Abstract
Although systemic inflammation and pulmonary complications increase the mortality rate in COVID-19, a broad spectrum of cardiovascular and neurological complications can also contribute to significant morbidity and mortality.
The molecular mechanisms underlying cardiovascular and neurological complications during and after SARS-CoV-2 infection are incompletely understood. Recently reported perturbations of the epitranscriptome of COVID-19 patients indicate that mechanisms including those derived from RNA modifications and non-coding RNAs may play a contributing role in the pathogenesis of COVID-19.
In this review paper, we gathered recently published studies investigating (epi)transcriptomic fluctuations upon SARS-CoV-2 infection, focusing on the brain-heart axis since neurological and cardiovascular events and their sequelae are of utmost prevalence and importance in this disease.
Keywords: Brain-heart axis, COVID-19, RNAs
Highlights
-
•
Respiratory, cardiac and neurological complications are substantial pathophysiological features of COVID-19.
-
•
Despite several types of vaccines available against COVID-19, it is too early to predict how useful they will be in eliminating SARS-CoV-2.
-
•
Rapidly evolving high-throughput next generation sequencing and mass spectrometry technologies are well positioned to allow discoveries that will increase our understanding of SARS-CoV-2 interaction with host immune response.
-
•
Identification of (epi)transcriptomic changes during SARS-CoV-2 infection in conjunction with other omics methodologies may lead to the discovery of novel prognostic and therapeutic approaches to combat COVID-19.
1. Introduction
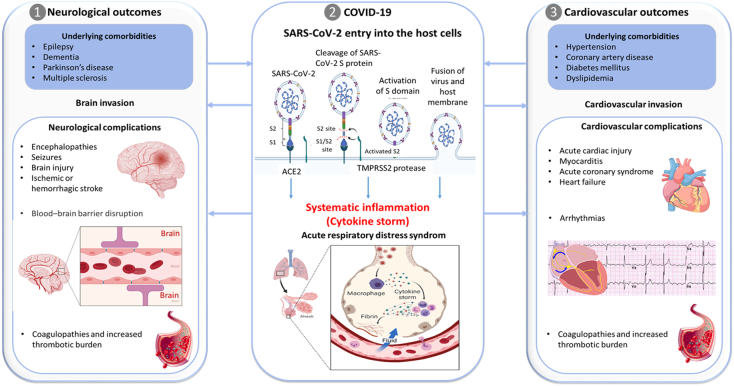
Coronavirus disease (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) demonstrating a wide spectrum of clinical manifestations, from asymptomatic, mild flulike illness to lethal acute respiratory distress syndrome. The SARS-CoV-2 enters human cells by binding to angiotensin-converting enzyme type 2 (ACE2), a transmembrane receptor expressed by pulmonary and inflammatory cells, but also by other cell types including cardiomyocytes, pericytes and neurons (Fig. 1). Functionally, ACE2 inactivates angiotensin II [1] and plays an important role in neuro-humoral regulation of the cardiovascular system. In severe cases, SARS-CoV-2 invasion of the host cells may result in progression toward the “COVID-19 cytokine storm” which is characterized by severe immune reaction and overwhelming systemic inflammation, haemodynamic instability and multiple organ failure with known and unknown clinical complications [2], [3]. Although, SARS-CoV-2 primarily affects the lungs, reports are emerging of the wide spectrum of cardiovascular and neurological manifestations and complications ranging from the de novo viral infection or its interplay with pre-existing comorbidities (Fig. 1). The underlying cardiovascular and neurological comorbidities seem to predispose the development of more severe cardiovascular and neurological complications in COVID-19 patients, which are in turn associated with higher mortality rates [4]. History of cardiovascular disease is associated with a nearly five-fold increased risk in fatality rates [5]. Most prevalent cardiovascular disease risk factors in COVID-19 patients are hypertension and diabetes mellitus, while most common cardiovascular complications observed in COVID-19 patients include arrhythmias, acute myocardial infarction, cardiac injury, fulminant myocarditis, pericarditis, cardiac arrhythmia, heart failure, and disseminated intravascular coagulation [6], [7]. The interaction between underlying cardiovascular comorbidities and the poor clinical outcome of COVID-19 may be multifaceted, including age, sex, cardiac dysfunction [8] and aorta ageing as defined by the estimated pulse wave velocity [9].
Fig. 1.
Interplay between SARS-CoV-2 infection, neurological and cardiovascular complications. 1) Underlying neurological and cardiovascular comorbidities are associated with high mortality in patients with COVID-19. 2) Multiple molecules at the cell surface are involved in the entry of SARS-CoV-2, including the major receptor ACE2, the membrane protease TMPRSS2, and other potential alternative/auxiliary receptors or cofactors such as cathepsin L, a transmembrane glycoprotein CD147, high-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) and neuropilin-1. The initial step of SARS-CoV-2 infection involves specific binding of spike protein (S) to the cellular entry receptor ACE2 and priming of S protein by TMPRSS2 at the cell surface or by cathepsin L in the endosomal compartment following ACE2-mediated endocytosis. After activation of the S2 domain on the spike, the virus enters the cell via membrane fusion. 3) Although the main presentation of COVID-19 is viral pneumonia, SARS-CoV-2 infection can also induce neurological and cardiovascular complications. Since the expression and tissue distribution of ACE2 dictates viral tropism and pathogenicity, ACE2 may facilitate direct invasion of neurons or myocardial cells leading to apoptosis and necrosis of neurons/cardiac and neighbouring cells. On the other hand, cytokine storm can damage an intact blood–brain barrier and disrupt the homeostasis of the central nervous system without the virus crossing the blood–brain barrier from the systemic circulation. In the cardiovascular system, an acute coronary syndrome can occur because of plaque rupture, coronary spasm or micro-thrombi owing to systemic inflammation or cytokine storm. In addition, the SARS-CoV-2 infection is associated with a pro-thrombotic state, which may lead to occlusion of blood vessels leading to injuries of both the heart and the brain. A part of this figure was created using “Mechanism of “SARS-CoV-2 Viral Entry” and “Cytokine storm” templates by BioRender.com (2020). Retrieved from https://app.biorender.com/biorender-templates.
Neurological and psychiatric sequelae of COVID-19 have been widely reported (Fig. 1) [10], [11]. A retrospective study among 236,379 patients diagnosed with COVID-19 estimated that the incidence of a neurological or psychiatric diagnosis in the following six months after infection was 33.62 % [12]. SARS-CoV-2 was recently detected in vivo in transgenic mice overexpressing human ACE2 and in post mortem cortical neurons of COVID-19 patients, showing the neuroinvasive capacity of SARS-CoV-2 [13]. However, the potential molecular mechanisms underlying cardiovascular and neurological complications during and post SARS-CoV-2 infection have not been fully elucidated.
Possible mechanisms include direct virus-mediated neuro- and cardiotoxicity, hypoxia-related injury, immune-mediated cytokine storm and systemic inflammation, and so forth. Recently reported perturbations of the epitranscriptome of COVID-19 patients indicate that mechanisms including those derived from RNA modifications and non-coding RNAs may play a contributing role in both short- and long-term cardiovascular and neurological outcomes.
2. Epitranscriptomic signature in the brain-heart axis of COVID-19 patients
2.1. Noncoding RNAs in the brain-heart axis
Eukaryotic cells produce different classes of non-protein coding RNA transcripts called ncRNAs participating in various cellular processes including, gene expression regulation, RNA maturation and protein synthesis. NcRNAs are transcribed by either RNA polymerase I, II or III, depending on the individual ncRNA [14]. According to their lengths, they can be divided into two main groups: (1) small or short ncRNAs including microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNA (piRNA) and small nucleolar RNAs (snoRNAs); and (2) long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) [15]. A rapidly growing number of studies has unravelled associations between aberrant ncRNA expression and human diseases. NcRNAs have emerged as promising candidates for the treatment of a variety of diseases and play a potential significant role in host-virus interactions [15]. Dysregulated noncoding RNAs (ncRNAs) expression levels in lung tissue and liquid biopsies from COVID-19 patients indicate that ncRNAs may play an important role in the pathogenesis, and hence clinical outcomes of COVID-19. For instance, altered levels of ncRNAs involved in T cell activation and differentiation have been found in peripheral blood mononuclear cell samples from COVID-19 patients at three different time points during their treatment, convalescence, and rehabilitation [16].
