) |
| Esophageal cancer | Akt/PCK1/Insig1/2 and nuclear SREBP-1 | Are higher in cancer specimens and positively correlated with tumor, node and metastasis stage and progression, and poor prognosis | (115) |
| Gastric cancer | SREBP-1c/FASN/SCD-1 | Is activated and promotes the expression of genes encoding fatty acid synthesis, such as SCD-1 and FASN for malignant phenotypes | (116) |
| Pancreatic cancer | Liver X receptors/SREBP-1 | Are significantly reduced in tumor tissues and control the transcription of polynucleotide kinase/phosphatase for regulating DNA repair and cell death | (117) |
| Pancreatic cancer | Transgelin-2/SREBP-1 | Is expressed in cancer tissues and indicates poor survival of patients, which induces SREBP-1-mediated transcription under the stimulation of insulin | (118) |
| Pancreatic cancer | High glucose microenvironment/SREBP-1 | Is associated with poor prognosis and regulates cancer proliferation, apoptosis, and autophagy by enhancing SREBP-1 expression | (119) |
| Pancreatic cancer | TNF-α/SREBP-1/FASN/ACC | Reduces cell migration and is associated with reduced SREBP-1, FASN, and ACC involved in lipogenesis, inflammation, and metastasis | (120) |
| Pancreatic cancer | SREBP-1/FASN/ACC/SCD-1 | Is significantly higher in cancer tissues and predicts poor prognosis and regulates lipid metabolism and tumor growth by regulating its downstream targets, FASN, ACC, SCD-1 | (41) |
| Endometrial cancer | SREBP-1 | Is significantly increased in cancer tissues and positively correlates with serum triglyceride | (121) |
| Endometrial cancer | SREBP-1 | Ten single-nucleotide polymorphisms are associated with increased risk and rs2297508 SNP with the C allele serves as a genetic factor for early detection | (122) |
| Endometrial cancer | SIRT1/FoxO1/SREBP-1 | Are changed in progestin-resistant cells and are involved in the development of progestin resistance | (123) |
| Endometrial cancer | FoxO1/SREBP-1 | Inhibits cell migration and invasion abilities and tumorigenesis in vitro and in vivo by directly targeting SREBP-1 | (124) |
| Ovarian cancer | Salt-inducible kinase-2/SREBP-1c/FASN | Promotes fatty acid synthesis by upregulating SREBP-1c expression and FASN, which is mediated by the PI3K/Akt pathway | (125) |
| Ovarian cancer | SREBP-1 | Is higher in cancer tissues compared to benign and borderline tumors and regulates cell growth, invasion, migration, and apoptosis in the cell and mouse xenograft models | (126, 127) |
| Clear cell renal cell carcinoma | E2F transcription factor 1/SREBP-1 | Is associated with poor prognosis and increases cell proliferation, epithelial–mesenchymal transition, and tumor progression by activating SREBP-1-mediated fatty acid biosynthesis | (128) |
| Bladder cancer | Pyruvate kinase 2/Akt/mTOR/SREBP-1c/FASN | Physically interacts with SREBP-1c to regulate FASN transcription for tumor growth by regulating Akt/mTOR signaling | (129) |
| Nasopharyngeal carcinoma | Long intergenic non-protein coding RNA 02570/miRNA-4649-3p/SREBP-1/FASN | Is upregulated in the late clinical stage and adsorbs microRNA-4649-3p to upregulate SREBP-1 and FASN for cancer progression | (130) |
| Nasopharyngeal carcinoma | Epstein–Barr virus-encoded latent membrane protein 1/SREBP-1 | Is overexpressed in cancer tissues and increases the expression, maturation, and activation of SREBP-1 for inducing de novo lipid synthesis and is involved in cancer pathogenesis driven by Epstein–Barr virus E | (131) |
| Oral squamous cell carcinoma | Glutathione peroxidase 4/SREBP-1/ferroptosis | Is higher in cancer cells, regulates cell proliferation and ferroptosis, and is correlated with p53 immunoreactivity | (132) |
| Thyroid cancer | SREBP-1 | Is significantly higher in invasive cancer tissues and associated with advanced disease stage and short survival | (28) |
| Thyroid cancer | SREBP-1/Hippo-YAP/CYR61/CTGF | Obviously increases oxygen consumption rate, invasion, and migration by upregulating the Hippo-YAP/CYR61/CTGF pathway | (133) |
| Sarcomas | Nuclear form of SREBP-1/PKR/PERK | Binds the promoter of protein kinase RNA(PKR)-like endoplasmic reticular kinase (PERK) to augment its expression and phosphorylation for malignant phenotypes | (134) |
GRP94, glucose-regulated protein 94; ACTO7, acyl-CoA thioesterase 7; SOAT, sterol O-acyltransferase.
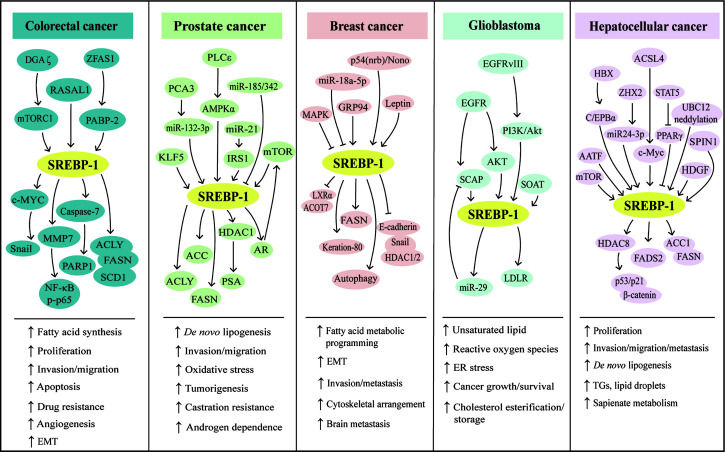
Figure 2.
SREBP-1-mediated lipogenesis in the five types of cancers. Multiple pathways can regulate SREBP-1 and its downstream targets to mediate aggressive characteristics, including proliferation, invasion, migration, EMT, tumorigenesis, metastasis, angiogenesis, and drug resistance in colorectal, prostate, breast, hepatocellular cancer, and glioblastoma. Meanwhile, in these cancers, SREBP-1 activation can increase de novo lipogenesis and regulate fatty acid metabolic programming, lipid droplets, and sapienate metabolism. EMT: epithelial–mesenchymal transition, ER: endoplasmic reticulum, TGs: triglycerides.
Targeting the SREBP-1 signaling pathway for cancer therapy
As reported, SREBP-1 has been a potential target and its inhibition by small molecules or natural products has been potential therapeutics for preventing and treating cancer (12). A specific inhibitor of SREBP activation, fatostatin is a diarylthiazole derivative and binds to SCAP to inhibit the translocations of SREBP-1 and SREBP-2 from the ER to Golgi (135, 136). In prostate cancer, fatostatin suppresses cell proliferation and colony formation in both androgen-responsive or -insensitive cancer cells and causes G2/M cell cycle arrest and cell death, which is mediated by the blockade of the SREBP-regulated metabolic pathway and AR signaling network (136). Fatostatin can inhibit SREBP activity to influence the assembly of mitotic microtubule spindle and cell division in various cancer cells (137). In breast cancer, fatostatin is more sensitive to estrogen receptor-positive cells in response with cell cycle arrest and apoptosis through accumulation of lipids under ER stress, such as polyunsaturated fatty acids (PUFAs), not the inhibition of SREBP activity (138). Moreover, fatostatin can reverse progesterone resistance through inhibition of the SREBP-1/NF-κB pathway in endometrial cancer (139). In pancreatic cancer, both fatostatin and PF429242 inhibit cell proliferation and the growth of xenograft tumor by reducing SREBP-1 and its downstream signaling cascades, such as FASN and SCD-1 (17). Additionally, the combination of fatostatin with tamoxifen has a synergistic effect on the inhibition of PI3K/Akt/mTOR signaling in ER-positive breast cancer (140). Together, fatostatin mainly regulates the activation of SREBP-1/2 to block different cancer progressions, while it also regulates the accumulations of PUFAs and the PI3K/Akt/mTOR pathway. Nelfinavir, a HIV protease inhibitor, induces ER stress and caspase-dependent apoptosis and increases SREBP-1 protein half-life by blocking intracellular trafficking of SREBP-1 and activating transcription factor 6 (ATF6) from site-2 protease (S2P) inhibition in liposarcoma cells and xenograft mouse models (141). In prostate cancer, nelfinavir inhibits AR activation and nuclear translocation of SREBP-1 and ATF6 by regulating S2P-mediated intramembrane proteolysis, which is associated with ER stress, inhibition of unfolded protein response, apoptosis, and autophagy for treating castration-resistant prostate cancer (142). In addition, nelfinavir and its analogs, #6, #7, and #8, have more potent effects on S2P cleavage than 1,10-phenanthroline, a metalloprotease-specific inhibitor. These molecules can block S2P cleavage activity to lead to the accumulations of precursor SREBP-1 and ATF6 against castration-resistant prostate cancer (143). These findings indicate that nelfinavir is a potential agent targeting S2P cleavage for cancer therapy. Currently, several targeted drugs targeting tyrosine kinase can regulate the SREBP-1 pathway to play anticancer effects. Osimertinib, the first-approved third-generation EGFR inhibitor, facilitates SREBP1 degradation and reduces the levels of its targets and lipogenesis in EGFR-mutant NSCLC cells and tumors, which suggests an effective strategy for overcoming acquired resistance of EGFR inhibitors by targeting SREBP-1 (144). Sorafenib, a multikinase inhibitor targeting RAS/MEK/ERK, VEGFR, and PDGFR, significantly affects SCD-1 expression to decrease the synthesis of monounsaturated fatty acids and suppresses ATP production to activate AMPK, which can reduce the levels of SREBP-1 and phosphorylate mTOR for the inhibition of liver cancer (145).
Furthermore, several small molecules or new dosage forms are reported as the regulators for SREBP-1-regulated lipogenesis. An AR degradation enhancer, ASC-J9 suppresses PCa cell growth and invasion by the AR/SREBP-1/FASN pathway in AR-positive cells and PI3K/Akt/SREBP-1/FASN signaling in AR-negative cells, which indicates that it mainly suppresses FASN-mediated PCa progression in both AR-dependent/independent manners (146). A newly developed AR antagonist, proxalutamide, significantly inhibits proliferation and migration, induces the caspase-dependent apoptosis, and diminishes the level of lipid droplets in PCa cells by regulating the levels of ACL, ACC, FASN, and SREBP-1. Moreover, proxalutamide can decrease AR expression in PCa cells, which may overcome the resistance of AR-targeted therapy (147). Leelamine, a pyruvate dehydrogenase kinase inhibitor, can downregulate the expressions of SREBP-1 and key fatty acid synthesis enzymes (ACLY, ACC1, FASN) at the mRNA and protein levels to suppress fatty acid synthesis against PCa progression (148). An HDAC inhibitor, valproic acid can inhibit prostate cancer cell viability and induce apoptosis by regulating the C/EBPα/SREBP-1 pathway based on the results of in vitro and in vivo experiments (149). A novel anti-beta2-microglobulin monoclonal antibody decreases cell proliferation, induces massive cell death, and decreases AR expression via inhibition of SREBP-1-mediated fatty acid synthesis in the models of multiple PCa cell lines and xenograft tumor (150). N-Arachidonoyl dopamine, a typical representative of N-acyl dopamines, can inhibit breast cancer cell migration, EMT, and stemness and cause decreased cholesterol biosynthesis by inhibiting SREBP-1, its key targets, and endoplasmic reticulum kinase 1/2 (ERK1/2) pathways (151). A novel small-molecule, SI-1, 1-(4-bromophenyl)-3-(pyridin-3-yl) urea inhibits aerobic glycolysis and enhances the antitumor effect of radiofrequency ablation in the HCC cells and xenograft tumors via inhibition of SREBP-1 activation (152). The treatment of docosahexaenoic acid, but neither n-6 PUFA arachidonic acid nor oleic acid, can inhibit the levels of the precursor of SREBP-1 and its mature form, and FASN is induced by estradiol and insulin to mediate breast cancer proliferation, which is the result from reduced phosphorylated Akt, not from ERK1/2 phosphorylation (153). In different GBM cells, phytol and retinol show cytotoxic effects at dose dependence, which might be mediated by the levels of SREBP-1, FASN, and farnesyl-diphosphate farnesyltransferase (FDFT1) to downregulate cholesterol and/or fatty acid biosynthetic pathways (154). In addition, it is reported that platinum complexes or micelles can regulate the SREBP-1 pathway to play a more effective function against cancer growth and progression. The pyridine co-ligand-functionalized cationic complexes, including C2, C6, and C8, can suppress cancer invasion, migration, and tumor spheroid formation through inhibition of SREBP-1-mediated lipid biogenesis (155). PB@LC/D/siR is synthesized by cross-linking for docetaxel and siSREBP1 delivery fused with PCa cell membranes and bone marrow mesenchymal stem cells, which show the enhanced antitumor effects in bone metastatic castration-resistant PCa, with the characteristics of deep tumor penetration, high safety, and bone protection via downregulation of SREBP-1 and SCD-1 at the mRNA level (156).
