Abstract
We report here the characterization of Tri10, a novel regulatory gene within the trichothecene gene cluster. Comparison of Tri10 genomic and mRNA sequences revealed that removal of a single 77-bp intron provided a 1,260-bp open reading frame, encoding a 420-amino-acid protein. Disruption of Tri10 in Fusarium sporotrichioides abolished T-2 toxin production and dramatically decreased the transcript accumulation for four trichothecene genes (Tri4, Tri5, Tri6, and Tri101) and an apparent farnesyl pyrophosphate synthetase (Fpps) gene. Conversely, homologous integration of a disruption vector by a single upstream crossover event significantly increased T-2 toxin production and elevated the transcript accumulation of the trichothecene genes and Fpps. Further analysis revealed that disruption of Tri10, and to a greater extent the disruption of Tri6, increased sensitivity to T-2 toxin under certain growth conditions. Although Tri10 is conserved in Fusarium graminearum and Fusarium sambucinum and clearly plays a central role in regulating trichothecene gene expression, it does not show any significant matches to proteins of known or predicted function or to motifs except a single transmembrane domain. We suggest a model in which Tri10 acts upstream of the cluster-encoded transcription factor TRI6 and is necessary for full expression of both the other trichothecene genes and the genes for the primary metabolic pathway that precedes the trichothecene biosynthetic pathway, as well as for wild-type levels of trichothecene self-protection. We further suggest the presence of a regulatory loop where Tri6 is not required for the transcription of Tri10 but is required to limit the expression of Tri10.
The trichothecenes represent a large family of toxic secondary metabolites produced by a variety of filamentous fungi, including Fusarium, Myrothecium, Stachybotrys, Trichoderma, and Trichothecium (16). They are primarily found as contaminants in food and animal feed, and consumption of these compounds by humans or livestock results in vomiting, alimentary hemorrhaging, and dermatitis (20). These toxins are potent inhibitors of eukaryotic protein synthesis (23) and induce apoptosis (24). In plants the trichothecenes are also phytotoxic and have been associated with virulence in specific plant-pathogen interactions (8, 9, 12, 25).
Significant progress has been made towards determining the trichothecene biosynthetic pathway and trichothecene gene organization and function. Trichothecenes are derived from farnesyl pyrophosphate, which is cyclized to form trichodiene (10). The biosynthetic sequence of events proceeding from trichodiene to complex trichothecenes, such as T-2 toxin, has been established by a combination of feeding experiments utilizing blocked mutants and heavy-isotope labeling of pathway precursors (3). To date, one regulatory gene, Tri6, one transporter gene, Tri12, and all but one of the known biosynthetic genes are clustered in Fusarium sporotrichioides (6), and similar gene clusters are present in Fusarium graminearum (6), Fusarium sambucinum (A. W. Peplow and M. N. Beremand, unpublished data), and Myrothecium roridum (17). Tri101, which encodes isotrichodermol 3-o-acetyltransferase, has been identified in both F. sporotrichioides and F. graminearum and appears to reside outside of the trichothecene gene cluster (19, 21). In addition to its acetyltransferase activity, the heterologous expression of Tri101 in yeast cells renders them resistant to trichothecenes (18, 21). However, disruption of Tri101 does not appear to cause a loss of toxin self-protection in F. sporotrichioides (21), suggesting that other genes can provide this function.
The regulation of trichothecene production is equally complex, and as with many secondary metabolites it can be controlled in liquid culture by the availability of certain nutrients, oxygen, pH, and temperature (33), as well as by the modulation of signal transduction pathways mediated by a G-protein Gα subunit (31). However, the complete genetic nature of the induction and repression of trichothecene biosynthesis is just beginning to be understood. Recently, the gene for TRI6, a zinc-finger DNA-binding protein, was isolated, characterized (15, 26), and shown to be required for induction of two trichothecene genes (Tri5 and Tri4), which encode, respectively, the enzymes for the first two steps in the T-2 toxin biosynthetic pathway (26). Here, we report the isolation and characterization of a second regulatory gene from within the trichothecene gene cluster, designated Tri10, which is required for trichothecene biosynthesis in F. sporotrichioides.
MATERIALS AND METHODS
Strains, plasmids, media, and culture conditions.
The F. sporotrichioides Sherb. wild-type strain NRRL 3299 was obtained from the ARS/USDA Culture Collection at the National Center for Agricultural Utilization Research in Peoria, Ill. The Fusarium sambucinum Fuckel (telomorph = Gibberella pulicaris [Fr.] Sacc.) wild-type strain R-6380 was obtained from the Fusarium Research Center, Pennsylvania State University. F. sporotrichioides strains MB5493 (Tri4 mutant) (3, 22) and NN4 (ΔTri6) (26) and the Fusarium graminearum Schwein. (telomorph = Gibberella zeae [Schwein.] Petch) wild-type strains GZ3639 (5) and W-8 (25) were described previously. Fresh stock cultures were routinely established from frozen glycerol stocks of conidia: transformants were inoculated onto V8 juice agar containing 300 μg of hygromycin B/ml and grown in the dark under alternating 12 h at 20°C and 12 h at 25°C conditions, while all other strains were inoculated onto V8 juice agar and incubated under alternating 12 h at 20°C dark, 12 h at 25°C light conditions (27). Strains were grown in liquid YEPD-2G medium (2% glucose, 0.3% yeast extract, 1% peptone) for DNA isolation and in liquid YEPD-5 medium (5% glucose, 0.1% yeast extract, and 0.1% peptone) for trichothecene analysis and RNA isolation (27). The vectors pT7Blue and pCR-Script were purchased from Novagen and Stratagene, respectively. The plasmid pGP53-1 was previously described (13).
RNA extraction and RT-PCR.