2.2. Small noncoding RNAs
Due to limited cardiac and brain tissues availability for research purposes, SARS-CoV-2-mediated neuro-cardiovascular transcriptome changes were investigated in liquid biopsies (plasma, serum and whole blood), human induced pluripotent stem cells (iPSC)-derived cardiomyocytes, human brain endothelial cells and human cholinergic neurons. Small ncRNAs (particularly miRNAs) associated with cardiovascular and neurological complications in COVID-19 patients are summarized in Table 1.
Table 1.
Aberrantly expressed miRNAs associated with the brain-heart axis and inflammation after SARS-CoV-2 infection.
| miRNA | Type of sample | Regulation | Validated targets in COVID-19 | Reference | |
|---|---|---|---|---|---|
| Brain | miR-24 | hBMECs | ↑ | NRP1 | [17] |
| Cardiac | miR-21-5p | Serum, RBC-depleted whole blood, | ↑ | MYC | [18], [19] |
| miR-200c | NRCMs, NRCFs, HCFs, HUVECS, cord blood derived hiPSC-CMs | ↓ | ACE | [20] | |
| miR-133a, miR-122 | Plasma | ↑↓ | – | [21] | |
| miR-208a | Serum | ↑ | – | [19] | |
| miR-499 | ↑ | FoxO1 | |||
| Inflammation | miR-155 | ↑ | – | ||
| miR-16-2-3p | Blood | ↑ | – | [22], [23] | |
| miR-6501-5p | ↑ | ||||
| miR-618 | ↑ | TLR-4, HAT1, TGF-β2 | |||
| miR-183-5p | ↓ | PTEN | |||
| miR-627-5p | ↓ | – | |||
| miR-144-3p | ↓ | FoxO1 | |||
| miR-423-5p | Plasma | ↑ | – | [24] | |
| miR-23a-3p | ↓ | – | |||
| miR-195-5p | ↑ | – | |||
| miR-1207-5p | Lung tissue | ↑ | CSF1 | [25] | |
| miR-146a-5p | Serum, RBC-depleted whole blood, | ↓ | IL-6, STAT1 | [18], [26] | |
| miR-429 | RBC-depleted whole blood | ↓ | – | [18] | |
| miR-142-3p | ↑ | IL6ST | |||
| miR-15b-5p | ↑ | IFNG, CD69 | |||
| miR-26a-5p | Post mortem lung tissue | ↓ | – | [27] | |
| miR-29b-3p | |||||
| miR-34a-5p |
hBMECs indicates human brain microvascular endothelial cells, HCFs – human cardiac fibroblasts, hiPSC-CMs - human induced pluripotent stem cell-derived cardiomyocytes, HUVECs – human umbilical vein endothelial cells, NRCFs - neonatal rat cardiac fibroblasts, NRCMs - neonatal rat cardiomyocytes, RBC – red blood cells.
In human brain microvascular endothelial cells (hBMECs), miR-24 targets cell surface receptor neuropilin-1 (NRP1), indicating its potential role in the neurological manifestation of COVID-19 [17]. Upregulation of miR-24 significantly reduced the permeability of hBMECs resulting in response of VEGF, an agonist of Neuropilin-1, which lead to the Neuropilin-1 overexpression and rescue of impaired cellular response [17]. Two independent studies have reported that NRP1 plays an important role in SARS-CoV-2 entry in human cells through interaction with S1 domain of viral Spike protein after its cleavage by furin protease [28], [29]. In silico analyses identified a set of hypothalamic miRNAs potentially playing a role in the regulation of expression of hypothalamic ACE2 and transmembrane serine protease 2 (TMPRSS2), essential proteins for SARS-CoV-2 cell entry [30]. Although hypothalamic circuits are known to be exposed to the entry of the virus via the olfactory bulb, the regulation of its interaction with SARS-CoV-2 is not yet fully understood.
Oxidative stress-induced miR-200c targets SARS-CoV-2 entry receptor ACE2 in rat primary cardiomyocytes and human iPSC-derived cardiomyocytes [20]. Two cardiometabolic miRNAs, miR-133a and miR-122 were associated with 28-day mortality of COVID-19 patients [21]. Serum levels of inflammation (miR-155) and cardiac myocyte-enriched miRNAs (miR-21, miR-208a and miR-499) were increased in COVID-19 patients compared to healthy controls, indicating an association between SARS-CoV-2 infection and cardiovascular issues [19]. A comprehensive analysis of transcriptomic expression profiles obtained from whole blood of moderate and severe COVID-19 patients has revealed several miRNAs such as miR-146a-5p, miR-21-5p, and miR-142-3p that may serve as potential biomarkers of disease severity [18]. The same study has highlighted three miRNAs, miR-146a-5p, miR-21-5p, and miR-142-3p, as potential therapeutic targets of COVID-19 [18].
2.3. Long noncoding RNAs
An increasing number of lncRNAs with potential neurological and immunological functions has been shown to be associated with the pathogenesis of COVID-19 (Table 2).
Table 2.
Aberrantly expressed lncRNAs associated with the brain-heart axis and inflammation after SARS-CoV-2 infection.
| ncRNA | Type of sample | Regulation | Pathways related to COVID-19 | Reference | |
|---|---|---|---|---|---|
| Brain-heart | DANCR | Lung tissue, brain | ↑ | mTOR | [31] |
| NEAT1 | Lung tissue, brain, PBMCs, HUVECs | ↓ | RUNX3, SPI1 | [31], [32], [33], [34] | |
| MALAT1 | PBMCs | ↓ | – | [32], [33] | |
| Inflammation | SNGH25 | PBMCs | ↓ | – | [32] |
| AC010904.2 | PBMCs | ↑ | – | [35] | |
| AC012065.4 | |||||
| AL365203.2 | |||||
| AC010175.1 | |||||
| LINC00562 | |||||
| AC010536.1 | |||||
| AP005671.1 | |||||
| SNHG1 | PBMCs | ↑ | IL-7 | [36] |
Up and down arrows, indicate an increase and decrease of expression with increasing disease severity, respectively. PBMCs – peripheral blood mononuclear cells.
Deep RNA sequencing of SARS-CoV-2-infected lung tissues and human-derived differentiated cholinergic neurons revealed several differentially expressed lncRNAs, reflecting their association with the inflammatory pathobiology related to COVID-19. For instance, transcriptomic analyses in lung has shown that two lncRNAs, DANCR and NEAT1, may predict the inflammatory profile of infected tissues and provide insights into adverse body and brain consequences of COVID-19 [31]. The lncRNA DANCR regulates inflammation and is responsible for the surveillance over cholinergic blockade of inflammation, thereby maintaining a balance between pro- and anti-inflammatory pathways in lung and brain tissues affected by SARS-CoV-2 [31]. In another study, expression levels of three lncRNAs, MALAT1, NEAT1, and SNGH25 were found to be downregulated in mild and severe COVID-19 patients [33]. Interestingly, MALAT1 and NEAT1 are implicated in both, cardiovascular and neurological complications. Recent findings on differential ncRNA expressions across SARS-CoV-2 infected tissues and bio-fluids of COVID-19 patients indicate that ncRNAs may constitute key players in the regulation of the immune response following infection [28], [29]. However, changes in ncRNAs landscape associated with cardiac and neurological manifestations of COVID-19 patients are yet to be fully characterized and their potential role in the course of the disease remains to be elucidated.
