Abstract
Aside from PD-L1 expression, biomarkers of response to immune checkpoint inhibitors (ICI) in non-small cell lung cancer (NSCLC) are needed. We previously reported in 100 NSCLC and kidney cancer patients that fecal Akkermansia muciniphila (Akk) correlated with ICI clinical success. The endpoint of this study was to prospectively validate the prognostic significance of fecal Akk in advanced NSCLC patients treated with first or second line ICI. We performed shotgun metagenomics-based microbiome profiling using two different pipelines in 338 NSCLC patients. Baseline stool Akk was associated with increased objective response rates and overall survival in multivariate analyses, independent of PD-L1 expression, antibiotics and performance status. Intestinal Akk was accompanied by a richer commensalism, including Eubacterium hallii and Bifidobacterium adolescentis and a more inflamed tumor microenvironment in a subset of patients. However, antibiotic use (20% of cases) coincided with a relative dominance of Akk above 4.8% accompanied with the genus Clostridium, both associated with resistance to ICI. Our study shows significant differences in relative abundance of Akk that could be useful for future studies as a tool to aid patient stratification.
Keywords: cancer, immuno-oncology, immune checkpoint blockade, PD-1, PD-L1, Akkermansia muciniphila, lung cancer
INTRODUCTION
The development of immune checkpoint inhibitors (ICI) targeting the PD-1/PD-L1 interaction has transformed the therapeutic landscape of patients with advanced non-small cell lung cancer (NSCLC)1–6. Landmark trials performed on previously treated patients with advanced NSCLC demonstrated superior overall survival (OS) with PD-1/PD-L1 blockade compared to standard chemotherapy1–3. Following randomized phase III trials on patients with previously untreated advanced NSCLC, ICI were approved in the first-line setting, either as monotherapy for patients with tumor PD-L1 expression ≥ 50 % on tumor cells or in combination with platinum-doublet chemotherapy irrespectively of PD-L1 expression4–7. However, only a minority (35%) of patients benefit from sustained response to ICI8. Most patients with NSCLC patients develop primary or secondary resistance, or occasional rapid acceleration of the disease called “hyper-progression”9. Therefore, understanding mechanisms of resistance to ICI to identify novel and robust biomarkers of resistance are urgently needed.
Primary resistance has been attributed to low tumor mutational burden and poor intrinsic antigenicity of tumor cells10,11, defective antigen presentation12, limited intratumoral lymphocyte infiltration related to T cell exhaustion13, and metabolic immunosuppressive pathways14,15. High dimensional omics technologies are currently developed to decipher the main regulators of the “cancer immune set-point” —the threshold beyond which an effective immune response can occur in the tumor bearer16.
Recent lines of evidence point to the biological significance of the composition of the gut microbiota in influencing peripheral immune tonus and effectiveness of ICI in patients with cancer17. The human gut microbiome, composed of 1013 micro-organisms, modulates many host processes including metabolism, inflammation, peristalsis, elimination of pathogens and xenobiotics, maturation of immune functions to maintain tolerance to microbial and food antigens as well as maintenance of intestinal epithelial barrier fitness. More recently, the gut microbiome has unexpectedly been shown to influence the effectiveness of ICI18. First, in pre-clinical models, experiments with germ-free or antibiotic (ATB)-treated mice unraveled that the antitumor activity of ICI requires the presence of gut microbial components19. Similarly, antibiotics (ATB) prior to ICI initiation drastically reduced the clinical benefit in several cohorts of patients across various cancer types (melanoma, advanced NSCLC, renal cell carcinoma (RCC), and urothelial cancer)18,20–22. This observation was confirmed in prospective trials and large meta-analyses, suggesting that gut microbiota may be instrumental for the immunostimulatory mode of action of ICI 23,24.
Supporting this contention, we and others reported that primary responses to anti-PD-1/PD-L1 antibodies in patients with epithelial tumours and melanoma could, at least in part, be attributed to the taxonomic composition of the gut microbial ecosystem18,20,25. A diverse microbiota and the presence of specific bacteria such as Akkermansia muciniphila, Ruminococcus or Bifidobacterium genera were associated with improved clinical response to ICI, correlating with increased systemic immune tonus18,20,25. We previously reported the stool metagenomic profiling of 100 patients diagnosed with refractory NSCLC or RCC receiving second or third line anti-PD-1 antibodies, concluding that the prevalence of A. muciniphila (Akk) was increased in patients presenting partial response or stable disease, as compared with patients in progressive disease (61 versus 34% respectively, p=0.007)18. Akk was also overrepresented in patients with progression free-survival (PFS) > 3 months, when analyzing a subgroup of 60 patients with NSCLC18. Another group performed 16S rRNA gene amplicon sequencing of 37 NSCLC patient feces, confirming that Akk was enriched in patients responding to ICI26. In patients treated with Abiraterone acetate, an inhibitor of androgen biosynthesis approved to treat metastatic castration-resistant prostate cancer, intestinal Akk also correlated with therapeutic responses27. Intestinal prevalence of Akk has also been associated with low body mass index, fitness, and successful aging (as indicated by its presence in disease-free centenarians)28. In animal models, Akk supplementation reduces obesity29 and its co-morbidities, palliates neurodegenerative disorders30 and counteracts progeria31. However, there are some epidemiological associations between this bacterium and inflammation-related pathologies (such as Parkinson disease32, colitis during graft-versus-host disease, 33 or diet-induced susceptibility to pathogenic Citrobacter rodentium34). Despite these reports, Akk may be viewed as a potential master regulator of homeostasis in the metaorganism.
Altogether, our earlier results obtained in small cohorts overlooking confounding factors allow to surmise that the gut microbiome composition, and more specifically the presence of Akk at diagnosis, could become a biomarker predicting objective responses and 12 month survival rates in advanced NSCLC treated with 1st and 2nd line ICI. To validate this hypothesis, we performed a prospective multicentric study and analyzed baseline microbiome profiles using two metagenomic taxonomic pipelines in 338 patients with advanced NSCLC patients to predict the clinical benefit to anti-PD-1 antibodies in multivariate analyses.
RESULTS
Association between the presence of Akkermansia muciniphila and clinical outcome
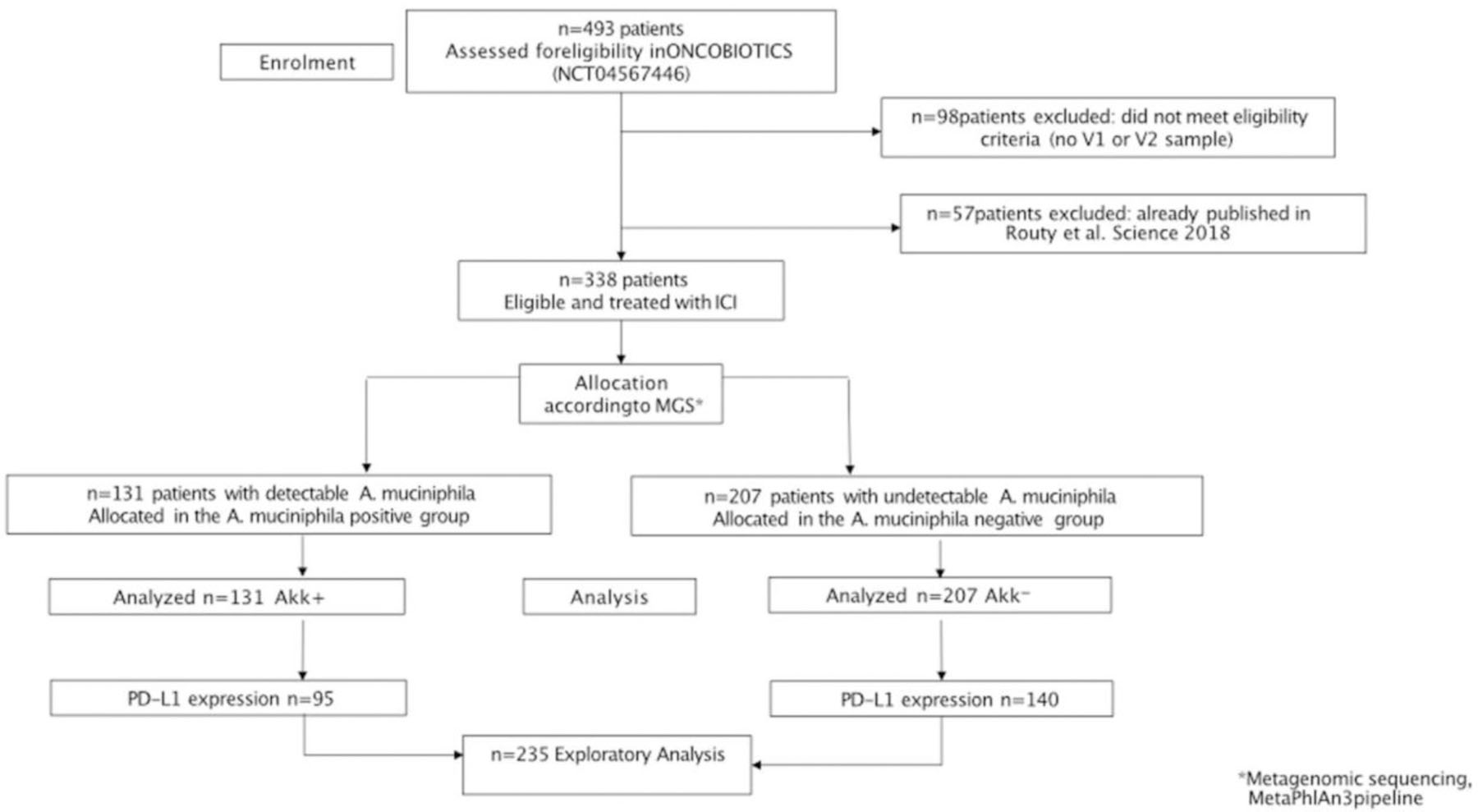
From December 2015 to November 2019, a total of 493 patients were screened for enrollment in this study and 338 patients met inclusion criteria, providing at least one baseline (V1 and/or V2) fecal sample for profiling (Extended Figure 1). Using shotgun metagenomic sequencing, we determined that Akk was detectable in 131 (39%) and absent in 207 (61%) patients using MetaPhlAn profiling expanded to identify the main Akk species-level genome bin (SGB9226 spp35, M&M Table 1). Baseline characteristics were well balanced between patients with detectable Akk (Akk+) and undetectable Akk (Akk−) groups with respect to sex, ECOG performance status, smoking history, tumor histology, body mass index (BMI), PD-L1 expression, and line of therapy (Table S1A). However, there was a non-statistically significant trend for older age in theAkk+ group (66 years versus 64 years, p=0.08). A total of 69 patients (20%) received antibiotics (ATB) 60 days prior to ICI initiation, with similar frequencies (p=0.68) and regimen in Akk+ and Akk− groups (Table S1A). Among ATB classes, beta-lactams were the most commonly prescribed in both groups (Table S1B, 80% in Akk+ versus 64% in Akk−, p=0.33).
M&M Table 1.
Prevalence of various substrains of Akkermansia in our cohort.
| SGB | |||||
|---|---|---|---|---|---|
|
| |||||
| 9226 – MucT – no. (%) | 9228 – no. (%) | 9224 – no. (%) | 9223 – (%) | None – no. (%) | |
| Prevalence of SGBs in all patients (n=338) | 131/338 (38.8) | 26/338 (7.7) | 5/338 (1.5) | 1/338 (0.3) | 175/338 (51.7) |
| Prevalence of SGBs in SGBs* patients (n=163) | 131/163 (80.4) | 26/163 (15.9) | 5/163 (3.1) | 1/163 (0.6) | - |
SGBs: Akkermansia candidate species or species-level genome bins; MucT: A. muciniphila.