For RNA isolation, conidia of F. sporotrichioides were washed from 7-day V8 juice agar cultures, filtered through sterile muslin, and inoculated into 100 ml of YEPD-5 to give a final concentration of 5.0 × 104 conidia per ml. Parallel cultures were grown for 15 h and 23 h (28°C and 200 rpm) and harvested by vacuum filtration through P-8 qualitative filter paper (Fisher). The mycelia were immediately frozen in liquid nitrogen, lyophilized, and stored at −80°C. Samples were pulverized in liquid nitrogen immediately prior to extraction. Total RNA was isolated using the Ultraspec II kit (Biotecx) according to the manufacturer's protocol with the addition of an acid phenol-chloroform-isoamyl alcohol (25:24:1, pH 4.2) extraction following the initial extraction. For reverse transcription (RT)-PCR, the purified 23-h total RNA was treated with RNase-free DNase I (Ambion), and first-strand cDNA was generated from 5 μg of total RNA and an oligo(dT) primer using the Superscript Preamplification kit (Gibco-BRL). The Tri10 cDNA was amplified from the first-strand cDNA reaction by PCR using oligonucleotides 320 (5′ CCACCCAGCAATCATCAG 3′) and A23 (5′ CTGTTGTCAATAGGCGAGTG 3′), which lie 22 bp upstream and 19 bp downstream of the putative start and stop codons, respectively (Fig. 1). A portion of the 3′ end of the Tri10 transcript was cloned by pairing primers 516 (5′ CTTCAGCTTATCGGGTTT 3′) and 356 (5′ GTACCTCGTTTCATGCC 3′), which lie 357 bp downstream of the putative start codon and 192 bp downstream of the putative stop codon, respectively. The PCR amplifications consisted of a single cycle of 5 min at 95°C; 25 cycles of 1 min at 95°C, 1 min at 50 to 55°C, and 1.5 min at 72°C; and 1 cycle of 10 min at 72°C. The amplified products were run on a 1% agarose gel and purified using the Qiaquick gel extraction kit (Qiagen). The purified products were then cloned into pCR-Script (Stratagene) and sequenced.
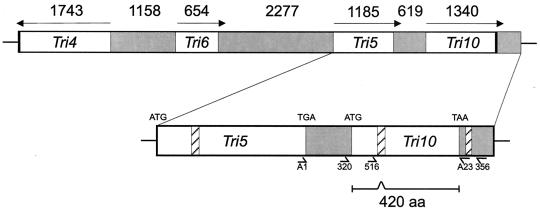
FIG. 1.
Graphic representation of the location of Tri10 and position of primers used for RT-PCR and for vector construction. Gray areas indicate intergenic regions, and the crosshatched areas represent the relative positions of introns within Tri5 and Tri10 in F. sporotrichioides. The numbers at the top of the figure indicate base pairs.
DNA sequencing.
Cloned RT-PCR products and genomic DNA clones were sequenced using specifically designed primers. Tri10 of F. sambucinum R-6380 was sequenced from the plasmid pGP53-1, which contains a 4.7-kb EcoRI fragment (harboring Tri5 and downstream sequences) isolated from a λgt11 library (13). Tri10 was also shown to be present and downstream from Tri5 in F. graminearum GZ3639 by Southern hybridization using the F. sporotrichioides NRRL 3299 Tri10 homolog as a probe. This downstream DNA, which is located on a 3.5-kb HindIII fragment, was amplified by chromosome crawling (28) via inverse PCR using two primers, B-1 (5′ GCGACGCTCGATACCGCCTCC 3′) and B-2 (5′ CGTGTCCATCACCTGAGGGTCC 3′), corresponding to the Tri5 gene from F. graminearum strain W-8 (25). All sequencing reactions were performed using the ABI PRISM Dye Terminator Cycle Sequencing Core Kit or the BigDye Terminator Cycle Sequencing Core Kit (Perkin-Elmer Corporation). Reactions were run on a Model 373A or Model 377 DNA Sequencer (Applied Biosystems) by the Gene Technologies Laboratory at Texas A&M University.
Plasmid construction and fungal transformation.
The Tri10 gene disruption plasmid, pTri10-1 (Fig. 2), was constructed in two steps. First, the PCR fragment (A1-356) (Fig. 1) containing the Tri10 coding region and flanking sequences was cloned into pT7-Blue. Then, the Tri10 amino acid coding region was disrupted by removing a 101-bp BstBI-BstEII fragment and replacing it with the Cochliobolus heterostrophus promoter 1 fused to the hygromycin B phosphotransferase coding region (32).
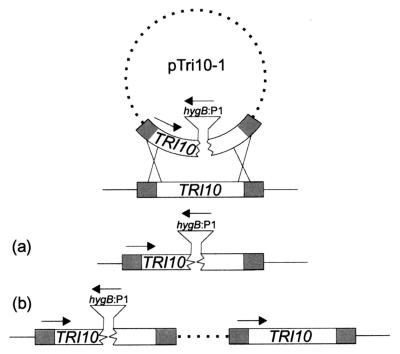
FIG. 2.
Results of integration of pTri10-1 into the Tri10 gene via double (a) and single (b) upstream homologous crossover events.
For fungal transformation, protoplasts were isolated by the following procedure: conidia (1 × 108 to 5 × 108) were inoculated into 100 ml of YEPD-2G broth and incubated for 7 h at 28°C with vigorous shaking (200 rpm). Germlings were harvested by filtration on a 0.2-μm-pore-size cellulose nitrate filter, washed once in 25 ml of 0.7 M NaCl, and resuspended in 20 ml of 0.7 M NaCl containing 0.1% Novozyme 234 (InterSpex Products), 1% driselase (Sigma), and 0.025% chitinase (Sigma). The germlings were then incubated at 28°C for 20 to 60 min with gentle shaking (75 to 90 rpm) until most of the culture had been converted to protoplasts. Protoplasts were collected by centrifugation (1,100 × g) for 5 min at room temperature and then washed once in 0.7 M NaCl and twice in STC buffer (1.2 M sorbitol, 10 mM Tris-HCl, 10 mM CaCl2). Transformation was performed according to the procedure described by Salch and Beremand (27).
DNA isolation and analysis.
Fungal genomic DNA was isolated as previously described (13). Southern blots were prepared according to standard techniques (28) using DNA digested with HindIII, which cuts the vector pTri10-1 once, and BglII, which does not cut the vector. The resulting blots were first hybridized with radiolabeled ([32P]dCTP) DNA probes prepared by nick translation (Gibco BRL) and then washed as recommended (Gene Images; USB).
RNA analysis.
For RNA blots, 5 μg of total RNA was separated by electrophoresis on formaldehyde-containing 1% agarose gels and transferred to Hybond N+ (Amersham) nylon membranes (28). Probes for RNA blots were gel-purified PCR fragments radiolabeled with [32P]dCTP by nick translation (Gibco BRL).