Importantly, natural products can regulate SREBP-1-regulated lipogenesis for the prevention and treatment of different cancers, including CRC, PCa, HCC, and breast cancer. In CRC, there are four compounds for regulating multiple signaling pathway-medicated SREBP-1 activation. One of ginger derivatives, 6-shogaol, can attenuate the adipocyte-conditioned medium effect by controlling the SREBP-1 level mediated by Akt, p70S6K, and AMPK signaling pathways in 5-FU-treated CRC (157). Ilexgenin A, the main bioactive compound from Ilex hainanensis Merr., significantly inhibits inflammatory colitis of mice induced by azoxymethane/dextran sulfate sodium and reverses the metabolites associated with colorectal cancer, which might be mediated by reprogramed lipid metabolism of the HIF-1α/SREBP-1 pathway (158). RA-XII, a natural cyclopeptide isolated from Rubia yunnanensis, decreases the motility of HCT 116 cells via inhibition of the β-catenin pathway and inhibits CRC growth and metastasis by downregulating the levels of SREBP-1, FASN, and SCD for restraining lipogenesis (159). Berberine inhibits colon cancer cell proliferation and induces cell cycle arrest of the G0/G1 phase via the Wnt/β-catenin pathway. Importantly, berberine blocks SREBP-1 activation and SCAP expression to downregulate the levels of lipogenic enzymes against tumor growth (160). The studies report that three compounds have effective anti-HCC effects by inhibiting SREBP-1 and its downstream targets. Cinobufotalin is extracted from the skin secretion of the giant toad and can effectively promote cell apoptosis, induce cell cycle G2/M arrest, and inhibit cell proliferation by downregulating SREBP-1 expression and the interaction with sterol regulatory elements for the inhibition of de novo lipid synthesis in HCC (161). Moreover, emodin, a main active component from Reynoutria multiflora (Thunb.) Moldenke, triggers apoptosis and reduces mitochondrial membrane potential by regulating intrinsic apoptosis signaling pathway in HCC cells. SREBP-1 and its downstream targets, such as ACLY, ACCα, FASN, and SCD-1, are inhibited to mediate the anticancer effect of emodin against HCC (162). Additionally, In HCC cells and xenograft tumors, betulin treatment inhibits cellular glucose metabolism to prevent metastatic potential and also facilitates the inhibitory effect of sorafenib on HCC through inhibition of SREBP-1 (163). In breast cancer cells, theanaphthoquinone, a member of the thearubigins generated by the oxidation of theaflavin, can block the EGF-induced nuclear translocation of SREBP-1 and also modulate the phosphorylation of ERK1/2, Akt, and EGFR/ErbB-2 induced by EGF, which can cause the blockade of FASN for the inhibition of cell viability and induction of cell death (164). Furthermore, in HER2-overexpressing breast cancer cells, piperineis strongly inhibits proliferation, induces apoptosis, and enhances paclitaxel sensitization through inhibition of SREBP-1/FASN signaling, which might be mediated by HER2 expression, ERK1/2, p38, and Akt signaling pathways (165). Additionally, vitexin and syringic acid from foxtail millet bran can inhibit breast cancer proliferation and block the conversion of saturated fatty acids to monounsaturated fatty acids, which is mediated by the decreases of glucose regulated protein 78 and SREBP-1, and its target, SCD-1 (166).
In NSCLC, ginsenoside Rh2 suppresses SREBP-1 expression and its nuclear translocation to disturb the interaction of SREBP-1 and FASN, which can enhance the immune effect and have a synergistic antitumor effect with cyclophosphamide (167). In gallbladder cancer, α-mangostin, a dietary xanthone, represses the proliferation, clone formation ability, and de novo lipogenesis; induces cell cycle arrest and apoptosis; and enhances gemcitabine sensitivity of gallbladder cancer cells, which might be mediated by AMPK activation and the inhibition of nuclear SREBP-1 translocation in cancer cells and tumor xenograft models (168). In cervical carcinoma, quercetin, a naturally occurring dietary flavonoid, decreases cell proliferation and induces cell death in Hela cells by reducing the O-GlcNAcylation of AMPK and the interaction of OGT and SREBP-1 (169). In both in vitro and in vivo models of pancreatic cancer, resveratrol induces gemcitabine chemosensitivity and suppresses sphere formation and markers of cancer stem cells by targeting SREBP-1, which indicates that resveratrol can be an effective sensitizer for chemotherapy (170). Timosaponin A3 can inhibit SREBP-1 and its targets, FASN and ACC, to reduce cell viability and increase cell cycle arrest and apoptosis of BxPC-3 cells and pancreatic cancer xenograft growth, which is independent in the Akt/GSK-3β pathway (171).
Besides these natural compounds, the extract, polysaccharides, or decoction from various herbs can also regulate the SREBP-1-mediated lipogenesis against PCa and pancreatic cancer growth and progression. In PCa, the ethanol extract of Ganoderma tsugae, a Chinese natural and herbal product, significantly inhibits the expressions of SREBP-1 and its downstream genes associated with lipogenesis and downregulates the levels of AR and PSA to block cancer growth and progression with androgen response and castration resistance (172). The results from the in vitro and in vivo experiments demonstrated that the ethanol extract of Davallia formosana can suppress proliferation, migration, and invasion in PCa cells by inhibiting the levels of SREBP-1, FASN, AR, and PSA, suggesting that SREBP-1/FASN/lipogenesis and the AR axis could be potential targets for the treatment of PCa (173). The cell suspension culture extract from Eriobotrya japonica significantly inhibits PCa cell growth, migration, and invasion by decreasing the SREBP-1/FASN-mediated lipid metabolism and AR signaling pathway in the cell and mouse models (174). Moreover, the polysaccharides from Astragalus membranaceus (APS) greatly inhibit the proliferation and invasion of PCa cells in a dose-dependent and time-dependent manner. Mechanistic studies demonstrate that APS treatment reduces the expressions of miR-138-5p, SIRT1, and SREBP-1 to block tumorigenesis and lipid metabolism in PCa (175). In addition, a traditional herbal decoction TJ001 has significant cytotoxicity, induces cell cycle arrest at the G1/S stage, and inhibits lipid accumulation in D145 and PCa cells with p53 mutation by regulating the ACC expression, SREBP-1 proteolytic cleavage, and the inhibition of AMPK/mTOR, which suggest that the combination of mutant p53 targeting and TJ001 can be considered as the potential strategy for PCa treatment (176). In pancreatic cancer, the CO2 supercritical extract of Yarrow (Achillea millefolium) can downregulate the levels of SREBP-1, FASN, and SCD to induce cytotoxicity in cancer cells and diminish tumor growth of xenograft mouse models, which can be developed as a complementary adjuvant or nutritional supplement (177). Taken together, the current findings of targeting SREBP-1-mediated lipogenesis by small molecules, natural products, or herb extracts are summarized in Table 3 and Figure 3 .
Table 3.
Targeting SREBP-1-mediated lipogenesis in different cancers.
| Treatment | Targets | Cancer type | Molecular mechanism | Refs |
|---|---|---|---|---|
| Fatostatin | SREBP-regulated metabolic pathway and AR signaling | Prostate cancer | Suppresses cell proliferation and colony formation and causes G2/M cell cycle arrest and cell death in both androgen-responsive or insensitive cancer cells | (136) |
| Fatostatin | SREBP activity | Various cancers | Possesses antitumor anti-mitotic properties by inhibiting tubulin polymerization and activating spindle assembly checkpoints | (137) |
| Fatostatin | Accumulation of polyunsaturated fatty acids | Breast cancer | Induces cell cycle arrest and apoptosis through the accumulation of lipids in response to ER stress, not by SREBP activity in estrogen receptor-positive cancer cells | (138) |
| Fatostatin | SREBP-1/NF-κB pathway | Endometrial cancer | Reverses progesterone resistance to inhibit proliferation and induces apoptosis both in vitro and in vivo models | (139) |
| Fatostatin and PF429242 | SREBP-1 and its downstream targets, FASN, SCD-1 | Pancreatic cancer | Inhibits cell viability and proliferation in a time- and dose-dependent manner | (17) |
| Fatostatin combined with tamoxifen | PI3K/Akt/mTOR pathway | Breast cancer | Significantly suppresses cell viability and invasion and regulates apoptosis and autophagy in the cell and mouse xenograft models | (140) |
| Nelfinavir | Intramembrane proteolysis of SREBP-1 and ATF6 | Liposarcoma | Induces the increases of SREBP-1 and ATF6 resulted from S2P inhibition against ER stress and caspase-mediated apoptosis | (141) |
| Nelfinavir | Intramembrane proteolysis of SREBP-1 and ATF6 | Prostate cancer | Inhibits androgen receptor activation and nuclear translocation of SREBP-1 to cause unprocessed SREBP-1 and ATF6 accumulations for the inhibition of proliferation and unfolded protein response | (142) |
| Nelfinavir and its analogs, #6, #7, and #8 | Intramembrane proteolysis of SREBP-1 and ATF6 | Prostate cancer | Induces the increases of SREBP-1 and ATF6 resulted from S2P inhibition against ER stress and apoptosis | (143) |
| Osimertinib | SREBP-1 | Non-small cell lung cancer | Facilitates SREBP-1 degradation, reduces the levels of its targets and lipogenesis to overcome the acquired resistance of EGFR inhibitors | (144) |
| Sorafenib | ATP/AMPK/mTOR/ SREBP-1 |
Hepatocellular carcinoma | Suppresses ATP production to activate AMPK and reduce SREBP-1 and phosphorated mTOR levels to disrupt SCD-1-mediated synthesis of monounsaturated fatty acids | (145) |
| ASC-J9 | AR/SREBP-1/FASN and PI3K/Akt/SREBP-1/FASN | Prostate cancer | Suppresses cell growth and invasion in both AR-dependent and AR-independent manner | (146) |
| Proxalutamide | SREBP-1/ACL/ACC/FASN | Prostate cancer | Inhibits the proliferation and migration, induces the caspase-dependent apoptosis, and diminishes the level of lipid droplets, which also overcome the resistance of AR-targeted therapy by decreasing AR expression | (147) |
| Leelamine | SREBP-1, ACLY, FASN, and SCD | Prostate cancer | Suppresses fatty acid synthesis in the cancer cells and tumor xenograft, not affected by AR status | (148) |
| Valproic acid | C/EBPα/SREBP-1 pathway (FASN, ACC1) | Prostate cancer | Inhibits cell viability and lipogenesis and induce apoptosis in the cancer cells | (149) |
| Beta2-microglobulin antibody | MAPK, SREBP-1 and AR | Prostate cancer | Decreases cell proliferation, induces apoptosis, and reduces AR expression to suppress tumor growth and progression | (150) |
| N-Arachidonoyl dopamine | SREBP-1/ERK1/2 pathways | Breast cancer | Inhibits cell migration, EMT, and stemness and causes decreased cholesterol biosynthesis | (151) |
| SI-1 | SREBP-1 activation | Hepatocellular carcinoma | Enhances the sensitivity to radiofrequency ablation in cancer cells, xenograft tumors, and the patients | (152) |