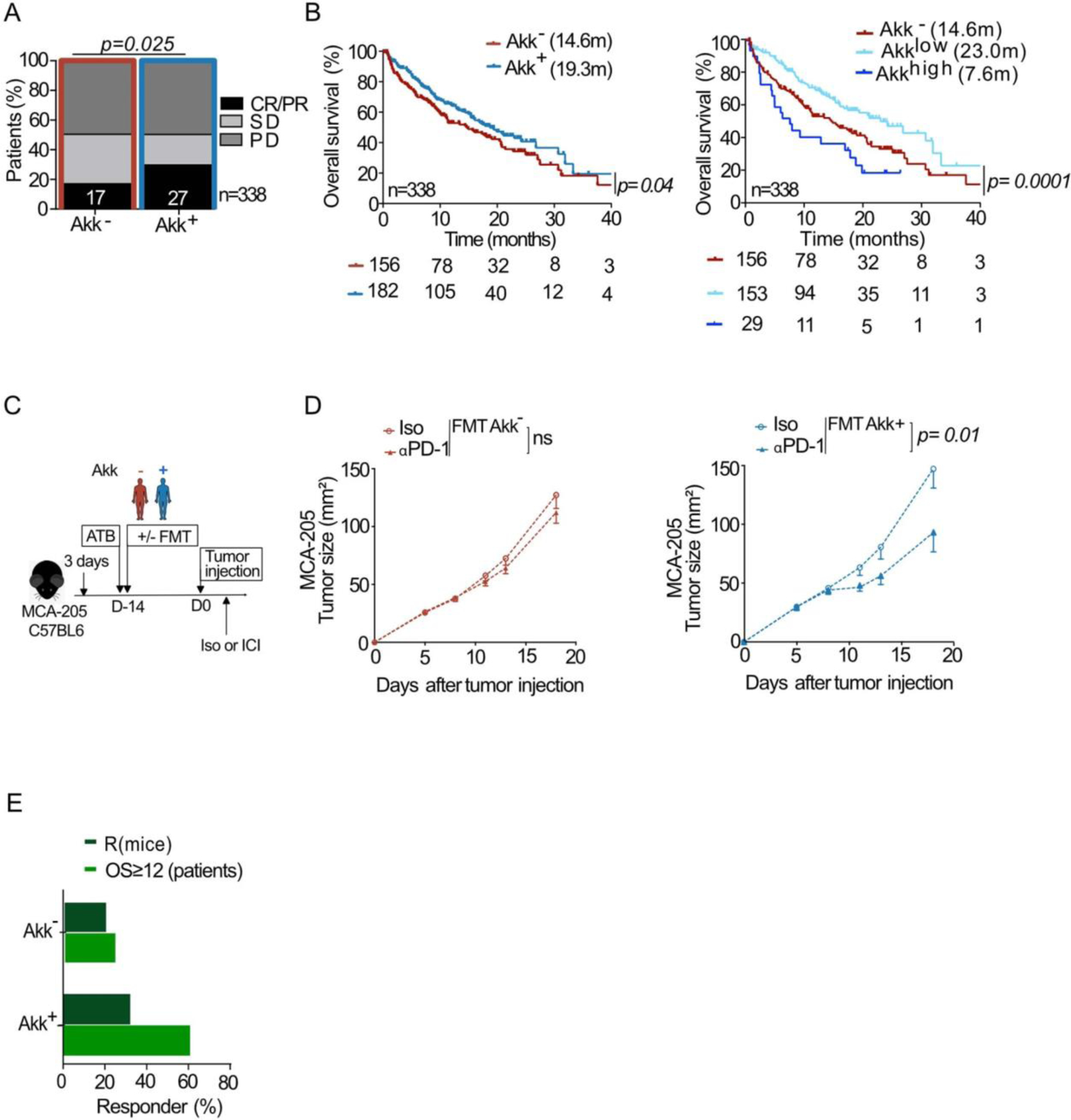
When considering Akk+ versus Akk− groups in our cohort, objective response rates (ORR) were 28% and 18% respectively (Figure 1A, p=0.04). Partial responses (PR), stable disease (SD) and progressive disease (PD) rates were 28%, 28% and 44%, respectively, for the Akk+ group versus 18%, 31% and 50% for the Akk− group. When considering the subgroup of patients who received immunotherapy alone as front-line treatment (1L IO, n=86), ORR were 41% and 19% in the Akk+ versus Akk− respectively (Figure 1B, p=0.016). Of note, for the whole cohort of patients (irrespective of the line of treatment), the median overall survival (mOS) was 18.8 months for the Akk+ group compared to 15.4 months in the Akk− group (HR=0.72; (95% CI :0.73–1.62); p=0.03, Figure 1C, Table S1A). The median OS of ≥ 2L NSCLC patients was 18.8 months for the Akk+ group compared to 13.4 months in the Akk− group (HR=0.70; (95% CI :0.50–0.98); p=0.04, Figure 1D, Table S1A). For the 1L IO patients, 59% of Akk+ patients were still alive after 12 months (OS ≥ 12) while only 35% of Akk− individuals were long term survivors (Figure 1E, p=0.04). We draw the same conclusions when using the MetaOMineR pipeline36 (Extended Figure 2A–B left panel).
Figure 1. Stool A. muciniphila (Akk) is associated with ICI clinical benefit.
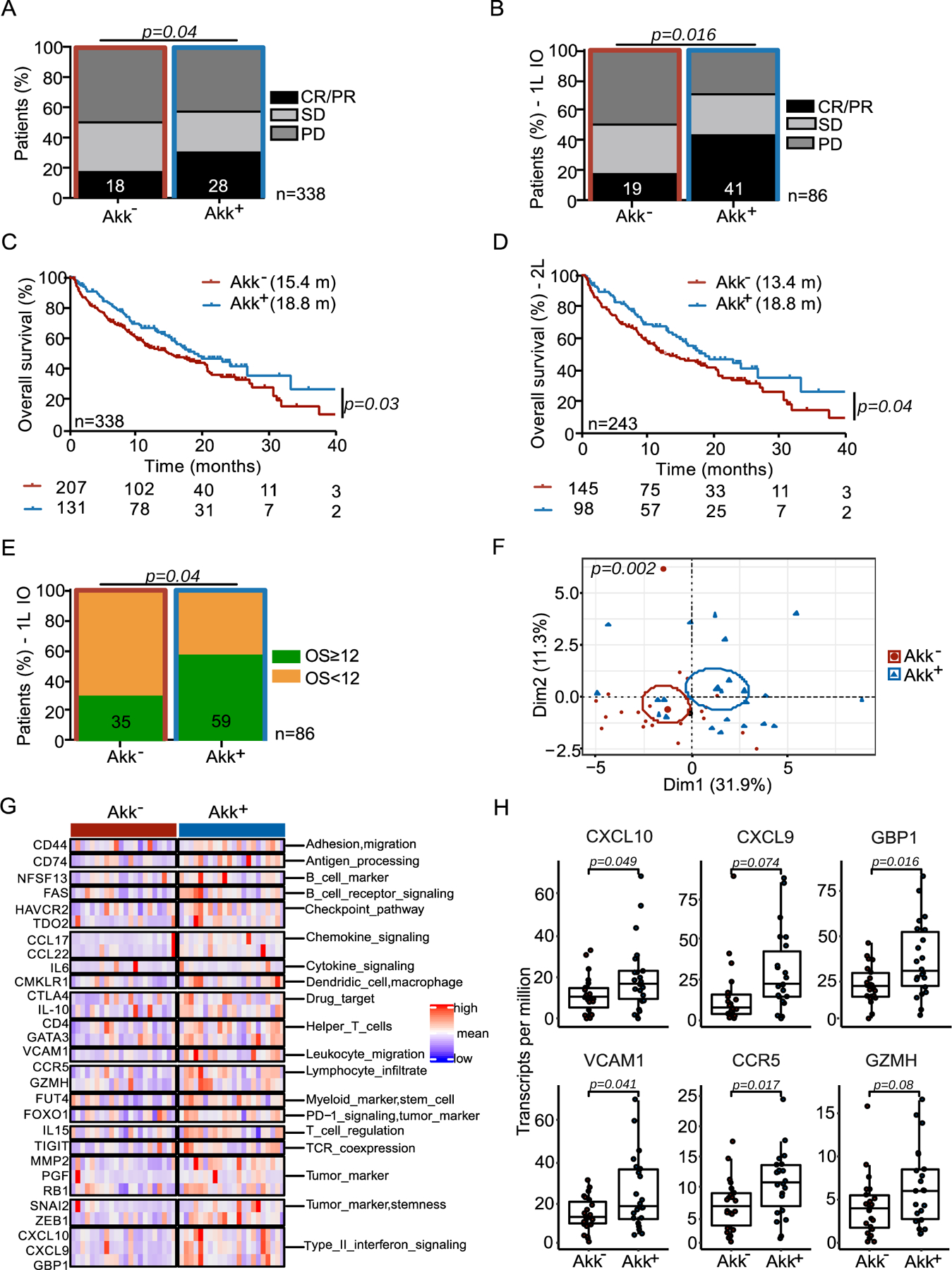
A-B Correlations between stool Akk and ORR in 1L+2L (n=338) and 1L immunotherapy NSCLC (n=86) patients. CR; complete response. PR; partial response, SD; stable disease, PD; progressive disease analyzed using Chi-square test. C-D Kaplan-Meier curves and Cox regression analyses of overall survival (OS) of 1L+2L (n=338) and 2L (n=243) according to Akk status. E. Difference of the intestinal prevalence of Akk between patients with OS < 12 months versus > 12 months in 1L immunotherapy (IO) analyzed using Chi-square test. F-H. RNA sequencing of tumor biopsies in a sub-group of 44 NSCLC patients (17 non-metastatic and 27 metastatic patients, Table S3). F. Principal Component Analysis (PCoA) of the differentially expressed genes according to intestinal prevalence of Akk, using the 395 immune-related gene selection of the Oncomine Immune Response Research Assay indicating significant differences using a Mann-Whitney p-value < 0.10 through PERMANOVA test using Euclidian distance. G. Heatmap of the differentially expressed gene products after normalization (between Akk+ vs Akk− patients) classified by category. H. Boxplot of selected gene expression values according to Akk groups. Differences between groups were assessed with Mann-Whitney tests. C-X-C motif chemokine ligand 10 (CXCL10), C-X-C motif chemokine ligand 9 (CXCL9), Guanylate binding protein 1 (GBP1), Vascular cell adhesion protein 1 (VCAM1), C-C chemokine receptor type 5 (CCR5), Granzyme H (GZMH).
To establish cause-effect relationships between the presence of Akk (and its ecosystem) with response to ICI, we retrospectively performed a preclinical meta-analysis gathering 29 experiments where the tumor growth kinetics were followed up in avatar mouse models17. These recipients were ATB-treated and then received fecal microbial transplants (FMT) from 26 different NSCLC patients (Table S2). Then, mice were implanted with syngeneic orthotopic MCA-205 sarcomas (representative tumor model for sensitivity to anti-PD-1 antibodies as previously described18,37) and later subjected to PD-1 blockade (Extended Figure 2C–E). As observed in our clinical cohort, resistance to therapy in mice was associated with the absence of detectable Akk in the FMT experiments (Extended Figure 2C–D).
Therefore, in this second independent and prospective study on 338 advanced NSCLC patients, we validated in humans using two different metagenomics pipelines as well as in mice that the presence of Akk is associated with higher ORR and longer OS in patients with NSCLC receiving ICI.
Links between fecal Akk, the intestinal ecosystem and the tumor microenvironment
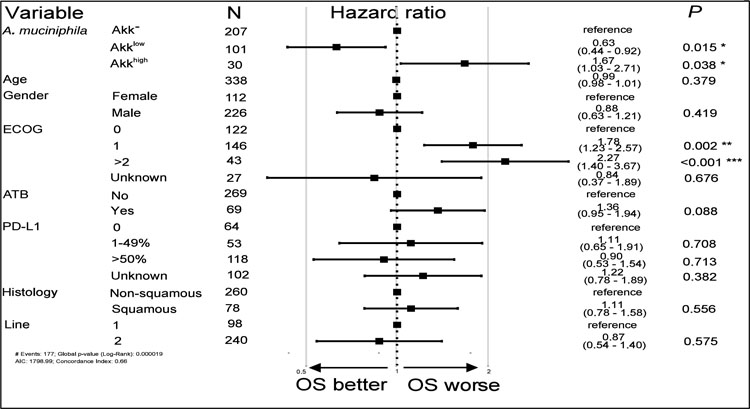
Given known correlations between compositional differences in the gut microbiota and tumor immune landscape18,20, that can vary across histological types38, we addressed the interactions between stool Akk detection and tumour histology (squamous versus non-squamous NSCLC). The presence of Akk in stools at diagnosis had no influence on histology (non-squamous vs squamous NSCLC (for multivariate analysis, p=0.556 in Table S1A, Table 1).
Table 1. Cox logistic regression multivariate analysis of overall survival in 338 NSCLC patients according to Akk relative abundance segregated in 3 groups (Akk−, Akklow and Akkhigh) and all the other relevant clinical parameters.
OS: overall survival. ECOG; eastern cooperative oncology group performance status. ATB: antibiotics.
|
In order to uncover intratumoral transcriptomic differences driven by Akk, we performed tumor RNA sequencing in a subset of patients with available tumor biopsies harvested from Akk+ (n=22) and Akk− (n=22) patients upon diagnosis of locally advanced or metastatic NSCLC (Table S3). The supervised analysis of significant gene expression differences between Akk+ and Akk− groups within a panel of 395 immune-related genes of the Oncomine Immune Response Research Assay39 revealed a set of differentally expressed genes associated with response to PD-1 blockade in lung cancer (Figure 1F) such as CD4+ T helper cells with activation (CD4, CD74, Vcam-1, associated with adhesion and trans-migration of T lymphocytes within tumor nests40, granzyme H serine protease Gzmh associated with cytolytic activity) and exhaustion (Ctla4, Fas, Tigit, Havcr2) markers and the IFN fingerprint (Ccr5, Cxcl9, Cxcl10, Tdo2, and IFN-inducible guanylate binding protein 1) (Figure 1G–H). These results support the possibility that Akk could promote the elicitation or recirculation of Th1 cells into the tumor microenvironment, as previously shown in mouse models18,19.
