This case-control study investigates the magnitude and predictive potential of univariate biological differences between individuals with depression and healthy individuals across neuroimaging modalities.
Key Points
Question
What is the neurobiological difference between healthy individuals and those with depression within common neuroimaging data modalities?
Findings
In this case-control study that included 1809 adults, the group differences in neuroimaging markers explained less than 2% variance, and the single-participant predictive utility was consistently below 56% accuracy. The distributional overlap between healthy individuals and those with depression even for the variables showing the largest difference was 87% to 95%.
Meaning
Study results suggest that patients with depression and healthy controls are remarkably similar regarding neural signatures of common neuroimaging modalities.
Abstract
Importance
Identifying neurobiological differences between patients with major depressive disorder (MDD) and healthy individuals has been a mainstay of clinical neuroscience for decades. However, recent meta-analyses have raised concerns regarding the replicability and clinical relevance of brain alterations in depression.
Objective
To quantify the upper bounds of univariate effect sizes, estimated predictive utility, and distributional dissimilarity of healthy individuals and those with depression across structural magnetic resonance imaging (MRI), diffusion-tensor imaging, and functional task-based as well as resting-state MRI, and to compare results with an MDD polygenic risk score (PRS) and environmental variables.
Design, Setting, and Participants
This was a cross-sectional, case-control clinical neuroimaging study. Data were part of the Marburg-Münster Affective Disorders Cohort Study. Patients with depression and healthy controls were recruited from primary care and the general population in Münster and Marburg, Germany. Study recruitment was performed from September 11, 2014, to September 26, 2018. The sample comprised patients with acute and chronic MDD as well as healthy controls in the age range of 18 to 65 years. Data were analyzed from October 29, 2020, to April 7, 2022.
Main Outcomes and Measures
Primary analyses included univariate partial effect size (η2), classification accuracy, and distributional overlapping coefficient for healthy individuals and those with depression across neuroimaging modalities, controlling for age, sex, and additional modality-specific confounding variables. Secondary analyses included patient subgroups for acute or chronic depressive status.
Results
A total of 1809 individuals (861 patients [47.6%] and 948 controls [52.4%]) were included in the analysis (mean [SD] age, 35.6 [13.2] years; 1165 female patients [64.4%]). The upper bound of the effect sizes of the single univariate measures displaying the largest group difference ranged from partial η2 of 0.004 to 0.017, and distributions overlapped between 87% and 95%, with classification accuracies ranging between 54% and 56% across neuroimaging modalities. This pattern remained virtually unchanged when considering either only patients with acute or chronic depression. Differences were comparable with those found for PRS but substantially smaller than for environmental variables.
Conclusions and Relevance
Results of this case-control study suggest that even for maximum univariate biological differences, deviations between patients with MDD and healthy controls were remarkably small, single-participant prediction was not possible, and similarity between study groups dominated. Biological psychiatry should facilitate meaningful outcome measures or predictive approaches to increase the potential for a personalization of the clinical practice.
Introduction
Major depressive disorder (MDD) is the single largest contributor to nonfatal health loss worldwide, annually affecting as many as 300 million people.1 The incremental economic burden of adults is estimated to be more than $320 billion in the US alone, including direct, suicide-related, and workplace costs. This represents a notable increase by 37.9% between 2010 and 2018.2,3 Driven by the discovery of efficient psychopharmacological medication and the insight that many mental disorders have a strong genetic component, the second half of the 20th century was dominated by biological psychiatry.4 With the emergence of cognitive neuroscience, neuroimaging, and neurogenetics, this paradigm evolved into a methodologically diverse systems-medicine approach aiming to explain mental disorders by dysfunctional neural systems at various levels.5 Correspondingly, identifying the neural and genetic basis of MDD to inform the improvement of treatments has been a mainstay of research in psychiatry for decades with more than 1500 neuroimaging studies listed on PubMed that investigate case-control differences between healthy individuals and those with depression (eMethods 1 in the Supplement).
Although the aim of large-scale projects and consortia that accumulate neuroimaging data from tens of thousands of patients is to consolidate and extend our understanding of mental disorders, there is growing concern regarding the replicability and prognostic utility of neural signatures derived from standard univariate analysis frameworks in psychiatry. Recent meta-analyses, including thousands of patients with depression, find either no or only very subtle spatial convergence of MDD vs healthy individual effects in unimodal paradigm-based and task-independent resting-state functional as well as structural magnetic resonance imaging (MRI) studies.6,7,8,9,10,11 Robust and convergent differences could only be revealed when combining functional hyperactivity of voxel-based physiological with morphometric modalities.7 In the same vein, a meta-analysis by the Enhancing Neuroimaging Genetics Through Meta-analysis (ENIGMA) consortium investigating subcortical brain structures showed a significant difference between 1728 patients with MDD and 7199 controls from 15 samples worldwide.12 This effect, however, was restricted to hippocampus volume and corresponds to a classification accuracy of merely 52.6%, leaving little hope for individualized prediction.13 Furthermore, it remains unclear how nonspecific volumetric changes advance our theoretical knowledge of the illness.14 This general notion of significant yet subtle differences was equally apparent in gray matter cortical and white matter disturbances in MDD.15,16 This lack of consistent findings and surprisingly small effects have been attributed to first, methodologic heterogeneity, including varying experimental designs, varying inclusion and exclusion criteria, or meta-analytic approaches, and second, to the heterogeneity of the clinical population and its assessment, including different severity, disease duration, or number of previous episodes.6,7,17
Methods
Participants
This case-control study was approved by the ethics committees of the medical faculties of the University of Marburg, Marburg, Germany, and the University of Münster, Münster, Germany. Participants received financial compensation and gave written and informed consent. Participants reported country of birth for their parents and grandparents. At the time of data analysis, 2036 healthy individuals and those with depression participated in the cross-sectional Marburg-Münster Affective Disorders Cohort Study (MACS).18,19 Data were collected at 2 different sites (Marburg and Münster, Germany). Exclusion criteria are available in eMethods 2 in the Supplement. For every data modality, all participants for whom data of the specific modality were available and passed quality checks were used. Patients with severe, moderate, mild or (partially) remitted MDD episodes were included irrespective of current treatment. Patients either fulfilled the DSM-IV criteria for an acute major depressive episode or had a lifetime history of a major depressive episode (eMethods 3, eFigures 1 and 2 in the Supplement). Secondary analysis included MDD subgroups of only patients with acute depression, chronic depression, or patients who received medication for MDD; inclusion criteria are available in eMethods 4 and 5 in the Supplement. A control analysis was conducted using a matched healthy sample for each modality (eMethods 6 in the Supplement). This study followed Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guidelines.
The present study quantified the magnitude and predictive potential of univariate biological differences between individuals with depression and healthy controls (HCs) across neuroimaging modalities in a harmonized study, minimizing methodological and clinical heterogeneity. To this end, we drew on data from the bicentric MACS, comprising major neuroimaging modalities including structural MRI, task-based functional MRI (fMRI), atlas-based connectivity, and voxel-based physiological and graph network parameters derived from resting-state fMRI and diffusion-tensor imaging (DTI).18,19 For comparison, we also investigated an MDD polygenic risk score (PRS) and environmental variables, including self-reported childhood maltreatment and social support.