2.4. Epitranscriptomics
RNA modifications, collectively termed the ‘epitranscriptome’, can effectively affect every aspect of RNA metabolism, including splicing, non-coding RNA biogenesis and maturation, as well as RNA stability and non-sense mediated decay, comprising thus an additional level of gene regulation [37]. Among over 100 epitranscriptomics modifications, A-to-I RNA editing and N6-methyladenosine (m6A) are the most studied and abundant modifications that control multiple biological functions in the cell, including in the cardiovascular system [38]. In addition, numerous less charted and abundant RNA modifications have been identified, including N1-methyladenosine (m1A), N5-methylcytosine (m5C), N7-methylguanosine (m7G), N6,2′-O-dimethyladenosine (m6Am), and pseudouridine (Ψ) [39]. In recent years, RNA modifications have been identified on coding RNA (messenger RNA, mRNA) as well as on ncRNA species including miRNAs, ribosomal (rRNAs) and transfer RNA (tRNAs), snoRNAs, lncRNAs and circRNAs [40], [41], [42], [43]. At the cellular level, dynamics of RNA modifications are finely regulated by different components of methylation machinery consisting of enzymes and proteins termed as writers, readers and erasers [44], [45].
Adenosine-to-inosine (A-to-I) RNA editing, mediated by the adenosine deaminase RNA specific (ADAR) family of enzymes, is the most widespread RNA modification, taking place mainly in the primate-specific, repetitive Alu elements [46], [47]. A recent study showed that ADAR1, the main RNA editing enzyme, was among the most upregulated genes in pulmonary alveolar type II cells of patients infected with SARS-CoV-2 [48], the main lung cells expressing the ACE2 receptor [49]. Similarly, ADAR1 expression was increased in peripheral blood cells of patients with severe COVID-19 compared to those with mild disease [50]. Increased ADAR1-induced RNA editing may affect the brain-heart axis in several ways. For instance, ADAR1 may prevent the excessive activation of innate immune receptors by the viral RNA [51], thus preventing the hyperinflammatory response and translational shutdown/apoptosis. Further, ADAR1 may prevent oxidative stress-induced inflammation and apoptosis in cardiomyocytes by inhibiting protein kinase R (PKR) hyperactivation [52]. Of interest, ADAR1 overexpression, which was also induced by oxidative stress, could limit PKR phosphorylation in cardiomyocytes [53], thus preventing the aberrant activation of innate immune sensors by viral double stranded RNA. On the other hand, extensive RNA editing of the serotonin 5-hydroxytryptamine receptor 2C (5-HT2CR) in the brain has been associated with aberrant sympathetic nervous activity [54]. 5-HT2CR has five editing sites in exon 5 leading to 3 amino acid substitutions, which can alter the coupling with downstream G proteins by 10–15-fold [55]. Of note, a persistent hypermetabolic state is observed in patients with COVID-19 [56], especially in those admitted to intensive care units [57] contributing to patient deterioration. Similar, yet unknown, possible editing events in other serotonin receptors leading to an aberrant activation of the sympathetic nervous system could potentially lead to arrhythmias or even sudden cardiac death especially in individuals with underlying heart conditions [58].
2.5. RNA editing of the viral RNA
Apart from its effects on host gene expression and cellular function under stress conditions including apoptosis, RNA editing may control SARS-CoV-2 propagation per se (reviewed in [59]). RNA editing has two major forms, deamination of adenosine-to-inosine (A-to-I) and deamination of cytosine-to-uracil (C-to-U), mediated by the ADAR and APOBEC family of enzymes, respectively [47], [60]. SARS-CoV-2 has a positive sense, single-stranded RNA genome of approximately 30-kilo bases length [61]. The high prevalence of C-to-U and A-to-G transitions [62], in sequence motifs compatible with ADAR/APOBEC-mediated deamination events observed in the human transcriptome [47], [60], [63], has suggested the involvement of host RNA editing machinery in SARS-CoV-2 mutagenesis. Editing of viral RNA can have either pro-viral or anti-viral effects depending on the host-virus interactions [64]; on one hand, editing events can affect viral protein synthesis by creating early stop codons (non-sense mutations). The complete lack of such A-to-G/C-to-U non-sense mutations in isolated genomic SARS-CoV-2 RNA from patient's broncho-alveolar lavage samples [63] suggests that such events may be incompatible with viral replication. On the other hand, a single point mutation, such as the D614G (A-to-G) substitution in the spike (S), can greatly affect viral transmissibility by: i) affecting loading of the spike protein into virions, ii) changing its conformation into a more ACE2-binding-competent state or iii) increasing binding affinity to ACE2 [65], [66]. Apart from the effects of site-specific editing events on viral propagation, extensive editing of the viral genome can affect its recognition by the host immune response. Heavily edited viral RNA can avoid recognition by innate immune receptors such as MDA-5 and PKR [64], thus avoiding the protective host immune response; on the other hand, hyper-editing of viral RNA can restrict viral replication or ‘mark’ viral RNA for degradation by endonucleases [67], [68]. Thus, RNA editing on the SARS-COV-2 genome could be a relevant mechanism controlling the dynamics of viral evolution, affecting virulence, pathogenicity and host response [69].
3. Crosstalk between RNAs and endothelial inflammation and vascular dysfunction in COVID-19
Endothelial inflammation is a central component of COVID-19 pathogenesis [70]. COVID-19 can lead to endothelial inflammation and dysfunction by two main mechanisms: 1) direct infection of endothelial cells by SARS-CoV-2 and 2) endothelial activation by the systemic inflammatory response. While an early analysis of organ autopsies using electron microscopy showed endothelial cells from multiple organs were infected with SARS-CoV-2 [71], a more recent study showed low capability of direct endothelial infection by SARS-CoV-2, mainly attributed to the very low expression levels of ACE2 [72]. Whether SARS-CoV-2 can enter endothelial cells via secondary receptors when at high titres [72], or the observed vascular inflammation in COVID-19 can be mainly attributed to the infection of neighbouring vascular cells, e.g. pericytes, and to the systemic inflammatory response remains to be elucidated.
SARS-CoV-2 shows a unique immunological profile compared to other viral infections such as Influenza (flu) [50]. In contrast to other viruses, which lead to the upregulation of the anti-viral part of the innate immune response (type I interferons), SARS-CoV-2 infection is characterized by the up-regulation of proinflammatory cytokines, predominantly IL-6 [50]. IL-6 activates the endothelium and upregulates the expression of adhesion molecules [E-selectin, vascular cell adhesion molecule 1 (VCAM-1), intercellular adhesion molecule 1 (ICAM-1)] as well as chemokines [interleukin-8, monocyte chemoattractant protein-1 (MCP-1)] which control the recruitment of immune cells into inflamed tissues [73]. Of note, circulating levels of endothelial adhesion molecules, including ICAM-1, VCAM-1, and vascular adhesion protein 1 (VAP-1), have been associated with more severe disease or increased mortality in patients with COVID-19 [74], [75]. [74], [75] Moreover, IL-6 trans-signalling can also increase plasminogen activator inhibitor-1 (PAI-1) production by endothelial cells [76], which may contribute to the thrombo-inflammatory cascade observed in COVID-19 patients [77].
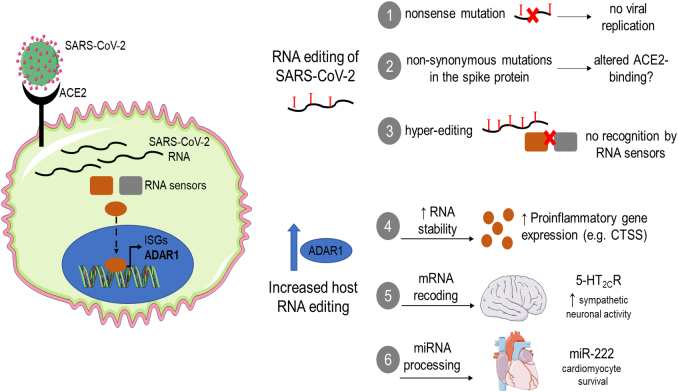
Apart from the transcriptional alterations induced by systemic inflammation in endothelial cells, a systematic upregulation of ADAR1-induced RNA editing in patients with COVID-19 could further propagate the inflammatory response by increasing stability of proinflammatory transcripts, as we have previously shown in chronic inflammatory diseases such as atherosclerosis and rheumatoid arthritis [78], [79]. For example, upregulation of cathepsin S (CTSS), an elastolytic enzyme that promotes the development of inflammatory, non-stable atherosclerotic plaques [80] could in turn destabilize existing plaques leading to acute coronary events or stroke. Cathepsin S is also involved in major histocompatibility complex (MHC)-II dependent antigen presentation [81], thus providing a link between innate and adaptive immune responses. Of interest, cathepsin S expression is increased in a subgroup of lung capillary endothelial cells along with genes involved in MHC class II-mediated antigen presentation [82], which could participate in initial recognition of SARS-CoV-2 and mounting of systemic inflammatory reaction [83]. Of note, CTSS is only one among thousands of mRNAs, as well as ncRNAs, including miRNAs and lncRNAs that can be affected by RNA editing [78], [80], [81], offering an additional level of gene regulation during the systematic inflammatory response induced by SARS-CoV-2 infection (Fig. 2).