T-2 toxin analysis.
Duplicate YEPD-5 liquid cultures (25 ml) were inoculated and grown in parallel as described above for RNA extraction. After 7 days of growth, a 5-ml aliquot of whole culture material was removed and frozen at −20°C until analyzed. T-2 toxin was extracted by vortexing 2 ml of whole culture material with 2 ml of ethyl acetate for 90 s. The phases were separated by centrifugation at 1,100 × g for 5 min. Supernatants were removed and diluted with ethyl acetate to various concentrations and then analyzed by gas chromatography-mass spectrometry (GC-MS) on a Hewlett Packard 5890 GC equipped with an HP 5972 MS engine. Samples were introduced via a heated injection port (260°C) with an HP 7673 autosampler (2-μl injection volume) into an HP5-MS bonded stationary phase capillary column (internal diameter, 30 m by 0.25 μm) with a film of 0.25-μm thickness. The oven temperature program began with an initial oven temperature of 90°C for 2.0 min, followed by a ramp (23°C/min) to 275°C for 2.0 min. The temperature was then increased (30°C/min) to 290°C and held for 4.7 min. Helium was used as the carrier gas at a constant flow rate of 0.75 ml/min. The mass selective detector was used in either ion-selective mode (T-2 toxin quantitation) or full-scan mode (to obtain a full spectrum) with an ionization energy of 70 eV and an ion source temperature of 180°C. Identification of T-2 toxin was based on comparison of retention times and spectra to those of standards. T-2 toxin was purchased from Sigma Chemical Co. (St. Louis, Mo.) and used without further purification. Toxin levels of transformants were compared to that of the wild type by a priori contrast using the JMP4 statistical software package.
T-2 toxin sensitivity assays.
F. sporotrichioides strains were tested for resistance or sensitivity to T-2 toxin by using three different assays. In the first assay, designated the drop-plate assay, T-2 toxin solutions were prepared in ethyl acetate to yield a concentration of 10, 25, 50, or 100 μg per 25 μl of solvent. A 25-μl sample of each concentration and a solvent control were individually spotted on the surface of a YEPD-2G agar plate (2% agar, 10 ml of agar per 100-mm-diameter plate) and allowed to evaporate. The plates were immediately inoculated with the desired strain by spreading 100 μl of a freshly prepared conidial suspension containing 1 × 106 to 2 × 106 conidia/ml and then incubated for 2 days in a growth chamber with an alternating 12 h at 25°C light,12 h at 20°C dark cycle. The second assay was conducted on GYEP agar (=YEPD-5 in this study) as previously described with the exceptions that plugs of mycelia were used for inoculation and that T-2 toxin was used instead of diacetoxyscirpenol and only at a 100-μg/ml concentration (1). The third T-2 toxin sensitivity assay was conducted in liquid media. T-2 toxin stock solutions of 40 and 20 mg/ml were prepared in ethyl acetate, and 25-μl aliquots of the appropriate concentration were added to make 1-ml aliquots of both YEPD-2G and YEPD-5, with final concentrations of 1,000 and 500 μg of T-2 toxin/ml. Aliquots of both media were also prepared with 25 μl of ethyl acetate as controls. Additionally, spent YEPD-5 media from 3-day cultures of NRRL 3299 and FsTri10-1–25 (this study) were analyzed by GC-MS to quantitate the amount of T-2 toxin present, and aliquots were sterilized through a nylon 0.2-μm-pore-size filter. All media were transferred to a 96-well microtiter plate (150 μl/well, in duplicate/strain) and inoculated with 7 μl of conidia of NRRL 3299, the ΔTri6 strain, or the ΔTri10 strain to bring the final concentration to 103 conidia/ml. Cultures were grown on a gyratory shaker with vigorous shaking at 28°C for 3 days. The response to T-2 toxin was determined by following optical density at λ = 595 at selected time points. A complete lack of increase in optical density corresponded to a complete lack of growth, as confirmed by visual examination of the cultures.
Nucleotide sequence accession numbers.
The nucleotide sequence and the predicted polypeptide sequence for Tri10 from F. sporotrichioides, F. graminearium, and F. sambucinum have been submitted to GenBank (accession no. AF364179, AF365969, and AF386074, respectively).
RESULTS
Identification and sequence analysis of Tri10.
Using mutant analysis and genomic DNA sequence from the trichothecene pathway gene cluster in F. sporotrichioides, Beremand and Hohn previously identified a potential 1,260-bp open reading frame (ORF) 619 bp downstream of the Tri5 stop codon which they designated Tri10 (4; M. N. Beremand, unpublished results) (Fig. 1). RNA blots probed with labeled DNA fragments from this region revealed the low-level production of a transcript from the predicted ORF, as seen in Fig. 3. In this study, the F. sporotrichioides Tri10 transcript was amplified by RT-PCR, cloned, and sequenced, confirming a 1,260-bp ORF encoding a putative protein of 420 amino acids and having a predicted molecular mass of 47,427 Da. Comparison of the genomic and cDNA nucleotide sequences indicated the excision of the predicted 77-bp intron from the coding region of all transcripts (bases 379 to 455) as well as the excision of a 69-bp intron from the 3′ untranslated region (bases 1424 to 1492) of some of the transcripts. Interestingly, at least one other trichothecene gene, Tri12, contains an intron in the 3′ untranslated region (1). Both Tri10 introns contain the consensus splicing signal sequence GT::AG for filamentous fungi, but only the 77-bp intron contains a consensus branch point (11).
FIG. 3.
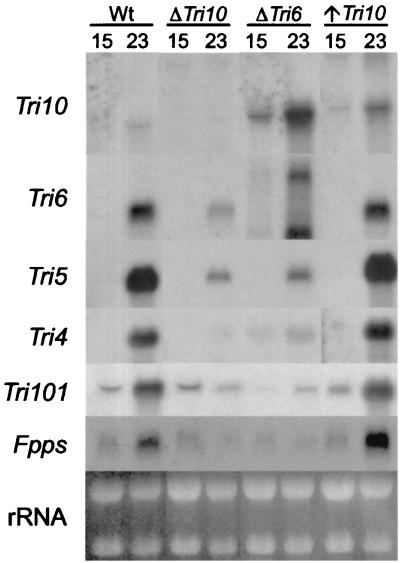
Northern analyses of Tri10, Tri6, Tri5, Tri4, Tri101, and Fpps in wild-type (Wt) (NRRL 3299), ΔTri10 (FsTri10-1–12), ΔTri6 (NN4), and ↑Tri10 (FsTri10-1-20) strains of F. sporotrichioides grown in parallel for 15 and 23 h. Five micrograms of total RNA was loaded per lane. Ribosomal RNAs were visualized in the gel by ethidium bromide staining.