In our analyses, we first assessed group difference effect sizes (η2) using established analysis standards for each modality (Figure 1). For all modalities, we reported results of the single variable (ie, score, voxel, graph metric, connectivity) displaying the largest difference between healthy individuals and those with depression, mirroring the mass-univariate statistical modeling most common in neuroimaging studies today. Second, to gauge potential predictive value of these variables showing the largest univariate difference, we estimated their predictive utility (ie, accuracy, sensitivity, specificity) in every modality. Third, we illustrated the similarity between individuals with depression and healthy participants with respect to the variable displaying the largest difference in every modality by calculating the overlapping coefficient, an intuitive measure of overlap between two populations.20 Although focusing on the single largest variable is prone to overestimating the true difference, the approach provides a solid upper bound for the true deviation between healthy individuals and patients with MDD in the respective modality. Explicitly investigating the substantial clinical heterogeneity often observed in MDD, we also conducted subgroup analyses including symptom severity, course of disease, sex, and scanner site.
Figure 1. Research Design and Analytical Procedure.
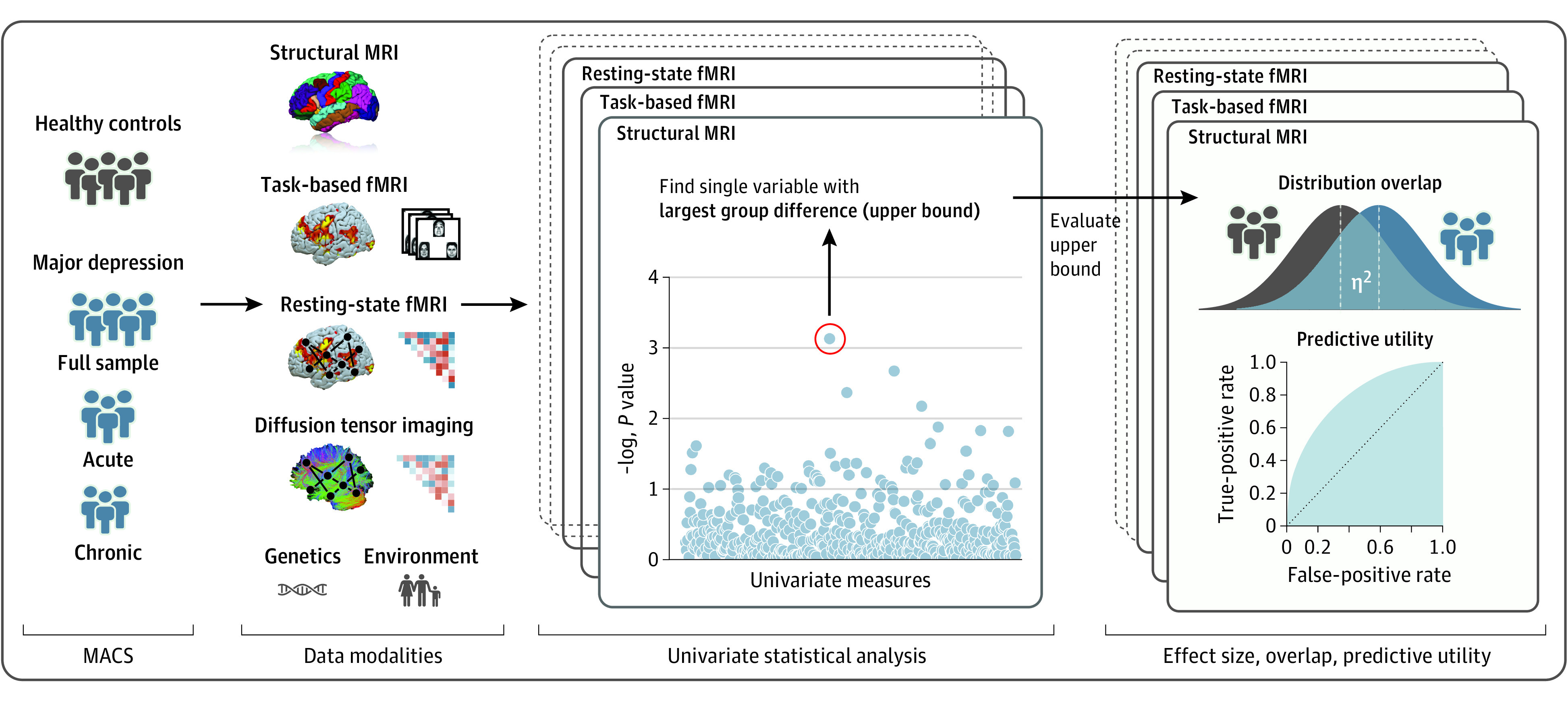
Schematic representation of the research design and analytical procedure. For all modalities, standard univariate models are calculated to find the single variable showing the largest difference between healthy individuals and those with depression, representing an upper bound for univariate group differences. Effect size (η2), distributional overlap, and predictive utility are estimated for these peak variables of the different modalities. fMRI, functional magnetic resonance imaging; MACS, Marburg-Münster Affective Cohort Study.
Data Modalities and Preprocessing
The established childhood trauma questionnaire was used to assess childhood maltreatment (eMethods 8 in the Supplement).21 Perceived social support was measured using the Social Support Questionnaire (eMethods 8 in the Supplement).22 A single PRS for major depression was calculated via bayesian regression and prior continuous shrinkage with a global scaling parameter (φ) of 1.30 × 10−4 using summary statistics from a recent genome-wide association study (eMethods 9 in the Supplement).23,24 Automated structural MRI segmentation was conducted using the cortical and subcortical parcellation stream of FreeSurfer Software Suite, version 5.3 (Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging), based on the Desikan-Killiany atlas (eMethods 10-12 in the Supplement).25 The CAT12 toolbox (Christian Gaser and Robert Dahnke, developers) was used to calculate voxel-based morphometry (VBM) from structural MRI (eMethods 11 in the Supplement).26 The Schaefer atlas with 100 parcels was used to derive connectivity matrices from 8 minutes of resting-state fMRI using the CONN toolbox (MIT Gabrieli Laboratory) (eMethods 13-14 in the Supplement).27,28 Local correlation as a measure of local coherence, the amplitude of low-frequency fluctuations (ALFF), and the fractional ALFF (fALFF) at each voxel were computed from resting-state fMRI.29,30,31 For the task-based fMRI data, an established emotional face-matching paradigm was used (eMethods 15 in the Supplement). The CATO toolbox (Dutch Connectome Laboratory) was used to reconstruct the anatomical connectome of the DTI data using a subdivision of the Desikan-Killiany atlas (eMethods 16 in the Supplement).32 Both DTI and resting-state connectivity matrices were binarized to calculate several representative graph network parameters such as global and local efficiency, betweenness centrality, or clustering coefficient (eMethods 17 in the Supplement).33
Statistical Analyses
An analysis of variance model predicting a single variable of interest was calculated for all variables of the different modalities with age, sex, and scanning site as a minimum set of covariates and a factor for HCs vs patients with MDD. Additional information on modality-specific covariates and the statistical procedure are available in eMethods 7 in the Supplement. This approach mirrors the traditional mass-univariate approach in neuroimaging by estimating an independent model for each variable (eg, voxel, connectivity). We, therefore, use the term univariate throughout the article. Correction for multiple comparisons was done within each data modality.