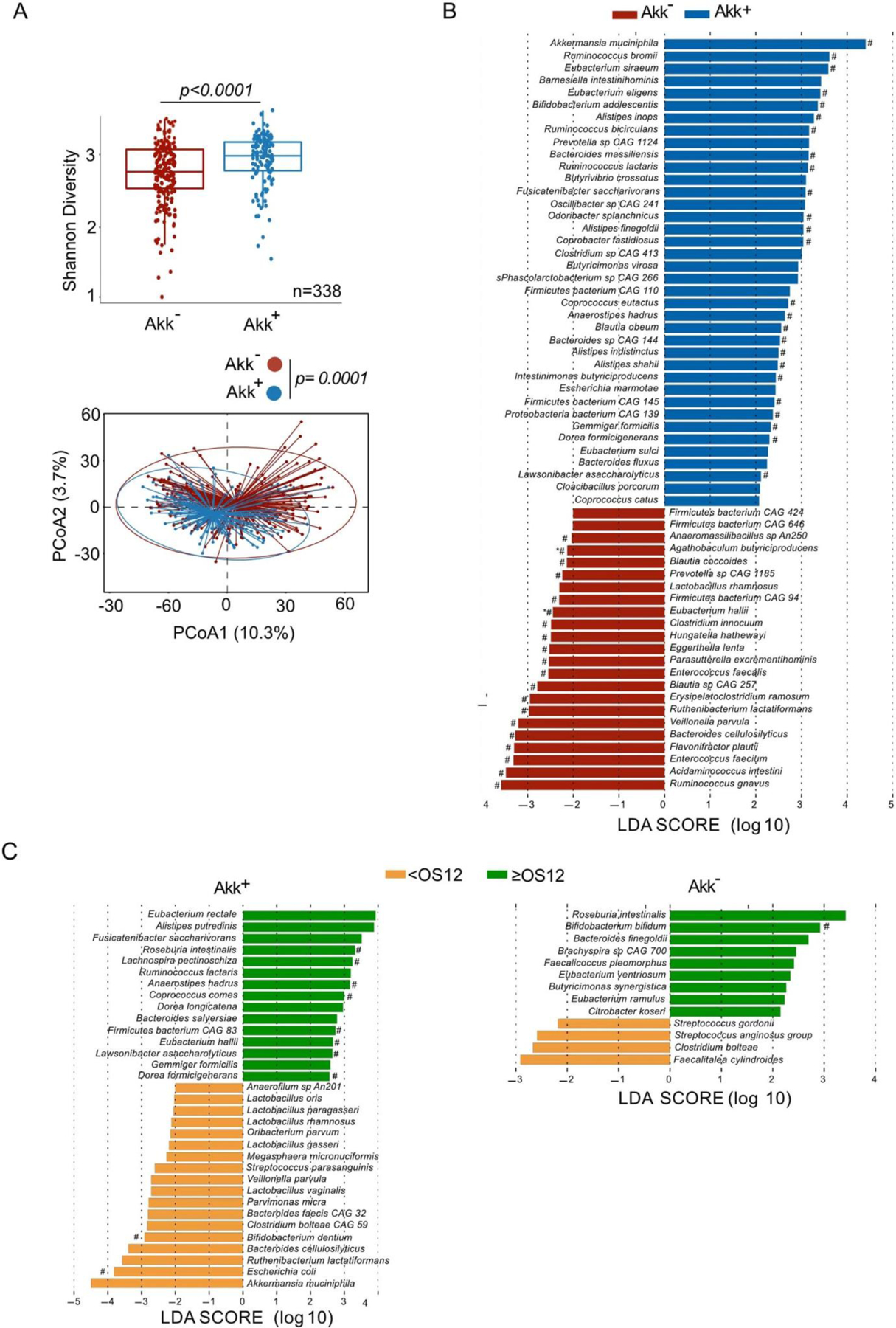
Next, we examined compositional taxonomic differences in the gut microbiota in Akk+ versus Akk− patients. We found a significant increase of the Shannon diversity index (Extended Figure 3A upper panel, p<0.0001) in Akk+ compared to Akk− patients, as well as differences in the overall microbial community composition between both groups (Extended Figure 3A lower panel, p=0.0001). Using LEfSe (Linear discriminant analysis Effect Size) analyses to investigate differences in species relative abundance between the 2 groups, we found Ruminococcacae (Ruminococcus bromii, R. bicirculans, R. lactaris), Lachnospiraceae (Eubacterium siraeum, E. eligens) family members and Alistipes spp. (A. inops, A. finegoldii, A. indistinctus, A. shahii) to be enriched in Akk+ stools as previously reported,41 while feces from Akk− patients were overabundant in Veillonella parvula, Actinomyces and genus Clostridium (C. innocuum, Hungatella hathewayi) (Extended Figure 3B) as already described in the lower airway microbiome of patients with NSCLC with poor prognosis 42 as well as patients with kidney cancer resistant to ICI37.
Furthermore, within the Akk+ group, we found significant differences in the overall microbial composition between patients with OS ≥ 12 versus OS < 12 months, but this difference was not observed in Akk− patients (Figure 2A, p=0.009 versus p=0.07). LEfSe analysis within the Akk+ group exhibiting OS ≥ 12 months versus those with OS < 12 months unveiled increased relative abundance of Lachnospiraceae family members (Dorea formicigenerans, D. longicatena, Eubacterium rectale, E. hallii, R. intestinalis, Coprococcus comes) in patients with OS ≥ 12. Conversely, species belonging to the Gammaproteobacteria (E. coli), Clostridia class (R. lactatiformans) or Bacilli class (such as Lactobacillus gasseri, L. paragasseri, L. oris, L. vaginalis, Streptococcus parasanguinis) and Veillonella parvula were dominant in patients with OS < 12 (Extended Figure 3C).
Figure 2. Akk relative abundance represents a prognostic marker of ICI.
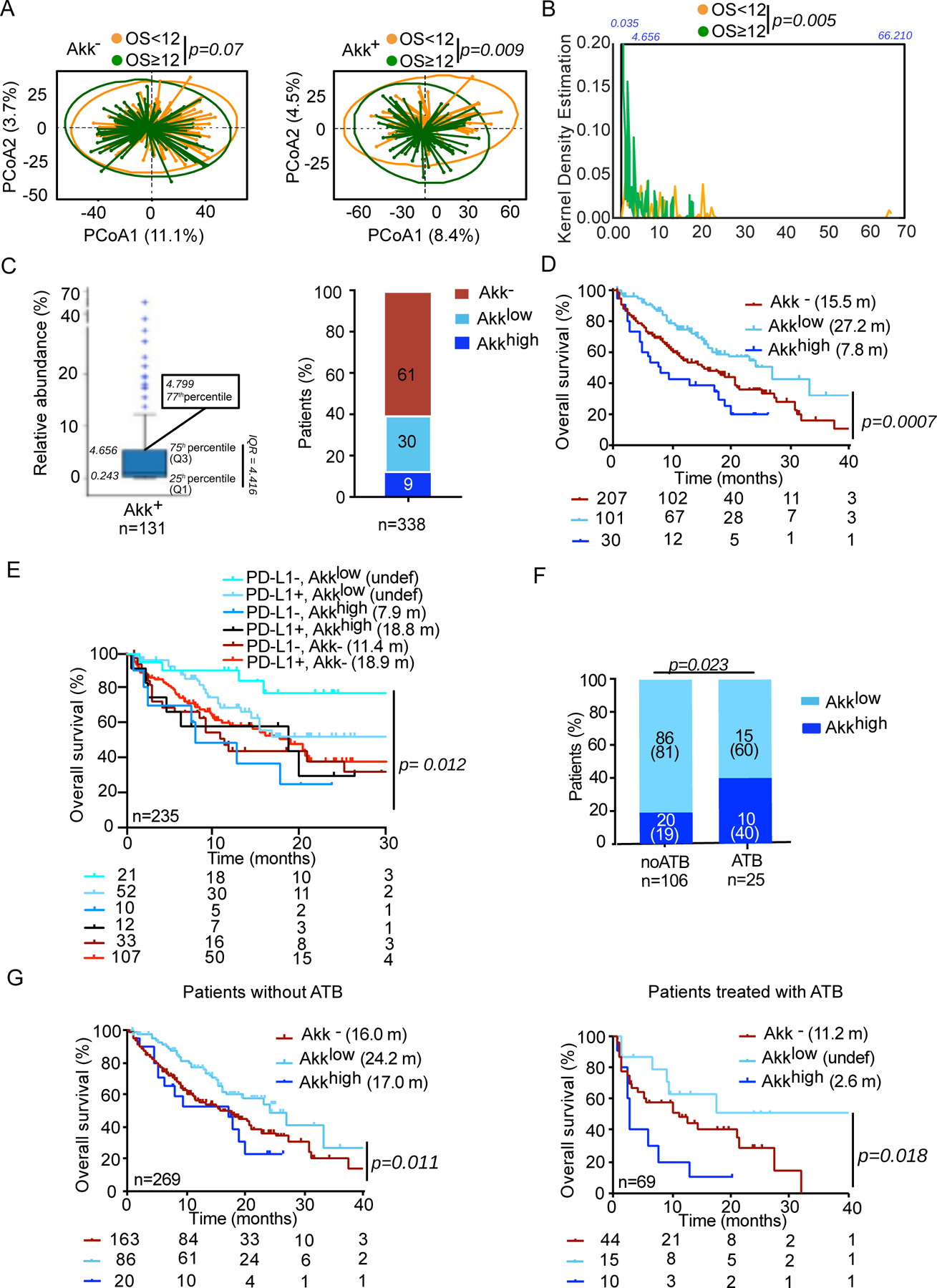
A. Beta-diversity measured by Bray-Curtis Index represented by Principal Coordinates analysis (PCoA) between Akk− versus Akk+ groups between OS < or ≥ 12 months within each subgroup. OS; overall survival. p-values were calculated using PERMANOVA with 999 permutations. B. Kernel density estimation aligning two variables, OS < or ≥ 12 and relative abundance of Akk in the entire cohort of 338 patients. C. Distribution of the relative abundance of Akk in patients with detectable Akk according to 77th percentile (left panel) and percentages of patients within each of the three groups of Akk relative abundance (right panel). Akk−: undetectable Akk, Akklow: Akk relative abundance between 0.035–4.799%, Akkhigh: >4.799% (77th percentile). D-E. Kaplan-Meier curve and Cox regression multivariate analysis of overall survival in 338 NSCLC patients according to Akk relative abundance segregated in 3 groups (Akk−, Akklow and Akkhigh) (D) and considering PD-L1 expression (E). F. Distribution of patients according to Akk relative abundance segregated in 2 groups (Akklow and Akkhigh) and ATB use (noATB: no exposure to ATB, ATB: antibiotics exposure within 2 months prior to ICI initiation). G. Kaplan-Meier curve and Cox regression multivariate analysis of overall survival in 338 NSCLC patients according to Akk relative abundance segregated in 3 groups (Akk−, Akklow and Akkhigh) and ATB use (noATB n=269, left panel and ATB n=69, right panel).
Taken together, these results indicate that the presence of Akk is associated with important, potentially prognosis-relevant shifts in the intestinal microbiota and tumor microenvironment of NSCLC patients.
Stratification of clinical outcome based on the relative abundance of Akk
We unexpectedly found an over-representation of Akk in patients with OS < 12 months within the Akk+ group, suggesting that the relative abundance of Akk may influence prognosis more than its absolute presence or absence. We next examined an ordinal rather than a categorical (Akk+ versus Akk−) variable to analyze the clinical significance of Akk. Indeed, the relative abundance of Akk within the Akk+ population ranged from 0.035% up to 66.210%. Using a Kernel density estimation of the relative abundance of Akk positionning patients with OS ≥ 12 or OS < 12, we noticed that patients harboring Akkhigh, at a relative abundance > 75th percentile (4.656%), did cluster within the OS < 12 months (Figure 2B–C left panel, p=0.005). To confirm this observation adjusting for the other risk factors, we used a supervised approach to statistically define an optimal cutoff for Akk to discriminate two hypothetical groups of Akk+ patients with different prognosis. We utilized a grid search algorithm based on the multivariable Cox model (adjusted for age, sex, ECOG performance status, ATB, histology, PD-L1 and line of treatment), with the Akaike information criterion as a performance index to determine this finest cutoff that corresponded to 4.799 (the 77th percentile of this cohort) (Figure 2C left panel). Among the whole cohort of 338 patients, we observed that only 9% of patients (23% of Akk+ subgroup) fell into the category of “relative overabundance” of Akk > 4.799 apostrophed “Akkhigh ” henceforth (Figure 2C right panel).
Moreover, we found a significant increase in Shannon diversity in Akklow compared to Akk− or Akkhigh specimens (Extended Figure 4A, p=0.00003), with the overall microbial community showing clear separation between Akk− versus Akklow as well as between Akklow versus Akkhigh patients (Extended Figure 4B–C, p=0.0001 and p=0.0003, respectively). Variable importance plot (VIP) discriminant analysis of taxonomic stool composition revealed that the ecosystem abnormally enriched in Akk (Akkhigh) was overrepresented by species of the genus Clostridium (Clostridium symbosium, C.innocuum, C. scindens, C. boltae, C. clostridioforme) at the expense of healthy commensals (F. prausntzii, C. comes, E. rectale, D. formicigenerans, D. longicatena, R. torques, Extended Figure 4D–E).
Kaplan Meier survival curves using the trichotomic stratification according to Akk relative abundance diverged (logrank test p=0.0007, Figure 2D, Table S4A–B), with a significantly longer median OS for Akklow patients compared to Akkhigh (27.2 months versus 7.8 months, Cox adjusted HR:0.38; 95%CI: 0.22–0.65, p=0.0005) and Akk− patients (27.2 months versus 15.5 months, adjusted HR:0.63; 95%CI:0.44–0.91, p=0.0150). The HR for the other risk factors considered in the multivariate Cox regression analysis are presented in Table 1 and Table S5. Not surprisingly, ECOG performance status ≥ 1 was another independent prognostic factor for this cohort31. The proportionality hazard assumption was questionable only for the ATB (Schoenfeld residuals trend test p=0.016), but no statistically significant interaction with time could be identified. In addition, ATB exposure was associated with shorter OS (in univariable analysis, p=0.009 (Table S4B, Table S5), and multivariate analysis, p=0.088 (Table 1, Table S5).
We also analyzed the interaction between Akk and PD-L1 in 235 advanced NSCLC patients with an available tumor expression of tumor PD-L1 (Table S1A, Extended Figure 1). We split the cohort into six groups according to PD-L1 expression and Akk fecal detection. The multivariate Cox regression analysis indicated that Akk dictated NSCLC prognosis, more than PD-L1 did (p=0.012 in 235 patients, Figure 2E).
Overall, we conclude that considering the trichotomic stratification of patients into Akk−, Akklow or Akkhigh individuals may be a more accurate independent prognostic factor of overall survival than the dichotomic (Akk− versus Akk+) division (likelihood ratio test of multivariable Cox models: p=0.0009). The presence of “normal levels” of Akk in the gut (Akklow) may be considered as a surrogate of host intestinal fitness. Akk overruled PD-L1 as a predictive biomarker of response to ICI in NSCLC patients.