Tri10 is transcribed in the same direction as Tri5. Two putative TATA boxes are present at positions −44 and −111 from the predicted translational start codon (Fig. 4). Five putative CAAT consensus sequences are located at positions −32, −48, −50, −107, and −207. A consensus eukaryotic ribosome-binding site exists beginning at position −6. No polyadenylation signals were found downstream of the stop codon. Comparisons of both the nucleotide sequence and the predicted amino acid sequence with multiple protein and DNA databases yielded only a single possible match (tBLASTn score = 93; Evalue = 4e−17) (2) to a pair of nearly identical expressed sequence tags (ESTs) (AL113997.1 and AL1116861.1) of unknown function from Botrytis cinerea grown under nitrogen-limiting conditions.
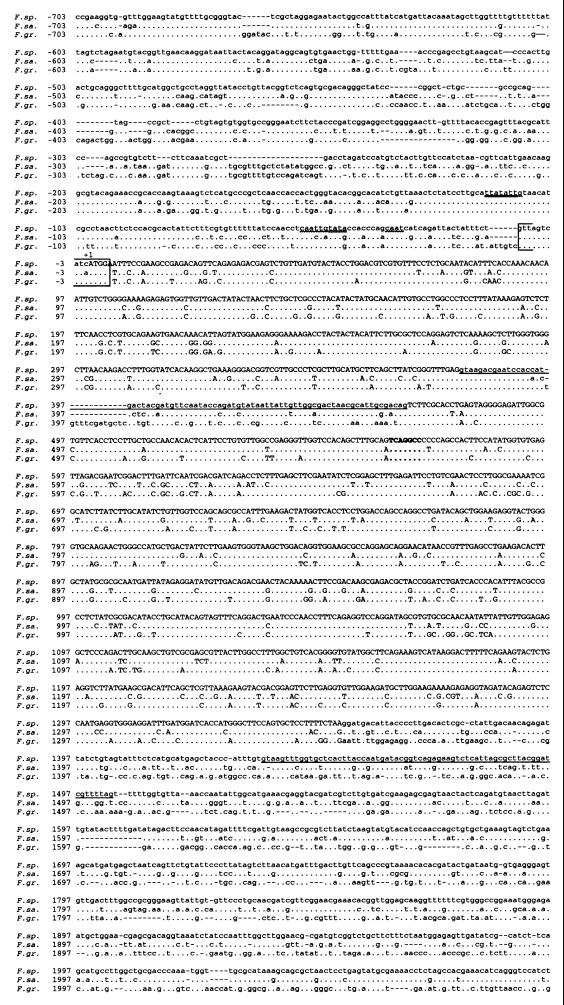
FIG. 4.
Sequence alignment of Tri10 from F. sporotrichioides (F. sp.) (NRRL 3299), F. sambucinum (F. sa.) (R-6380), and F. graminearum (F. gr.) (GZ3639) produced using Multiple Sequence Alignment (Baylor College of Medicine [http://dot.imgen.bcm.tmc.edu:9331/multi-align/Options/map.html]). Dots indicate conserved bases with F. sporotrichioides, and dashes indicate gaps between species. Capital letters indicate the putative amino acid coding region. A single underline indicates intron sequences. A double underline indicates conserved putative TATA elements, and a wavy underline represents conserved putative CAAT elements. The boxed region is the conserved ribosome binding site. The conserved TRI6 binding site is indicated by boldface type.
Transformation with pTri10-1 leading to the disruption and displacement of Tri10.
We wanted to examine the effect of Tri10 gene disruption on Tri gene expression and toxin production. However, in previous work the homologous integration of a Tri5 gene replacement vector into the Tri5 downstream sequence resulted in an increase in trichothecene gene expression and toxin production (M. N. Beremand, unpublished data). Since this vector contained the downstream region of Tri5 beginning with the last few codons of Tri5 and extended into what we now know to be the middle of the Tri10 gene, we designed the pTri10-1 vector so that it could either disrupt Tri10 as a result of homologous integration via a double crossover event or essentially duplicate the homologous integration event described above via a single crossover event (Fig. 2). Accordingly, the pTri10-1 vector carried a 2.1-kb DNA fragment which began at the end of the Tri5 amino acid coding region and included the entire Tri10 gene except for a 101-bp amino acid coding segment which was replaced by a hygromycin resistance cassette. Southern blot analysis (data not shown) of the hygromycin-resistant transformants obtained following transformation of the wild-type F. sporotrichioides NRRL 3299 strain with pTri10-1 revealed that both classes of transformants were produced.
Tri10 plays a major role in controlling toxin production and the transcription of known trichothecene cluster genes and Tri101.
To investigate if disruption of Tri10 had an effect on T-2 toxin production, we grew YEPD-5 liquid shake cultures for 7 days and measured the culture filtrate for T-2 toxin using GC-MS. Transformants which contain a single disrupted copy of Tri10 (ΔTri10) consistently produced no T-2 toxin, as seen in Table 1, and do not appear to accumulate trichothecene pathway intermediates (data not shown).
TABLE 1.
Production of T-2 toxin by Tri10 transformants
| Groupa | Strain | Amt of T-2 toxin (μg/ml)b |
|---|---|---|
| Wild type | NRRL 3299 | 267 ± 5 |
| ΔTri10 | FsTri10-1-12 | 0 |
| ↑Tri10 | FsTri10-1-8 | 566 ± 5 |
| FsTri10-1-16 | 604 ± 46 | |
| FsTri10-1-20 | 633 ± 14 | |
| FsTri10-1-23 | 418 ± 7 | |
| FsTri10-1-24 | 644 ± 6 | |
| FsTri10-1-25 | 999 ± 137 |
Each group was shown to be significantly different by a priori contrast.
Values shown represent the means of duplicate extractions of duplicate cultures ± the standard deviations.