For each modality, the variable showing the strongest effect (largest F value) was selected and partial η2 was calculated as measure of effect size for the group factor (HC vs MDD). Bootstrap CIs were calculated using the bias-corrected and accelerated bootstrap method including group stratification.34
For further analyses, the covariates were regressed out of the variables showing the largest group effect. To quantify their predictive potential, a logistic regression was trained to classify between patients and controls, and balanced accuracy, sensitivity, and specificity were calculated. Lastly, we calculated the overlapping coefficient for the maximum effect variables to illustrate the similarity between depressive and healthy participants.20 All code implementing the statistical analyses and figures is publicly available.35
Additional sensitivity analyses were performed on more homogeneous subgroups to test whether group differences between healthy individuals and those with depression increase when some clinical and methodological variance is removed. To that end, we analyzed female and male participants as well as samples acquired in Münster, Germany, and Marburg, Germany, separately. In addition, we analyzed a subgroup of patients with acute MDD, chronic MDD, or patients who received medication for MDD. All P values were 2-sided, and significance was set at P < .05. Data were analyzed from October 29, 2020, to April 7, 2022, using Python, version 3.7 (Python Software Foundation).
Results
Effect Sizes, Distributional Overlap, and Classification Performance for HC vs MDD
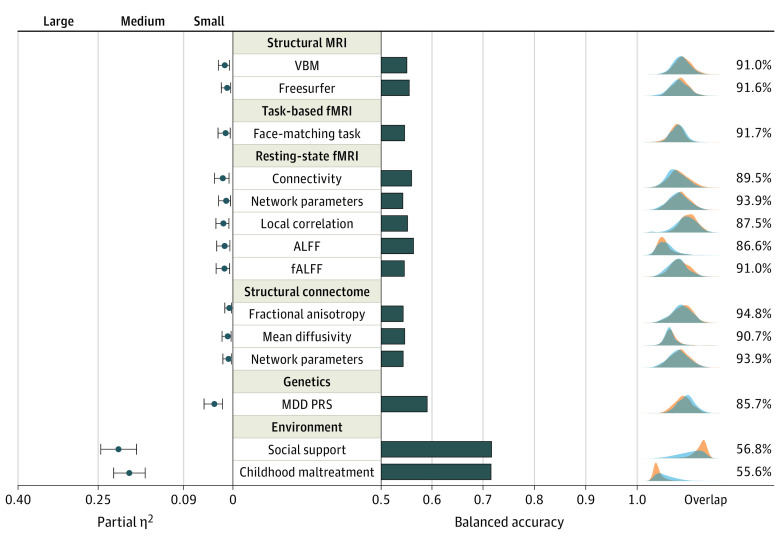
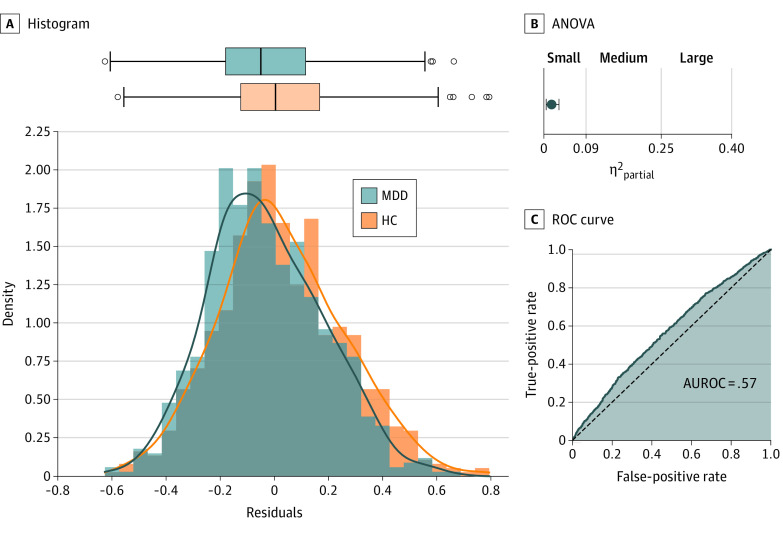
A total of 1809 individuals (861 patients [47.6%] and 948 controls [52.4%]) were included in the analysis (mean [SD] age, 35.6 [13.2] years; 1165 female patients [64.4%]; 644 male patients [35.6%]) (Table 1). For the single variables displaying the largest difference between HCs and patients with MDD, analysis results of variance effect sizes were small in all neuroimaging modalities. They ranged from partial η2 of 0.004 for the largest effect in DTI data to partial η2 of 0.017 for the largest effect in resting-state connectivity (Figure 2, Table 2). For structural MRI, the greatest difference between healthy individuals and those with depression could be observed in a voxel in the left gyrus rectus (VBM: F1, 1737 = 22.82; partial η2 = 0.013; uncorrected P < .001; corrected P = .03) (eFigure 3 in the Supplement) and for the total cortical volume of the right hemisphere (FreeSurfer: F1, 1735 = 14.92; partial η2 = 0.009; uncorrected P < .001; corrected P = .06) (eFigure 4 in the Supplement). For task-based fMRI, the greatest difference in brain activation during a face-matching task between healthy individuals and those with depression was observed in a voxel within the left superior frontal region (F1, 1235 = 14.3; partial η2 = 0.011; uncorrected P < .001; corrected P = .40) (eFigure 5 in the Supplement). For resting-state fMRI, the greatest difference between healthy individuals and those with depression was measured for the connectivity between a region of the right peripheral visual network and a region of the somatomotor network A (F1, 1330 = 22.53; partial η2 = 0.017; uncorrected P < .001; corrected P = .07) (Figure 3) and the degree centrality of region 54 (peripheral visual network) of the Schaefer atlas (F1, 1339 = 13.94; partial η2 = .01; uncorrected P < .001; corrected P = .28) (eFigure 6 in the Supplement). For local correlation of resting-state fMRI, the greatest difference between healthy individuals and those with depression was found for a voxel in the right paracentral lobule (F1, 1330 = 20.98; partial η2 = 0.016; uncorrected P < .001; corrected P = .07) (eFigure 7 in the Supplement). For ALFF, the greatest effect was found for a voxel in the right parahippocampal region (F1, 1330 = 17.86; partial η2 = 0.013; uncorrected P < .001; corrected P = .10) (eFigure 8 in the Supplement). For fALFF, the greatest effect was found for a voxel in the right medial orbitofrontal cortex (F1, 1330 = 18.29; partial η2 = 0.014; uncorrected P < .001; corrected P = .24) (eFigure 9 in the Supplement). For DTI, the greatest effect was found between the right pars triangularis and right rostral middle frontal region for fractional anisotropy (F1, 1496 = 6.71; partial η2 = 0.004; uncorrected P = .01; corrected P < .99) (eFigure 10 in the Supplement), mean diffusivity (F1, 1494 = 11.20; partial η2 = 0.007; uncorrected P = .001; corrected P < .99) (eFigure 11 in the Supplement), and the average degree centrality network parameter (F1, 1502 = 8.93; partial η2 = 0.006; uncorrected P = .003; corrected P < .99) (eFigure 12 in the Supplement).