The impact of ATB use on Akk relative abundance and survival outcome
Akkhigh levels as well as ATB exposure were considered standalone variables associated with shorter OS in NSCLC patients treated with ICI. Given these observations, we first combined the dichotomic classification of patients with respect to Akk (Akk− versus Akk+) with their history of prior antibiotic exposure to segregate patients into four groups. The Akk+ group without ATB exposure showed the strongest clinical benefit (median OS of 23.0 months) compared to the three other groups (Extended Figure 5A). The next favorable prognostic group fell into Akk− without ATB exposure, with a median OS of 16.0 months. In contrast, exposure to ATB showed the shortest OS (mOS around 9 months, Extended Figure 5A, p=0.017). Accordingly, ATB tended to reduce the alpha diversity of the Akk+ group (Extended Figure 5B–C). Moreover, ATB exposure enriched the Akk+ group in Gammaproteobacteria (E. coli), Clostridia class (Clostridium bolteae, Ruthenibacterium lactatiformens), and H2S producing bacteria (Bilophila wadsworthia) (Extended Figure 5D), as already described 37 at the expense of health-associated bacteria (C. aerofaciens, D. longicatena, D. formicigenerans, Eubacterium sp. CAG 38)17,43 also over-represented in the Akk+ group who did not take ATB (Extended Figure 5E).
In an attempt to establish an association between ATB use and the relative overabundance of Akk, we compared the percentages of Akkhigh stools in NSCLC patients with ATB use versus those that were ATB-free (Figure 2F) and performed a Kaplan Meier OS curve using the trichotomic classification (Akk−, Akklow versus Akkhigh) in two subgroups according to ATB exposure before the first administration of ICI (Figure 2G). We found a drastic increase of the proportion of Akkhigh patients among ATB users versus non users (40% versus 19%, p=0.023, Figure 2F). We confirmed that the overabundance of stool Akk at diagnosis in patients exposed to ATB was associated with lower overall survival despite ICI therapy (Extended Figure 5C, Figure 2G right panel). In ATB-free subjects, Akkhigh patients exhibited a reduced benefit to ICI compared with Akklow individuals but not worse than the Akkneg subgroup (Figure 2G left panel).
Altogether, these results demonstrate that ATB exposure is a negative predictor of survival to ICI, associated with overabundance of Akk (Akkhigh) and the relative dominance of Clostridium spp (C. bolteae, Lachnoclostridium).
Stratification of clinical outcome based on other components of the Akk - associated ecosystem
We used various statistical methods such as Linear discriminant analysis effect size (Extended Figure 3), Volcano plot from ANOVA model (Figure 3A), and MaAsLin multivariate statistical framework (Table S6) to compare taxonomic compositional variations between Akk+ versus Akk− stools in order to identify 16 bacterial species, among which 14 were positively and 2 were negatively associated with Akk fecal prevalence independently of other confounding factors such as age, gender, BMI, lines of therapy (Figure 3A, Table S6). Next, we performed Cox logistic regression multivariate analyses to determine the impact of each bacterium on patient clinical outcome using a trichotomic distribution of their respective relative abundances (according to a cut-off defined above). Apart from Akk, very few bacteria (except Eubacterium hallii, Bifidobacterium adolescentis, Parasutterella excrementihominis, Intestinimonas butyriciproducens, or C. innocuum already described to mediate immunomodulatory functions43–45 were also associated with prolonged (Figure 3B, Figure 3D) or reduced survival following ICI therapy (Figure 3F, Table S6) with a “linear dose” response in contrast to what was observed for Akk. There was an added value to consider the coordinated presence of both, Akk as well as E. hallii or B. adolescentis, to accurately predict ORR and/or mOS in this cohort of 338 NSCLC patients (Figure 3C,E). Hence, these findings suggest that Akk and its collateral commensalism may participate in the ICI-mediated clinical benefit of patients.
Figure 3. Stratification of clinical outcome based on other components of the Akk - associated ecosystem.
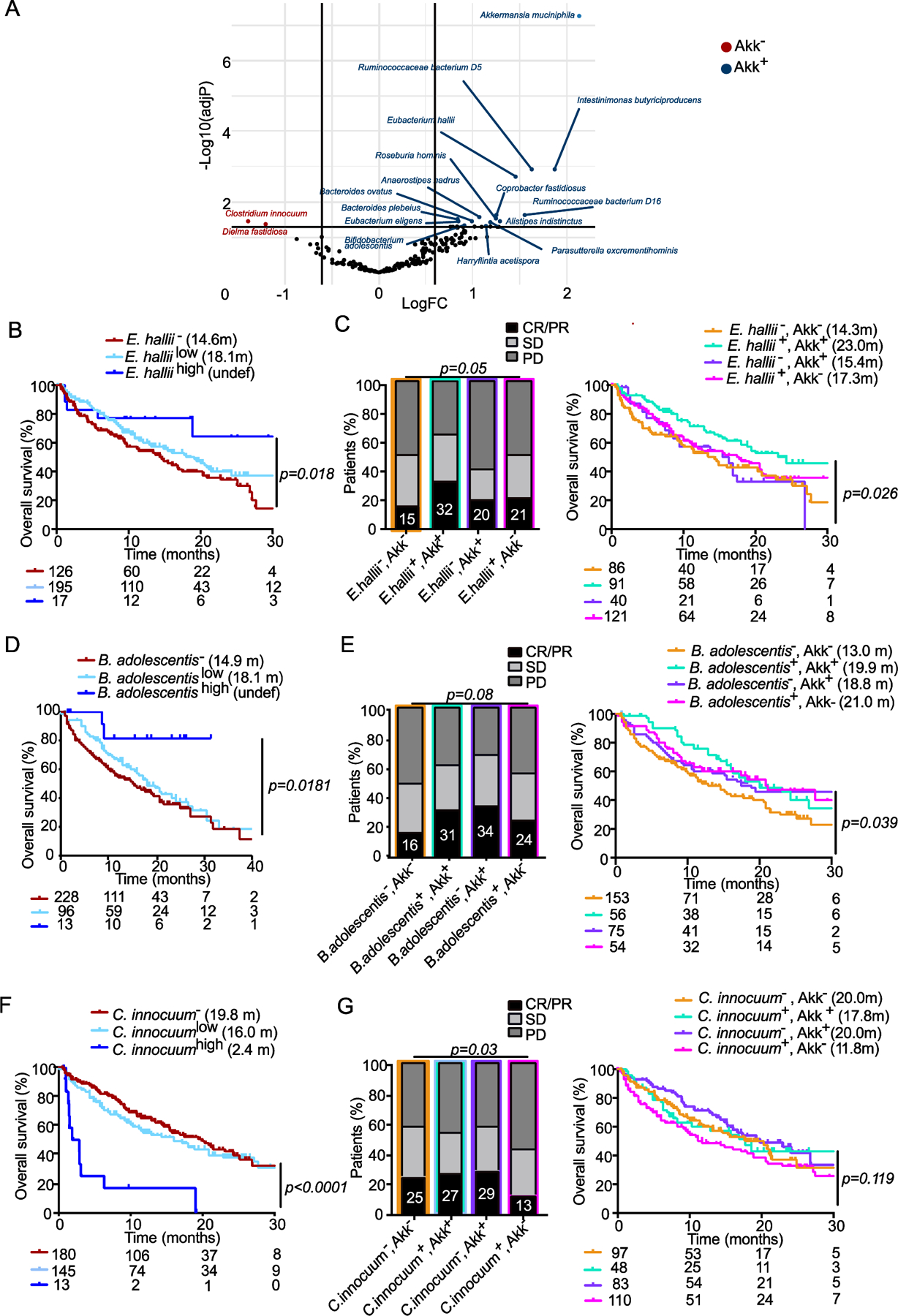
A. Volcano plot (indicating Fold Change (FC) and p-values in ANOVA statistical analyses) to segregate taxonomic species (with a prevalence> 2.5%) according to their relative abundance in baseline fecal specimen of 338 patients based on their association with Akk: species significantly associated with or excluded from Akk− enriched ecosystems (Akk+, blue dots, Akk−, red dots) B, D, F. Kaplan Meier overall survival curves in 338 NSCLC patients according to the trichotomic distribution of the relative abundance of beneficial or harmful bacteria (undetectable bacterium: −, low versus high) retained in the LEfSe model, MaAsLin2 and the Volcano plot/ANOVA Table S6). C, E, G. Influence of collateral bacteria associated with Akk (retained in Table S6) in the Akk-associated impact on ORR and OS in a dichotomic pattern (presence/absence) using Chi-square test (for ORR, left panels) and Cox regression multivariate analysis for Kaplan Meier curves (right panels). CR; complete response. PR; partial response, SD; stable disease, PD; progressive disease.
Akk rescued response to PD-1 blockade by shifting the microbiome towards species associated with good prognosis
Trying to establish a link between Akk and/or its collateral ecosystem and patient clinical outcome, we turned to our microbiota humanized avatar tumor bearing mouse models described above. As depicted and commented above (Extended Figure 2C–E), mouse resistance to anti-PD-1 antibodies was associated with FMT bereft of Akk. Next, we analyzed the compensatory effects brought up by oral supplementation with an immunogenic strain of Akk, Akkp226118 in both groups of animals, receiving Akk+ versus Akk− FMT. Mice reared in specific pathogen-free conditions (SPF), indicated as FMT-in Extended Figure 6A–B) and inoculated with the same tumor cell line were used as controls of “murine eubiosis”. Indeed, we could restore responsiveness to PD-1 blockade, only in the setting of Akk− FMT (Extended Figure 6B–C). Next, the objective was to analyze whether oral supplementation with Akkp2261 would change the host microbiome of the recipient mice and whether the remodeling of the host ecosystem would correlate with preclinical benefit to exogenous Akkp2261. For this purpose, we concatenated all tumor models syngeneic of BALB/c (CT26, 4T1) and C57BL/6 (B16F10, MCA-205-, MC38) mice that were first transferred with stools from 29 individual NSCLC patients (Table S2) and then treated with anti-PD-1 antibodies (Extended Figure 6A) and collected recipient feces pre-and post-oral feeding with Akkp2261. The non-supervised hierarchical clustering of the ratios between the tumor growth kinetics with anti-PD-1 antibodies with or without exogenous Akkp2261 normalized on the effect of PD-1 blockade in eubiotic conditions (SPF versus FMT) revealed that exogenous Akkp2261 was effective in about 50% cases (Extended Figure 6D, Extended Figure 6E left panel). We detected taxonomic differences in the recipient microbiota in responders (R) versus non responders (NR) to exogenous Akkp2261 (Extended Figure 6E right panel). R mice harbored a relative overrepresentation of metagenomics species annotated in Figure 3A in humans harboring Akk+ stools namely Intestinimonas butyriciproducens, Parasuterrella excrementihominis compared with NR mice, that in contrast, tended to be enriched in Bacteroides spp (Extended Figure 6E right panel).
Altogether, avatar mice transferred with Akk− human fecal material exhibited a phenotype of tumor resistance to PD-1 blockade but were rescued by Akkp2261 when Akkp2261 could shift the microbiome towards the favorable Akk associated collateral ecosystem.
DISCUSSION
Here, we report the results of a prospective, multicentric study based on the profiling of the gut microbiota of patients with advanced NSCLC treated with PD-1 blockade. The relative abundance of Akk was associated with clinical benefit, defined by an increase in ORR and survival, taking into accounting the main microbiota-relevant confounding factors (age, gender, BMI, lines of therapy). The prognostic significance of this gut bacterium was validated by multivariate analyses and interaction studies indicating that Akk is markedly associated to the prognosis of advanced NSCLC treated with ICI, independently from age, gender, ECOG PS, ATB use and PD-L1. The intestinal residence of Akk was a proxy of richness of the gut ecosystem, as shown by the association of Akk at a relative abundance within the 77th percentile (Akklow < 4.799%), with stool alpha diversity (Shannon diversity index). These results expand on previous observations that have been made in smaller cohorts of patients with NSCLC 18,41 and provide evidence that gut microbiome diversity and composition, specifically the relative abundance of Akk, offer relevant information to predict survival of patients with NSCLC amenable to ICI.
Our study is the largest metagenomics prospective analysis that attempted to validate Akk as a new prognostic factor for ICI. Our study meet the pre-specified criterium of statistical significance which was set at 10% ORR increase between Akk− and Akk+ patients when considering all (mostly 2L) 338 NSCLC patients (from 18.2% to 28.2% in ORR, Figure 1A). Moreover, we also observed a > 10% ORR increase between Akk− and Akk+ (from 19% to 41% ORR, Figure 1B) with a mOS advantage in 1L NSCLC patients (Figure 1E). Moreover, our study linked the gut microbiota composition to the tumor microenvironmental landscape, highlighting increased transcription of gene products of the adaptive immunity and the IFN fingerprint. Finally, we validated in mice that Akk− stools confered resistance to PD-1 blockade.
In addition to prospectively validating the hypothesis in a larger and homogeneous cohort, we report that Akk was associated not only with increased alpha diversity but also with a distinct bacterial community associated with a health or immunogenic status represented by Ruminococcacae (Faecalibacterium prausnitzii, R. lactaris) and Lachnospiraceae (Dorea formicigenerans & D. longicatena, Eubacterium rectale & E. hallii, Roseburia faecis & R. intestinalis) family members as well as Bifidobacterium adolescentis, Intestinimonas butyricyproducens and others. These findings reconcile the results across several works, geographical distributions and sequencing technologies since Faecalibacterium, Ruminococcus and Bifidobacterium were previously reported to be enriched in North-American, Japanese and South Korean patients with melanoma and NSCLC cancers and have favorable outcome 17,20,41,44. Moreover, even in avatar gut humanized mouse models, responders to exogenous Akkp2261 shifted their microbiome towards an over-representation of some of the above mentionned species belonging to the Akk+ ecosystem.