Fig. 2.
RNA editing in COVID-19. SARS-CoV-2 enters the cell through its interaction with host ACE2 receptor and is subsequently recognized by cytosolic RNA sensors eliciting an innate immune response. Recognition of SARS-CoV-2 leads to the upregulation of interferon-stimulated genes, including ADAR1. ADAR1-induced RNA editing can in turn affect viral propagation by: 1) creating nonsense mutations, that may inhibit viral protein synthesis and replication, 2) creating non-synonymous mutations in the spike that can alter binding to ACE2 receptor and 3) destabilizing double-stranded RNA structures created during viral replication to prevent recognition of SARS-CoV-2 by innate immune sensors. Moreover, a systematic increase of RNA editing in the host can affect the brain-heart axis by: 4) increasing RNA stability of proinflammatory genes, such as CTSS, and thus propagating the systematic, as well as the tissue-specific (atherosclerotic plaque destabilization) inflammatory response, 5) creating recoding events in 5-HT2CR in the brain leading to a hypermetabolic state, 6) increasing miRNA processing by DICER and thus mature miR-222 expression, which prevents apoptosis of infected myocardial cells.
3.1. Brain-heart crosstalk
It is projected that 0.04 % of the overall COVID-19 patients have been affected with neurological diseases (e.g. ischemic strokes, encephalopathy, psychosis) at the central nervous system (CNS) [84]. On another hand, it is estimated that 20 % to 30 % of hospitalized patients develop cardiac injury [85], [86]. Clinical data indicate that both the susceptibility to and the outcomes of COVID-19 are strongly associated with cardiovascular disease (reviewed in refs [86], [87]). Currently, it is relatively unknown what is the level of communication between the brain and the heart in the context of COVID-19 and which cellular or molecular mechanisms play major roles in this communication [88].
Inflammation may be a potential link between heart and brain communication in the context of COVID-19. It is well accepted that the proteome signature is affected significantly in the acute-phase response in severe COVID-19 patients [89], [90]. Results obtained in different studies indicate the up-regulation of IL-6 signalling pathway [89], [91], [92], which is more affected than other inflammatory pathways such as the tumor necrosis factor and interferon gamma pathways. Both the classical complement pathway and the complement modulators are also activated, C-reactive protein and serum amyloid proteins are up-regulated, and modulators of inflammation such as gelsolin that is part of the extracellular actin scavenger system which removes toxic F-actin filaments are dysregulated [89].
A second important route of communication between heart and brain are the extracellular vesicles (EVs). EVs are lipid vesicles secreted by cells containing several biomolecules (miRNAs, mRNA, proteins) [93] which are involved in inter-organ communication [94]. These EVs can be classified in three different categories according to their biogenesis: (i) exosomes, (ii) microvesicles and (iii) apoptotic bodies. EVs secreted by the brain or the heart have the potential to carry biological information in the brain-heart axis. For example, EVs released by the CNS activate the acute-phase response leading to the hepatic release of acute-phase proteins (e.g. TNF, CXCL1, serum amyloid proteins) which are released into the circulation and influence the activity of the cardiovascular system [95], [96]. Recently, it has been reported that COVID-19 patients have high levels of plasmatic EVs containing tissue factor (TF) activity, a main activator of the coagulation cascade, and thus a trigger of thrombotic events [97]. It has been speculated that the EVs containing TF are derived from activated endothelial cells or perivascular cells. Further studies are required to investigate in more detail the role of EVs in the brain-heart axis.
Stem/progenitor cells are also a route of communication between heart and brain. Circulating levels of endothelial progenitor cells (CD45−CD31+CD34+CD146−) have been reported to be increased in COVID-19 patients with mild and severe symptoms compared to healthy controls [98]. Interestingly, some endothelial progenitor cells had the phenotype CD34+KDR+CD19+, indicative of their lymphocyte lineage [99].
3.2. Diagnostic potential of ncRNAs in COVID-19
The major hallmarks of severe COVID-19 are respiratory (pulmonary embolism), cardiac (ischemia, tachyarrhythmia, myocarditis and pericarditis), and neurological (transient ischemic attack and stroke). Up to date, sensitive and reliable markers for the early risk stratification of cardio-neurological complications and mortality risk assessment in COVID-19 patients have not been established yet. Over the past decade, (epi)transcriptome network has emerged as important player in regulation of virtually all cellular processes and indicators of outcomes of a plethora of disease states including cardiovascular, neurodegenerative disease, cancers and infectious diseases. Clinical applications of RNAs in different human pathologies are on the horizon. Identification of differentially expressed ncRNAs as well as changes in RNA modification profiles and their functional annotation in biofluids of COVID-19 patients may open up new avenues for developing biomarkers against this deadly disease. For instance, several properties of ncRNAs particularly miRNAs suggest their potential value as biomarkers for COVID-19 outcome prognosis. They are present and stable in the circulation, they demonstrate tissue-specificity, participate in disease evolution, they are measurable using reliable and sensitive techniques and they are easily accessible from biofluids (“liquid biopsies”). In addition to high sensitivity and specificity, RNA analysis is cheaper compared to protein analysis and offers a greater overview of cell regulation and states compared to DNA analysis.
3.3. Future of COVID-19 diagnostics and therapy
The scientific community around the globe is working frenetically to support emergency responses to the ongoing pandemic by implementing innovative research strategies aiming to develop diagnostic, prognostic and therapeutic solutions for COVID-19, as well as to prepare for future similar issues. The battle against COVID-19 and preparedness for future pandemics rely on the development of innovative approaches to identify risk stratification biomarkers and therapeutic targets capable to predict disease evolution and improve clinical outcomes. Although several types of vaccines are currently administered against COVID-19, it is too early to predict how efficient they will be to eliminate SARS-CoV-2, notably due to the capacity of the virus to mutate and generate novel and potentially more dangerous variants. Taking into account high transmissibility of new viral variants and heterogeneity of clinical symptoms among COVID-19 patients, there is a critical need to foster multi-omics approaches to discover novel diagnostics and therapeutic solutions for COVID-19.
Recent intensified interest in (epi)transcriptomic research by scientific community worldwide, pharmaceutical industries and biotechnology companies and the lessons learned from COVID-19 pandemic should be applied for the future benefit of patients and the healthcare system.
A comprehensive understanding of the complex and multilayer interplay between SARS-CoV-2 and host immune responses in COVID-19 patients is fundamental to defining the effectiveness of treatments, predicting disease evolution, and for understanding heterogeneity of disease severities and outcomes [100]. Two recently published studies demonstrate the power of multi-omics approach to characterize a sharp disease-state shift between mild and moderate illness [101], [102]. A wider application of multi-omics high-throughput technologies may contribute to refine our knowledge of SARS-CoV-2 pathogenicity and decipher fundamental changes at genomic, transcriptomic, proteomics and metabolomics levels. In this review paper, we gathered recently published studies investigating transcriptomic fluctuations upon SARS-CoV-2 infection, with a focus on the brain-heart axis since neurological and cardiovascular events and sequelae are of utmost prevalence and importance. Identification of dysregulated ncRNAs and epitranscriptomics changes during SARS-CoV-2 infection may constitute a source of biomarkers for early identification of patients at risk of cardiovascular and neurological events. Additionally, a deep mining of the interplay between (epi)transcriptome affecting both the brain and the heart and interacting with SARS-CoV-2 may provide significant mechanistic insights and catalyse the discovery of novel drugs to limit adverse events, thereby impacting on healthcare and improving patients' outcomes.