Table 1. Social Demographics, Clinical Characteristics, and Neurobiological and Genetic Data of All Participants.
| Sociodemographic and clinical variables | Participant | |||
|---|---|---|---|---|
| Healthy | Major depression | |||
| No. | Mean (SD) [range] | No. | Mean (SD) [range] | |
| Sex | ||||
| Male | 339 | NA | 305 | NA |
| Female | 609 | 556 | ||
| Age, y | NA | 34.41 (13.00) [18-65] | NA | 36.81 (13.26) [18-65] |
| Hamilton Depression Rating Scale | 1.46 (2.18) [0-17] | 9.36 (7.17) [0-34] | ||
| Beck Depression Inventory | 4.12 (4.27) [0-31] | 17.54 (11.02) [0-52] | ||
| Childhood Trauma Questionnaire | 32.61 (8.61) [25-77] | 44.96 (15.88) [25-125] | ||
| Social support | 4.51 (0.54) [2-5] | 3.77 (0.87) [1-5] | ||
| Medication index | NA | 1.31 (1.49) [0-11] | ||
| No. of previous inpatient treatments | 1.58 (2.08) [0-17] | |||
| No. of previous depressive episodes | 4.02 (6.78) [1-90] | |||
| Total duration of previous inpatient treatments, wk | 12.16 (18.58) [0-187] | |||
| Total duration of all previous depressive episodes, mo | 43.91 (62.66) [1-480] | |||
| Time since first psychiatric treatment, mo | 86.15 (95.45) [0-552] | |||
| Neurobiological and genetic data | ||||
| Structural MRI | ||||
| Voxel-based morphometry | 926 | NA | 818 | NA |
| Cortical and subcortical surface, thickness, volume | 923 | 818 | ||
| Functional MRI | ||||
| Resting state | 702 | NA | 634 | NA |
| Face-matching task | 656 | 585 | ||
| Diffusion tensor imaging | 819 | 689 | ||
| Polygenic risk score | 850 | 771 | ||
Abbreviations: MRI, magnetic resonance imaging; NA, not applicable.
Figure 2. Effect Size, Classification Accuracy, and Distributional Overlap.
Partial effect size (η2) of single variables displaying the overall largest effect in each modality. Error bars indicate upper and lower bounds for bootstrapped CIs for partial η2. Balanced classification accuracy for all modalities based on logistic regression of single variables displaying the largest effect. Kernel density estimation plots of deconfounded values, including distributional overlap for healthy participants and those with depression are plotted on the right side of the figure. DTI indicates diffusion-tensor imaging; FA, fractional anisotropy; fALFF, fractional amplitude of low-frequency fluctuation; fMRI, functional magnetic resonance imaging; MDD, major depressive disorder; PRS, polygenic risk score; VBM, voxel-based morphometry.
Table 2. Neurobiological, Genetic, and Environmental Differences Between Healthy Individuals and Those With Depression.
| HC vs MDD | No. (HC, MDD) | df 1 | df 2 | F value | Uncorrected P value | Corrected P value | Partial η2 [range] | No. (%) | Covariatesa | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overlap | BACC | Sensitivity | Specificity | AUROC | |||||||||
| Structural MRI | |||||||||||||
| VBM | 1744 (926, 818) | 1 | 1737 | 22.82 | <.001 | .03 | 0.013 [0.004-0.025] | 90.96 | 54.30 | 53.35 | 55.26 | 56.19 | a |
| Cortical and subcortical surface, thickness, volume | 1741 (923, 818) | 1 | 1735 | 14.92 | <.001 | .06 | 0.009 [0.003-0.019] | 91.64 | 54.71 | 54.28 | 55.13 | 55.12 | b |
| Task-based fMRI | |||||||||||||
| Face-matching task | 1241 (656, 585) | 1 | 1235 | 14.25 | <.001 | .40 | 0.011 [0.003-0.026] | 91.75 | 53.83 | 52.29 | 55.38 | 55.71 | b |
| Resting-state fMRI | |||||||||||||
| Bivariate connectivity | 1336 (702, 634) | 1 | 1330 | 22.53 | <.001 | .07 | 0.017 [0.005-0.032] | 89.46 | 55.20 | 51.57 | 58.83 | 57.16 | b |
| Network parameters | 1336 (702, 634) | 1 | 1330 | 13.94 | <.001 | .28 | 0.010 [0.002-0.024] | 91.84 | 53.51 | 49.29 | 57.73 | 55.47 | b |
| Local correlation | 1336 (702, 634) | 1 | 1330 | 20.98 | <.001 | .07 | 0.016 [0.005-0.030] | 87.51 | 54.43 | 55.70 | 53.15 | 56.31 | b |
| ALFF | 1336 (702, 634) | 1 | 1330 | 17.86 | <.001 | .10 | 0.013 [0.003-0.028] | 86.60 | 55.58 | 63.53 | 47.63 | 55.98 | b |
| fALFF | 1336 (702, 634) | 1 | 1330 | 18.29 | <.001 | .24 | 0.014 [0.004-0.029] | 90.97 | 53.80 | 52.71 | 54.89 | 56.59 | b |
| Structural connectome | |||||||||||||
| FA | 1502 (815, 687) | 1 | 1496 | 6.71 | .01 | <.99 | 0.004 [0.001-0.013] | 94.77 | 53.52 | 55.21 | 51.82 | 53.88 | b |
| MD | 1500 (814, 686) | 1 | 1494 | 11.20 | <.001 | <.99 | 0.007 [0.001-0.018] | 90.67 | 53.85 | 46.19 | 61.52 | 54.63 | b |
| Network parameters | 1508 (819, 689) | 1 | 1502 | 8.93 | .003 | <.99 | 0.006 [0.001-0.017] | 93.91 | 53.57 | 53.72 | 53.41 | 54.18 | b |
| Genetics | |||||||||||||
| Polygenic risk score | 1621 (850, 771) | 1 | 1613 | 54.17 | <.001 | <.001 | 0.032 [0.017-0.052] | 85.68 | 58.27 | 57.53 | 59.01 | 60.14 | c |
| Environment | |||||||||||||
| Social support | 1797 (945, 852) | 1 | 1792 | 481.92 | <.001 | <.001 | 0.212 [0.178-0.245] | 56.76 | 70.80 | 79.15 | 62.44 | 76.61 | d |
| Childhood maltreatment | 1799 (947, 852) | 1 | 1794 | 425.58 | <.001 | <.001 | 0.192 [0.162-0.221] | 55.61 | 70.70 | 80.25 | 61.15 | 76.01 | d |
Abbreviations: AUROC, area under the receiver operating curve; BACC, balanced accuracy; FA, fractional anisotropy; fALFF, fractional amplitude of low-frequency fluctuations; HC, healthy control; MD, mean diffusivity; MDD, major depressive disorder; MDS, multidimensional scaling; VBM, voxel-based morphometry.
Covariates in the statistical models: a = age + sex + dummy scanner + total intracranial volume; b = age + sex + dummy scanner; c = age + sex + dummy site + MDS 1 + MDS 2 + MDS 3, d = age + sex + dummy site.
Figure 3. Effect Size, Predictive Utility, and Distributional Overlap of the Best Modality.