In contrast, transformants resulting from a single homologous integration event in the upstream portion of Tri10 displayed a significant (P = 0.0003) increase in T-2 toxin production over that of the wild-type strain (Table 1). The observation that T-2 toxin production is substantially elevated in these transformants parallels our previous and subsequent observations with integration events in this region of the gene cluster. Thus, we have designated this group of pTri10-1 transformants as Tri10 overproducers (↑Tri10).
RNA blot analysis was employed to examine the effect of the disruption of Tri10 on Tri gene expression. Comparison of transcript levels of representative Tri cluster genes between the ΔTri10 transformant (FsTri10-1-12) and the wild type showed that the expression of the trichothecene biosynthetic genes Tri5 and Tri4 and the regulatory gene Tri6 was greatly reduced in the ΔTri10 strain (Fig. 3). The Tri5, Tri4, and Tri6 gene transcripts are not detectable in the wild-type strain at 15 h but reach a high level of accumulation by 23 h. In contrast, only low levels of these gene transcripts appeared in the ΔTri10 strain. Full expression of Tri101, a Tri gene which putatively lies outside of the Tri gene cluster, was also dependent upon Tri10. While Tri101 was expressed at the same level at 15 h in both the wild-type and the ΔTri10 strains, the level of Tri101 transcript increased at 23 h in the wild-type strain but not in the ΔTri10 strain (Fig. 3). The transcription of Tri10 is also effectively blocked in the ΔTri10 strain. The trace amount of hybridizing material observed in the 23-h sample is most likely from a fusion transcript with the 5′ end of Tri10 joined to the P1:hygB sequence inserted in the Tri10 coding region (Fig. 2).
Examination of this same set of Tri gene transcripts in the ↑Tri10 transformant FsTri10-1–20 revealed that all of these transcripts are at least slightly elevated over wild-type levels at 23 h (Fig. 3). Notably, the Tri10 transcript is obviously elevated. Collectively, these data further demonstrate that T-2 toxin production parallels Tri gene expression and support the hypothesis that Tri10 plays a major regulatory role in coordinately regulating Tri gene expression and T-2 toxin production.
Tri6 is not required for transcription of Tri10.
Interestingly, Tri10 appears to be overexpressed at both time points in the Tri6 disruption mutant NN4 relative to the wild-type strain, as seen in Fig. 3. This is in sharp contrast to the other Tri genes, which show a marked decrease in the amount of transcript in response to the disruption of Tri6. These data suggest that the transcription of Tri10 is not positively regulated by TRI6 but instead may be negatively regulated either directly or indirectly when TRI6 is present.
Tri10 gene expression indirectly controls Fpps gene expression.
To determine if Tri10 gene expression affects the expression of genes in the primary metabolic pathway that directly feeds into the trichothecene biosynthetic pathway, we examined the expression of Fpps. FPPS catalyzes the formation of farnesyl pyrophosphate, the last intermediate in the isoprenoid primary metabolic pathway and the immediate precursor to trichodiene, the first intermediate of the trichothecene biosynthetic pathway. As seen in Fig. 3, Fpps transcript levels are reduced in the ΔTri10 transformant and slightly elevated at 23 h in the ↑Tri10 transformant. It is also noteworthy that the Fpps transcripts are reduced in the ΔTri6 transformant even though Tri10 transcripts are elevated. This suggests that Tri10 does not directly control Fpps transcript levels but rather does so indirectly via its control of the expression of Tri6, another gene(s), or trichothecene toxin levels.
Tri10 expression affects self-protection from T-2 toxin.
The results shown in the drop plate assay (Fig. 5) indicate that the deletion of Tri10 reduces the level of T-2 toxin self-protection and suggest that the expression of Tri10 may be required for wild-type levels of T-2 toxin self-protection. Unlike the wild-type parent, the ΔTri10 strain clearly displayed reduced growth in the presence of 100 and 50 μg of T-2 toxin. To determine if this response was mediated through the lack of Tri6 gene expression and hence the lack of expression of a gene(s) under the control of Tri6 or was simply due to the lack of T-2 toxin production, the abilities of a ΔTri6 strain and a Tri4 mutant strain to grow in the presence of T-2 toxin were examined. Although both strains fail to make T-2 toxin, the ΔTri6 strain does so because it is blocked in the transcription of the known Tri cluster genes, and the Tri4 mutant strain does so because it does not make an active trichodiene oxygenase enzyme and is therefore blocked in the second enzymatic step (feeding experiments have shown that this Tri4 mutant strain expresses all of the other pathway enzymes and therefore the genes) (22). As seen in Fig. 5, the ΔTri6 strain showed an even greater sensitivity to T-2 toxin than did the ΔTri10 strain, while the Tri4 mutant strain looked like the wild-type parent. We also examined T-2 toxin sensitivity using the YEPD-5 plate assay described by Alexander et al. (1) with the exceptions that T-2 toxin and mycelial plugs were used. Under these test conditions we saw no difference between the growth of the wild-type strain and either the ΔTri6 or ΔTri10 transformants on media containing 100 μg of T-2 toxin/ml (data not shown). Because we had observed a discernible difference between these strains and the wild-type strain in the drop-plate assay, we performed a third assay utilizing both YEPD-2G and YEPD-5 liquid media as well as spent YEPD-5 media containing known amounts of T-2 toxin. The results of this assay are shown in Fig. 6. Both the ΔTri10 and ΔTri6 transformants can be distinguished from the wild-type parent. However, in contrast to the ΔTri6 strain, which demonstrated no growth at all in the YEPD-2G (1,000 μg of T-2 toxin/ml) and the spent media, the ΔTri10 strain was able to grow under these conditions, albeit in an altered fashion, as measured by optical density.
FIG. 5.
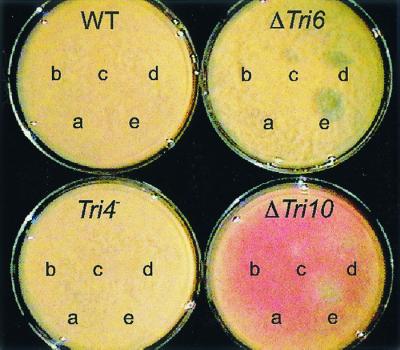
Drop-plate T-2 toxin sensitivity assay. Twenty-five microliters of ethyl acetate containing 0 (a), 10 (b), 25 (c), 50 (d), or 100 (e) μg of T-2 toxin was spotted onto YEPD-2G plates, and the ethyl acetate was allowed to evaporate. The plates were then spread with 1 × 105 to 2 × 105 conidia and allowed to grow for 2 days.