| Docosahexaenoic acid | Precursor and mature SREBP-1, FASN | Breast cancer | Inhibits cancer cell proliferation induced by estradiol and insulin, which is dependent on Akt phosphorylation, not ERK1/2 phosphorylations | (153) |
| Phytol and retinol | SREBP-1, FASN, and farnesyl-diphosphate farnesyltransferase | Glioblastoma | Have cytotoxic effects in a dose-dependent manner and inhibit cholesterol and/or fatty acid biosynthetic pathways | (154) |
| Platinum complexes, C2, C6, C8 | SREBP-1-regulated metabolic pathway (LDLR, FASN, HMGCR) | Breast, liver, and lung cancer | Inhibits invasion, migration, and cancer stem cell formation and induce apoptosis in vitro | (155) |
| PB@LC/D/siR | SREBP-1 and SCD-1 | Bone metastatic castration-resistant PCa | Shows the enhanced antitumor effects with the characteristics of deep tumor penetration, high safety, and bone protection | (156) |
| 6-Shogaol | Akt, p70S6K, and AMPK-mediated SREBP-1 levels | Colorectal cancer | Attenuates the effect of adipocyte-conditioned medium on 5-FU resistance | (157) |
| Ilexgenin A | HIF-1α/SREBP-1 pathway | Colorectal cancer | Inhibits azoxymethane/dextran sulfate sodium-induced inflammatory colitis and reverses colorectal cancer-associated metabolites by reprogramed lipid metabolism | (158) |
| RA-XII | SREBP-1, FASN, and SCD | Colorectal cancer | Decreases cell motility, tumor growth, and metastasis by restraining lipogenesis | (159) |
| Berberine | SCAP/SREBP-1 pathway | Colon cancer | Inhibits cell proliferation and induces G0/G1 cell cycle arrest and regulates the levels of lipogenic enzymes in the in vitro and in vivo studies | (160) |
| Cinobufotalin | SREBP-1 expression and the interaction with sterol regulatory elements | Hepatocellular carcinoma | Induces cell cycle G2-M arrest and apoptosis and inhibits cell proliferation via the inhibition of de novo lipid synthesis | (161) |
| Emodin | SREBP-1 and its downstream targets, ACLY, ACCα, FASN, and SCD-1 | Hepatocellular carcinoma | Triggers apoptosis and reduces mitochondrial membrane potential to play an anticancer effect | (162) |
| Betulin | SREBP-1 | Hepatocellular carcinoma | Inhibits cellular glucose metabolism to prevent metastatic potential and facilitates the inhibitory effect of sorafenib | (163) |
| Theanaphthoquinone | EGF-induced nuclear translocation of SREBP-1 and FASN expression | Breast cancer | Decreases cell viability and induces cell death by regulating ERK1/2 and Akt phosphorylation and EGFR/ErbB-2 pathways | (164) |
| Piperine | SREBP-1 and FASN expression | HER2-overexpressing breast cancer | Inhibits proliferation, induces apoptosis, and enhances sensitization to paclitaxel by regulating ERK1/2, p38 MAPK, and Akt signaling pathways | (165) |
| Vitexin and syringic acid | GRP78/SREBP-1/SCD-1 | Breast cancer | Inhibits cell proliferation and impairs tumor growth by regulating cellular membrane and lipid droplet | (166) |
| Ginsenoside Rh2 | SREBP-1 nuclear translocation and FASN | Non-small cell lung cancer | Enhances the immune effect and has a synergistic antitumor effect with cyclophosphamide | (167) |
| α-Mangostin | AMPK activation and nuclear SREBP-1 translocation | Gallbladder cancer | represses cell proliferation, clone formation ability, and de novo lipogenesis; induces cell cycle arrest and apoptosis; and enhances gemcitabine sensitivity | (168) |
| Quercetin | SREBP-1 and the interaction with O-linked N-acetylglucosamine transferase | Cervical cancer | In cervical carcinoma, quercetin, a naturally occurring dietary flavonoid, decreases cell proliferation and induces cell death in Hela cells by reducing the O-GlcNAcylation of AMPK | (169) |
| Resveratrol | SREBP-1 | Pancreatic cancer | Induces gemcitabine chemosensitivity and suppresses sphere formation and the markers of cancer stem cells in both in vitro and in vivo models | (170) |
| Timosaponin A3 | SREBP-1 and its downstream targets, FASN, ACC | Pancreatic cancer | Inhibits cell viability and induces cell cycle arrest, apoptosis in the cancer cells and xenograft model, which is independent in the Akt/GSK-3β pathway | (171) |
| Ethanol extract of Ganoderma tsugae | SREBP-1 and its downstream genes, AR, PSA | Prostate cancer | Inhibits cancer growth and activates apoptosis by blocking the SREBP-1/AR axis | (172) |
| Ethanol extract of Davallia formosana | SREBP-1/FASN/lipogenesis and AR axis | Prostate cancer | Suppresses cancer proliferation, migration, and invasion by inhibiting the levels of SREBP-1, FASN, AR, and PSA in in vitro and in vivo experiments | (173) |
| Cell suspension culture extract from Eriobotrya japonica | SREBP-1/FASN and AR signaling pathways | Prostate cancer | Inhibits cell growth, migration, and invasion by decreasing the SREBP-1/FASN-mediated lipid metabolism and AR signaling pathway in the cell and mouse models | (174) |
| Astragalus polysaccharides | miR-138-5p/SIRT1/SREBP-1 pathway | Prostate cancer | Inhibits proliferation and invasion in a dose- and time-dependent manner and blocks tumorigenesis and lipid metabolism by the miR-138-5p/SIRT1/SREBP-1 pathway | (175) |
| TJ001 | ACC expression, SREBP-1 proteolytic cleavage, | Prostate cancer | Has significant cytotoxicity, induces cell cycle arrest at the G1/S stage, and inhibits lipid accumulation in D145 cells by regulating the AMPK/mTOR pathway | (176) |
| CO2 supercritical extract of Yarrow | SREBP-1, FASN, and SCD | Pancreatic cancer | Induces cytotoxicity in cancer cells and diminishes tumor growth of the xenograft mouse model | (177) |
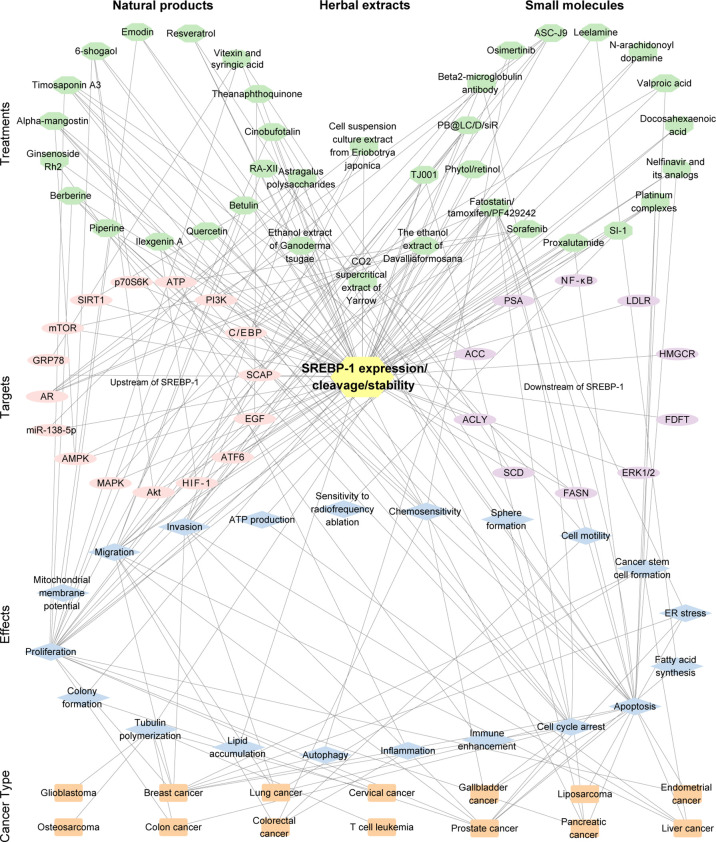
Figure 3.
The interaction of the treatment by small molecules, natural products, or the extract of herbs, the targets (SREBP-1 expression/cleavage/stability), effects, and cancer types. The pink/purple circles represent upstream and downstream targets of SREBP-1. The blue diamonds represent the effects from the inhibition of the SREBP-1 pathway. The orange rectangles represent cancer types. The number of edges connected between the nodes in the network represents the count of their connections.
Conclusions
This review mainly summarized the recent findings of SREBP-1-mediated lipogenesis in different cancer and as potential targets by small molecules, natural products, or the extracts of herbs against cancer growth and progression. Multiple signaling pathways, such as EGFR and PI3K/Akt/mTOR, control SREBP-1 expression and activation to regulate the transcription of multiple genes for fatty acid synthesis (ACLY, ACC, FASN, SCD-1/5) and lipid uptake (LDLR) in human cancer. Multiple signaling molecules can regulate SREBP-1 expression, activation, stability, and binding. Furthermore, SREBP-1 can regulate downstream signaling pathways to mediate cancer proliferation, apoptosis, endoplasmic reticulum stress, and epithelial–mesenchymal transition for tumor growth and metastasis of different cancers, including colon, prostate, breast, lung, and hepatocellular cancer. Additionally, many studies have demonstrated that fatostatin, nelfinavir, AR antagonist, natural compounds, or herbal extracts can target SREBP-1 and its downstream targets for the inhibition of lipid biosynthesis in tumor growth and progression of various cancers. This review could provide new insights into the critical function of the SREBP-1-regulated lipogenesis in different cancers and its potential for cancer therapy for targeting SREBP-1.
Critically, several issues and future perspectives are concerned, as in the following: 1) Targeting SREBP-1 can inhibit multiple enzymes in lipogenesis to significantly block cancer growth and aggressive progression, which is a promising strategy of multitarget therapeutics. 2) Current studies of targeting the SREBP-1 pathway against cancer are preclinical studies for investigating their effects and molecular mechanisms. It is necessary to conduct a clinical evaluation of SREBP-1-targeted therapy against cancer in future. 3) Except its role in cancer, SREBP-1 also plays a critical role in lipid metabolism of metabolic-related diseases. At present, no research is reported showing that those treatments targeting SREBP-1 have side effects, which might be related with the lower level of SREBP-1 in normal tissues, compared to that in cancer. The development of new and specific drugs targeting SREBP-1 with less or no toxic should be potential research directions.
Author Contributions
QZ, XL, and GW: conceptualization, writing for original preparation. XL and GW: supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (82100076). The funder had no role in the decision to publish or in the preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- SREBPs
sterol regulatory element-binding proteins
- bHLH-LZ
basic helix–loop–helix leucine zipper
- GBM
glioblastoma
- EGFR
epidermal growth factor receptor
- PI3K
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- mTOR
mammalian target of rapamycin
- SCAP
SREBP cleavage-activating protein
- SREs
sterol regulatory elements
- ACLY
ATP citrate lyase
- ACC
acetyl-CoA carboxylase
- FASN
fatty acid synthase
- SCD-1/5
stearoyl-CoA desaturase 1 and 5
- LDLR
low-density lipoprotein receptor
- mTORC1
mammalian/mechanistic target of rapamycin complex 1
- GS
glutamine synthetase
- O-GlcNAcylation
O-linked N-acetylglucosaminylation
- Sp1
specificity protein 1
- OGT
O-GlcNAc transferase
- AMPK
AMP-activated protein kinase
- sCLU
secretory clusterin
- TDG
thymine DNA glycosylase
- MAP
mitogen-activated protein
- CRC
colorectal cancer
- SNAIL
snail family transcriptional repressor 1
- AR
androgen receptor
- KLF5
Kruppel-like factor 5
- HMGCR
hydroxy-3-methylglutaryl CoA reductase
- GRP94
glucose-regulated protein 94
- EGFRvIII
EGFR-activating mutation
- HDGF
hepatoma-derived growth factor
- AATF
apoptosis-antagonizing transcription factors
- ZHX2
zinc fingers and homeoboxes 2
- ACS
acyl-CoA synthetases
- ACSL4
acyl CoA synthetase 4
- SPIN1
spindlin 1
- HBV
hepatitis B virus
- HBx
hepatitis B virus-encoded X protein
- STAT5
signal transducers and activators of transcription 5
- GEO
Gene Expression Omnibus
- TNM
tumor-node metastasis
- PCK1
phosphoenolpyruvate carboxykinase 1
- NSCLC
non-small cell lung cancer
- PTMI5
protein arginine methyltransferase 5
- DNFA
de novo fatty acid biosynthesis
- SCCs
squamous cell carcinomas
- TP63
tumor protein 63
- ZEB1
zinc finger E-box-binding homeobox 1
- LXRs
liver X receptors
- PNKP
polynucleotide kinase/phosphatase
- SNPs
single-nucleotide polymorphisms
- SIRT1
sirtuin 1
- FoxO1
forkhead transcription factor 1
- ccRCC
clear cell renal cell carcinoma
- E2F1
E2F transcription factor 1
- EMT
epithelial–mesenchymal transition
- NPC
nasopharyngeal carcinoma
- GPX4
glutathione peroxidase 4
- nSREBP-1
nuclear form of SREBP-1
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- ER
endoplasmic reticulum
- PUFAs
polyunsaturated fatty acids
- ATF6
activating transcription factor 6
- S2P
site-2 protease
- ERK1/2
endoplasmic reticulum kinase 1/2
- FDFT1
farnesyl-diphosphate farnesyltransferase
- APS
polysaccharides from Astragalus membranaceus.