Distributional overlap, effect size (η2), and classification performance for the single variable displaying the largest effect across all neuroimaging modalities (resting-state functional magnetic resonance imaging [fMRI] connectivity between a region of the right peripheral visual network and a region of the somatomotor network). A, Histogram with Gaussian kernel density estimation as a solid line and box plot of the confound-corrected values of the resting-state region of interest (ROI) to ROI connectivity displaying the largest effect. Middle line of the box plot represents the median value; the end of the boxes represents the lower and upper quartile. The boundaries of the whiskers are found within 1.5 IQR. B, Partial η2 analysis of variance (ANOVA) effect size for the strongest resting-state connectivity. Light blue indicates the upper bound of a bootstrapped 95% CI. C, Receiver operating characteristic (ROC) curve for logistic regression classification based on the single variable displaying the largest effect. AUROC indicates area under the ROC curve; HC, healthy control; MDD, major depressive disorder.
In comparison to the neuroimaging data, individuals and those with depression differed significantly in the PRS for major depression (F1, 1613 = 20.56; partial η2 = 0.032; P < .001), social support (F1, 1792 = 481.93; partial η2 = 0.211; P < .001), and childhood maltreatment (F1, 1794 = 425.59; partial η2 = 0.192; P < .001).
Distributions of the variables displaying the largest difference between HCs and MDD overlapped between 86.6% and 94.8% across all neuroimaging modalities (Figure 2). Even under ideal statistical conditions, this corresponds to classification accuracies between 53.5% and 55.6%. Resting-state ALFF displayed the highest overall classification accuracy. In comparison, MDD PRS was found to have an overlap of 85.7% (balanced accuracy = 58.3%). In contrast, environmental variables showed an overlap of 55.6% and 56.8%, corresponding to a classification accuracy of 70.7% and 70.8%.
To further analyze the effect of heterogeneity owing to research site or sex, we repeated all analyses for the 2 study sites in Marburg, Germany, and Münster, Germany, as well as for male and female participants separately. Although methodologic and biological homogeneity were expected to increase within the respective subsamples, results did not fundamentally change (eTables 4-5 in the Supplement). A control analysis using a matched healthy sample also showed highly similar results (eFigure 16 in the Supplement).
Analysis of Subgroups With Acute and Chronic Depression and Those Who Received Medication for MDD
Results do not fundamentally change when considering only those with acute or chronic depression as well as subgroups of patients with MDD who received medication (eTables 1-3 and eFigures 13-15 in the Supplement). For the variables displaying the largest group difference in each modality, distributions of healthy individuals and those with acute depression overlapped between 86.2% and 94.1% for all neuroimaging modalities. Classification accuracies ranged between 53.9% and 55.8% for those variables displaying the maximum effect. Largest effect size was found within resting-state connectivity (partial η2 = 0.021). Comparably, distributions of maximum difference variables for healthy individuals and those with chronic depression overlapped between 79.1% and 92.0% for all neuroimaging modalities. Classification accuracies ranged between 53.4% and 59.0%. Largest effect size was found within resting-state ALFF (partial η2 = 0.029). Individuals with depression who received medication showed overlap rates between 84.6% and 93.9%. Classification accuracies ranged between 53.4% and 59.2%. The largest effect size was found within resting-state local correlation (partial η2 = 0.027).
Discussion
In this case-control study, results suggest that healthy individuals and those with depression are strikingly similar with regard to univariate neurobiological and genetic measures. Even when considering the upper bound of the deviation in each modality, none could be considered informative from a personalized psychiatry perspective with both groups being nearly indistinguishable on a single-participant level. This is true despite near-ideal harmonization of study protocols, quality control, neuroimaging data acquisition, and clinical assessment, employing standard processing and analysis pipelines frequently used in the scientific community. Overall, no modality explained more than approximately 2% of the variance between healthy individuals and those with depression. Our results for structural MRI data are in line with Schmaal et al12 who reported an explained variance of approximately 1% for their largest effect of hippocampal volume reduction (Cohen d = 0.21; η2 = 0.011; statistical transformation from d to η2).36 Importantly, however, we extend this finding to a comprehensive set of neuroimaging modalities and show that results are similar also for task-based and resting-state fMRI as well as DTI. Crucially, as this large data set was acquired by only 2 research sites, we showed that the observed low effect sizes cannot be explained by a lack of harmonization of studies as previously suggested.17
Likewise, extensive subgroup analyses revealed that clinical heterogeneity alone is also not concealing potentially relevant differences. Nominally, patients with chronic depression showed slightly larger effect sizes and less overlap with healthy participants. Although this could indicate that depression severity increases neurobiological deviation, this association does not seem to be particularly strong. In contrast to previous reports, our study leaves little room to attribute the lack of substantial differences between HC and MDD to small sample size or heterogeneity in study protocols and assessments.
If the informational and predictive value of cross-sectional, univariate group differences is negligible, we must first explain why we see such a similarity between neurobiological measurements of healthy individuals and those with depression despite a substantial behavioral difference. Second, we need to derive ways to deliver accurate predictions that can change the clinical practice, finally improving the well-being of patients.37
When trying to explain the surprising subtlety of neurobiological deviations in depression, 2 major reasons can be identified. In principle, it could be possible that clinical neuroscience may simply be measuring properties of the brain irrelevant to MDD. Given the consistently small effects across all investigated modalities, including brain structure, function, and genetics, our results would then suggest to direct research efforts toward brain measurements that are temporally and spatially more finely grained and could thus provide more clinically relevant information. Work on magnetoencephalography (MEG), electroencephalography (EEG), and high-field MRI in neuroimaging and psychiatry substantiates this research direction and demonstrates that these methods are already finding its way into the core toolbox of clinical neuroimaging (review38 on 7-T MRI in depression,39 for a review on MEG and for EEG in treatment-response prediction).40 Although more attainable, EEG and MEG studies must build platforms and consortia to obtain similar sample sizes as current large-scale structural MRI studies. Regarding fMRI, increasing evidence points to low reliability values particularly of task-based fMRI, and efforts have been suggested to address these issues.41 Although structural MRI does not encounter such reliability issues, standard volumetric analysis pipelines, such as FreeSurfer, may not capture variance with direct relevance for disease-related mechanisms.12,16 Owing to their high level of standardization, these structural data pipelines have, however, dominated large consortia such as ENIGMA lately. Voxel-based morphometry methods provide a similar level of standardization but do not rely upon a specific parcellation; therefore, disease-specific regions identified by large meta-analyses such as Gray et al7 can be targeted in future research.