FIG. 6.
Microtiter plate T-2 toxin sensitivity assay. Wild-type (NRRL 3299) (♦), ΔTri10 (▪), and ΔTri6 (▴) strains were grown in YEPD-2G and YEPD-5 containing either 1,000, 500, or 0 μg of T-2 toxin/ml. These strains were also grown in spent YEPD-5 media containing 264 or 534 μg of T-2 toxin/ml. All strains were grown for 64 h, and the optical density was measured at λ = 595 (O.D. 595). Error bars represent the range of readings observed per isolate per time point.
The above-described experiments indicate that the assay conditions are critical for observing the increased sensitivities of both the ΔTri10 and the ΔTri6 strains to T-2 toxin. Overall, the ΔTri6 transformant was more sensitive, showing clearly inhibited growth in two of the three assay formats, while the ΔTri10 transformant was intermediate between ΔTri6 and the wild type, showing some clearly inhibited growth only in the drop plate assay. These results, together with the observation that a low level of the Tri6 transcript was present in the ΔTri10 transformant, imply that Tri10 via Tri6 controls the expression of one or more genes that contribute to T-2 toxin self-protection and that the wild-type levels of expression of these genes become essential only under certain growth conditions.
Tri10 is conserved in other trichothecene-producing Fusarium species
To determine if Tri10 was present in other trichothecene-producing species and also to ask what motifs within Tri10 might be conserved, we examined the genomes of F. sambucinum R-6380 (G. pulicaris), which produces primarily diacetoxyscirpenol, and F. graminearum GZ3639 (Gibberella zeae), which produces primarily deoxynivalenol. We isolated and sequenced Tri10 from both of these strains as described in Materials and Methods and determined that the placement and direction of transcription relative to Tri5 are conserved. Likewise the amino acid coding sequence of Tri10 in F. sambucinum and F. graminearum is of identical length to that found in F. sporotrichioides, and all three species share >85% nucleotide identity (Fig. 4) and approximately 88% amino acid identity. All three species share at least one putative TATA and CAAT element (Fig. 4). They also share the conserved eukaryotic ribosome-binding site (with one nucleotide substitution), conserved 5′ and 3′ splice sites surrounding the intron within the coding region, and the conserved branch point within the intron (14) (Fig. 4). However, the removal of this intron has not been confirmed by cDNA sequencing in F. sambucinum or F. graminearum. In addition, F. sambucinum and F. graminearum appear to lack the consensus splice sites for the intron in the 3′ untranslated region.
DISCUSSION
In this study we define a key regulatory gene, Tri10, which controls trichothecene production and related gene expression. As depicted in the model in Fig. 7, the data suggest that Tri10 exerts this control, at least in part, by regulating the transcription of Tri6. Tri10 appears to be a positive regulator of Tri6, since Tri10 transcripts appear before Tri6 transcripts in the Tri10 overproducing strain and deletion of Tri10 severely inhibits trichothecene gene expression and blocks trichothecene production. However, because disruption of Tri10 does not cause a complete loss of Tri6, Tri5, Tri4, and Tri101 gene transcripts and the disruption of Tri6 likewise causes a severe reduction only of the last three gene transcripts, each of these genes must also have an additional basal route or mechanism by which its expression is activated. Furthermore, in order to account for the lack of T-2 toxin production by the ΔTri10 strain, the loss of Tri10 must completely block the transcription of some additional gene(s) required for T-2 toxin production. Alternatively, a combined marked reduction in both the Tri gene transcript levels and the farnesyl pyrophosphate pool, as a result of the parallel inhibition of Fpps and other isoprenoid genes (described below), could be sufficient to prevent T-2 toxin production in the ΔTri10 strain. These possibilities can be addressed by future experiments.
FIG. 7.
Proposed regulatory model for trichothecene biosynthesis. Solid arrows indicate known positive activators. Dotted arrows indicate possible activation. Blocked arrows indicate known inhibitory activities, and dotted blocked arrows indicate possible inhibitory activities. Question marks indicate other hypothesized but unknown regulatory signals or factors.
As shown in the model in Fig. 7, this study provides evidence for a regulatory circuit that links the primary and secondary metabolic pathways involved in trichothecene production by the coordinate regulation of transcript levels for these pathway enzymes. A deletion of either Tri10 or Tri6 causes a severe reduction in Fpps transcripts, while overexpression of Tri10 causes an increase. Based on additional work with our cDNA library (A. W. Peplow, A. G. Tag, G. Garifullina, and M. Beremand, unpublished data), EST database (Fusarium sporotrichioides Sequencing Project; B. A. Roe, Q. Ren, D. Kupfer, Hong-Shing Lai, M. Beremand, A. Peplow, and A. Tag [http: //www.genome.ou.edu/fsporo.html]), and cDNA microarray studies (A. G. Tag, A. W. Peplow, T.-F. Hsieh, T. L. Thomas, and M. N. Beremand, unpublished data) of members of our group, we have determined that this regulation extends to other genes in the isoprenoid biosynthetic pathway. We are currently investigating the underlying regulatory mechanisms that mediate this control.
The regulatory relationship between Tri10 and Tri6 appears to be further linked. The elevation of Tri10 transcripts in the Tri6 deletion strain compared to the wild-type strain raises the interesting possibility of a regulatory loop whereby activation of Tri10 upregulates Tri6 transcription and the activation of Tri6 in turn directly or indirectly downregulates Tri10 transcription. However, there must also be an independent mechanism which turns off Tri10 gene expression; otherwise, the Tri10 transcript levels would be constitutive in the Tri6 deletion strain.