References
- 1. Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, et al. Srebp-1, a Basic-Helix-Loop-Helix-Leucine Zipper Protein That Controls Transcription of the Low Density Lipoprotein Receptor Gene. Cell (1993) 75(1):187–97. doi: 10.1016/S0092-8674(05)80095-9 [DOI] [PubMed] [Google Scholar]
- 2. Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, et al. Srebp-2, a Second Basic-Helix-Loop-Helix-Leucine Zipper Protein That Stimulates Transcription by Binding to a Sterol Regulatory Element. Proc Natl Acad Sci USA (1993) 90(24):11603–7. doi: 10.1073/pnas.90.24.11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown MS, Goldstein JL. The Srebp Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor. Cell (1997) 89(3):331–40. doi: 10.1016/S0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- 4. Amemiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, et al. Transcriptional Activities of Nuclear Srebp-1a, -1c, and -2 to Different Target Promoters of Lipogenic and Cholesterogenic Genes. J Lipid Res (2002) 43(8):1220–35. doi: 10.1194/jlr.M100417-JLR200 [DOI] [PubMed] [Google Scholar]
- 5. Horton JD, Goldstein JL, Brown MS. Srebps: Activators of the Complete Program of Cholesterol and Fatty Acid Synthesis in the Liver. J Clin Invest (2002) 109(9):1125–31. doi: 10.1172/JCI15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harada N, Yonemoto H, Yoshida M, Yamamoto H, Yin Y, Miyamoto A, et al. Alternative Splicing Produces a Constitutively Active Form of Human Srebp-1. Biochem Biophys Res Commun (2008) 368(3):820–6. doi: 10.1016/j.bbrc.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 7. Rome S, Lecomte V, Meugnier E, Rieusset J, Debard C, Euthine V, et al. Microarray Analyses of Srebp-1a and Srebp-1c Target Genes Identify New Regulatory Pathways in Muscle. Physiol Genomics (2008) 34(3):327–37. doi: 10.1152/physiolgenomics.90211.2008 [DOI] [PubMed] [Google Scholar]
- 8. Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. Srebp Transcription Factors: Master Regulators of Lipid Homeostasis. Biochimie (2004) 86(11):839–48. doi: 10.1016/j.biochi.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 9. Shimano H, Sato R. Srebp-Regulated Lipid Metabolism: Convergent Physiology - Divergent Pathophysiology. Nat Rev Endocrinol (2017) 13(12):710–30. doi: 10.1038/nrendo.2017.91 [DOI] [PubMed] [Google Scholar]
- 10. Ferre P, Foufelle F. Srebp-1c Transcription Factor and Lipid Homeostasis: Clinical Perspective. Horm Res (2007) 68(2):72–82. doi: 10.1159/000100426 [DOI] [PubMed] [Google Scholar]
- 11. Sun L, Halaihel N, Zhang W, Rogers T, Levi M. Role of Sterol Regulatory Element-Binding Protein 1 in Regulation of Renal Lipid Metabolism and Glomerulosclerosis in Diabetes Mellitus. J Biol Chem (2002) 277(21):18919–27. doi: 10.1074/jbc.M110650200 [DOI] [PubMed] [Google Scholar]
- 12. Guo D, Bell EH, Mischel P, Chakravarti A. Targeting Srebp-1-Driven Lipid Metabolism to Treat Cancer. Curr Pharm Design (2014) 20(15):2619–26. doi: 10.2174/13816128113199990486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of Androgen Receptor, Lipogenesis, and Oxidative Stress Converged by Srebp-1 Is Responsible for Regulating Growth and Progression of Prostate Cancer Cells. Mol Cancer Res (2012) 10(1):133–42. doi: 10.1158/1541-7786.MCR-11-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao J, Zhu L, Zhu Q, Su J, Liu M, Huang W. Srebp-1 Is an Independent Prognostic Marker and Promotes Invasion and Migration in Breast Cancer. Oncol Lett (2016) 12(4):2409–16. doi: 10.3892/ol.2016.4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, et al. An Lxr Agonist Promotes Glioblastoma Cell Death Through Inhibition of an Egfr/Akt/Srebp-1/Ldlr-Dependent Pathway. Cancer Discovery (2011) 1(5):442–56. doi: 10.1158/2159-8290.CD-11-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Porstmann T, Griffiths B, Chung YL, Delpuech O, Griffiths JR, Downward J, et al. Pkb/Akt Induces Transcription of Enzymes Involved in Cholesterol and Fatty Acid Biosynthesis Via Activation of Srebp. Oncogene (2005) 24(43):6465–81. doi: 10.1038/sjrnc.1208802 [DOI] [PubMed] [Google Scholar]
- 17. Siqingaowa, Sekar S, Gopalakrishnan V, Taghibiglou C. Sterol Regulatory Element-Binding Protein 1 Inhibitors Decrease Pancreatic Cancer Cell Viability and Proliferation. Biochem Biophys Res Commun (2017) 488(1):136–40. doi: 10.1016/j.bbrc.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 18. Kanagasabai T, Li G, Shen TH, Gladoun N, Castillo-Martin M, Celada SI, et al. Microrna-21 Deficiency Suppresses Prostate Cancer Progression Through Downregulation of the Irs1-Srebp-1 Signaling Pathway. Cancer Lett (2022) 525:46–54. doi: 10.1016/j.canlet.2021.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of the Human Gene Encoding Sterol Regulatory Element Binding Protein-1 (Srebf1) and Localization of Srebf1 and Srebf2 to Chromosomes 17p11.2 and 22q13. Genomics (1995) 25(3):667–73. doi: 10.1016/0888-7543(95)80009-B [DOI] [PubMed] [Google Scholar]
- 20. Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential Expression of Exons 1a and 1c in Mrnas for Sterol Regulatory Element Binding Protein-1 in Human and Mouse Organs and Cultured Cells. J Clin Invest (1997) 99(5):838–45. doi: 10.1172/JCI119247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of Sterol Regulatory Element Binding Protein Is Less Active Than Isoform 1a in Livers of Transgenic Mice and in Cultured Cells. J Clin Invest (1997) 99(5):846–54. doi: 10.1172/JCI119248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes AL, Todd BL, Espenshade PJ. Srebp Pathway Responds to Sterols and Functions as an Oxygen Sensor in Fission Yeast. Cell (2005) 120(6):831–42. doi: 10.1016/j.cell.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 23. Guillet-Deniau I, Mieulet V, Le Lay S, Achouri Y, Carre D, Girard J, et al. Sterol Regulatory Element Binding Protein-1c Expression and Action in Rat Muscles: Insulin-Like Effects on the Control of Glycolytic and Lipogenic Enzymes and Ucp3 Gene Expression. Diabetes (2002) 51(6):1722–8. doi: 10.2337/diabetes.51.6.1722 [DOI] [PubMed] [Google Scholar]
- 24. Ruiz R, Jideonwo V, Ahn M, Surendran S, Tagliabracci VS, Hou Y, et al. Sterol Regulatory Element-Binding Protein-1 (Srebp-1) Is Required to Regulate Glycogen Synthesis and Gluconeogenic Gene Expression in Mouse Liver. J Biol Chem (2014) 289(9):5510–7. doi: 10.1074/jbc.M113.541110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferre P, Phan F, Foufelle F. Srebp-1c and Lipogenesis in the Liver: An Update1. Biochem J (2021) 478(20):3723–39. doi: 10.1042/BCJ20210071 [DOI] [PubMed] [Google Scholar]
- 26. Im SS, Yousef L, Blaschitz C, Liu JZ, Edwards RA, Young SG, et al. Linking Lipid Metabolism to the Innate Immune Response in Macrophages Through Sterol Regulatory Element Binding Protein-1a. Cell Metab (2011) 13(5):540–9. doi: 10.1016/j.cmet.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu S, Naar AM. Srebp1-Dependent De Novo Fatty Acid Synthesis Gene Expression Is Elevated in Malignant Melanoma and Represents a Cellular Survival Trait. Sci Rep (2019) 9(1):10369. doi: 10.1038/s41598-019-46594-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li C, Peng X, Lv J, Zou H, Liu J, Zhang K, et al. Srebp1 as a Potential Biomarker Predicts Levothyroxine Efficacy of Differentiated Thyroid Cancer. Biomed Pharmacother = Biomed Pharmacotherapie (2020) 123:109791. doi: 10.1016/j.biopha.2019.109791 [DOI] [PubMed] [Google Scholar]
- 29. Wang X, Sato R, Brown MS, Hua X, Goldstein JL. Srebp-1, a Membrane-Bound Transcription Factor Released by Sterol-Regulated Proteolysis. Cell (1994) 77(1):53–62. doi: 10.1016/0092-8674(94)90234-8 [DOI] [PubMed] [Google Scholar]
- 30. DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-Dependent Proteolysis of Srebp: Relocation of Site-1 Protease From Golgi to Er Obviates the Need for Srebp Transport to Golgi. Cell (1999) 99(7):703–12. doi: 10.1016/s0092-8674(00)81668-2 [DOI] [PubMed] [Google Scholar]
- 31. Goldstein JL, DeBose-Boyd RA, Brown MS. Protein Sensors for Membrane Sterols. Cell (2006) 124(1):35–46. doi: 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- 32. Hegarty BD, Bobard A, Hainault I, Ferre P, Bossard P, Foufelle F. Distinct Roles of Insulin and Liver X Receptor in the Induction and Cleavage of Sterol Regulatory Element-Binding Protein-1c. Proc Natl Acad Sci United States America (2005) 102(3):791–6. doi: 10.1073/pnas.0405067102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin Activates the Rat Sterol-Regulatory-Element-Binding Protein 1c (Srebp-1c) Promoter Through the Combinatorial Actions of Srebp, Lxr, Sp-1 and Nf-Y Cis-Acting Elements. Biochem J (2005) 385(Pt 1):207–16. doi: 10.1042/BJ20040162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakakuki M, Kawano H, Notsu T, Imada K, Mizuguchi K, Shimano H. A Novel Processing System of Sterol Regulatory Element-Binding Protein-1c Regulated by Polyunsaturated Fatty Acid. J Biochem (2014) 155(5):301–13. doi: 10.1093/jb/mvu019 [DOI] [PubMed] [Google Scholar]
- 35. Nakakuki M, Shimano H, Inoue N, Tamura M, Matsuzaka T, Nakagawa Y, et al. A Transcription Factor of Lipid Synthesis, Sterol Regulatory Element-Binding Protein (Srebp)-1a Causes G(1) Cell-Cycle Arrest After Accumulation of Cyclin-Dependent Kinase (Cdk) Inhibitors. FEBS J (2007) 274(17):4440–52. doi: 10.1111/j.1742-4658.2007.05973.x [DOI] [PubMed] [Google Scholar]
- 36. Motallebipour M, Enroth S, Punga T, Ameur A, Koch C, Dunham I, et al. Novel Genes in Cell Cycle Control and Lipid Metabolism With Dynamically Regulated Binding Sites for Sterol Regulatory Element-Binding Protein 1 and Rna Polymerase Ii in Hepg2 Cells Detected by Chromatin Immunoprecipitation With Microarray Detection. FEBS J (2009) 276(7):1878–90. doi: 10.1111/j.1742-4658.2009.06914.x [DOI] [PubMed] [Google Scholar]