If we assume that clinically relevant information is contained in our data, the way we statistically and methodologically map depressive behavior to neuroimaging data must be inadequate. This can relate to the traditional mass-univariate statistical modeling as well as the clinical definition of the disease itself and the resulting heterogeneity of the clinical population. Standard univariate analysis approaches may not be able to adequately model the complexity of the depressive phenotype and underlying biological causalities. Network neuroscience continues to have a considerable effect on the field, applying methods from mathematical graph theory to model the functional integration of brain regions using functional and effective connectivity. Although the graph metrics used in this study did not increase the difference between healthy individuals and those with depression, the increased capacity of network neuroscience approaches is more likely to be able to model complex clinical phenotypes.33,42,43,44 To reach a higher level of personalization in psychiatry, multivariate machine learning methods with their clear focus on predictive and clinical utility as well as their ability to model complex relationships should become an even more significant part of the neuroimaging tool kit.45,46,47,48,49
Complementary to the possibility of inadequate neurobiological measurements and modeling approaches is the possibility that we may be considering ill-defined phenotypes. Although numerous attempts such as the Research Domain Criteria have been suggested to assess phenotype characteristics in a manner as to make them more accessible to neuroscientific investigation, none have yielded substantial progress thus far.50 Along the same lines, many have argued that depression is not a consistent syndrome with a fixed set of symptoms identical for all patients. In an investigation of patient’s symptoms in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study, Fried and Nesse51 identified over 1000 unique symptoms in about 3700 patients with depression, irrespective of depression severity. With these symptoms potentially differing from each other with respect to their underlying biology, severity, or effect on functioning, the common notion of aggregating across these diverse symptom profiles and focusing on MDD as homogeneous phenomena has likely hampered the development of clinically useful biomarkers of depression.52 Still, depressive core symptoms shared across patients clearly point to the existence of at least some common dysfunctions that also need to have a neurobiological basis. Thus, investigating the neurobiological basis of individual symptoms or dysfunctions is a promising research direction that receives increased attention.53 From a statistical point of view, normative modeling may be another way of parsing the clinical heterogeneity within and across disorders.54,55 With increasing sample sizes, such approaches will become increasingly more important in the future.
Limitations
This study had several limitations. Although the current study comprised a wide range of neuroimaging modalities, these were analyzed separately. The combination of multiple sources of information may decrease overlap between patients and controls and should be further explored. Machine learning methods in particular provide a sound basis for modality integration while controlling in-sample overfitting. Another important limitation of this work was the cross-sectional nature of this study. Within-participant longitudinal measurements may be more suitable to reveal mechanistic insights or predictive potential. To this end, especially outcome-based, longitudinal research designs are key to advancing our understanding of causal mechanisms with a direct effect on the clinical practice. More ecologically valid and easy to administer symptom measurements, eg, via smartphone applications, may aid this endeavor.56,57
Conclusions
Results of this case-control study suggest that even for maximum univariate biological differences, deviations between healthy individuals and patients with MDD were remarkably small. For future research, we recommend the following: (1) all researchers should clearly communicate the relevance of their findings by reporting measures of predictive utility or distributional overlap in addition to P values; if predictive utility cannot be demonstrated, researchers should precisely state in what way a significant effect advances the development of a quantitative neurobiological theory of depression, and stake holders may want to consider novel approaches to fMRI paradigm design58; (2) the community should prioritize more comprehensive phenotyping, including deep phenotyping of existing cohorts, the systematic assessment of novel digital phenotypes, moving beyond simple case-control designs, as well as longitudinal assessments of symptom dynamics and life events; and (3) the major issue of poor predictive performance needs to be addressed; machine learning approaches are increasingly used to investigate multivariate patterns of deviations and map high-dimensional biological information to complex phenotypes.47 Although these methods can also be useful in the context of advancing or falsifying theories, this clear shift from explanation to prediction might be more likely to have a direct effect on clinical practice in the short term.59
eMethods 1. PubMed Search Terms
eMethods 2. Exclusion Criteria
eMethods 3. Diagnosis and Remission Status
eMethods 4. Calculation of Medication Load Index
eMethods 5. Analysis of the Depressive Subgroups
eMethods 6. Matching Procedure for an Age- and Sex-Matched Healthy Sample
eMethods 7. Statistical Covariates and Correction
eMethods 8. Assessment of Childhood Maltreatment and Social Support
eMethods 9. Polygenic Risk Scores
eMethods 10. Magnetic Resonance Imaging
eMethods 11. T1-Weighted MRI
eMethods 12. Freesurfer Measures
eMethods 13. Functional MRI Image Acquisition
eMethods 14. Resting-State fMRI
eMethods 15. Task-Based fMRI
eMethods 16. DTI Image Acquisition and Preprocessing
eMethods 17. Graph Network Parameters
eTable 1. Statistical Analyses of Differences Between Healthy and Acutely Depressed Individuals
eTable 2. Statistical Analyses of Differences Between Healthy and Chronically Depressed Individuals
eTable 3. Statistical Analyses of Differences Between Healthy and Medicated Depressive Individuals
eTable 4. Statistical Analyses of Differences Between Healthy and Depressive Individuals Analyzed Separately for Females and Males
eTable 5. Statistical Analyses of Differences Between Healthy and Depressive Individuals Analyzed Separately for Marburg and Münster
eFigure 1. Depressive Symptom Severity of Lifetime and Acute MDD
eFigure 2. Depressive Symptom Severity of Chronically Depressed and Medicated MDD
eFigure 3. Predictive Utility—VBM—Healthy Individuals vs Those With Depression
eFigure 4. Predictive Utility—Freesurfer—Healthy Individuals vs Those With Depression
eFigure 5. Predictive Utility—Task fMRI—Healthy Individuals vs Those With Depression
eFigure 6. Predictive Utility—Resting-State fMRI Network Parameters—Healthy Individuals vs Those With Depression
eFigure 7. Predictive Utility—Resting-State fMRI local correlation—Healthy Individuals vs Those With Depression
eFigure 8. Predictive Utility—Resting-State fMRI ALFF—Healthy Individuals vs Those With Depression
eFigure 9. Predictive Utility—Resting-State fMRI fALFF—Healthy Individuals vs Those With Depression
eFigure 10. Predictive Utility—DTI FA—Healthy Individuals vs Those With Depression
eFigure 11. Predictive Utility—DTI MD—Healthy Individuals vs Those With Depression
eFigure 12. Predictive Utility—DTI Network Parameters—Healthy Individuals vs Those With Depression
eFigure 13. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Acute Depression
eFigure 14. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Long-term Depression