The regulation of Tri10 and toxin production is also controlled by a regulatory region upstream of Tri10. In this study we show that homologous integration of pTri10-1 upstream of Tri10 increased trichothecene toxin production and Tri10, Tri6, Tri5, Tri4, Tri101, and Fpps gene expression. This integration event produced a break in the Tri gene cluster approximately 700 bp upstream of Tri10 at the very end of the Tri5 amino acid-coding region. We have conducted additional experiments which have revealed that this coordinate increase in toxin production and gene expression is defined by a regulatory region which extends upstream of the Tri10 coding region to the Tri5 promoter region and that overproduction can be caused by the disruption of this contiguous sequence (G. Garifullina, A. Tag, A. Peplow, and M. Beremand, unpublished data) (7). These data suggest that the overproduction phenotype is due to the interruption of a cluster regulatory region that normally downregulates Tri10 gene expression. This downregulation could be mediated in part through the Tri10-Tri6 regulatory loop proposed in the model above (Fig. 7) and further discussed below.
Interestingly, both regulatory genes Tri6 and Tri10 flank the gene for the first biosynthetic step of the trichothecene pathway, Tri5, and this topography as well as the direction of transcription are conserved in F. sambucinum (A. W. Peplow and M. N. Beremand, unpublished data) and F. graminearum (6). All three species also lack a TRI6 binding site (15) immediately upstream from Tri10, which is consistent with our data and the observation that the transcription of Tri10 is not dependent on TRI6 (this study). Conversely, Tri10 is overexpressed in the ΔTri6 strain, and notably, it contains a TRI6 binding site motif in the middle of its amino-acid coding region which could potentially be instrumental in mediating a reduction in Tri10 gene expression. Although this internal binding site has not been shown to bind TRI6 in vitro (15), it could be utilized in vivo by TRI6 alone or associated with another inhibitory factor(s) to effectively block Tri10 gene expression. It is also possible that the Tri6-dependent downregulation of Tri10 is directly associated with the downregulation of Tri10 exerted by the regulatory region upstream of Tri10. For example, the enhanced transcription of Tri5 by Tri6 may interfere with the transcription of Tri10. Chromatin remodeling could also be involved, independently or in conjunction with Tri6 activity. However, regardless of the underlying mechanism(s) involved, the dependency of the wild-type regulation of Tri10 gene expression and toxin production on the DNA sequence extending upstream of Tri10 to in front of the Tri5 ORF suggests that the relative positions of Tri5 and Tri10 may impose some evolutionary constraints on this region of the cluster, such that it may be more stable than other regions within the cluster.
The regulatory controls exerted by Tri10 and imposed on Tri10 are complex and intimately intertwined, with Tri6 and cluster topography playing important roles. As discussed above, these controls extend beyond the Tri gene cluster to a noncluster Tri gene (Tri101) and to genes for primary metabolism. As discussed below, the expression of Tri10 and Tri6 also impact some aspects of trichothecene self-protection.
While both Tri6 and to a lesser extent Tri10 are required for wild-type levels of T-2 toxin self-protection, the loss of self-protection in the ΔTri6 and ΔTri10 strains is partial and dependent upon the culture conditions. Thus, Tri6 and Tri10 appear to play roles in self-protection which only become critical under certain conditions. Tri6, and Tri10 via Tri6, may help to mediate the expression of one or more self-protection genes. To date, two genes that potentially contribute to trichothecene self-protection have been identified: Tri12, which is a major facilitator superfamily transporter gene located within the Tri gene cluster, and Tri101, which is a 3-o-acetyltransferase gene located outside of the Tri gene cluster. Tri101 was first identified as a potential self-protection gene as it conferred trichothecene toxin resistance on otherwise toxin-sensitive yeast cells (18, 21). However, additional genes for self-protection are likely to be present in F. sporotrichioides, since the deletion of Tri12 only partially decreased toxin self-protection and the deletion of Tri101 had no apparent effect on sensitivity to toxin (1, 21). Nonetheless, it is possible that Tri101 may contribute to self-protection under certain conditions, including those when other self-protection genes are repressed. In this regard, it is interesting that even though Tri101 displays what appears to be a background constitutive level of transcript accumulation in the ΔTri10 and ΔTri6 strains, the full expression of both Tri101 (this study) and presumably Tri12 is dependent on the expression of both Tri6 and Tri10. Consequently, the increased sensitivity to T-2 toxin displayed by the ΔTri10 and ΔTri6 strains could be due to the simultaneous reduction in the expression of both Tri12 and Tri101 as well as of one or more additional genes that contribute to toxin self-protection.
Finally, we also addressed whether the observed reduction in self-protection in the ΔTri10 and ΔTri6 strains was due to an inability to make T-2 toxin. Examination of three different T-2 toxin mutant strains revealed that only the ΔTri10 and ΔTri6 strains and not the mutant unable to make a functional trichodiene oxygenase (Tri4 mutant) displayed an increased sensitivity to T-2 toxin. Thus, the lack of T-2 toxin production itself does not lead to the reduction in self-protection, and consequently the expression of self-protection in F. sporotrichioides is not dependent upon the self-production of T-2 toxin.
The features of Tri10 strongly suggest a role as a regulatory gene in trichothecene biosynthesis. The mRNA expression pattern of Tri10 is consistent with that of a regulatory gene. The level of Tri10 transcript accumulation is low in wild-type cultures, and the transcripts appear before Tri6 transcripts in the Tri10 overproducing strain. Although Tri10 is highly conserved among F. sporotrichioides, F. graminearum, and F. sambucinum, only one other gene (an EST, function unknown) in GenBank had similarity to Tri10. Likewise, a single motif, a putative transmembrane domain (amino acids 340 to 360), was identified within TRI10 (Simple Modular Architecture Research Tool) (29, 30). Curiously, TRI10 is predicted to contain 16% leucine, but it does not contain any conserved motifs consistent with leucine repeats or zipper structures. While the precise mode of action of TRI10 remains a mystery, its function is clear as an essential control point in trichothecene production and gene expression. We are currently utilizing other molecular approaches in parallel with DNA microarrays to elucidate the function of Tri10 and further investigate the extent of the regulatory circuits defined by Tri10.
ACKNOWLEDGMENTS
We gratefully acknowledge Tom McDonald for use of the GC-MS instrumentation.
This work was supported in part by USDA/CSREES NRICGP grant no. 9503702. A.G.T. and A.W.P. were supported by a National Science Foundation grant to the Program for the Biology of Filamentous Fungi.