- 37. Lecomte V, Meugnier E, Euthine V, Durand C, Freyssenet D, Nemoz G, et al. A New Role for Sterol Regulatory Element Binding Protein 1 Transcription Factors in the Regulation of Muscle Mass and Muscle Cell Differentiation. Mol Cell Biol (2010) 30(5):1182–98. doi: 10.1128/MCB.00690-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y, Chen K, Liu X, Huang L, Zhao D, Li L, et al. Srebp-1 Interacts With C-Myc to Enhance Somatic Cell Reprogramming. Stem Cells (2016) 34(1):83–92. doi: 10.1002/stem.2209 [DOI] [PubMed] [Google Scholar]
- 39. Varghese JF, Patel R, Yadav UCS. Sterol Regulatory Element Binding Protein (Srebp) -1 Mediates Oxidized Low-Density Lipoprotein (Oxldl) Induced Macrophage Foam Cell Formation Through Nlrp3 Inflammasome Activation. Cell signalling (2019) 53:316–26. doi: 10.1016/j.cellsig.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 40. Guo D, Prins RM, Dang J, Kuga D, Iwanami A, Soto H, et al. Egfr Signaling Through an Akt-Srebp-1-Dependent, Rapamycin-Resistant Pathway Sensitizes Glioblastomas to Antilipogenic Therapy. Sci Signaling (2009) 2(101):ra82. doi: 10.1126/scisignal.2000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, et al. Srebp1 Regulates Tumorigenesis and Prognosis of Pancreatic Cancer Through Targeting Lipid Metabolism. Tumour Biol (2015) 36(6):4133–41. doi: 10.1007/s13277-015-3047-5 [DOI] [PubMed] [Google Scholar]
- 42. Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu K, et al. Srebp-1 Has a Prognostic Role and Contributes to Invasion and Metastasis in Human Hepatocellular Carcinoma. Int J Mol Sci (2014) 15(5):7124–38. doi: 10.3390/ijms15057124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kikuchi K, Tsukamoto H. Stearoyl-Coa Desaturase and Tumorigenesis. Chemico-biological Interact (2020) 316:108917. doi: 10.1016/j.cbi.2019.108917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Icard P, Wu Z, Fournel L, Coquerel A, Lincet H, Alifano M. Atp Citrate Lyase: A Central Metabolic Enzyme in Cancer. Cancer Lett (2020) 471:125–34. doi: 10.1016/j.canlet.2019.12.010 [DOI] [PubMed] [Google Scholar]
- 45. Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules (2020) 25(17):3935. doi: 10.3390/molecules25173935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang C, Ma J, Zhang N, Yang Q, Jin Y, Wang Y. The Acetyl-Coa Carboxylase Enzyme: A Target for Cancer Therapy? Expert Rev Anticancer Ther (2015) 15(6):667–76. doi: 10.1586/14737140.2015.1038246 [DOI] [PubMed] [Google Scholar]
- 47. Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, et al. Glucose-Mediated N-Glycosylation of Scap Is Essential for Srebp-1 Activation and Tumor Growth. Cancer Cell (2015) 28(5):569–81. doi: 10.1016/j.ccell.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic Activation of Pi3k-Akt-Mtor Signaling Suppresses Ferroptosis Via Srebp-Mediated Lipogenesis. Proc Natl Acad Sci United States America (2020) 117(49):31189–97. doi: 10.1073/pnas.2017152117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bakan I, Laplante M. Connecting Mtorc1 Signaling to Srebp-1 Activation. Curr Opin Lipidology (2012) 23(3):226–34. doi: 10.1097/MOL.0b013e328352dd03 [DOI] [PubMed] [Google Scholar]
- 50. de la Cruz Lopez KG, Toledo Guzman ME, Sanchez EO, Garcia Carranca A. Mtorc1 as a Regulator of Mitochondrial Functions and a Therapeutic Target in Cancer. Front Oncol (2019) 9:1373. doi: 10.3389/fonc.2019.01373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jhu JW, Yan JB, Lin ZH, Lin SC, Peng IC. Srebp1-Induced Glutamine Synthetase Triggers a Feedforward Loop to Upregulate Srebp1 Through Sp1 O-Glcnacylation and Augments Lipid Droplet Formation in Cancer Cells. Int J Mol Sci (2021) 22(18):9814. doi: 10.3390/ijms22189814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sodi VL, Bacigalupa ZA, Ferrer CM, Lee JV, Gocal WA, Mukhopadhyay D, et al. Nutrient Sensor O-Glcnac Transferase Controls Cancer Lipid Metabolism Via Srebp-1 Regulation. Oncogene (2018) 37(7):924–34. doi: 10.1038/onc.2017.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim MJ, Choi MY, Lee DH, Roh GS, Kim HJ, Kang SS, et al. O-Linked N-Acetylglucosamine Transferase Enhances Secretory Clusterin Expression Via Liver X Receptors and Sterol Response Element Binding Protein Regulation in Cervical Cancer. Oncotarget (2018) 9(4):4625–36. doi: 10.18632/oncotarget.23588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yan JB, Lai CC, Jhu JW, Gongol B, Marin TL, Lin SC, et al. Insulin and Metformin Control Cell Proliferation by Regulating Tdg-Mediated DNA Demethylation in Liver and Breast Cancer Cells. Mol Ther Oncolytics (2020) 18:282–94. doi: 10.1016/j.omto.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, et al. Treg Cells Promote the Srebp1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages Via Repression of Cd8(+) T Cell-Derived Interferon-Gamma. Immunity (2019) 51(2):381–97.e6. doi: 10.1016/j.immuni.2019.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koizume S, Takahashi T, Yoshihara M, Nakamura Y, Ruf W, Takenaka K, et al. Cholesterol Starvation and Hypoxia Activate the Fvii Gene Via the Srebp1-Gilz Pathway in Ovarian Cancer Cells to Produce Procoagulant Microvesicles. Thromb Haemostasis (2019) 119(7):1058–71. doi: 10.1055/s-0039-1687876 [DOI] [PubMed] [Google Scholar]
- 57. Yang YA, Han WF, Morin PJ, Chrest FJ, Pizer ES. Activation of Fatty Acid Synthesis During Neoplastic Transformation: Role of Mitogen-Activated Protein Kinase and Phosphatidylinositol 3-Kinase. Exp Cell Res (2002) 279(1):80–90. doi: 10.1006/excr.2002.5600 [DOI] [PubMed] [Google Scholar]
- 58. Choi WI, Jeon BN, Park H, Yoo JY, Kim YS, Koh DI, et al. Proto-Oncogene Fbi-1 (Pokemon) and Srebp-1 Synergistically Activate Transcription of Fatty-Acid Synthase Gene (Fasn). J Biol Chem (2008) 283(43):29341–54. doi: 10.1074/jbc.M802477200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y, Li C, Hu C, Wu Q, Cai Y, Xing S, et al. Lin28 Enhances De Novo Fatty Acid Synthesis to Promote Cancer Progression Via Srebp-1. EMBO Rep (2019) 20(10):e48115. doi: 10.15252/embr.201948115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gao Y, Zhao Q, Mu X, Zhu H, Liu B, Yao B, et al. Srebp1 Promotes 5-Fu Resistance in Colorectal Cancer Cells by Inhibiting the Expression of Caspase7. Int J Clin Exp Pathol (2019) 12(3):1095–100. [PMC free article] [PubMed] [Google Scholar]
- 61. Gao Y, Nan X, Shi X, Mu X, Liu B, Zhu H, et al. Srebp1 Promotes the Invasion of Colorectal Cancer Accompanied Upregulation of Mmp7 Expression and Nf-Kappab Pathway Activation. BMC Cancer (2019) 19(1):685. doi: 10.1186/s12885-019-5904-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li JN, Mahmoud MA, Han WF, Ripple M, Pizer ES. Sterol Regulatory Element-Binding Protein-1 Participates in the Regulation of Fatty Acid Synthase Expression in Colorectal Neoplasia. Exp Cell Res (2000) 261(1):159–65. doi: 10.1006/excr.2000.5054 [DOI] [PubMed] [Google Scholar]
- 63. Qiu Z, Deng W, Hong Y, Zhao L, Li M, Guan Y, et al. Biological Behavior and Lipid Metabolism of Colon Cancer Cells Are Regulated by a Combination of Sterol Regulatory Element-Binding Protein 1 and Atp Citrate Lyase. Onco Targets Ther (2021) 14:1531–42. doi: 10.2147/OTT.S282906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wen YA, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, et al. Downregulation of Srebp Inhibits Tumor Growth and Initiation by Altering Cellular Metabolism in Colon Cancer. Cell Death Dis (2018) 9(3):265. doi: 10.1038/s41419-018-0330-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhai D, Cui C, Xie L, Cai L, Yu J. Sterol Regulatory Element-Binding Protein 1 Cooperates With C-Myc to Promote Epithelial-Mesenchymal Transition in Colorectal Cancer. Oncol Lett (2018) 15(4):5959–65. doi: 10.3892/ol.2018.8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shen W, Xu T, Chen D, Tan X. Targeting Srebp1 Chemosensitizes Colorectal Cancer Cells to Gemcitabine by Caspase-7 Upregulation. Bioengineered (2019) 10(1):459–68. doi: 10.1080/21655979.2019.1676485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jin Y, Chen Z, Dong J, Wang B, Fan S, Yang X, et al. Srebp1/Fasn/Cholesterol Axis Facilitates Radioresistance in Colorectal Cancer. FEBS Open Bio (2021) 11(5):1343–52. doi: 10.1002/2211-5463.13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang G, Li Z, Li X, Zhang C, Peng L. Rasal1 Induces to Downregulate the Scd1, Leading to Suppression of Cell Proliferation in Colon Cancer Via Lxralpha/Srebp1c Pathway. Biol Res (2019) 52(1):60. doi: 10.1186/s40659-019-0268-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang H, Chen Y, Liu Y, Li Q, Luo J, Wang L, et al. The Lncrna Zfas1 Regulates Lipogenesis in Colorectal Cancer by Binding Polyadenylate-Binding Protein 2 to Stabilize Srebp1 Mrna. Mol Ther Nucleic Acids (2022) 27:363–74. doi: 10.1016/j.omtn.2021.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Torres-Ayuso P, Tello-Lafoz M, Merida I, Avila-Flores A. Diacylglycerol Kinase-Zeta Regulates Mtorc1 and Lipogenic Metabolism in Cancer Cells Through Srebp-1. Oncogenesis (2015) 4:e164. doi: 10.1038/oncsis.2015.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ettinger SL, Sobel R, Whitmore TG, Akbari M, Bradley DR, Gleave ME, et al. Dysregulation of Sterol Response Element-Binding Proteins and Downstream Effectors in Prostate Cancer During Progression to Androgen Independence. Cancer Res (2004) 64(6):2212–21. doi: 10.1158/0008-5472.can-2148-2 [DOI] [PubMed] [Google Scholar]
- 72. O'Malley J, Kumar R, Kuzmin AN, Pliss A, Yadav N, Balachandar S, et al. Lipid Quantification by Raman Microspectroscopy as a Potential Biomarker in Prostate Cancer. Cancer Lett (2017) 397:52–60. doi: 10.1016/j.canlet.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Suh JH, Gong EY, Kim JB, Lee IK, Choi HS, Lee K. Sterol Regulatory Element-Binding Protein-1c Represses the Transactivation of Androgen Receptor and Androgen-Dependent Growth of Prostatic Cells. Mol Cancer Res (2008) 6(2):314–24. doi: 10.1158/1541-7786.MCR-07-0354 [DOI] [PubMed] [Google Scholar]