eFigure 15. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Depression Taking Medication
eFigure 16. Effect Size and Classification Accuracy—Matched Healthy Individuals vs Those With Depression
eReferences
References
- 1.World Health Organization . Depression and other common mental disorders. Accessed November 11, 2021. https://apps.who.int/iris/bitstream/handle/10665/254610/WHO-MSD-MER-2017.2-eng.pdf
- 2.Greenberg PE, Fournier AA, Sisitsky T, Pike CT, Kessler RC. The economic burden of adults with major depressive disorder in the US (2005 and 2010). J Clin Psychiatry. 2015;76(2):155-162. doi: 10.4088/JCP.14m09298 [DOI] [PubMed] [Google Scholar]
- 3.Greenberg PE, Fournier AA, Sisitsky T, et al. The economic burden of adults with major depressive disorder in the US (2010 and 2018). Pharmacoeconomics. 2021;39(6):653-665. doi: 10.1007/s40273-021-01019-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shorter E. A History of Psychiatry: From the Era of the Asylum to the Age of Prozac by Edward Shorter. 2nd ed. Wiley; 1998. [Google Scholar]
- 5.Walter H. The third wave of biological psychiatry. Front Psychol. 2013;4:582. doi: 10.3389/fpsyg.2013.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller VI, Cieslik EC, Serbanescu I, Laird AR, Fox PT, Eickhoff SB. Altered brain activity in unipolar depression revisited: meta-analyses of neuroimaging studies. JAMA Psychiatry. 2017;74(1):47-55. doi: 10.1001/jamapsychiatry.2016.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray JP, Müller VI, Eickhoff SB, Fox PT. Multimodal abnormalities of brain structure and function in major depressive disorder: a meta-analysis of neuroimaging studies. Am J Psychiatry. 2020;177(5):422-434. doi: 10.1176/appi.ajp.2019.19050560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprooten E, Rasgon A, Goodman M, et al. Addressing reverse inference in psychiatric neuroimaging: meta-analyses of task-related brain activation in common mental disorders. Hum Brain Mapp. 2017;38(4):1846-1864. doi: 10.1002/hbm.23486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodkind M, Eickhoff SB, Oathes DJ, et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry. 2015;72(4):305-315. doi: 10.1001/jamapsychiatry.2014.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanasse TJ, Fox PM, Barron DS, et al. BrainMap VBM: an environment for structural meta-analysis. Hum Brain Mapp. 2018;39(8):3308-3325. doi: 10.1002/hbm.24078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sha Z, Xia M, Lin Q, et al. Metaconnectomic analysis reveals commonly disrupted functional architectures in network modules and connectors across brain disorders. Cereb Cortex. 2018;28(12):4179-4194. doi: 10.1093/cercor/bhx273 [DOI] [PubMed] [Google Scholar]
- 12.Schmaal L, Veltman DJ, van Erp TGM, et al. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol Psychiatry. 2016;21(6):806-812. doi: 10.1038/mp.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fried EI, Kievit RA. The volumes of subcortical regions in depressed and healthy individuals are strikingly similar: a reinterpretation of the results by Schmaal et al. Mol Psychiatry. 2016;21(6):724-725. doi: 10.1038/mp.2015.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malhi GS, Das P, Outhred T. Size matters; but so does what you do with it! Mol Psychiatry. 2016;21(6):725-726. doi: 10.1038/mp.2015.200 [DOI] [PubMed] [Google Scholar]
- 15.van Velzen LS, Kelly S, Isaev D, et al. White matter disturbances in major depressive disorder: a coordinated analysis across 20 international cohorts in the ENIGMA MDD working group. Mol Psychiatry. 2020;25(7):1511-1525. doi: 10.1038/s41380-019-0477-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmaal L, Hibar DP, Sämann PG, et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry. 2017;22(6):900-909. doi: 10.1038/mp.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmaal L, Veltman DJ, van Erp TGM, et al. Response to Dr Fried & Dr Kievit, and Dr Malhi et al. Mol Psychiatry. 2016;21(6):726-728. doi: 10.1038/mp.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogelbacher C, Möbius TWD, Sommer J, et al. The Marburg-Münster Affective Disorders Cohort Study (MACS): a quality assurance protocol for MR neuroimaging data. Neuroimage. 2018;172:450-460. doi: 10.1016/j.neuroimage.2018.01.079 [DOI] [PubMed] [Google Scholar]
- 19.Kircher T, Wöhr M, Nenadic I, et al. Neurobiology of the major psychoses: a translational perspective on brain structure and function-the FOR2107 consortium. Eur Arch Psychiatry Clin Neurosci. 2019;269(8):949-962. doi: 10.1007/s00406-018-0943-x [DOI] [PubMed] [Google Scholar]
- 20.Inman HF, Bradley EL. The overlapping coefficient as a measure of agreement between probability distributions and point estimation of the overlap of 2 normal densities. Commun Stat Theory Methods. 1989;18(10):3851-3874. doi: 10.1080/03610928908830127 [DOI] [Google Scholar]
- 21.Bernstein DP, Fink L, Handelsman L, et al. Initial reliability and validity of a new retrospective measure of child abuse and neglect. Am J Psychiatry. 1994;151(8):1132-1136. doi: 10.1176/ajp.151.8.1132 [DOI] [PubMed] [Google Scholar]
- 22.Fydrich T, Sommer G, Tydecks S, Brähler E. Fragebogen zur sozialen Unterstützung (F-SozU): Normierung der Kurzform (K-14). Z Med Psychol. 2009;18(1):43-48. [Google Scholar]
- 23.Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343-352. doi: 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. doi: 10.1038/s41467-019-09718-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781. doi: 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaser C, Kurth F. A computational anatomy toolbox for SPM. Accessed June 24, 2021. http://www.neuro.uni-jena.de/cat/
- 27.Schaefer A, Kong R, Gordon EM, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28(9):3095-3114. doi: 10.1093/cercor/bhx179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2(3):125-141. doi: 10.1089/brain.2012.0073 [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Long XY, Yang Y, et al. Amplitude of low frequency fluctuation within visual areas revealed by resting-state functional MRI. Neuroimage. 2007;36(1):144-152. doi: 10.1016/j.neuroimage.2007.01.054 [DOI] [PubMed] [Google Scholar]
- 30.Zou QH, Zhu CZ, Yang Y, et al. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods. 2008;172(1):137-141. doi: 10.1016/j.jneumeth.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshpande G, LaConte S, Peltier S, Hu X. Integrated local correlation: a new measure of local coherence in fMRI data. Hum Brain Mapp. 2009;30(1):13-23. doi: 10.1002/hbm.20482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lange SC, van den Heuvel MP. Structural and functional connectivity reconstruction with CATO—a connectivity analysis toolbox. bioRxiv. Preprint posted online May 31, 2021. doi: 10.1101/2021.05.31.446012 [DOI] [PubMed]
- 33.Farahani FV, Karwowski W, Lighthall NR. Application of graph theory for identifying connectivity patterns in human brain networks: a systematic review. Front Neurosci. 2019;13:585. doi: 10.3389/fnins.2019.00585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diciccio TJ, Romano JP. A review of bootstrap confidence intervals. J Royal Stat Soc Series B Stat Methodol. 1988;50(3):338-354. doi: 10.1111/j.2517-6161.1988.tb01732.x [DOI] [Google Scholar]
- 35.Github . Code for analyses and figures for Quantifying Deviations of Brain Structure and Function in Major Depressive Disorder across Neuroimaging Modalities. Accessed June 27, 2022. https://github.com/wwu-mmll/more-alike-than-different-paper2021 [DOI] [PMC free article] [PubMed]
- 36.Cohen J. Statistical Power Analysis for the Behavioral Sciences. Academic Press; 1988. [Google Scholar]
- 37.Paulus MP, Thompson WK. The challenges and opportunities of small effects: the new normal in academic psychiatry. JAMA Psychiatry. 2019;76(4):353-354. doi: 10.1001/jamapsychiatry.2018.4540 [DOI] [PubMed] [Google Scholar]