3.4. Extracellular vesicles based COVID-19 therapeutics
Several EVs based therapies for the treatment of COVID-19 are under clinical evaluation due to their immunomodulatory properties. According to ClinicalTrials.gov there are four clinical trials (phase I) and one pilot clinical study (NCT04276987) running or completed for the therapeutic evaluation of EVs. The EVs originated either from mesenchymal stem cells (MSC) (NCT04602442; NCT04491240; NCT04276987), T cells (NCT04389385) or bone marrow cells (NCT04493242). One of the completed randomized trials (NCT04491240; total number of COVID-19 patients untreated and treated with EVs were 10 and 20, respectively) reported no adverse reactions of MSC derived-EVs administered in patients by inhalation. These clinical trials are justified by the encouraging results obtained by the use of EVs in pre-clinical lung injured models such as acute respiratory distress syndrome, lipopolysaccharide-induced lung injury and pneumonia (reviewed in reference [103]).
4. Concluding remarks and future perspectives
Although the rate of hospitalization of SARS-CoV-2 infected individuals is diminishing, the COVID-19 pandemic remains a significant public health threat worldwide, mostly due to emerging long-term sequelae (“long COVID”). An in-depth characterization of the omics contributors of the disease would allow refining diagnostic strategies, identify markers of disease severity and progression, and finally identify novel therapeutic approaches.
Rapidly evolving high-throughput technologies are well positioned to allow discoveries that will increase our understanding of SARS-CoV-2 interaction with host immune response. This may lead to new biomarkers and treatments that can, if possible at the same time, identify and treat patients at risk of developing severe forms of COVID-19 or long-term sequelae affecting for instance the cardiovascular of neurological systems. For complex traits such as COVID-19 involving multiple biological pathways and organs, omics approaches may allow the identification of novel disease mechanisms to further understand and better characterize SARS-CoV-2 pathophysiology. In line with that, dynamic monitoring of cardiovascular and neurological symptoms and organ dysfunction with laboratory markers might help unravel the underlying pathways linking cardiovascular and neurological disorders to poor clinical outcome.
Integrative (epi)transcriptomic approaches have the potential to aid personalizing healthcare for optimal outcomes. This holds true not only for COVID-19 but also for any other disease with an (epi)transcriptomic component. After the demonstration of their — until recently doubtable — usefulness as vaccines, RNAs are now entering a new era of RNAs for biomarkers and therapeutic purposes. Hence, more examples of the use of RNAs in precision medicine are expected in the few upcoming years.
Funding
AJ is funded by the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement 893435.
YD is funded by the EU Horizon 2020 project COVIRNA (Grant Agreement # 101016072), the National Research Fund (grants # C14/BM/8225223, C17/BM/11613033 and COVID-19/2020-1/14719577/miRCOVID), the Ministry of Higher Education and Research, and the Heart Foundation-Daniel Wagner of Luxembourg.
KS is supported by grants from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (MODVASC, grant agreement No 759248) and the German Research Foundation DFG (SFB834 B12, project number 75732319).
LF is funded by the European Union's Horizon 2020 research and innovation program under grant agreement numbers 764958 (project NANOSTEM), 952266 (project RESETageing) and 101016072 (project COVIRNA), INTERREG program funding (Projects “NeuroAtlantic” and 2IQBioneuro, Ref: EAPA_791/2018 and Ref: 0624_2IQBIONEURO_6_E, respectively) and Portuguese Research Funds (project POCI-01-0145-FEDER-029229).
A.H.B is supported by the BHF Chair of Translational Cardiovascular Sciences (CH/11/2/28733), BHF programme grants (RG/14/3/30706, RG/20/5/34796 and RG/19/3/34265), BHF Centre for Vascular Regeneration and BHF Research Excellence Award RE/18/5/34216. He is also funded by the EU Horizon 2020 project COVIRNA (Grant Agreement # 101016072).
Author contributions
All authors contributed substantially to all aspects of the article.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Yvan Devaux reports was provided by Luxembourg Institute of Health. Yvan Devaux holds patents related to diagnostic and therapeutic applications of RNAs.
Acknowledgements
This article is based upon work from EU-CardioRNA COST Action CA17129 (www.cardiorna.eu) supported by COST (European Cooperation in Science and Technology).
Glossary
- Angiotensin-converting enzyme type 2 (ACE2) - a transmembrane receptor engaged as SARS-CoV-2 entry door for cell infection.
- Epitranscriptome – the ensemble of biochemical RNA modifications taking place throughout the transcriptome including coding and non-coding regions of mRNA and small and long non-coding RNAs.
- Extracellular vesicles (EVs) - lipid vesicles secreted by cells containing several biomolecules (miRNAs, mRNA, proteins).
- COVID-19 cytokine storm - a severe immune reaction characterized by systematic hyper-inflammation, haemodynamic instability and multiple organ failure.
- MicroRNAs (miRNAs) – small endogenous noncoding RNAs of ~22 nucleotides in length involved in the regulation of gene expression at the posttranscriptional level and implicated in many fundamental physiologic processes.
- Noncoding RNAs (ncRNAs) - a heterogeneous group of non-protein coding transcripts (RNAs) in terms of length, structure, function, localization and biogenesis.
- Long noncoding RNAs (lncRNAs) – most prevalent and functionally versatile class of ncRNA transcripts longer than > 200 nucleotides.
- Severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) - is a member of a large family of viruses called coronaviruses.
References
- 1.Gao Y.M., Xu G., Wang B., Liu B.C. Cytokine storm syndrome in coronavirus disease 2019: a narrative review. J Intern Med. 2021;289(2):147–161. doi: 10.1111/joim.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., U.K. Hlh across speciality collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazaridis C., Vlachogiannis N.I., Bakogiannis C., Spyridopoulos I., Stamatelopoulos K., Kanakakis I., Vassilikos V., Stellos K. Involvement of cardiovascular system as the critical point in coronavirus disease 2019 (COVID-19) prognosis and recovery. Hellenic J Cardiol. 2020;61(6):381–395. doi: 10.1016/j.hjc.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long B., Brady W.J., Koyfman A., Gottlieb M. Cardiovascular complications in COVID-19. Am J Emerg Med. 2020;38(7):1504–1507. doi: 10.1016/j.ajem.2020.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H., Rhee J.W., Cheng P., Waliany S., Chang A., Witteles R.M., Maecker H., Davis M.M., Nguyen P.K., Wu S.M. Cardiovascular complications in patients with COVID-19: consequences of viral toxicities and host immune response. Curr Cardiol Rep. 2020;22(5):32. doi: 10.1007/s11886-020-01292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patone M., Mei X.W., Handunnetthi L., Dixon S., Zaccardi F., Shankar-Hari M., Watkinson P., Khunti K., Harnden A., Coupland C.A.C., Channon K.M., Mills N.L., Sheikh A., Hippisley-Cox J. Risks of myocarditis, pericarditis, and cardiac arrhythmias associated with COVID-19 vaccination or SARS-CoV-2 infection. Nat Med. 2021;28(2):410–422. doi: 10.1038/s41591-021-01630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan W.J., Liang W.H., He J.X., Zhong N.S. Cardiovascular comorbidity and its impact on patients with COVID-19. Eur Respir J. 2020;55(6) doi: 10.1183/13993003.01227-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamatelopoulos K., Georgiopoulos G., Baker K.F., Tiseo G., Delialis D., Lazaridis C., Barbieri G., Masi S., Vlachogiannis N.I., Sopova K., Mengozzi A., Ghiadoni L., van der Loeff I.S., Hanrath A.T., Ajdini B., Vlachopoulos C., Dimopoulos M.A., Duncan C.J.A., Falcone M., Stellos K., Pisa C.-R.G., Newcastle C.-R.G. Estimated pulse wave velocity improves risk stratification for all-cause mortality in patients with COVID-19. Sci Rep. 2021;11(1):20239. doi: 10.1038/s41598-021-99050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komaroff A.L., Lipkin W.I. Insights from myalgic encephalomyelitis/chronic fatigue syndrome may help unravel the pathogenesis of postacute COVID-19 syndrome. Trends Mol Med. 2021;27(9):895–906. doi: 10.1016/j.molmed.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosseini N., Nadjafi S., Ashtary B. Overview of COVID-19 and neurological complications. Rev Neurosci. 2021;32(6):671–691. doi: 10.1515/revneuro-2020-0116. [DOI] [PubMed] [Google Scholar]