- 74. Audet-Walsh E, Vernier M, Yee T, Laflamme C, Li S, Chen Y, et al. Srebf1 Activity Is Regulated by an Ar/Mtor Nuclear Axis in Prostate Cancer. Mol Cancer Res (2018) 16(9):1396–405. doi: 10.1158/1541-7786.MCR-17-0410 [DOI] [PubMed] [Google Scholar]
- 75. Zheng Y, Jin J, Gao Y, Luo C, Wu X, Liu J. Phospholipase Cepsilon Regulates Prostate Cancer Lipid Metabolism and Proliferation by Targeting Amp-Activated Protein Kinase (Ampk)/Sterol Regulatory Element-Binding Protein 1 (Srebp-1) Signaling Pathway. Med Sci Monitor (2020) 26:e924328. doi: 10.12659/MSM.924328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee MY, Moon JS, Park SW, Koh YK, Ahn YH, Kim KS. Klf5 Enhances Srebp-1 Action in Androgen-Dependent Induction of Fatty Acid Synthase in Prostate Cancer Cells. Biochem J (2009) 417(1):313–22. doi: 10.1042/BJ20080762 [DOI] [PubMed] [Google Scholar]
- 77. Li L, Hua L, Fan H, He Y, Xu W, Zhang L, et al. Interplay of Pkd3 With Srebp1 Promotes Cell Growth Via Upregulating Lipogenesis in Prostate Cancer Cells. J Cancer (2019) 10(25):6395–404. doi: 10.7150/jca.31254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li X, Chen YT, Josson S, Mukhopadhyay NK, Kim J, Freeman MR, et al. Microrna-185 and 342 Inhibit Tumorigenicity and Induce Apoptosis Through Blockade of the Srebp Metabolic Pathway in Prostate Cancer Cells. PloS One (2013) 8(8):e70987. doi: 10.1371/journal.pone.0070987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guo S, Zhang Y, Wang S, Yang T, Ma B, Li X, et al. Lncrna Pca3 Promotes Antimony-Induced Lipid Metabolic Disorder in Prostate Cancer by Targeting Mir-132-3 P/Srebp1 Signaling. Toxicol Lett (2021) 348:50–8. doi: 10.1016/j.toxlet.2021.05.006 [DOI] [PubMed] [Google Scholar]
- 80. Perone Y, Farrugia AJ, Rodriguez-Meira A, Gyorffy B, Ion C, Uggetti A, et al. Srebp1 Drives Keratin-80-Dependent Cytoskeletal Changes and Invasive Behavior in Endocrine-Resistant Eralpha Breast Cancer. Nat Commun (2019) 10(1):2115. doi: 10.1038/s41467-019-09676-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yang Y, Morin PJ, Han WF, Chen T, Bornman DM, Gabrielson EW, et al. Regulation of Fatty Acid Synthase Expression in Breast Cancer by Sterol Regulatory Element Binding Protein-1c. Exp Cell Res (2003) 282(2):132–7. doi: 10.1016/s0014-4827(02)00023-x [DOI] [PubMed] [Google Scholar]
- 82. Zhang N, Zhang H, Liu Y, Su P, Zhang J, Wang X, et al. Srebp1, Targeted by Mir-18a-5p, Modulates Epithelial-Mesenchymal Transition in Breast Cancer Via Forming a Co-Repressor Complex With Snail and Hdac1/2. Cell Death Differ (2019) 26(5):843–59. doi: 10.1038/s41418-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pham DV, Tilija Pun N, Park PH. Autophagy Activation and Srebp-1 Induction Contribute to Fatty Acid Metabolic Reprogramming by Leptin in Breast Cancer Cells. Mol Oncol (2021) 15(2):657–78. doi: 10.1002/1878-0261.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Santana-Codina N, Marce-Grau A, Muixi L, Nieva C, Marro M, Sebastian D, et al. Grp94 Is Involved in the Lipid Phenotype of Brain Metastatic Cells. Int J Mol Sci (2019) 20(16):3883. doi: 10.3390/ijms20163883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu Z, Zhao X, Zhao L, Yang H, Liu L, Li J, et al. P54(Nrb)/Nono Regulates Lipid Metabolism and Breast Cancer Growth Through Srebp-1a. Oncogene (2016) 35(11):1399–410. doi: 10.1038/onc.2015.197 [DOI] [PubMed] [Google Scholar]
- 86. Li X, Wu C, Chen N, Gu H, Yen A, Cao L, et al. Pi3k/Akt/Mtor Signaling Pathway and Targeted Therapy for Glioblastoma. Oncotarget (2016) 7(22):33440–50. doi: 10.18632/oncotarget.7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Guo D, Bell EH, Chakravarti A. Lipid Metabolism Emerges as a Promising Target for Malignant Glioma Therapy. CNS Oncol (2013) 2(3):289–99. doi: 10.2217/cns.13.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ru P, Hu P, Geng F, Mo X, Cheng C, Yoo JY, et al. Feedback Loop Regulation of Scap/Srebp-1 by Mir-29 Modulates Egfr Signaling-Driven Glioblastoma Growth. Cell Rep (2016) 16(6):1527–35. doi: 10.1016/j.celrep.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ru P, Guo D. Microrna-29 Mediates a Novel Negative Feedback Loop to Regulate Scap/Srebp-1 and Lipid Metabolism. RNA Dis (2017) 4(1):e1525. doi: 10.14800/rd.1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, et al. Sterol Regulatory Element Binding Protein-Dependent Regulation of Lipid Synthesis Supports Cell Survival and Tumor Growth. Cancer Metab (2013) 1(1):3. doi: 10.1186/2049-3002-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Geng F, Cheng X, Wu X, Yoo JY, Cheng C, Guo JY, et al. Inhibition of Soat1 Suppresses Glioblastoma Growth Via Blocking Srebp-1-Mediated Lipogenesis. Clin Cancer Res (2016) 22(21):5337–48. doi: 10.1158/1078-0432.CCR-15-2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Min X, Wen J, Zhao L, Wang K, Li Q, Huang G, et al. Role of Hepatoma-Derived Growth Factor in Promoting De Novo Lipogenesis and Tumorigenesis in Hepatocellular Carcinoma. Mol Oncol (2018) 12(9):1480–97. doi: 10.1002/1878-0261.12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kumar DP, Santhekadur PK, Seneshaw M, Mirshahi F, Uram-Tuculescu C, Sanyal AJ. A Regulatory Role of Apoptosis Antagonizing Transcription Factor in the Pathogenesis of Nonalcoholic Fatty Liver Disease and Hepatocellular Carcinoma. Hepatology (2019) 69(4):1520–34. doi: 10.1002/hep.30346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yu X, Lin Q, Wu Z, Zhang Y, Wang T, Zhao S, et al. Zhx2 Inhibits Srebp1c-Mediated De Novo Lipogenesis in Hepatocellular Carcinoma Via Mir-24-3p. J Pathol (2020) 252(4):358–70. doi: 10.1002/path.5530 [DOI] [PubMed] [Google Scholar]
- 95. Chen J, Ding C, Chen Y, Hu W, Yu C, Peng C, et al. Acsl4 Reprograms Fatty Acid Metabolism in Hepatocellular Carcinoma Via C-Myc/Srebp1 Pathway. Cancer Lett (2021) 502:154–65. doi: 10.1016/j.canlet.2020.12.019 [DOI] [PubMed] [Google Scholar]
- 96. Zhao M, Bu Y, Feng J, Zhang H, Chen Y, Yang G, et al. Spin1 Triggers Abnormal Lipid Metabolism and Enhances Tumor Growth in Liver Cancer. Cancer Lett (2020) 470:54–63. doi: 10.1016/j.canlet.2019.11.032 [DOI] [PubMed] [Google Scholar]
- 97. Zhang X, Zhang X, Zhang JJ, Qiao L. [Hepatitis B Virus X Protein Regulates Lipid Metabolism and Promotes the Proliferation of Liver Cancer Cells Via the C/Ebpa/Srebp-1 Pathway]. Zhonghua gan zang bing za zhi = Zhonghua Ganzangbing zazhi = Chin J Hepatol (2020) 28(12):1036–41. doi: 10.3760/cma.j.cn501113-20190923-00346 [DOI] [PubMed] [Google Scholar]
- 98. Tian Y, Wong VW, Wong GL, Yang W, Sun H, Shen J, et al. Histone Deacetylase Hdac8 Promotes Insulin Resistance and Beta-Catenin Activation in Nafld-Associated Hepatocellular Carcinoma. Cancer Res (2015) 75(22):4803–16. doi: 10.1158/0008-5472.CAN-14-3786 [DOI] [PubMed] [Google Scholar]
- 99. Mueller KM, Kornfeld JW, Friedbichler K, Blaas L, Egger G, Esterbauer H, et al. Impairment of Hepatic Growth Hormone and Glucocorticoid Receptor Signaling Causes Steatosis and Hepatocellular Carcinoma in Mice. Hepatology (2011) 54(4):1398–409. doi: 10.1002/hep.24509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Heo MJ, Kang SH, Kim YS, Lee JM, Yu J, Kim HR, et al. Ubc12-Mediated Srebp-1 Neddylation Worsens Metastatic Tumor Prognosis. Int J Cancer (2020) 147(9):2550–63. doi: 10.1002/ijc.33113 [DOI] [PubMed] [Google Scholar]
- 101. Triki M, Rinaldi G, Planque M, Broekaert D, Winkelkotte AM, Maier CR, et al. Mtor Signaling and Srebp Activity Increase Fads2 Expression and Can Activate Sapienate Biosynthesis. Cell Rep (2020) 31(12):107806. doi: 10.1016/j.celrep.2020.107806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, et al. Co-Ordinate Activation of Lipogenic Enzymes in Hepatocellular Carcinoma. Eur J Cancer (2005) 41(9):1316–22. doi: 10.1016/j.ejca.2004.12.037 [DOI] [PubMed] [Google Scholar]
- 103. Jiang H, Zhu L, Xu D, Lu Z. A Newly Discovered Role of Metabolic Enzyme Pck1 as a Protein Kinase to Promote Cancer Lipogenesis. Cancer Commun (2020) 40(9):389–94. doi: 10.1002/cac2.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shao F, Bian X, Wang J, Xu D, Guo W, Jiang H, et al. Prognostic Impact of Pck1 Protein Kinase Activity-Dependent Nuclear Srebp1 Activation in Non-Small-Cell Lung Carcinoma. Front Oncol (2021) 11:561247. doi: 10.3389/fonc.2021.561247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu L, Yan H, Ruan M, Yang H, Wang L, Lei B, et al. An Akt/Prmt5/Srebp1 Axis in Lung Adenocarcinoma Regulates De Novo Lipogenesis and Tumor Growth. Cancer Sci (2021) 112(8):3083–98. doi: 10.1111/cas.14988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Luo D, Xiao H, Dong J, Li Y, Feng G, Cui M, et al. B7-H3 Regulates Lipid Metabolism of Lung Cancer Through Srebp1-Mediated Expression of Fasn. Biochem Biophys Res Commun (2017) 482(4):1246–51. doi: 10.1016/j.bbrc.2016.12.021 [DOI] [PubMed] [Google Scholar]
- 107. Ruiz CF, Montal ED, Haley JA, Bott AJ, Haley JD. Srebp1 Regulates Mitochondrial Metabolism in Oncogenic Kras Expressing Nsclc. FASEB J (2020) 34(8):10574–89. doi: 10.1096/fj.202000052R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xu C, Zhang L, Wang D, Jiang S, Cao D, Zhao Z, et al. Lipidomics Reveals That Sustained Srebp-1-Dependent Lipogenesis Is a Key Mediator of Gefitinib-Acquired Resistance in Egfr-Mutant Lung Cancer. Cell Death Discovery (2021) 7(1):353. doi: 10.1038/s41420-021-00744-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lin L, Bao YX, Tian M, Ren Q, Zhang W. Mir-29 Family Inhibited the Proliferation and Migration of Lung Cancer Cells by Targeting Srebp-1. Mol Cell Toxicol (2022) 18(2):165–75. doi: 10.1007/s13273-021-00180-3 [DOI] [Google Scholar]
- 110. Talebi A, Dehairs J, Rambow F, Rogiers A, Nittner D, Derua R, et al. Sustained Srebp-1-Dependent Lipogenesis as a Key Mediator of Resistance to Braf-Targeted Therapy. Nat Commun (2018) 9(1):2500. doi: 10.1038/s41467-018-04664-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yamauchi Y, Furukawa K, Hamamura K, Furukawa K. Positive Feedback Loop Between Pi3k-Akt-Mtorc1 Signaling and the Lipogenic Pathway Boosts Akt Signaling: Induction of the Lipogenic Pathway by a Melanoma Antigen. Cancer Res (2011) 71(14):4989–97. doi: 10.1158/0008-5472.CAN-10-4108 [DOI] [PubMed] [Google Scholar]