REFERENCES
- 1.Alexander N J, McCormick S P, Hohn T M. Tri12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol Gen Genet. 1999;261:977–984. doi: 10.1007/s004380051046. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beremand M N. Isolation and characterization of mutants blocked in T-2 toxin biosynthesis. Appl Environ Microbiol. 1987;53:1855–1859. doi: 10.1128/aem.53.8.1855-1859.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beremand M N, Hohn T M. Tri10: a new gene in the trichothecene gene cluster in Fusarium sporotrichioides. Fungal Genet News. 1995;42A:100. [Google Scholar]
- 5.Bowden R L, Leslie J F. Sexual recombination in Gibberella zeae. Phytopathology. 1998;89:182–188. doi: 10.1094/PHYTO.1999.89.2.182. [DOI] [PubMed] [Google Scholar]
- 6.Brown D W, McCormick S P, Alexander N J, Proctor R H, Desjardins A E. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet Biol. 2001;32:121–133. doi: 10.1006/fgbi.2001.1256. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, McCormick S P, Hohn T M. Altered regulation of 15-acetyldeoxynivalenol production in Fusarium graminearum. Appl Environ Microbiol. 2000;66:2062–2065. doi: 10.1128/aem.66.5.2062-2065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjardins A E, Gardner H W, Weltring K M. Detoxification of sesquiterpene phytoalexins by Gibberella pulicaris (Fusarium sambucinum) and its importance for virulence on potato tubers. J Ind Microbiol. 1992;9:201–211. [Google Scholar]
- 9.Desjardins A E, Spencer G F, Plattner R D, Beremand M N. Furanocoumarin phytoalexins, trichothecene toxins, and infection of Pastinaca sativa by Fusarium sporotrichioides. Phytopathology. 1989;79:170–175. [Google Scholar]
- 10.Evans R, Holtom A M, Hanson J R. Biosynthesis of 2-cis-farnesol. J Chem Soc Chem Commun. 1973;1973:465. [Google Scholar]
- 11.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 12.Harris L J, Desjardins A E, Plattner R D, Nicholson P G, Butler G, Young J C, Weston G, Proctor R H, Hohn T M. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 1999;83:954–960. doi: 10.1094/PDIS.1999.83.10.954. [DOI] [PubMed] [Google Scholar]
- 13.Hohn T M, Desjardins A E. Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol Plant-Microbe Interact. 1992;5:249–256. doi: 10.1094/mpmi-5-249. [DOI] [PubMed] [Google Scholar]
- 14.Hohn T M, Desjardins A E, McCormick S P, Proctor R H. Biosynthesis of trichothecenes, genetic and molecular aspects. In: Eklund M, Richard J L, Mise K, editors. Molecular approaches to food safety issues involving toxic microorganisms. Fort Collins, Colo: Alaken, Inc.; 1995. pp. 239–248. [Google Scholar]
- 15.Hohn T M, Krishna R, Proctor R H. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet Biol. 1999;26:224–235. doi: 10.1006/fgbi.1999.1122. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis B B, Trapp S C, Hohn T M. Fungal toxins produced by Brazilian Baccharis species: a case for horizontal gene transfer? Proc. Int. Compositae Conf. Kew—Biol. Utilization. 1996;20:261–267. [Google Scholar]
- 17.Keller N P, Hohn T M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 18.Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins: cloning and characterization of Tri101. J Biol Chem. 1998;273:1654–1661. doi: 10.1074/jbc.273.3.1654. [DOI] [PubMed] [Google Scholar]
- 19.Kimura M, Shingu Y, Yoneyama K, Yamaguchi I. Features of Tri101, the trichothecene 3-o-acetyltransferase gene, related to the self-defense mechanism in Fusarium graminearum. Biosci Biotechnol Biochem. 1998;62:1033–1036. doi: 10.1271/bbb.62.1033. [DOI] [PubMed] [Google Scholar]
- 20.Marasas W F O, Nelson P E, Toussoun T A. Toxigenic Fusarium species: identity and mycotoxicology. University Park, Pa: The Pennsylvania State University Press; 1984. [Google Scholar]
- 21.McCormick S P, Alexander N J, Trapp S E, Hohn T M. Disruption of TRI101, the gene encoding trichothecene 3-o-acetyltransferase, from Fusarium sporotrichioides. Appl Environ Microbiol. 1999;65:5252–5256. doi: 10.1128/aem.65.12.5252-5256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick S P, Taylor S L, Plattner R D, Beremand M N. Bioconversion of possible T-2 toxin precursors by a mutant strain of Fusarium sporotrichioides NRRL 3299. Appl Environ Microbiol. 1990;56:702–706. doi: 10.1128/aem.56.3.702-706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin C S, Vaughn M H, Campbell I M, Wei C M, Stafford M E, Hansen B S. Inhibition of protein synthesis by trichothecenes. In: Rodricks J V, Hesseltine C W, Mehlman M A, editors. Mycotoxins in human and animal health. College Park, Md: Patholox Publishers; 1977. pp. 263–273. [Google Scholar]
- 24.Okumwai H, Yoshino N, Suglura Y, Sugamata M, Hintikka E-L, Jarvis B, Ueno Y. Trichothecenes as potent inducers of apoptosis. In: Johanning E, editor. Bioaerosols, fungi and mycotoxins: health effects, assessment, prevention and control. Albany, N.Y: Boyd Printing Co., Inc.; 1999. pp. 221–231. [Google Scholar]
- 25.Proctor R H, Hohn T M, McCormick S P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant-Microbe Interact. 1995;8:593–601. doi: 10.1094/mpmi-8-0593. [DOI] [PubMed] [Google Scholar]
- 26.Proctor R H, Hohn T M, McCormick S P, Desjardins A E. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl Environ Microbiol. 1995;61:1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salch Y P, Beremand M N. Gibberella pulicaris transformants: state of transforming DNA during asexual and sexual growth. Curr Genet. 1993;23:343–350. doi: 10.1007/BF00310897. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tag A, Hicks J, Garifullina G, Ake C, Jr, Phillips T D, Beremand M, Keller N. G-protein signalling mediates differential production of toxic secondary metabolites. Mol Microbiol. 2000;38:658–665. doi: 10.1046/j.1365-2958.2000.02166.x. [DOI] [PubMed] [Google Scholar]
- 32.Turgeon B G, Garber R C, Yoder O C. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987;7:3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueno Y, Sawano M, Ishi K. Production of trichothecene mycotoxins by Fusarium species in shake culture. Appl Microbiol. 1975;30:4–9. doi: 10.1128/am.30.1.4-9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]