Confirming the clinical significance of bacterial diversity and commensals associated with responses, our results validate the growing body of evidence linking ATB use and poor clinical outcome 23,45. In addition to depleting favorable genera associated with survival (such as Ruminococcus)41, ATB use tends to deviate the gut microbiome composition towards harmful bacteria previously associated with proinflammatory or immunoregulatory pathways (such as E. coli, and Clostridium bolteae) 46, supporting previous findings in renal cell carcinomas amenable to ICI 37. Surprisingly, in addition to acting as an independent negative prognostic factor, ATB promoted the overabundance of Akk (Akkhigh) above the 77th percentile level associated with poor prognosis. Indeed, ATB use doubled the proportion of individuals presenting a stool Akkhigh phenotype. This phenotypic trait of overabundance of Akk > 4.799 was associated with a dominance of the Clostridium species (C. bolteae, C. innocuum, C. asparagiforme, C. scindens, C. symbosium) belonging to clusters IV and XIVa of the genus Clostridium, known to maintain IL-10 producing Treg in colonic lamina propria47.
Aside from ATB use, overabundance of Akk > 4.799 (Akkhigh) was associated with a shorter overall survival than “normal” relative abundance of Akk < 4.799, possibly reflecting an underlying pathophysiological disorder of the intestinal barrier in these advanced cancer patients. High relative proportions or subdominance of A. muciniphila in the ecosystem has been associated with pathophysiological failures (such as anorexia nervosa48, GVHD33, Aging49, dysmetabolism50, HIV infection 51, pathobionts52 or liver injury53). Hence, in the context of gut injury by, or conducive to, ATB use, Akk might constitute a biomarker of ongoing but imperfect intestinal repair. Of note, we failed to observe a similar trichotomic distribution correlating with opposite clinical outcome investigating other bacteria (Figure 3).
Hence, we conclude that Akk relative abundance could represent a reliable biomarker of favorable or dismal prognosis for patients receiving immunotherapy with PD-1 blockade. It may be of utmost importance to risk-stratified I-O patients based on shot-gun metagenomics (rather than by 16S rRNA) sequencing to precisely quantify the relative abundance of Akk in addition to ATB use, and PD-L1 expression in prospective trials including NSCLC patients and designed to discover optimal biomarkers.
Therapeutic strategies modulating the microbiome such as FMT or commensals are currently being evaluated to boost ICI responses or circumvent primary resistance to ICI, though without patient stratification based on their degree of dysbiosis54. Here, we provide preclinical data suggesting that Akk could therapeutically bypass the resistance to anti-PD-1 blockade conferred by FMT bereft of endogenous Akk (Extended Figure 6 and17,18). The oral supplementation with exogenous Akk (Akkp2261) was particularly effective when Akk shifted the recipient microbiome towards a healthy status (Extended Figure 6D–E). A recent preclinical study indicates that the anticancer effects of recombinant interleukin-2 could be improved by Akk in a Toll-like receptor-2 dependent fashion55. In patients, food supplementation with Akk is safe and reduces insulinoresistance and dyslipidemia in the context of pre-diabetes56,57. Altogether, we surmise that therapeutic supplementation with a lyophilized encapsulated Akkp2261 would benefit the subgroups of patients not exposed to ATB, and devoid of endogenous Akk. In contrast, complex polymicrobial consortia or fecal microbial transplantation may be best suited for patients with prior ATB exposure.
The limitations of this study are the following ones. First, it did not evaluate the predictive value of Akk in 1L chemo-immunotherapy combinations (only considering 1L IO presenting with PD-L1 >50% tumor expression). Second, it could not consider the interactions between the tumor mutational burden and stool Akk prevalence. Moreover, this study failed to investigate the functional and clinical relevance of more rare Akkermansia spp.58 different from the SGB9226 monitored in this study using MetaPhlAn 3 pipeline with a custom marker database (see Methods). Nevertheless, our study provides a strong rationale for the development of diagnostic tools assessing gut dysbiosis for routine oncological management, as well as framework for the design of microbiota-centered interventions to circumvent primary resistance to ICI in patients with NSCLC.
METHODS
Study design and treatment
Patients eligibility.
The ONCOBIOTICS trial (NCT04567446), a multicentric prospective observational study was designed to evaluate the impact of the microbiome composition in the clinical outcome of patients with advanced NSCLC treated with anti-PD-(L)1. We enrolled patients across 12 academic centers in France and two centers in Canada. Adult patients with pathologically confirmed advanced non-squamous or squamous NSCLC and an Eastern Cooperative Oncology Group (ECOG) performance-status score of 0–2, amenable to ICI as standard-of-care and compelling to provide a stool sample were eligible. Eligible patients received ICI following progression on platinum-based chemotherapy regimens, either with nivolumab or atezolizumab regardless of PD-L1 expression or with pembrolizumab if PD-L1 ≥ 1%. Given the subsequent approval of first-line ICI in the first-line setting during the study accrual period, patients who received pembrolizumab monotherapy or in combination with platinum-based chemotherapy, depending on PD-L1 expression were also included. Standard-of- are treatment was continued until disease progression, unacceptable adverse effects, or completion as per protocol (2 years of ICI). Full eligibility criteria are listed in the trial protocol (available at NCT04567446). Baseline characteristics including detailed listing of concurrent medications received the last two months prior to ICI initiation, and date of last follow-up were entered at each center in an electronic case report form.
Hypothesis.
Sample size calculation was performed based on the primary end-point defined as investigator-assessed ORR from the hypothesis proposed in Routy et al18 that in a population with metagenomics detectable Akk (Akk+) in the gut microbiome, the response rate would be higher than in the population with undetectable Akk (Akk−).We considered that a meaningful clinical difference would correlate to an ORR incremental from 10% in the Akk− to 20% in the Akk+ group. Given the superiority hypothesis, power was set at 80% with a two-sided alpha level of 5%, using EAST® program. Hence, we determined that at least 292 patients—with equal distribution of 146 patients in both Akk+ and Akk− groups –would be necessary to confirm our primary objective.
Study end-point and assessments.
Computed tomography (CT) scans were performed at baseline and every 8–12 weeks for the first year and every 12–15 weeks thereafter until disease progression. Tumor response was assessed using the Response Evaluation Criteria in Solid Tumors version (RECIST) 1.159. The primary end-point was investigator-assessed objective response rate (ORR) which was defined as the number and percentage of patients with a Best Overall Response (BOR) of confirmed complete response (CR) or partial response (PR). Best overall response (BOR) was defined as the best response designation, recorded between the date of first treatment dose and the date of the initial objectively documented tumor progression per RECIST v1.1 or the date of subsequent therapy, whichever occurs first. For patients without documented progression or subsequent therapy, all available response designations contributed to the BOR determination. Secondary end-points included overall survival (OS), and microbiome variables such as alpha and beta diversity and differential abundance analyses at the genus-level. Overall survival was defined as the time from trial inclusion until death from any cause. The follow-up of patients alive at the database lock was censored to the date of last record of contact.
Treatment modalities
Treatment modalities: the number of Pembrolizumab (every other 21 days) or Nivolumab (every other 15 days) or Atezolizumab (every other 21 days) injections received was 4+/− 2 at 8–12 weeks and was 20+/− 4 at 12 months.
Ethical issues.
The ancillary studies have been designed according to an IRB approved-study (Oncobiotics* Sponsor Protocol N : CSET 2017/2619, ID-RCB N : 2017-A02010–53 https://clinicaltrials.gov/ct2/show/NCT04567446). The trial was conducted in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki. All patients provided written informed consent. General Data Protection Regulation procedures and anonymization rules have been applied according to Oncobiome H2020 model system already in place in the ClinicoBiome, Gustave Roussy. All data and sample collection were performed in compliance with regulatory, ethical, and European GDPR requirements.
Human stool samples and metagenomics analyses
Fecal samples were prospectively collected at different time points (V1: pre-ICI, V2: before the second ICI injection, V3: at 3 months post-ICI and V4: at 6 months post-ICI) at each center following the International Human Microbiome Standards (IHMS) guidelines, but only the baseline V1 sample was considered for this analysis. Indeed, patients were included in the analysis only if they provided a V1 sample, or for patients where such timely collection was not feasible, V2 samples were also considered as “baseline” as in Routy et al. Science 2018. Of note, the 60 patients with NSCLC already described in our previous report18 were excluded from the principal analysis. For metagenomic analysis, the stools were processed for total DNA extraction and sequencing with Ion Proton technology following MetaGenoPolis (INRA) France, as previously reported18,60,61. Cleaning, filtering and classification of reads were performed with two different pipelines: MetaOMineR and MetaPhlAn 362. In order to determine Akkermansia muciniphila presence/absence, we used a total of 463 genetic markers identified from four Akkermansia candidate species-level genome bins (SGBs) (SGB9223 – 38 markers, SGB9224 – 54 markers, SGB9226 – 171 markers and SGB9228 – 200 markers)35 in MetaPhlAn. As outlined in the M&M Table 1, the type of strain of A. muciniphila (MucT) delineated as SGB9226, was the most prevalent species in our cohort (>80% of Akkermansia positive subset) and was therefore used for the calculation of the relative abundance of Akk in the main figures of this article.
We found msp_0025 to correspond to SGB9226, and used its relative abundance values as a proxy for A. muciniphila in MetaOMineR. A full description of both DNA purification and metagenomic pipelines is available in Derosa et al 37. Starting from abundance matrices, only taxa that were present in at least 2.5% of all samples were considered, and then raw data were normalized and standardized (Sci-Kit-learn version 0.20.3).
Tumor RNA sequencing
Using previously published technique63 total RNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumors from patients with advanced NSCLC included in the main analysis as well as from patients with limited stages (baseline characteristics included in Table S4). Libraries were prepared from 12 μl of total RNA with the TruSeq Stranded Total RNA using Ribo-Zero (Illumina) following manufacturer instructions. BBMAP v38.87 was used to trim the sequencing adapters and filtered the low quality and too short reads. Kallisto software64 was used for quantifying transcript abundance from RNA-seq data against GRCh38 cDNA reference transcriptome from the Ensembl database, release 101. Only protein-coding transcripts and genes were included in the downstream analysis. Transcript Per Million values have been used for downstream analysis. Mann-Whitney tests have been performed to compare gene expression according to Akkermansia groups. PERMANOVA test with Euclidian distance has been used to assess the difference between groups on the subset of differentially expressed genes.
Pre-clinical study details:
Mice.
All animal experiments were carried out in compliance with French and European laws and regulations. The local institutional animal ethics board and French Ministère de la Recherche approved all mice experiments (permission numbers: 2016–049-4646, 2018–020-510263031v3). Mice avatar studies have been approved by the regulatory animal facility local and national committees and the French Ministry (Everimmune #13366–2018020510263031 v3, APAFIS#17530–201811413352738 v2 (03/2019–03/2024). APAFIS# 21378–201907080848483459). Female C57Bl/6 and BALB/c were purchased from Harlan (France) and Janvier (France), respectively. Mice were used between 8 and 16 weeks of age housed in specific pathogen-free conditions (SPF). All mouse experiments were performed at the animal facility in Gustave Roussy Cancer Campus where animals were housed in specific pathogen-free conditions.
Cell culture, reagents and tumor cell lines.
MC38, MCA-205 and B16F10 (syngeneic from C57BL/6 mice), and 4T1 cell lines (syngeneic from BALB/c mice) were purchased from ATCC. 4T1, MCA-205 and MC38 cells were cultured in RPMI 1640 containing 10% FCS, 2mM L-glutamine, 100 UI/ml penicillin/streptomycin, 1 mM sodium pyruvate and MEM non-essential amino. All reagents were purchased from Gibco-Invitrogen (Carlsbad, CA, USA). B16F10 and CT26 cells were cultured in DMEM containing containing 10% FCS, + 100 UI/ml penicillin/streptomycin + non-essential amino acid. All cell lines were cultured at 37°C with 5% CO2 and regularly tested to be free of mycoplasma contamination.
Subcutaneous model of MCA-205, MC38 and B16F10 and 4T1.
Syngeneic C57BL/6 mice were respectively implanted with 0.8 × 106 MCA-205, 1.0 × 106 MC38/CT26 or 3 × 105 B16F10 cells subcutaneously. Syngeneic BALB/c mice were implanted with 3 × 105 4T1 cells subcutaneously. For tumor growth experiments, tumor-implanted mice were treated intraperitoneally (i.p.) when tumors reached 20 to 40 mm² in size with anti-PD-1 mAbs (250µg/mouse; clone RMP1–14) or isotype control (clone 2A3). Mice were injected 4 times at 3-day intervals with anti-PD-1 mAbs. Tumor length and width were routinely monitored every 3 times per week by means of a caliper. All antibodies were purchased from BioXcell, NH, US.
Antibiotic treatments.
Mice were treated with an antibiotic solution (ATB) containing ampicillin (1 mg/ml), streptomycin (5mg/ml), and colistin (1 mg/ml) (Sigma-Aldrich) added in the drinking water of mice. Antibiotic activity was confirmed by cultivating fecal pellets resuspended in BHI+15% glycerol at 0.1 g/ml on COS (Columbia Agar with 5% Sheep Blood) plates for 48h at 37°C in aerobic and anaerobic conditions. In brief, in the context of fecal microbial transplantation experiments, mice received 3 days of ATB before undergoing fecal microbial transplantation the next day by oral gavage using animal feeding needles.
FMT experiments.
Fecal microbiota transfer (FMT) was performed by thawing fecal material. Two hundred µL of the suspension was then transferred by oral gavage into ATB pre-treated recipient (as described above). In addition, another 100µL was applied on the fur of each animal. Two weeks after FMT, tumor cells were injected subcutaneously and mice were treated with anti-PD-1 mAbs or isotype control as previously explained.
We used MCA-205 fibrosarcomas because it is normally -in SPF eubiotic mice-sensitive to anti-PD-1 Ab and has been used as a reference mouse model in our previous avatar experiments reported in 18 and 37, both papers showing that results obtained with MCA-205 were recapitulated in orthotopic TC1 lung cancer or RENCA RENal CArcinoma models, respectively. So, we can trust the biological relevance and suitability of this MCA-205 model system to probe FMT or taxonomic fecal composition in future experiments.
Detailed experimental setting of the murine meta-analysis (Extended Figure 3E–F).