- 112. Li LY, Yang Q, Jiang YY, Yang W, Jiang Y, Li X, et al. Interplay and Cooperation Between Srebf1 and Master Transcription Factors Regulate Lipid Metabolism and Tumor-Promoting Pathways in Squamous Cancer. Nat Commun (2021) 12(1):4362. doi: 10.1038/s41467-021-24656-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang J, Ling R, Zhou Y, Gao X, Yang Y, Mao C, et al. Srebp1 Silencing Inhibits the Proliferation and Motility of Human Esophageal Squamous Carcinoma Cells Via the Wnt/Beta-Catenin Signaling Pathway. Oncol Lett (2020) 20(3):2855–69. doi: 10.3892/ol.2020.11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Huang CM, Huang CS, Hsu TN, Huang MS, Fong IH, Lee WH, et al. Disruption of Cancer Metabolic Srebp1/Mir-142-5p Suppresses Epithelial-Mesenchymal Transition and Stemness in Esophageal Carcinoma. Cells (2019) 9(1):7. doi: 10.3390/cells9010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shao F, Bian X, Jiang H, Zhao G, Zhu L, Xu D, et al. Association of Phosphoenolpyruvate Carboxykinase 1 Protein Kinase Activity-Dependent Sterol Regulatory Element-Binding Protein 1 Activation With Prognosis of Oesophageal Carcinoma. Eur J Cancer (2021) 142:123–31. doi: 10.1016/j.ejca.2020.09.040 [DOI] [PubMed] [Google Scholar]
- 116. Sun Q, Yu X, Peng C, Liu N, Chen W, Xu H, et al. Activation of Srebp-1c Alters Lipogenesis and Promotes Tumor Growth and Metastasis in Gastric Cancer. Biomed Pharmacother = Biomed Pharmacotherapie (2020) 128:110274. doi: 10.1016/j.biopha.2020.110274 [DOI] [PubMed] [Google Scholar]
- 117. Yang B, Zhang B, Cao Z, Xu X, Huo Z, Zhang P, et al. The Lipogenic Lxr-Srebf1 Signaling Pathway Controls Cancer Cell DNA Repair and Apoptosis and Is a Vulnerable Point of Malignant Tumors for Cancer Therapy. Cell Death Differ (2020) 27(8):2433–50. doi: 10.1038/s41418-020-0514-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, et al. Role of Transgelin-2 in Diabetes-Associated Pancreatic Ductal Adenocarcinoma. Oncotarget (2017) 8(30):49592–604. doi: 10.18632/oncotarget.17519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhou C, Qian W, Li J, Ma J, Chen X, Jiang Z, et al. High Glucose Microenvironment Accelerates Tumor Growth Via Srebp1-Autophagy Axis in Pancreatic Cancer. J Exp Clin Cancer Res (2019) 38(1):302. doi: 10.1186/s13046-019-1288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Al-Zoubi M, Chipitsyna G, Saxena S, Sarosiek K, Gandhi A, Kang CY, et al. Overexpressing Tnf-Alpha in Pancreatic Ductal Adenocarcinoma Cells and Fibroblasts Modifies Cell Survival and Reduces Fatty Acid Synthesis Via Downregulation of Sterol Regulatory Element Binding Protein-1 and Activation of Acetyl Coa Carboxylase. J Gastrointestinal Surg (2014) 18(2):257–68. doi: 10.1007/s11605-013-2370-7 [DOI] [PubMed] [Google Scholar]
- 121. Shafiee MN, Mongan N, Seedhouse C, Chapman C, Deen S, Abu J, et al. Sterol Regulatory Element Binding Protein-1 (Srebp1) Gene Expression Is Similarly Increased in Polycystic Ovary Syndrome and Endometrial Cancer. Acta Obstetricia Gynecologica Scandinavica (2017) 96(5):556–62. doi: 10.1111/aogs.13106 [DOI] [PubMed] [Google Scholar]
- 122. Qiu CP, Lv QT, Dongol S, Wang C, Jiang J. Single Nucleotide Polymorphism of Srebf-1 Gene Associated With an Increased Risk of Endometrial Cancer in Chinese Women. PloS One (2014) 9(3):e90491. doi: 10.1371/journal.pone.0090491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang Y, Zhang L, Che X, Li W, Liu Z, Jiang J. Roles of Sirt1/Foxo1/Srebp-1 in the Development of Progestin Resistance in Endometrial Cancer. Arch Gynecol Obstetrics (2018) 298(5):961–9. doi: 10.1007/s00404-018-4893-3 [DOI] [PubMed] [Google Scholar]
- 124. Zhang Y, Zhang L, Sun H, Lv Q, Qiu C, Che X, et al. Forkhead Transcription Factor 1 Inhibits Endometrial Cancer Cell Proliferation Via Sterol Regulatory Element-Binding Protein 1. Oncol Lett (2017) 13(2):731–7. doi: 10.3892/ol.2016.5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhao J, Zhang X, Gao T, Wang S, Hou Y, Yuan P, et al. Sik2 Enhances Synthesis of Fatty Acid and Cholesterol in Ovarian Cancer Cells and Tumor Growth Through Pi3k/Akt Signaling Pathway. Cell Death Dis (2020) 11(1):25. doi: 10.1038/s41419-019-2221-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yang J, Stack MS. Lipid Regulatory Proteins as Potential Therapeutic Targets for Ovarian Cancer in Obese Women. Cancers (2020) 12(11):3469. doi: 10.3390/cancers12113469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nie LY, Lu QT, Li WH, Yang N, Dongol S, Zhang X, et al. Sterol Regulatory Element-Binding Protein 1 Is Required for Ovarian Tumor Growth. Oncol Rep (2013) 30(3):1346–54. doi: 10.3892/or.2013.2575 [DOI] [PubMed] [Google Scholar]
- 128. Shen D, Gao Y, Huang Q, Xuan Y, Yao Y, Gu L, et al. E2f1 Promotes Proliferation and Metastasis of Clear Cell Renal Cell Carcinoma Via Activation of Srebp1-Dependent Fatty Acid Biosynthesis. Cancer Lett (2021) 514:48–62. doi: 10.1016/j.canlet.2021.05.012 [DOI] [PubMed] [Google Scholar]
- 129. Tao T, Su Q, Xu S, Deng J, Zhou S, Zhuang Y, et al. Down-Regulation of Pkm2 Decreases Fasn Expression in Bladder Cancer Cells Through Akt/Mtor/Srebp-1c Axis. J Cell Physiol (2019) 234(3):3088–104. doi: 10.1002/jcp.27129 [DOI] [PubMed] [Google Scholar]
- 130. Liu F, Wei J, Hao Y, Lan J, Li W, Weng J, et al. Long Intergenic Non-Protein Coding Rna 02570 Promotes Nasopharyngeal Carcinoma Progression by Adsorbing Microrna Mir-4649-3p Thereby Upregulating Both Sterol Regulatory Element Binding Protein 1, and Fatty Acid Synthase. Bioengineered (2021) 12(1):7119–30. doi: 10.1080/21655979.2021.1979317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lo AK, Lung RW, Dawson CW, Young LS, Ko CW, Yeung WW, et al. Activation of Sterol Regulatory Element-Binding Protein 1 (Srebp1)-Mediated Lipogenesis by the Epstein-Barr Virus-Encoded Latent Membrane Protein 1 (Lmp1) Promotes Cell Proliferation and Progression of Nasopharyngeal Carcinoma. J Pathol (2018) 246(2):180–90. doi: 10.1002/path.5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Fukuda M, Ogasawara Y, Hayashi H, Okuyama A, Shiono J, Inoue K, et al. Down-Regulation of Glutathione Peroxidase 4 in Oral Cancer Inhibits Tumor Growth Through Srebp1 Signaling. Anticancer Res (2021) 41(4):1785–92. doi: 10.21873/anticanres.14944 [DOI] [PubMed] [Google Scholar]
- 133. Kuo CY, Chang YC, Chien MN, Jhuang JY, Hsu YC, Huang SY, et al. Srebp1 Promotes Invasive Phenotypes by Upregulating Cyr61/Ctgf Via the Hippo-Yap Pathway. Endocr Relat Cancer (2021) 29(2):47–58. doi: 10.1530/ERC-21-0256 [DOI] [PubMed] [Google Scholar]
- 134. Hu Q, Mao Y, Liu M, Luo R, Jiang R, Guo F. The Active Nuclear Form of Srebp1 Amplifies Er Stress and Autophagy Via Regulation of Perk. FEBS J (2020) 287(11):2348–66. doi: 10.1111/febs.15144 [DOI] [PubMed] [Google Scholar]
- 135. Shao W, Machamer CE, Espenshade PJ. Fatostatin Blocks Er Exit of Scap But Inhibits Cell Growth in a Scap-Independent Manner. J Lipid Res (2016) 57(8):1564–73. doi: 10.1194/jlr.M069583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Li X, Chen YT, Hu P, Huang WC. Fatostatin Displays High Antitumor Activity in Prostate Cancer by Blocking Srebp-Regulated Metabolic Pathways and Androgen Receptor Signaling. Mol Cancer Ther (2014) 13(4):855–66. doi: 10.1158/1535-7163.MCT-13-0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gholkar AA, Cheung K, Williams KJ, Lo YC, Hamideh SA, Nnebe C, et al. Fatostatin Inhibits Cancer Cell Proliferation by Affecting Mitotic Microtubule Spindle Assembly and Cell Division. J Biol Chem (2016) 291(33):17001–8. doi: 10.1074/jbc.C116.737346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Brovkovych V, Izhar Y, Danes JM, Dubrovskyi O, Sakallioglu IT, Morrow LM, et al. Fatostatin Induces Pro- and Anti-Apoptotic Lipid Accumulation in Breast Cancer. Oncogenesis (2018) 7(8):66. doi: 10.1038/s41389-018-0076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ma X, Zhao T, Yan H, Guo K, Liu Z, Wei L, et al. Fatostatin Reverses Progesterone Resistance by Inhibiting the Srebp1-Nf-Kappab Pathway in Endometrial Carcinoma. Cell Death Dis (2021) 12(6):544. doi: 10.1038/s41419-021-03762-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Liu Y, Zhang N, Zhang H, Wang L, Duan Y, Wang X, et al. Fatostatin in Combination With Tamoxifen Induces Synergistic Inhibition in Er-Positive Breast Cancer. Drug Des Devel Ther (2020) 14:3535–45. doi: 10.2147/DDDT.S253876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Guan M, Fousek K, Jiang C, Guo S, Synold T, Xi B, et al. Nelfinavir Induces Liposarcoma Apoptosis Through Inhibition of Regulated Intramembrane Proteolysis of Srebp-1 and Atf6. Clin Cancer Res (2011) 17(7):1796–806. doi: 10.1158/1078-0432.CCR-10-3216 [DOI] [PubMed] [Google Scholar]
- 142. Guan M, Fousek K, Chow WA. Nelfinavir Inhibits Regulated Intramembrane Proteolysis of Sterol Regulatory Element Binding Protein-1 and Activating Transcription Factor 6 in Castration-Resistant Prostate Cancer. FEBS J (2012) 279(13):2399–411. doi: 10.1111/j.1742-4658.2012.08619.x [DOI] [PubMed] [Google Scholar]
- 143. Guan M, Su L, Yuan YC, Li H, Chow WA. Nelfinavir and Nelfinavir Analogs Block Site-2 Protease Cleavage to Inhibit Castration-Resistant Prostate Cancer. Sci Rep (2015) 5:9698. doi: 10.1038/srep09698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chen Z, Yu D, Owonikoko TK, Ramalingam SS, Sun SY. Induction of Srebp1 Degradation Coupled With Suppression of Srebp1-Mediated Lipogenesis Impacts the Response of Egfr Mutant Nsclc Cells to Osimertinib. Oncogene (2021) 40(49):6653–65. doi: 10.1038/s41388-021-02057-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liu G, Kuang S, Cao R, Wang J, Peng Q, Sun C. Sorafenib Kills Liver Cancer Cells by Disrupting Scd1-Mediated Synthesis of Monounsaturated Fatty Acids Via the Atp-Ampk-Mtor-Srebp1 Signaling Pathway. FASEB J (2019) 33(9):10089–103. doi: 10.1096/fj.201802619RR [DOI] [PubMed] [Google Scholar]