- 12.Taquet M., Geddes J.R., Husain M., Luciano S., Harrison P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry. 2021;8(5):416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.E., Liu F., Dai Y., Szigeti-Buck K., Yasumoto Y., Wang G., Castaldi C., Heltke J., Ng E., Wheeler J., Alfajaro M.M., Levavasseur E., Fontes B., Ravindra N.G., Van Dijk D., Mane S., Gunel M., Ring A., Kazmi S.A.J., Zhang K., Wilen C.B., Horvath T.L., Plu I., Haik S., Thomas J.L., Louvi A., Farhadian S.F., Huttner A., Seilhean D., Renier N., Bilguvar K., Iwasaki A. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218(3) doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich J.A., Kugel J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7(8):612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 15.Badimon L., Robinson E.L., Jusic A., Carpusca I., deWindt L.J., Emanueli C., Ferdinandy P., Gu W., Gyongyosi M., Hackl M., Karaduzovic-Hadziabdic K., Lustrek M., Martelli F., Nham E., Potocnjak I., Satagopam V., Schneider R., Thum T., Devaux Y. Cardiovascular RNA markers and artificial intelligence may improve COVID-19 outcome: a position paper from the EU-CardioRNA COST action CA17129. Cardiovasc Res. 2021;117(8):1823–1840. doi: 10.1093/cvr/cvab094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H.Y., Xu M., Yang C.X., Tian R.R., Zhang M., Li J.J., Wang X.C., Ding Z.L., Li G.M., Li X.L., He Y.Q., Dong X.Q., Yao Y.G., Zheng Y.T. Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct Target Ther. 2020;5(1):294. doi: 10.1038/s41392-020-00457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mone P., Gambardella J., Wang X., Jankauskas S.S., Matarese A., Santulli G. miR-24 targets the transmembrane glycoprotein neuropilin-1 in human brain microvascular endothelial cells. Noncoding RNA. 2021;7(1):9. doi: 10.3390/ncrna7010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H., Gao Y., Li Z., Miao Y., Huang Z., Liu X., Xie L., Li H., Wen W., Zheng Y., Su W. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin Transl Med. 2020;10(6) doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., Sonnenschein K., Pink I., Hoeper M.M., Welte T., Bauersachs J., David S., Bar C., Thum T. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur J Heart Fail. 2021;23(3):468–475. doi: 10.1002/ejhf.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., Bar C., Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J Mol Cell Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutmann C., Khamina K., Theofilatos K., Diendorfer A.B., Burnap S.A., Nabeebaccus A., Fish M., McPhail M.J.W., O'Gallagher K., Schmidt L.E., Cassel C., Auzinger G., Napoli S., Mujib S.F., Trovato F., Sanderson B., Merrick B., Roy R., Edgeworth J.D., Shah A.M., Hayday A.C., Traby L., Hackl M., Eichinger S., Shankar-Hari M., Mayr M. Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc Res. 2022;118(2):461–474. doi: 10.1093/cvr/cvab338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C., Hu X., Li L., Li J.H. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal. 2020;34(10) doi: 10.1002/jcla.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck B.E., Wood C.D., Whenham G.R. Triaryl phosphate poisoning in cattle. Vet Pathol. 1977;14(2):128–137. doi: 10.1177/030098587701400205. [DOI] [PubMed] [Google Scholar]
- 24.Farr R.J., Rootes C.L., Rowntree L.C., Nguyen T.H.O., Hensen L., Kedzierski L., Cheng A.C., Kedzierska K., Au G.G., Marsh G.A., Vasan S.S., Foo C.H., Cowled C., Stewart C.R. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021;17(7) doi: 10.1371/journal.ppat.1009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertolazzi G., Cipollina C., Benos P.V., Tumminello M., Coronnello C. miR-1207-5p can contribute to dysregulation of inflammatory response in COVID-19 via targeting SARS-CoV-2 RNA. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.586592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabbatinelli J., Giuliani A., Matacchione G., Latini S., Laprovitera N., Pomponio G., Ferrarini A., Svegliati Baroni S., Pavani M., Moretti M., Gabrielli A., Procopio A.D., Ferracin M., Bonafe M., Olivieri F. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech Ageing Dev. 2021;193 doi: 10.1016/j.mad.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centa A., Fonseca A.S., Ferreira S., Azevedo M.L.V., Vaz de Paula C.B., Nagashima S., Machado-Souza C., Miggiolaro A., Baena C.P., de Noronha L., Cavalli L.R. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am J Physiol Lung Cell Mol Physiol. 2020;320(3):L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D., Mussa B.M. Identification of novel hypothalamic MicroRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in silico analysis. Brain Sci. 2020;10(10) doi: 10.3390/brainsci10100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meydan C., Madrer N., Soreq H. The neat dance of COVID-19: NEAT1, DANCR, and Co-modulated cholinergic RNAs link to inflammation. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.590870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaath H., Alajez N.M. Identification of PBMC-based molecular signature associational with COVID-19 disease severity. Heliyon. 2021;7(5) doi: 10.1016/j.heliyon.2021.e06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaath H., Vishnubalaji R., Elkord E., Alajez N.M. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. 2020;9(11) doi: 10.3390/cells9112374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachogiannis N.I., Sachse M., Georgiopoulos G., Zormpas E., Bampatsias D., Delialis D., Bonini F., Galyfos G., Sigala F., Stamatelopoulos K., Gatsiou A., Stellos K. Adenosine-to-inosine alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease. J Mol Cell Cardiol. 2021;160:111–120. doi: 10.1016/j.yjmcc.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng J., Zhou X., Feng W., Jia M., Zhang X., An T., Luan M., Pan Y., Zhang S., Zhou Z., Wen L., Sun Y., Zhou C. Risk stratification by long non-coding RNAs profiling in COVID-19 patients. J Cell Mol Med. 2021;25(10):4753–4764. doi: 10.1111/jcmm.16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Li B., Bai Q., Wang P., Wei G., Li Z., Hu L., Tian Q., Zhou J., Huang Q., Wang Z., Yue S., Wu J., Yang L., Zhou X., Jiang L., Ni T., Ye L., Wu Y. The lncRNA Snhg1-Vps13D vesicle trafficking system promotes memory CD8 T cell establishment via regulating the dual effects of IL-7 signaling. Signal Transduct Target Ther. 2021;6(1):126. doi: 10.1038/s41392-021-00492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatsiou A., Stellos K. Dawn of epitranscriptomic medicine. Circ Genom Precis Med. 2018;11(9) doi: 10.1161/CIRCGEN.118.001927. [DOI] [PubMed] [Google Scholar]