We conducted 29 individual experiments over 2 years comprising 6–8 groups (including 6 mice/group) where the growth kinetics of orthotopic MCA-205 sarcomas (and other tumors such as MC38 colon cancer (syngeneic from C57BL/6 mice), or 4T1 breast or CT26 colon tumors (syngeneic from BALB/c mice) or B16 (melanoma) were monitored in avatar mouse models 17.
These recipients were ATB-treated and then received fecal microbial transplants (FMT) from 26 different NSCLC patients (phenotype of “Responder, R “versus “Non Responder, NR” to PD1 blockade, and relative abundance of stool Akk, both described in Table S5). Then, the mice were implanted with syngeneic tumors and later subjected to immunotherapy with anti-PD-1 antibodies. FMT could confer sensitivity (when stools were Akk+) or resistance (when stools were Akk neg) to anti-PD-1 Ab, when compared with animals in eubiosis reared in SPF conditions (without FMT). Next, we analyzed the benefit and compensatory effects of oral supplementation of Akkp2261, according to stool Akk +/− in the original FMT. Of note, exogenous Akkp2261 was not detectable more than 30 hrs in the recipient intestines (as shown in qRT-PCR using A. muciniphila specific probe sets). To better scrutinize whether the exogenous Akkp2261 could shift the microbiome of the recipient tumor bearers differently in R versus NR mice, we concatanated all tumor models syngeneic of BALB/c mice (CT26, 4T1) and C57BL/6 mice (B16F10, MCA-205, MC38) that were treated by FMT from 29 different NSCLC patients and then treated with anti-PD-1 mAbs +/− Akkp2261 prior to stool harvesting for 16S rRNA based-sequencing of fecal amplicons. We calculated the relative benefit of exogenous Akk by dividing the ratio of tumor size between anti-PD-1+ Akkp2261/anti-PD-1+ water ) normalized on the ratio of tumor sizes between anti-PD-1/Ctl mAbs in SPF mice for each time points for the whole kinetics. The non-supervised hierarchical clustering of the ratios between the tumor growth kinetics with anti-PD-1 mAbs with or without exogenous Akkp2261 normalized on the effect of PD-1 blockade in eubiotic conditions revealed that exogenous Akkp2261 was effective in about 50% cases (heatmap in the Extended Figure S3E). To delineate which Akk-associated ecosystem was associated with responses (ratios> mean of the cohort), LEfSe was used to identify the taxonomic changes segregating R versus NR to exogenous Akk. These species were compared with the bacteria featuring in human stools described in Extended Figure 1 and Figure 3A of the revised manuscript.
Gut colonization with Akk CSUR p2261 Akk CSUR p2261
Gut colonization with Akk CSUR p2261 Akk CSUR p2261 was provided by the Institut hospitalo-universitaire Méditerranée Infection, Marseille, France. Akkp2261 was grown on 5% sheep blood enriched Columbia agar (COS) plates in an anaerobic atmosphere created using 3 anaerobic generators (BioMerieux) at 37°C for at least 72h. Itentification of the bacterium was performed using a Matrix-Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) mass spectrometer (Microflex LT analyser, Bruker Daltonics, Germany). Colonization of ATB pre-treated mice was performed by oral gavage with 100 µl of suspension containing 1 × 108 bacteria obtained from a suspensions of 109 CFU/mL using a fluorescence spectrophotometer (Eppendorf) at an optical density of 600 nm in PBS. Five bacterial gavages were performed for each mouse: the first 24h before the first injection of anti-PD-1 mAbs and, subsequently, four times on the same day of ICI.
Mouse fecal DNA extraction and microbiota characterization.
Feces were harvested in each mouse and group for metagenomics between 7 and 14 days after start of immunotherapy. Samples were stored at −80°C until processing. Preparation and sequencing of mouse fecal samples was performed at IHU Méditerranée Infection, Marseille, France. Briefly, DNA was extracted using two protocols. The first protocol consisted of physical and chemical lysis, using glass powder and proteinase K respectively, then processing using the Macherey-Nagel DNA Tissue extraction kit (Duren, Germany). The second protocol was identical to the first protocol, with the addition of glycoprotein lysis and deglycosylation steps. The resulting DNA was sequenced, targeting the V3–V4 regions of the 16S rRNA gene. Raw FASTQ files were analyzed with Mothur pipeline v.1.39.5 for quality check and filtering (sequencing errors, chimerae) on a Workstation DELL T7910 (Round Rock, Texas, United States). Raw reads were filtered and clustered into Operational Taxonomic Units (OTUs), followed by elimination of low-populated OTUs (till 5 reads) and by de novo OTU picking at 97% pair-wise identity using standardized parameters and SILVA rDNA Database v.1.19 for alignment. A prevalence threshold of ≥ 2.5% was implemented for statistical analyses on recognized OTUs, performed with Python v3.8.2. The most representative and abundant read within each OTU (as evidenced in the previous step with Mothur v.1.39.5) underwent a nucleotide Blast using the National Center for Biotechnology Information (NCBI) Blast software (ncbi-blast-2.9.0) and the latest NCBI 16S Microbial Database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/). A matrix of bacterial relative abundances was built at each taxon level (phylum, class, order, family, genus, species) for subsequent multivariate statistical analyses.
Statistical analyses
In humans, data matrices were firstly normalized then standardized using QuantileTransformer and StandardScaler methods from Sci-Kit learn package v0.20.3. Normalization using the output_distribution=‘normal’ option transforms each variable to a strictly Gaussian-shaped distribution, whilst the standardization results in each normalized variable having a mean of zero and variance of one. These two steps of normalization followed by standardization ensure the proper comparison of variables with different dynamic ranges, such as bacterial relative abundances. For microbiota analysis, measurements of α diversity (within sample diversity) such as Richness and Shannon index, were calculated at species level using the SciKit-learn package v.0.4.1. Exploratory analysis of β-diversity (between sample diversity) was calculated using the Bray-Curtis measure of dissimilarity and represented in Principal Coordinate Analyses (PCoA), along with methods to compare groups of multivariate sample units (analysis of similarities - ANOSIM, permutational multivariate analysis of variance - PERMANOVA) to assess significance in data points clustering. ANOSIM and PERMANOVA were automatically calculated after 999 permutations, as implemented in SciKit-learn package v0.4.1. We implemented Partial Least Square Discriminant Analysis (PLS-DA) and the subsequent Variable Importance Plot (VIP) as a supervised analysis wherein the VIP values (order of magnitude) are used to identify the most discriminant bacterial species. All the analyses were performed within a Python v3.8.2 environment. Univariate differential abundance analysis was performed via linear discriminant analysis of effect size (LEfSe)65. We added further support of differentially abundant species using two different multivariate differential abundance methods; ANCOM-BC66 and MaAsLin267, that included covariates such as age, sex, BMI, cohort and sequencing batch. France’s Data Protection Article 8 legislation (Commission Nationale Informatique et Libertés [CNIL]) prohibits the analysis of the racial and ethnic origins. Raw sequencing counts were estimated from species-level MetaPhlAn 3 relative abundances by multiplying these values by the total number of reads for each sample and these were used in ANCOM-BC (v.1.0.1) with default parameters, a library size cutoff of 500 reads and no structural zero detection. Masslin2 (v.1.4.0) was run using Logit transformed relative abundances that were normalized with total-sum-scaling (TSS) and using the variable of interest as a fixed effect.
Survival curves were estimated using the Kaplan-Meier method and compared with the log-rank test (Mantel-Cox method) in a univariate analysis. Multivariate analyses were performed using Cox regression models to determine HRs and 95% confidence intervals (CIs) for OS adjusting for other clinicopathologic features. The proportionality hazard assumption was checked testing the trend of the Schoenfeld residuals with the cox.zph R function.When the test was statistically significant for a variable, its interaction with time was introduced in the model and tested using the tt (time transformation) function with different functional forms (linear, exponential, logarithmic, and penalized spline). The optimal cutoff for each bacterial species to define different prognosis groups was obtained with grid search algorithm based on the multivariate Cox model to take into account the potential confounding factors (age, sexe,…). The grid was defined for each species by the percentiles of the distribution of the non-zero prevalence values. The cutoff corresponding to the model with the better Akaike information criterion (AIC, lower is better) was selected as the optimal cutoff.
All tests were two-sided and statistical significance was set at a p-value <0.05. Statistical analyses were conducted using the GraphPad Prism 7 and R software (http://www.R-project.org/).
In mice, all tumor growth curves were analyzed using software developed in Professor Guido Kroemer’s laboratory and information about statistical analyses can be found at this following link: https://kroemerlab.shinyapps.io/TumGrowth/. Between-group comparisons of mice, global comparison were performed using Kruskall-Wallis test, post-hoc multiple comparisons using Dunn’s test.
Finally, natural tumor growth data deriving fom mice experiments (at least 6 mice per experiment) were averaged for each timepoint (T0 to T8), then longitudinally normalized on the first timepoint, in order to have a common starting value of 1. All averaged and normalized tumor values were then expressed with Fold Ratios (FR, as reported in Extended Figure 3E), and underwent base 2 logaritmization in order to enhance augmentation (red) or diminution (blue) of the tumor growth. A Hierarchical Clusterization Analysis (HCA) based on Bray-Curtis distance was implemented on longitudinal FR data in order to define a branch of responder (R) to Akkp2261(green cluster), or a group of non-responder (NR) to Akkp2261 (red cluster). All reported p-values underwent Benjamini-Hochberg two-stages False Discovery Rate (FDR) at 10%.
Extended Data
Extended Data Fig. 1:
Consort diagram describing the stool collection in the whole NSCLC ONCOBIOTICS cohort.
Extended Data Fig. 2:
MetaOMineR-based analysis of the association between stool A. muciniphila (Akk) and clinical benefit to ICI in patients.
A-B. Correlations between stool prevalence of Akk (MetaOMineR pipeline) and ORR (A) or OS (B) in 1+2L NSCLC patients (N=338, A-B) based on MGS identification of Akk in the MetaOMineR algorithm (INRAE). Chi-square test (A) and Cox regression analysis for median overall survival (OS) depicted in Kaplan Meier curves according to detectable or undetectable Akk (Akk+ or Akk) analyzed in 2 groups (B, left panel) or segregated in 3 groups (Akk−, Akklow and Akkhigh) (B, right panel). Chi-square test P-values are two-sided, with no adjustments made for multiple comparisons (A). The Akk status was compared using the stratified log-rank test. P-values are one-sided with no adjustment (B). C. Experimental setting of avatar mice. FMT of NSCLC patients (Supplementary Table S3) segregated according to the presence or absence of Akk into MCA-205 tumor bearing C57BL/6 mice. Treatments are indicated by arrows (ATB, FMT, anti-PD-1 (ICI) mAbs, or isotype control mAbs (Iso)). D. MCA-205 tumor growth kinetics in each group of FMT according to the prevalence of Akk. in isotype Ctl versus anti-PD-1 mAbs treated mice. Data are presented as mean values +/−SEM of tumor sizes within 6 animals/group. Concatenation of at least n=8 experiments (using a different stool of NSCLC patient) containing 6 mice/group. Tumor sizes according to FMT Akk− (D, left panel) versus Akk+ (D, right panel) are depicted, each dot representing one mouse. Statistics were mixed-effect modeling with specific software ((https://kroemerlab.shinyapps.io/TumGrowth/) for longitudinal tumor growth analysis. P-value are indicated. E. Percentages of responding mice (tumor reduction of>25 % compared with means of controls in the anti-PD-1 mAbs -treated group) and patients (ORR) in each category of stools used for FMT (derived from patients in Supplementary Table S3). CR; complete response. PR; partial response, SD; stable disease, PD; progressive disease.
Extended Data Fig. 3:
Metagenomic species characterizing Akk+ stools and patient survival.
Shannon diversity index representing stool alpha diversity in Akk+ and Akk− groups of fecal specimens (N=338) (A, upper panel). Beta-diversity measured by Bray-Curtis Index represented by Principal Coordinates analysis (PCoA) between Akk+ versus Akk− groups in the whole cohort of 1+2L (A, lower panel). p-values were calculated using PERMANOVA with 999 permutations. The lower and the upper hinges of boxplots corresponds to the 25th and 75th percentiles, respectively. The midline is the median. The upper and lower whiskers extend from the hinges to the largest (or smallest) value no further than 1.5 interquartile range from the hinge, defined as the distance between the 25th and 75th percentiles. P-values were calculated testing the null hypothesis and using a two-sided test. Exact p-value: 3.84573e-05. B-C. Differential abundance of metagenomic species measured by linear discriminant analysis of effect size (LEfSe) according to the presence of A. muciniphila (Akk) (B) and the OS at 12 months (C) within Akk+ group (C, left panel) and Akk− group (C, right panel). LDA; Linear discriminant analysis. OS: overall survival. P-values were calculated using a two-sided nonparametric factorial Kruskal-Wallis (KW) sum-rank test. # Multivariate analysis (ANCOM-BC/Maaslin2) with a false discovery rate (FDR) adjusted p-value <0.2.