- 146. Wen S, Niu Y, Lee SO, Yeh S, Shang Z, Gao H, et al. Targeting Fatty Acid Synthase With Asc-J9 Suppresses Proliferation and Invasion of Prostate Cancer Cells. Mol Carcinog (2016) 55(12):2278–90. doi: 10.1002/mc.22468 [DOI] [PubMed] [Google Scholar]
- 147. Gu Y, Xue M, Wang Q, Hong X, Wang X, Zhou F, et al. Novel Strategy of Proxalutamide for the Treatment of Prostate Cancer Through Coordinated Blockade of Lipogenesis and Androgen Receptor Axis. Int J Mol Sci (2021) 22(24):13222. doi: 10.3390/ijms222413222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Singh KB, Hahm ER, Pore SK, Singh SV. Leelamine Is a Novel Lipogenesis Inhibitor in Prostate Cancer Cells in Vitro and in Vivo . Mol Cancer Ther (2019) 18(10):1800–10. doi: 10.1158/1535-7163.MCT-19-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pang B, Zhang J, Zhang X, Yuan J, Shi Y, Qiao L. Inhibition of Lipogenesis and Induction of Apoptosis by Valproic Acid in Prostate Cancer Cells Via the C/Ebpalpha/Srebp-1 Pathway. Acta Biochim Biophys Sin (Shanghai) (2021) 53(3):354–64. doi: 10.1093/abbs/gmab002 [DOI] [PubMed] [Google Scholar]
- 150. Huang WC, Zhau HE, Chung LW. Androgen Receptor Survival Signaling Is Blocked by Anti-Beta2-Microglobulin Monoclonal Antibody Via a Mapk/Lipogenic Pathway in Human Prostate Cancer Cells. J Biol Chem (2010) 285(11):7947–56. doi: 10.1074/jbc.M109.092759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bandyopadhayaya S, Akimov MG, Verma R, Sharma A, Sharma D, Kundu GC, et al. N-Arachidonoyl Dopamine Inhibits Epithelial-Mesenchymal Transition of Breast Cancer Cells Through Erk Signaling and Decreasing the Cellular Cholesterol. J Biochem Mol Toxicol (2021) 35(4):e22693. doi: 10.1002/jbt.22693 [DOI] [PubMed] [Google Scholar]
- 152. Zou XZ, Hao JF, Zhou XH. Inhibition of Srebp-1 Activation by a Novel Small-Molecule Inhibitor Enhances the Sensitivity of Hepatocellular Carcinoma Tissue to Radiofrequency Ablation. Front Oncol (2021) 11:796152. doi: 10.3389/fonc.2021.796152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Huang LH, Chung HY, Su HM. Docosahexaenoic Acid Reduces Sterol Regulatory Element Binding Protein-1 and Fatty Acid Synthase Expression and Inhibits Cell Proliferation by Inhibiting Pakt Signaling in a Human Breast Cancer Mcf-7 Cell Line. BMC Cancer (2017) 17(1):890. doi: 10.1186/s12885-017-3936-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Facchini G, Ignarro RS, Rodrigues-Silva E, Vieira AS, Lopes-Cendes I, Castilho RF, et al. Toxic Effects of Phytol and Retinol on Human Glioblastoma Cells Are Associated With Modulation of Cholesterol and Fatty Acid Biosynthetic Pathways. J Neuro-Oncol (2018) 136(3):435–43. doi: 10.1007/s11060-017-2672-9 [DOI] [PubMed] [Google Scholar]
- 155. Bai X, Ali A, Wang N, Liu Z, Lv Z, Zhang Z, et al. Inhibition of Srebp-Mediated Lipid Biosynthesis and Activation of Multiple Anticancer Mechanisms by Platinum Complexes: Ascribe Possibilities of New Antitumor Strategies. Eur J Med Chem (2022) 227:113920. doi: 10.1016/j.ejmech.2021.113920 [DOI] [PubMed] [Google Scholar]
- 156. Chen J, Wu Z, Ding W, Xiao C, Zhang Y, Gao S, et al. Srebp1 Sirna Enhance the Docetaxel Effect Based on a Bone-Cancer Dual-Targeting Biomimetic Nanosystem Against Bone Metastatic Castration-Resistant Prostate Cancer. Theranostics (2020) 10(4):1619–32. doi: 10.7150/thno.40489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lee KC, Wu KL, Yen CK, Chen CN, Chang SF, Huang WS. 6-Shogaol Antagonizes the Adipocyte-Conditioned Medium-Initiated 5-Fluorouracil Resistance in Human Colorectal Cancer Cells Through Controlling the Srebp-1 Level. Life (Basel) (2021) 11(10):1067. doi: 10.3390/life11101067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhang L, Qiao X, Chen M, Li P, Wen X, Sun M, et al. Ilexgenin a Prevents Early Colonic Carcinogenesis and Reprogramed Lipid Metabolism Through Hif1alpha/Srebp-1. Phytomedicine (2019) 63:153011. doi: 10.1016/j.phymed.2019.153011 [DOI] [PubMed] [Google Scholar]
- 159. Wang Y, Guo D, He J, Song L, Chen H, Zhang Z, et al. Inhibition of Fatty Acid Synthesis Arrests Colorectal Neoplasm Growth and Metastasis: Anti-Cancer Therapeutical Effects of Natural Cyclopeptide Ra-Xii. Biochem Biophys Res Commun (2019) 512(4):819–24. doi: 10.1016/j.bbrc.2019.03.088 [DOI] [PubMed] [Google Scholar]
- 160. Liu Y, Hua W, Li Y, Xian X, Zhao Z, Liu C, et al. Berberine Suppresses Colon Cancer Cell Proliferation by Inhibiting the Scap/Srebp-1 Signaling Pathway-Mediated Lipogenesis. Biochem Pharmacol (2020) 174:113776. doi: 10.1016/j.bcp.2019.113776 [DOI] [PubMed] [Google Scholar]
- 161. Meng H, Shen M, Li J, Zhang R, Li X, Zhao L, et al. Novel Srebp1 Inhibitor Cinobufotalin Suppresses Proliferation of Hepatocellular Carcinoma by Targeting Lipogenesis. Eur J Pharmacol (2021) 906:174280. doi: 10.1016/j.ejphar.2021.174280 [DOI] [PubMed] [Google Scholar]
- 162. Yang N, Li C, Li H, Liu M, Cai X, Cao F, et al. Emodin Induced Srebp1-Dependent and Srebp1-Independent Apoptosis in Hepatocellular Carcinoma Cells. Front Pharmacol (2019) 10:709. doi: 10.3389/fphar.2019.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Yin F, Feng F, Wang L, Wang X, Li Z, Cao Y. Srebp-1 Inhibitor Betulin Enhances the Antitumor Effect of Sorafenib on Hepatocellular Carcinoma Via Restricting Cellular Glycolytic Activity. Cell Death Dis (2019) 10(9):672. doi: 10.1038/s41419-019-1884-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Weng MS, Ho CT, Ho YS, Lin JK. Theanaphthoquinone Inhibits Fatty Acid Synthase Expression in Egf-Stimulated Human Breast Cancer Cells Via the Regulation of Egfr/Erbb-2 Signaling. Toxicol Appl Pharmacol (2007) 218(2):107–18. doi: 10.1016/j.taap.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 165. Do MT, Kim HG, Choi JH, Khanal T, Park BH, Tran TP, et al. Antitumor Efficacy of Piperine in the Treatment of Human Her2-Overexpressing Breast Cancer Cells. Food Chem (2013) 141(3):2591–9. doi: 10.1016/j.foodchem.2013.04.125 [DOI] [PubMed] [Google Scholar]
- 166. Zhang LC, La XQ, Tian JM, Li HQ, Li AP, Liu YZ, et al. The Phytochemical Vitexin and Syringic Acid Derived From Foxtail Fillet Bran Inhibit Breast Cancer Cells Proliferation Via Grp78/Srebp-1/Scd1 Signaling Axis. J Funct Foods (2021) 85(26):104620. doi: 10.1016/j.jff.2021.104620 [DOI] [Google Scholar]
- 167. Qian Y, Huang R, Li S, Xie R, Qian B, Zhang Z, et al. Ginsenoside Rh2 Reverses Cyclophosphamide-Induced Immune Deficiency by Regulating Fatty Acid Metabolism. J Leukoc Biol (2019) 106(5):1089–100. doi: 10.1002/JLB.2A0419-117R [DOI] [PubMed] [Google Scholar]
- 168. Shi Y, Fan Y, Hu Y, Jing J, Wang C, Wu Y, et al. Alpha-Mangostin Suppresses the De Novo Lipogenesis and Enhances the Chemotherapeutic Response to Gemcitabine in Gallbladder Carcinoma Cells Via Targeting the Ampk/Srebp1 Cascades. J Cell Mol Med (2020) 24(1):760–71. doi: 10.1111/jcmm.14785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Ali A, Kim MJ, Kim MY, Lee HJ, Roh GS, Kim HJ, et al. Quercetin Induces Cell Death in Cervical Cancer by Reducing O-Glcnacylation of Adenosine Monophosphate-Activated Protein Kinase. Anat Cell Biol (2018) 51(4):274–83. doi: 10.5115/acb.2018.51.4.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zhou C, Qian W, Ma J, Cheng L, Jiang Z, Yan B, et al. Resveratrol Enhances the Chemotherapeutic Response and Reverses the Stemness Induced by Gemcitabine in Pancreatic Cancer Cells Via Targeting Srebp1. Cell Prolif (2019) 52(1):e12514. doi: 10.1111/cpr.12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Kim Y, Jee W, An EJ, Ko HM, Jung JH, Na YC, et al. Timosaponin A3 Inhibits Palmitate and Stearate Through Suppression of Srebp-1 in Pancreatic Cancer. Pharmaceutics (2022) 14(5):945. doi: 10.3390/pharmaceutics14050945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Huang SY, Huang GJ, Wu HC, Kao MC, Huang WC. Ganoderma Tsugae Inhibits the Srebp-1/Ar Axis Leading to Suppression of Cell Growth and Activation of Apoptosis in Prostate Cancer Cells. Molecules (2018) 23(10):2539. doi: 10.3390/molecules23102539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hsieh PF, Jiang WP, Huang SY, Basavaraj P, Wu JB, Ho HY, et al. Emerging Therapeutic Activity of Davallia Formosana on Prostate Cancer Cells Through Coordinated Blockade of Lipogenesis and Androgen Receptor Expression. Cancers (2020) 12(4):914. doi: 10.3390/cancers12040914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Hsieh PF, Jiang WP, Basavaraj P, Huang SY, Ruangsai P, Wu JB, et al. Cell Suspension Culture Extract of Eriobotrya Japonica Attenuates Growth and Induces Apoptosis in Prostate Cancer Cells Via Targeting Srebp-1/Fasn-Driven Metabolism and Ar. Phytomedicine (2021) 93:153806. doi: 10.1016/j.phymed.2021.153806 [DOI] [PubMed] [Google Scholar]
- 175. Guo S, Ma B, Jiang X, Li X, Jia Y. Astragalus Polysaccharides Inhibits Tumorigenesis and Lipid Metabolism Through Mir-138-5p/Sirt1/Srebp1 Pathway in Prostate Cancer. Front Pharmacol (2020) 11:598. doi: 10.3389/fphar.2020.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Kang S, Kim HI, Choi YJ, Lee SK, Kim JH, Cheon C, et al. Traditional Herbal Formula Taeeumjowi-Tang (Tj001) Inhibits P53-Mutant Prostate Cancer Cells Growth by Activating Ampk-Dependent Pathway. Evidence-Based Complementary Altern Med (2019) 2019:2460353. doi: 10.1155/2019/2460353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Mouhid L, Gomez de Cedron M, Garcia-Carrascosa E, Reglero G, Fornari T, Ramirez de Molina A. Yarrow Supercritical Extract Exerts Antitumoral Properties by Targeting Lipid Metabolism in Pancreatic Cancer. PloS One (2019) 14(3):e0214294. doi: 10.1371/journal.pone.0214294 [DOI] [PMC free article] [PubMed] [Google Scholar]