- 38.Sweaad W.K., Stefanizzi F.M., Chamorro-Jorganes A., Devaux Y., Emanueli C., E.U.-C.C.A. CA Relevance of N6-methyladenosine regulators for transcriptome: implications for development and the cardiovascular system. J Mol Cell Cardiol. 2021;160:56–70. doi: 10.1016/j.yjmcc.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 40.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coker H., Wei G., Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):310–318. doi: 10.1016/j.bbagrm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Kwast R., Quax P.H.A., Nossent A.Y. An emerging role for isomiRs and the microRNA epitranscriptome in neovascularization. Cells. 2019;9(1) doi: 10.3390/cells9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chanfreau G.F. Impact of RNA modifications and RNA-modifying enzymes on eukaryotic ribonucleases. Enzymes. 2017;41:299–329. doi: 10.1016/bs.enz.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatsiou A., Vlachogiannis N., Lunella F.F., Sachse M., Stellos K. Adenosine-to-inosine RNA editing in health and disease. Antioxid Redox Signal. 2018;29(9):846–863. doi: 10.1089/ars.2017.7295. [DOI] [PubMed] [Google Scholar]
- 47.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17(2):83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe A., Abbondanza D., Fleming S.J., Subramanian A., Montoro D.T., Jagadeesh K.A., Dey K.K., Sen P., Slyper M., Pita-Juarez Y.H., Phillips D., Biermann J., Bloom-Ackermann Z., Barkas N., Ganna A., Gomez J., Melms J.C., Katsyv I., Normandin E., Naderi P., Popov Y.V., Raju S.S., Niezen S., Tsai L.T., Siddle K.J., Sud M., Tran V.M., Vellarikkal S.K., Wang Y., Amir-Zilberstein L., Atri D.S., Beechem J., Brook O.R., Chen J., Divakar P., Dorceus P., Engreitz J.M., Essene A., Fitzgerald D.M., Fropf R., Gazal S., Gould J., Grzyb J., Harvey T., Hecht J., Hether T., Jane-Valbuena J., Leney-Greene M., Ma H., McCabe C., McLoughlin D.E., Miller E.M., Muus C., Niemi M., Padera R., Pan L., Pant D., Pe'er C., Pfiffner-Borges J., Pinto C.J., Plaisted J., Reeves J., Ross M., Rudy M., Rueckert E.H., Siciliano M., Sturm A., Todres E., Waghray A., Warren S., Zhang S., Zollinger D.R., Cosimi L., Gupta R.M., Hacohen N., Hibshoosh H., Hide W., Price A.L., Rajagopal J., Tata P.R., Riedel S., Szabo G., Tickle T.L., Ellinor P.T., Hung D., Sabeti P.C., Novak R., Rogers R., Ingber D.E., Jiang Z.G., Juric D., Babadi M., Farhi S.L., Izar B., Stone J.R., Vlachos I.S., Solomon I.H., Ashenberg O., Porter C.B.M., Li B., Shalek A.K., Villani A.C., Rozenblatt-Rosen O., Regev A. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202(5):756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., Koltsida O., Mentis A., Koulouris N., Tsiodras S., Koutsoukou A., Andreakos E. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol. 2021;22(1):32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 51.Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellaker C., Vesely C., Ponting C.P., McLaughlin P.J., Jantsch M.F., Dorin J., Adams I.R., Scadden A.D., Ohman M., Keegan L.P., O'Connell M.A. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9(4):1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z., Li Y., Sheng C., Yang C., Chen L., Sun J. Tanshinone IIA inhibits apoptosis in the myocardium by inducing microRNA-152-3p expression and thereby downregulating PTEN. Am J Transl Res. 2016;8(7):3124–3132. [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Men M., Xie B., Shan J., Wang C., Liu J., Zheng H., Yang W., Xue S., Guo C. Inhibition of PKR protects against H2O2-induced injury on neonatal cardiac myocytes by attenuating apoptosis and inflammation. Sci Rep. 2016;6:38753. doi: 10.1038/srep38753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara Y., Grimberg A., Teegarden S., Mombereau C., Liu S., Bale T.L., Blendy J.A., Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28(48):12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 56.Lakenman P.L.M., van der Hoven B., Schuijs J.M., Eveleens R.D., van Bommel J., Olieman J.F., Joosten K.F.M. Energy expenditure and feeding practices and tolerance during the acute and late phase of critically ill COVID-19 patients. Clin Nutr ESPEN. 2021;43:383–389. doi: 10.1016/j.clnesp.2021.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whittle J., Molinger J., MacLeod D., Haines K., Wischmeyer P.E., L.-C.S. Group Persistent hypermetabolism and longitudinal energy expenditure in critically ill patients with COVID-19. Crit Care. 2020;24(1):581. doi: 10.1186/s13054-020-03286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zucker I.H. Novel mechanisms of sympathetic regulation in chronic heart failure. Hypertension. 2006;48(6):1005–1011. doi: 10.1161/01.HYP.0000246614.47231.25. [DOI] [PubMed] [Google Scholar]
- 59.Vlachogiannis N.I., Verrou K.M., Stellos K., Sfikakis P.P., Paraskevis D. The role of A-to-I RNA editing in infections by RNA viruses: possible implications for SARS-CoV-2 infection. Clin Immunol. 2021;226 doi: 10.1016/j.clim.2021.108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lerner T., Papavasiliou F.N., Pecori R., RNA Editors Cofactors, and mRNA targets: an overview of the C-to-U RNA editing machinery and its implication in human disease. Genes (Basel) 2018;10(1) doi: 10.3390/genes10010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R., Hozumi Y., Zheng Y.H., Yin C., Wei G.W. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12(10) doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Giorgio S., Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv. 2020;6(25):eabb5813. doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samuel C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411(2):180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V., Okuda K., Markmann A.J., Bartelt L., de Silva A., Margolis D.M., Boucher R.C., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun. 2021;12(1):848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scadden A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12(6):489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 68.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picardi E., Mansi L., Pesole G. Detection of A-to-I RNA editing in SARS-COV-2. Genes (Basel) 2021;13(1) doi: 10.3390/genes13010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Libby P., Luscher T. COVID-19 is, in the end, an endothelial disease. Eur Heart J. 2020;41(32):3038–3044. doi: 10.1093/eurheartj/ehaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCracken I.R., Saginc G., He L., Huseynov A., Daniels A., Fletcher S., Peghaire C., Kalna V., Andaloussi-Mae M., Muhl L., Craig N.M., Griffiths S.J., Haas J.G., Tait-Burkard C., Lendahl U., Birdsey G.M., Betsholtz C., Noseda M., Baker A.H., Randi A.M. Lack of evidence of angiotensin-converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation. 2021;143(8):865–868. doi: 10.1161/CIRCULATIONAHA.120.052824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romano M., Sironi M., Toniatti C., Polentarutti N., Fruscella P., Ghezzi P., Faggioni R., Luini W., van Hinsbergh V., Sozzani S., Bussolino F., Poli V., Ciliberto G., Mantovani A. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;6(3):315–325. doi: 10.1016/s1074-7613(00)80334-9. [DOI] [PubMed] [Google Scholar]
- 74.Spadaro S., Fogagnolo A., Campo G., Zucchetti O., Verri M., Ottaviani I., Tunstall T., Grasso S., Scaramuzzo V., Murgolo F., Marangoni E., Sega F.Vieceli Dalla, Fortini F., Pavasini R., Rizzo P., Ferrari R., Papi A., Volta C.A., Contoli M. Markers of endothelial and epithelial pulmonary injury in mechanically ventilated COVID-19 ICU patients. Crit Care. 2021;25(1):74. doi: 10.1186/s13054-021-03499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tong M., Jiang Y., Xia D., Xiong Y., Zheng Q., Chen F., Zou L., Xiao W., Zhu Y. Elevated expression of serum endothelial cell adhesion molecules in COVID-19 patients. J Infect Dis. 2020;222(6):894–898. doi: 10.1093/infdis/jiaa349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang S., Tanaka T., Inoue H., Ono C., Hashimoto S., Kioi Y., Matsumoto H., Matsuura H., Matsubara T., Shimizu K., Ogura H., Matsuura Y., Kishimoto T. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci U S A. 2020;117(36):22351–22356. doi: 10.1073/pnas.2010229117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Connors J.M., Levy J.H. Thromboinflammation and the hypercoagulability of COVID-19. J Thromb Haemost. 2020;18(7):1559–1561. doi: 10.1111/jth.14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jae N., Rossbach O., Amrhein C., Sigala F., Boon R.A., Furtig B., Manavski Y., You X., Uchida S., Keller T., Boeckel J.N., Franco-Cereceda A., Maegdefessel L., Chen W., Schwalbe H., Bindereif A., Eriksson P., Hedin U., Zeiher A.M., Dimmeler S. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med. 2016;22(10):1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 79.Vlachogiannis N.I., Gatsiou A., Silvestris D.A., Stamatelopoulos K., Tektonidou M.G., Gallo A., Sfikakis P.P., Stellos K. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J Autoimmun. 2020;106 doi: 10.1016/j.jaut.2019.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sukhova G.K., Zhang Y., Pan J.H., Wada Y., Yamamoto T., Naito M., Kodama T., Tsimikas S., Witztum J.L., Lu M.L., Sakara Y., Chin M.T., Libby P., Shi G.P. Deficiency of cathepsin S reduces atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2003;111(6):897–906. doi: 10.1172/JCI14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi G.P., Villadangos J.A., Dranoff G., Small C., Gu L., Haley K.J., Riese R., Ploegh H.L., Chapman H.A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10(2):197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 82.Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L.C., Pircher A., Geldhof V., de Rooij L., Kalucka J., Sokol L., Garcia-Caballero M., Zheng Y., Qian J., Teuwen L.A., Khan S., Boeckx B., Wauters E., Decaluwe H., De Leyn P., Vansteenkiste J., Weynand B., Sagaert X., Verbeken E., Wolthuis A., Topal B., Everaerts W., Bohnenberger H., Emmert A., Panovska D., De Smet F., Staal F.J.T., McLaughlin R.J., Impens F., Lagani V., Vinckier S., Mazzone M., Schoonjans L., Dewerchin M., Eelen G., Karakach T.K., Yang H., Wang J., Bolund L., Lin L., Thienpont B., Li X., Lambrechts D., Luo Y., Carmeliet P. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37(3):421. doi: 10.1016/j.ccell.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 83.Teuwen L.A., Geldhof V., Pasut A., Carmeliet P. Author correction: COVID-19: the vasculature unleashed. Nat Rev Immunol. 2020;20(7):448. doi: 10.1038/s41577-020-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellul M.A., Benjamin L., Singh B., Lant S., Michael B.D., Easton A., Kneen R., Defres S., Sejvar J., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19(9):767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5(7):811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akhmerov A., Marban E. COVID-19 and the heart. Circ Res. 2020;126(10):1443–1455. doi: 10.1161/CIRCRESAHA.120.317055. [DOI] [PubMed] [Google Scholar]
- 87.Nishiga M., Wang D.W., Han Y., Lewis D.B., Wu J.C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol. 2020;17(9):543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beis D., Zerr I., Martelli F., Doehner W., Devaux Y. RNAs in brain and heart diseases. Int J Mol Sci. 2020;21(10) doi: 10.3390/ijms21103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris-Taube K., Vernardis S.I., Egger A.S., Kreidl M., Ludwig D., Kilian C., Agostini F., Zelezniak A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., von Kalle C., Campbell A., Hayward C., Porteous D.J., Marioni R.E., Langenberg C., Lilley K.S., Kuebler W.M., Mulleder M., Drosten C., Suttorp N., Witzenrath M., Kurth F., Sander L.E., Ralser M. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24. doi: 10.1016/j.cels.2020.05.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruan Q., Yang K., Wang W., Jiang L., Song J. Correction to: clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intens Care Med. 2020;46(6):1294–1297. doi: 10.1007/s00134-020-06028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Abreu R.C., Fernandes H., da Costa Martins P.A., Sahoo S., Emanueli C., Ferreira L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat Rev Cardiol. 2020;17(11):685–697. doi: 10.1038/s41569-020-0389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verweij F.J., Revenu C., Arras G., Dingli F., Loew D., Pegtel D.M., Follain G., Allio G., Goetz J.G., Zimmermann P., Herbomel P., Bene F.Del, Raposo G., van Niel G. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell. 2019;48(4):573–589. doi: 10.1016/j.devcel.2019.01.004. e4. [DOI] [PubMed] [Google Scholar]
- 95.Couch Y., Akbar N., Roodselaar J., Evans M.C., Gardiner C., Sargent I., Romero I.A., Bristow A., Buchan A.M., Haughey N., Anthony D.C. Circulating endothelial cell-derived extracellular vesicles mediate the acute phase response and sickness behaviour associated with CNS inflammation. Sci Rep. 2017;7(1):9574. doi: 10.1038/s41598-017-09710-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson B.D., Kip K.E., Marroquin O.C., Ridker P.M., Kelsey S.F., Shaw L.J., Pepine C.J., Sharaf B., Bairey Merz C.N., Sopko G., Olson M.B., Reis S.E., L. National Heart, I. Blood, serum amyloid a as a predictor of coronary artery disease and cardiovascular outcome in women: the National Heart, Lung, and Blood Institute-sponsored Women's ischemia syndrome evaluation (WISE) Circulation. 2004;109(6):726–732. doi: 10.1161/01.CIR.0000115516.54550.B1. [DOI] [PubMed] [Google Scholar]
- 97.Rosell A., Havervall S., von Meijenfeldt F., Hisada Y., Aguilera K., Grover S.P., Lisman T., Mackman N., Thalin C. Patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality. Arterioscler Thromb Vasc Biol. 2021;41(2):878–882. doi: 10.1161/ATVBAHA.120.315547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mancuso P., Gidaro A., Gregato G., Raveane A., Cremonesi P., Quarna J., Caccia S., Gusso L., Rusconi S., Giacomelli A., Cogliati C., Bertolini F. Circulating endothelial progenitors are increased in COVID-19 patients and correlate with SARS-CoV-2 RNA in severe cases. J Thromb Haemost. 2020;18(10):2744–2750. doi: 10.1111/jth.15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guerin C.L., Guyonnet L., Goudot G., Revets D., Konstantinou M., Chipont A., Chocron R., Blandinieres A., Khider L., Rancic J., Peronino C., Debuc B., Cras A., Knosp C., Latremouille C., Capel A., Ollert M., Diehl J.L., Jansen P., Planquette B., Sanchez O., Gaussem P., Mirault T., Carpentier A., Gendron N., Smadja D.M. Multidimensional proteomic approach of endothelial progenitors demonstrate expression of KDR restricted to CD19 cells. Stem Cell Rev Rep. 2021;17(2):639–651. doi: 10.1007/s12015-020-10062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin B., Liu J., Liu Y., Qin X. Progress in understanding COVID-19: insights from the omics approach. Crit Rev Clin Lab Sci. 2021;58(4):242–252. doi: 10.1080/10408363.2020.1851167. [DOI] [PubMed] [Google Scholar]
- 101.Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., Baloni P., Qin G., Smith B., Kornilov S.A., Rostomily C., Xu A., Li J., Dong S., Rothchild A., Zhou J., Murray K., Edmark R., Hong S., Heath J.E., Earls J., Zhang R., Xie J., Li S., Roper R., Jones L., Zhou Y., Rowen L., Liu R., Mackay S., O'Mahony D.S., Dale C.R., Wallick J.A., Algren H.A., Zager M.A., Unit I.S.-S.C.B., Wei W., Price N.D., Huang S., Subramanian N., Wang K., Magis A.T., Hadlock J.J., Hood L., Aderem A., Bluestone J.A., Lanier L.L., Greenberg P.D., Gottardo R., Davis M.M., Goldman J.D., Heath J.R. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6):1479–1495. doi: 10.1016/j.cell.2020.10.037. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Overmyer K.A., Shishkova E., Miller I.J., Balnis J., Bernstein M.N., Peters-Clarke T.M., Meyer J.G., Quan Q., Muehlbauer L.K., Trujillo E.A., He Y., Chopra A., Chieng H.C., Tiwari A., Judson M.A., Paulson B., Brademan D.R., Zhu Y., Serrano L.R., Linke V., Drake L.A., Adam A.P., Schwartz B.S., Singer H.A., Swanson S., Mosher D.F., Stewart R., Coon J.J., Jaitovich A. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12(1):23–40. doi: 10.1016/j.cels.2020.10.003. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Al-Khawaga S., Abdelalim E.M. Potential application of mesenchymal stem cells and their exosomes in lung injury: an emerging therapeutic option for COVID-19 patients. Stem Cell Res Ther. 2020;11(1):437. doi: 10.1186/s13287-020-01963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]