Extended Data Fig. 4:
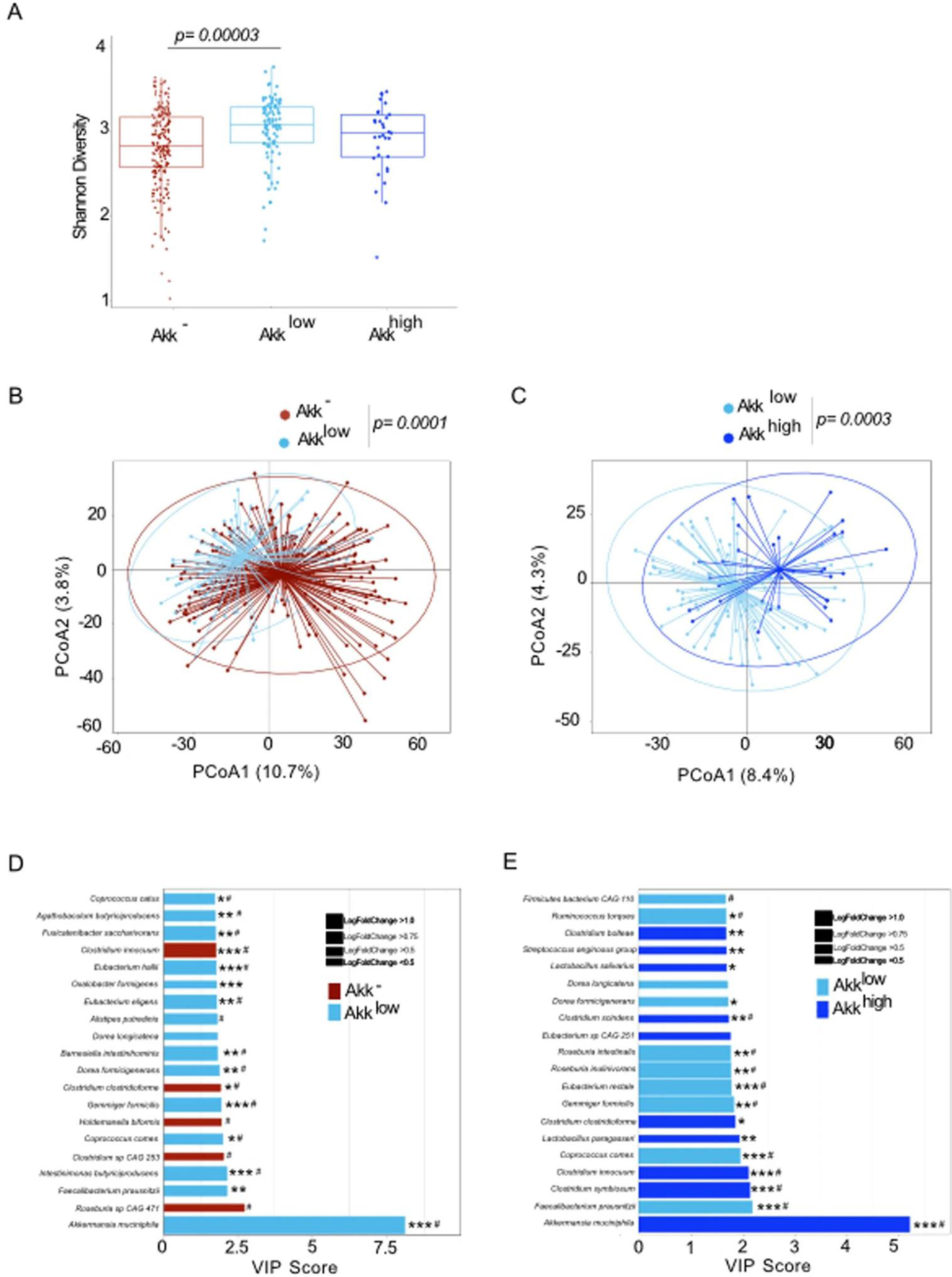
Compositional taxonomic differences in stools of NSCLC patients segregated according to Akk relative abundance. A. Alpha diversity according to Akk relative abundance segregated in 3 groups Akk−: undetectable Akk, Akklow: A. muciniphila relative abundance between 0.035–4.799% (<77th percentile of positive samples), and Akkhigh: 4.799% (> 77th percentile) (N=338). The lower and upper hinges of boxplots correspond to the 25th and 75th percentiles, respectively. The midline is the median. The upper and lower whiskers extend from the hinges to the largest (or smallest) value no further than ×1.5 interquartile range from the hinge, defined as the distance between the 25th and 75th percentiles. P-values were calculated using a two-sided nonparametric Wilcoxon sum-rank test. B-C. Beta-diversity using PCoA between Akk− and Akklow (B) and between Akklow and Akkhigh (C) p-values were calculated using PERMANOVA with 999 permutations. The PERMANOVA test compares groups of objects and tests the null hypothesis that the centroids and dispersion of the groups are equivalent. The P-value is calculated by comparing the actual F test to that gained from (in this case 999) random permutations of the objects between the groups. If p <0.05, the null hypothesis is disregarded and we conclude that the centroids and dispersion between the groups are not equivalent. D-E. Variable importance plot (VIP) discriminant analysis of taxonomic stool composition according to Akk relative abundance, between Akk− versus Akklow (D) and Akklow versus Akkhigh (E). Differences in bacterial prevalence and abundance in fold ratios are indicated in these VIP plots. VIP: Variable importance plot. * p <0.05,** p <0.01, *** p <0.001. P-values were calculated using a two-sided nonparametric Wilcoxon sum-rank test. # Multivariate analysis (ANCOM-BC/Maaslin2) with a false discovery rate (FDR) adjusted p-value <0.2.
Extended Data Fig. 5:
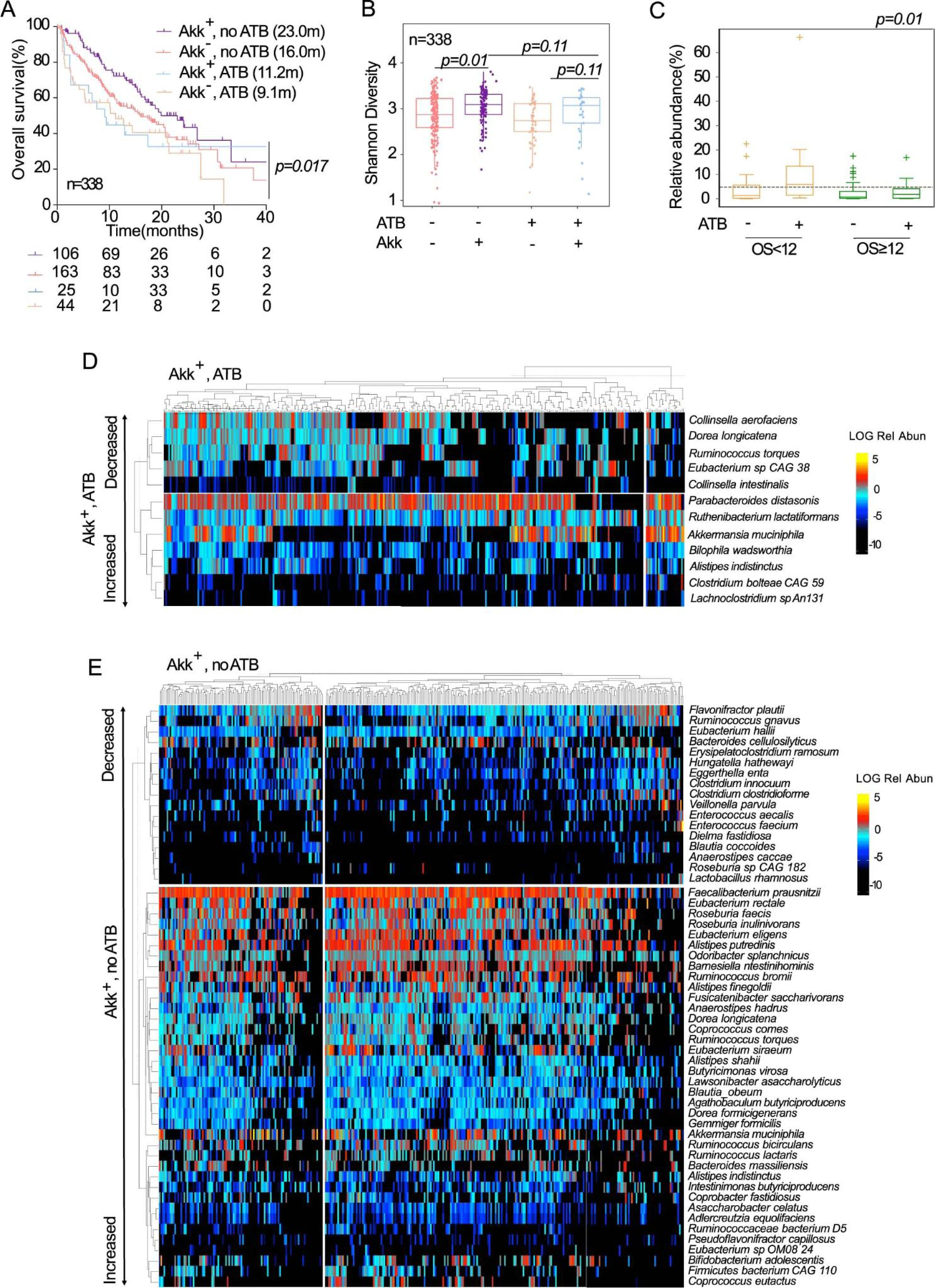
Interaction between ATB and A.muciniphila on survival and microbiome composition. A. Kaplan-Meier curve and Cox regression analysis of overall survival in the n=338 patients according to detectable versus undetectable Akk (Akk+ and Akk−) and ATB use (noATB: no exposure to ATB, ATB: antibiotics exposure within 2 months prior to ICI initiation). The Akk status and ATB use were compared using the stratified log-rank test. P-values are one-sided with no adjustment. B. Shannon diversity index representing stool alpha diversity in Akk+ and Akk− groups of fecal specimen from patients exposed or not to ATB (N=338). The lower and upper hinges of boxplots correspond to the 25th and 75th percentiles, respectively. The midline is the median. The upper and lower whiskers extend from the hinges to the largest (or smallest) value no further than ×1.5 interquartile range from the hinge, defined as the distance between the 25th and 75th percentiles. P-values were calculated using a two-sided nonparametric Wilcoxon sum-rank test. C. Box Plots representing the relative abundance (mean+/−SEM) of Akk according to overall survival at 12 months and exposure or not to ATB in n=338 patients. The lower and upper hinges of boxplots correspond to the 25th and 75th percentiles, respectively. The midline is the median. The upper and lower whiskers extend from the hinges to the largest (or smallest) value no further than ×1.5 interquartile range from the hinge, defined as the distance between the 25th and 75th percentiles. The test used was Kruskal-Wallis, two-sided, 5% level of significance. No adjustments were made for multiple comparisons. D. Heatmap showing differentially abundant species identified in stools with detectable Akk (Akk+) in patients exposed to (D) or not exposed to (E) ATB within 2 months prior to ICI initiation. Species were identified using a non-parametric Kruskall-Wallis test comparing 4 groups made up of 2 variables: Akkermansia muciniphila presence/absence and antibiotic use. The figure shows species’ abundances across samples whose False Discovery Rate (FDR) was <0.2 in the KW test and whose Wilcoxon Rank Sum Test p-value was <0.05 when comparing the highlighted group to the rest.
Extended Data Fig. 6:
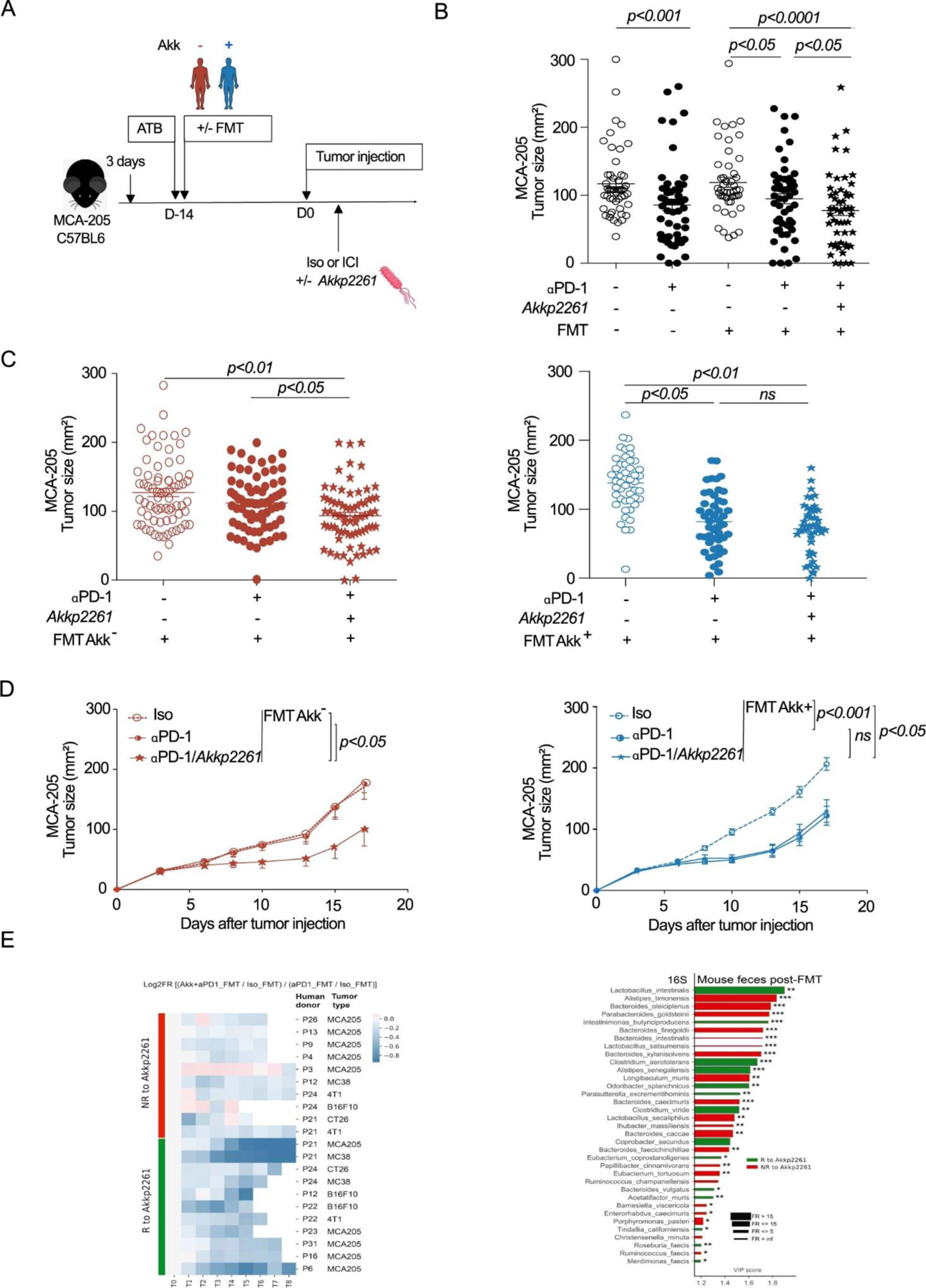
Akkp2261 modulated the murine microbiome composition, rescuing responsiveness to PD-1 blockade. A. Experimental setting. After 3 days of ATB, FMT was performed in mice by oral gavage using patient stools classified according to Akk (Akk+ and Akk−). 14 days later, MCA-205 tumors were i.d inoculated, and mice were treated with anti-PD-1 or iso-control mAbs 4 times every 3 days concomitantly with oral supplementation of Akkp2261 four times every 3 days. B-D. Mean MCA-205 tumor sizes+/−SEM are depicted at day 12 after 4 therapeutic injections of anti-PD-1 mAbs, in each FMT groups (Akk+ and Akk−) supplemented or not with Akkp2261 as well as in animals reared in SPF conditions (FMT-). Concatenation of>25 experiments using n=53 mice in Iso group, n=51 in Iso FMT+ group, n=56 in anti-PD-1 and anti-PD-1 FMT+ groups. Each experiment comprising 6 mice/group and was performed at least 2 times for each FMT (Supplementary Table S6) (B). Tumor sizes according to FMT Akk− (C left, n=72/group; C right, n=49 in Iso group and n=48 in other groups) versus Akk+ (D left, n=6/group, D right, n=12 in Iso and anti-PD-1 groups, n=14 in anti-PD-1 with Akkp2261) are depicted, each dot representing one mouse. Statistics were mixed-effect modeling with specific software ((https://kroemerlab.shinyapps.io/TumGrowth/) for longitudinal tumor growth analysis (D) and Mann–Whitney U-test (B-C) to compare two independent groups (after Kruskal–Wallis test was implemented using Dunn’s test for multiple groups). ns=not significant. E. Clustermap of ratios of Akkp2261-related tumor reduction at day 12–15 following PD-1 mAbs in FMT normalized onto ratios obtained in SPF mice. The relative tumor size reduction follows a blue color code (the darker the greater; R, Responders)). 29 FMT were performed according to A. N=29–30 mice/group in total. Each experiment contained 6 mice/group and was performed 2–3 times for each tumor model (E, left panel). 16S rRNA sequencing of gene amplicons of stools harvested in recipient avatar tumor bearers at day 12 post-4 injections of anti-PD-1 Abs and 4 oral gavages with Akkp2261 divided into green (R) and red (NR) groups. VIP plot repartition of discriminant metagenomic species segregating groups of mice that responded to oral Akkp2261 (R, green bars) or not (NR, red bars). (E, right panel). Asterisks represent significant Mann-Whitney U test without FDR at 10%. * p <0.05, ** p <0.01, *** p <0.001. P-values were calculated using a two-sided nonparametric Wilcoxon sum-rank test. Adjustments for multiple comparisons were not made.