- 38.Cattarinussi G, Delvecchio G, Maggioni E, Bressi C, Brambilla P. Ultrahigh-field imaging in major depressive disorder: a review of structural and functional studies. J Affect Disord. 2021;290:65-73. doi: 10.1016/j.jad.2021.04.056 [DOI] [PubMed] [Google Scholar]
- 39.Uhlhaas PJ, Liddle P, Linden DEJ, Nobre AC, Singh KD, Gross J. Magnetoencephalography as a tool in psychiatric research: current status and perspective. Biol Psychiatry Cogn Neurosci Neuroimaging. 2017;2(3):235-244. doi: 10.1016/j.bpsc.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolle CE, Fonzo GA, Wu W, et al. Cortical connectivity moderators of antidepressant vs placebo treatment response in major depressive disorder: secondary analysis of a randomized clinical trial. JAMA Psychiatry. 2020;77(4):397-408. doi: 10.1001/jamapsychiatry.2019.3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott ML, Knodt AR, Ireland D, et al. What is the test-retest reliability of common task-functional MRI measures? new empirical evidence and a meta-analysis. Psychol Sci. 2020;31(7):792-806. doi: 10.1177/0956797620916786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park HJ, Friston K. Structural and functional brain networks: from connections to cognition. Science. 2013;342(6158):1238411. doi: 10.1126/science.1238411 [DOI] [PubMed] [Google Scholar]
- 43.Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017;20(3):353-364. doi: 10.1038/nn.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cauda F, Nani A, Costa T, et al. The morphometric co-atrophy networking of schizophrenia, autistic, and obsessive spectrum disorders. Hum Brain Mapp. 2018;39(5):1898-1928. doi: 10.1002/hbm.23952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bzdok D, Meyer-Lindenberg A. Machine learning for precision psychiatry: opportunities and challenges. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3(3):223-230. doi: 10.1016/j.bpsc.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 46.Bzdok D, Varoquaux G, Steyerberg EW. Prediction, not association, paves the road to precision medicine. JAMA Psychiatry. 2021;78(2):127-128. doi: 10.1001/jamapsychiatry.2020.2549 [DOI] [PubMed] [Google Scholar]
- 47.Winter NR, Cearns M, Clark SR, et al. From multivariate methods to an AI ecosystem. Mol Psychiatr. 2021;26(11):6116-6120. doi: 10.1038/s41380-021-01116-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn T, Nierenberg AA, Whitfield-Gabrieli S. Predictive analytics in mental health: applications, guidelines, challenges and perspectives. Mol Psychiatry. 2017;22(1):37-43. doi: 10.1038/mp.2016.201 [DOI] [PubMed] [Google Scholar]
- 49.Janssen RJ, Mourão-Miranda J, Schnack HG. Making individual prognoses in psychiatry using neuroimaging and machine learning. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3(9):798-808. doi: 10.1016/j.bpsc.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 50.Insel T, Cuthbert B, Garvey M, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167(7):748-751. doi: 10.1176/appi.ajp.2010.09091379 [DOI] [PubMed] [Google Scholar]
- 51.Fried EI, Nesse RM. Depression is not a consistent syndrome: An investigation of unique symptom patterns in the STAR*D study. J Affect Disord. 2015;172:96-102. doi: 10.1016/j.jad.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fried EI, Nesse RM. Depression sum-scores don’t add up: why analyzing specific depression symptoms is essential. BMC Med. 2015;13(1):72. doi: 10.1186/s12916-015-0325-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fried EI, Epskamp S, Nesse RM, Tuerlinckx F, Borsboom D. What are “good” depression symptoms? comparing the centrality of DSM and non-DSM symptoms of depression in a network analysis. J Affect Disord. 2016;189:314-320. doi: 10.1016/j.jad.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 54.Marquand AF, Rezek I, Buitelaar J, Beckmann CF. Understanding heterogeneity in clinical cohorts using normative models: beyond case-control studies. Biol Psychiatry. 2016;80(7):552-561. doi: 10.1016/j.biopsych.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF. From estimating activation locality to predicting disorder: a review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci Biobehav Rev. 2015;57:328-349. doi: 10.1016/j.neubiorev.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 56.Goltermann J, Emden D, Leehr EJ, et al. Smartphone-based self-reports of depressive symptoms using the Remote Monitoring Application in Psychiatry (ReMAP): Interformat Validation Study. JMIR Ment Health. 2021;8(1):e24333. doi: 10.2196/24333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torous J, Staples P, Onnela JP. Realizing the potential of mobile mental health: new methods for new data in psychiatry. Curr Psychiatry Rep. 2015;17(8):602. doi: 10.1007/s11920-015-0602-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eickhoff SB, Milham M, Vanderwal T. Toward clinical applications of movie fMRI. Neuroimage. 2020;217:116860. doi: 10.1016/j.neuroimage.2020.116860 [DOI] [PubMed] [Google Scholar]
- 59.Paulus MP. Pragmatism instead of mechanism: a call for impactful biological psychiatry. JAMA Psychiatry. 2015;72(7):631-632. doi: 10.1001/jamapsychiatry.2015.0497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods 1. PubMed Search Terms
eMethods 2. Exclusion Criteria
eMethods 3. Diagnosis and Remission Status
eMethods 4. Calculation of Medication Load Index
eMethods 5. Analysis of the Depressive Subgroups
eMethods 6. Matching Procedure for an Age- and Sex-Matched Healthy Sample
eMethods 7. Statistical Covariates and Correction
eMethods 8. Assessment of Childhood Maltreatment and Social Support
eMethods 9. Polygenic Risk Scores
eMethods 10. Magnetic Resonance Imaging
eMethods 11. T1-Weighted MRI
eMethods 12. Freesurfer Measures
eMethods 13. Functional MRI Image Acquisition
eMethods 14. Resting-State fMRI
eMethods 15. Task-Based fMRI
eMethods 16. DTI Image Acquisition and Preprocessing
eMethods 17. Graph Network Parameters
eTable 1. Statistical Analyses of Differences Between Healthy and Acutely Depressed Individuals
eTable 2. Statistical Analyses of Differences Between Healthy and Chronically Depressed Individuals
eTable 3. Statistical Analyses of Differences Between Healthy and Medicated Depressive Individuals
eTable 4. Statistical Analyses of Differences Between Healthy and Depressive Individuals Analyzed Separately for Females and Males
eTable 5. Statistical Analyses of Differences Between Healthy and Depressive Individuals Analyzed Separately for Marburg and Münster
eFigure 1. Depressive Symptom Severity of Lifetime and Acute MDD
eFigure 2. Depressive Symptom Severity of Chronically Depressed and Medicated MDD
eFigure 3. Predictive Utility—VBM—Healthy Individuals vs Those With Depression
eFigure 4. Predictive Utility—Freesurfer—Healthy Individuals vs Those With Depression
eFigure 5. Predictive Utility—Task fMRI—Healthy Individuals vs Those With Depression
eFigure 6. Predictive Utility—Resting-State fMRI Network Parameters—Healthy Individuals vs Those With Depression
eFigure 7. Predictive Utility—Resting-State fMRI local correlation—Healthy Individuals vs Those With Depression
eFigure 8. Predictive Utility—Resting-State fMRI ALFF—Healthy Individuals vs Those With Depression
eFigure 9. Predictive Utility—Resting-State fMRI fALFF—Healthy Individuals vs Those With Depression
eFigure 10. Predictive Utility—DTI FA—Healthy Individuals vs Those With Depression
eFigure 11. Predictive Utility—DTI MD—Healthy Individuals vs Those With Depression
eFigure 12. Predictive Utility—DTI Network Parameters—Healthy Individuals vs Those With Depression
eFigure 13. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Acute Depression
eFigure 14. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Long-term Depression
eFigure 15. Effect Size and Classification Accuracy—Healthy Individuals vs Those With Depression Taking Medication
eFigure 16. Effect Size and Classification Accuracy—Matched Healthy Individuals vs Those With Depression
eReferences