Supplementary Material
Acknowledgements:
LZ and BB were funded by the RHU Torino Lumière (ANR-16-RHUS-0008), while LZ was supported by H2020 ONCOBIOME and ANR – Frecnh-German Ileobiome - 19-CE15–0029-01. LZ and GK received a donation from Seerave Foundation. LZ, LD and ED were supported by the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE). LD and LZ were supported by SIGN’IT ARC foundation. LZ and GK were supported by the Ligue contre le Cancer (équipe labelisée); ANR Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Bristol-Myers Squibb Company (International Immuno-Oncology Network), Cancéropôle Ile-de-France; Chancellerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; the European Commission (ArtForce); the European Research Council (ERC); Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; FHU CARE, Dassault, and Badinter Philantropia, and the Paris Alliance of Cancer Research Institutes (PACRI). BR is supported by the Canadian Institutes of Health Research (CIHR), Institut du cancer de Montreal and the Prefontaine family. AC is supported by the CPRIT Research Training Program (RP170067). NS was supported by the European Research Council (ERC-STG project MetaPG-716575); MIUR ‘Futuro in Ricerca’ (grant No. RBFR13EWWI_001) to NS; the European H2020 program (ONCOBIOME-825410 project and MASTER-818368 project); the National Cancer Institute of the National Institutes of Health (1U01CA230551); by the Premio Internazionale Lombardia e Ricerca 2019 to GK. AM has been or is currently an investigator in clinical trials sponsored by BMS, MSD, GSK/Tesaro, Janssen, Roche/Genentech, Pfizer, Astra Zeneca (AZ), Amgen. AM has been or is currently a member of Clinical Trial Scientific Steering Committee for AZ and GSK. AM has been or is currently a member of the scientific advisory board of the following companies: Merck Serono, Novartis, BMS, Symphogen, Amgen, Tesaro/GSK, Pfizer, Astra Zeneca/Medimmune, Servier, Sanofi. AM has provided Scientific & Medical Consulting to the following companies: Roche, Sanofi. AM is a member of the Data Safety and Monitoring Board for the following trial NCT02423863 (TLR3 agonist; Oncovir). AM has received research funding and or drug supply for pre-Clinical & clinical research projects from: BMS, Boehringer Ingelheim, Idera, MSD, Fondation MSD Avenir, SIRIC (INCa-DGOS-Inserm_12551).
Abbreviations:
- Akk
Akkermansia muciniphila
- Akk−
A. muciniphila undetectable in stool
- Akk+
A. muciniphila detectable in stool
- Akklow
A. muciniphila < cut-off value
- Akkhigh
A. muciniphila > cut-off value
- ATB
antibiotics
- BMI
body mass index
- FMT
Fecal Microbial Transplantation
- ICI
immune checkpoint inhibitor
- I-O
immuno-oncology
- NSCLC
non-small cell lung cancer
- ORR
objective response rate
- OS
overall survival
- PD
progressive disease
- PFS
progression-free survival
- RCC
renal cell carcinoma
- SD
stable disease
Footnotes
Code availability
No unique software or computational code was created for this study. Code detailing implementation of established tools/pipelines are described in details in the Method section and available upon request to the corresponding author.
CONFLICTS OF INTEREST
LZ received research contract from Kaleido and Innovate Pharma and Pilege. LD had consulting, and advisory role for BMS, Sanofi and was supported by Philantropia Fondation Gustave Roussy. GZ received a research grant from Fondation Roche, received fees from Roche, MSD, BMS, Astra-Zeneca and is consultant for Da Volterra & Inventiva. ED reports grants and personal fees from Roche Genentech, grants from Boehringer, grants from Astrazeneca, grants and personal fees from Merck Serono, grants from BMS, and grants from MSD. PD had consulting, and advisory role for AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Daiichi Sankyo, Eli Lilly, Merck, Novartis, Pfizer, prIME Oncology, Peer CME, Roche, Samsung, as well as honoraria from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Eli Lilly, Merck, Novartis, Pfizer, prIME Oncology, Peer CME, Roche, Samsung. PD ran clinical trials as principal or co-investigator for AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Eli Lilly, Merck, Novartis, Pfizer, Roche, Medimmun, Sanofi-Aventis, Taiho Pharma, Novocure, Daiichi Sankyo, and received Travel, Accommodation, Expenses: from AstraZeneca, Roche, Novartis, prIME Oncology, Pfizer. FG received honoraria from Amgen,Sanofi, Merk Serono, MSD, BMS, Astra Zeneca, had a consultancy or advisory role for Roche, Enterome and received direct research fundings from Roche, Enterome, Astra Zeneca, Servier and traveling supports from Servier, Amgen, Roche. JCS In the last 2 years consultancy fees from Relay Therapeutics, Gritstone are holds shares from Hookipa, Gritstone, AstraZeneca, Daiichi Sankyo and was a full time employee for AstraZeneca 2017–2019.
Data availability
The data generated or analyzed during this study are included within the paper, its Supplementary Information files and public repositories. Detailed information on the cohort is available in Supplementary Table 7, raw metagenomic sequences are available in the SRA under the Bioproject accession PRJNA751792 and raw RNA sequencing are available in the SRA under accession the NCBI accession GSE182328.
REFERENCES
- 1.Herbst RS et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. The Lancet 387, 1540–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Brahmer J et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-cell Lung Cancer. N. Engl. J. Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borghaei H et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-cell Lung Cancer. N. Engl. J. Med 373, 1627–1639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi L et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med 378, 2078–2092 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Paz-Ares L et al. Pembrolizumab plus Chemotherapy for Squamous Non–Small-Cell Lung Cancer. N. Engl. J. Med 379, 2040–2051 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Reck M et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Elkrief A et al. Therapeutic landscape of metastatic non-small-cell lung cancer in Canada in 2020. Curr. Oncol 27, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadgeel S et al. Updated Analysis From KEYNOTE-189: Pembrolizumab or Placebo Plus Pemetrexed and Platinum for Previously Untreated Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 38, 1505–1517 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Ferrara R et al. Hyperprogressive Disease in Patients With Advanced Non-Small Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or With Single-Agent Chemotherapy. JAMA Oncol 4, 1543–1552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riaz N et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 171, 934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spranger S, Bao R & Gajewski TF Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Smyth MJ, Ngiow SF, Ribas A & Teng MWL Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol 13, 143–158 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Koyama S et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun 7, 10501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young A et al. Co-inhibition of CD73 and A2AR Adenosine Signaling Improves Anti-tumor Immune Responses. Cancer Cell 30, 391–403 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Chen DS & Mellman I Elements of cancer immunity and the cancer–immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Routy B et al. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol 15, 382–396 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Routy B et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Vétizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan V et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derosa L et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol 29, 1437–1444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derosa L & Zitvogel L Antibiotics impair immunotherapy for urothelial cancer. Nat. Rev. Urol 17, 605–606 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Lurienne L et al. NSCLC Immunotherapy Efficacy and Antibiotic Use: A Systematic Review and Meta-Analysis. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 15, 1147–1159 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Derosa L et al. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov 11, 2396–2412 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Matson V et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y et al. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 14, 1378–1389 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Daisley BA et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat. Commun 11, 4822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro A et al. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell. Mol. Life Sci. CMLS 75, 129–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q et al. Gut bacteria Akkermansia is associated with reduced risk of obesity: evidence from the American Gut Project. Nutr. Metab 17, 90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blacher E et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572, 474–480 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Bárcena C et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med 25, 1234–1242 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Romano S et al. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. Npj Park. Dis 7, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shono Y et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med 8, 339ra71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai MS et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karcher N et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol 22, 209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Chatelier E et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Derosa L et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol 78, 195–206 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Meng X et al. Immune Microenvironment Differences Between Squamous and Non-squamous Non-small-cell Lung Cancer and Their Influence on the Prognosis. Clin. Lung Cancer 20, 48–58 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Hwang S et al. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci. Rep 10, 643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakajima K et al. IAP inhibitor, Embelin increases VCAM-1 levels on the endothelium, producing lymphocytic infiltration and antitumor immunity. Oncoimmunology 9,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hakozaki T et al. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. Cancer Immunol. Res 8, 1243–1250 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Tsay J-CJ et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov (2020) doi: 10.1158/2159-8290.CD-20-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benevides L et al. New Insights into the Diversity of the Genus Faecalibacterium. Front. Microbiol 8, 1790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S-H et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol 6, 277–288 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Elkrief A, Derosa L, Kroemer G, Zitvogel L & Routy B The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Ann. Oncol. Off. J. Eur. Soc. Med. Oncol 30, 1572–1579 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Seo SW, Kim D, Szubin R & Palsson BO Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep 12, 1289–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Atarashi K et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruusunen A, Rocks T, Jacka F & Loughman A The gut microbiome in anorexia nervosa: relevance for nutritional rehabilitation. Psychopharmacology (Berl.) 236, 1545–1558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Lugt B et al. Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1−/Δ7 mice. Immun. Ageing 16, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Depommier C et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med 25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouyang J et al. The Bacterium Akkermansia muciniphila: A Sentinel for Gut Permeability and Its Relevance to HIV-Related Inflammation. Front. Immunol 11, 645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huck O et al. Akkermansia muciniphila reduces Porphyromonas gingivalis-induced inflammation and periodontal bone destruction. J. Clin. Periodontol 47, 202–212 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Wu W et al. Protective Effect of Akkermansia muciniphila against Immune-Mediated Liver Injury in a Mouse Model. Front. Microbiol 8, 1804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baruch EN et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2020) doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 55.Shi L et al. Combining IL-2-based immunotherapy with commensal probiotics produces enhanced antitumor immune response and tumor clearance. J. Immunother. Cancer 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Depommier C et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med 25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plovier H et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med 23, 107–113 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Cani PD & de Vos WM Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front. Microbiol 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenhauer EA et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer Oxf. Engl 1990 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Li J et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol 32, 834–841 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Nielsen HB et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol 32, 822–828 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Beghini F et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. bioRxiv 2020.11.19.388223 (2020) doi: 10.1101/2020.11.19.388223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fumet J-D et al. Prognostic and predictive role of CD8 and PD-L1 determination in lung tumor tissue of patients under anti-PD-1 therapy. Br. J. Cancer 119, 950–960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Segata N et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin H & Peddada SD Analysis of compositions of microbiomes with bias correction. Nat. Commun 11, 3514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mallick R et al. Subduction initiation and the rise of the Shillong Plateau. Earth Planet. Sci. Lett 543, 116351 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated or analyzed during this study are included within the paper, its Supplementary Information files and public repositories. Detailed information on the cohort is available in Supplementary Table 7, raw metagenomic sequences are available in the SRA under the Bioproject accession PRJNA751792 and raw RNA sequencing are available in the SRA under accession the NCBI accession GSE182328.


