Abstract
Human peripheral blood mononuclear cells (PBMCs) represent a highly responsive primary tissue that is composed of innate and adaptive immune cells. In this study, we compared modulation of the transcriptome of PBMCs by the vitamin D metabolites 25-hydroxyvitamin D3 (25(OH)D3), 25(OH)D2 and 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3). Saturating concentrations of 1,25(OH)2D3, 25(OH)D3 and 25(OH)D2 resulted after 24 h stimulation in a comparable number and identity of target genes, but below 250 nM 25(OH)D3 and 25(OH)D2 were largely insufficient to affect the transcriptome. The average EC50 values of 206 common target genes were 322 nM for 25(OH)D3 and 295 nM for 25(OH)D2 being some 600-fold higher than 0.48 nM for 1,25(OH)2D3. The type of target gene, such as primary/secondary, direct/indirect or up-/down-regulated, had no significant effect on vitamin D metabolite sensitivity, but individual genes could be classified into high, mid and lower responders. Since the 1α-hydroxylase CYP27B1 is very low expressed in PBMCs and early (4 and 8 h) transcriptome responses to 25(OH)D3 and 25(OH)D2 were as prominent as to 1,25(OH)2D3, both vitamin D metabolites may directly control gene expression. In conclusion, at supra-physiological concentrations 25(OH)D3 and 25(OH)D2 are equally potent in modulating the transcriptome of PBMCs possibly by directly activating the vitamin D receptor.
Keywords: vitamin D, transcriptome, PBMCs, target genes, 1α, 25-dihydroxyvitamin D3, 25-hydroxyvitamin D3, 25-hydroxyvitamin D2
Introduction
Although humans can produce vitamin D3 endogenously when they expose their skin to UV-B (1), predominant indoor lifestyle as well as insufficient UV indices in Northern latitudes (above 38°) at winter times (2), suggest the supplementation of the vitamin for at least a part of the year (3). Humans and animals use the cholesterol precursor 7-dehydrocholesterol as the substrate for vitamin D3 synthesis, while fungi and yeast produce vitamin D2 on the basis of the sterol ergosterol (4). Vitamin D2 and vitamin D3 differ only in the structure of their side chain, but in human intestine the uptake vitamin D3 seems to be more effective (5). Nevertheless, both forms of vitamin D are used for food fortification and direct supplementation (6).
Vitamin D2 and vitamin D3 are metabolized in the liver by the CYP (cytochrome P450) enzymes CYP2R1 and CYP27A1 to 25(OH)D2 and 25(OH)D3, respectively (7). The sum of the serum concentrations of both metabolites [or sometimes only 25(OH)D3] is traditionally used as biomarker for the vitamin D status of an individual (8, 9). A vitamin D status below 50 nM increases the risk for musculoskeletal disorders, such as rickets in children and osteomalacia as well as fractures in adults (10). Moreover, insufficient 25(OH)D serum levels are associated with a number of immune-related diseases, such as rheumatoid arthritis (11), multiple sclerosis (12), type I diabetes (13) and inflammatory bowel disease (14). In addition, vitamin D deficiency raises the risk for severe consequence of microbe infections in tuberculosis, influenza or COVID-19 (coronavirus disease) (15–17). Therefore, one should aim for serum 25(OH)D levels in the range of 75–100 nM, i.e., 30–40 ng/ml (18). In contrast, a vitamin D status of above 250 nM should be avoided, in order to prevent deleterious effects of hypercalcemia (19).
The biologically most active form of vitamin D3 and D2 are 1,25(OH)2D3 and 1,25(OH)2D2, respectively, which function in sub-nanomolar concentrations as nuclear hormones (20). For endocrine purposes 1,25(OH)2D is synthesized in the kidneys by the enzyme CYP27B1 using 25(OH)D as a substrate (21), while for paracrine use 1,25(OH)2D is produced also in CYP27B1 expressing keratinocytes and immune cells (22). Since 1,25(OH)2D is the natural, high affinity (KD = 0.1 nM) ligand of the nuclear receptor VDR (vitamin D receptor) (23, 24), vitamin D affects the activity of hundreds of genes in VDR expressing tissues (25). Thus, the physiological functions of vitamin D are associated with changes of the transcriptome of multiple tissues and cell types by ligand-activated VDR (26). The vitamin D-triggered transcriptome has been studied in vitro in a number of cell culture models, such as THP-1 monocytic leukemia cells (27–29), as well as in PBMCs (30, 31). Primary cells like PBMCs are a natural mixture of innate and adaptive immune cells like monocytes, natural killer cells, T and B cells. They are far closer to the human in vivo situation than cell lines and can be obtained with minimal harm to the donor (32).
The affinity of VDR for 25(OH)D3 is 100- to 1,000-fold lower than for 1,25(OH)2D3 (33, 34), which parallels with the fact that the serum concentration of 25(OH)D3 is some 1,000-fold higher than that of 1,25(OH)2D3 (0.05–0.15 nM) (35). This relation raised already earlier the question, whether 25(OH)D3 has the potential to act as a nuclear hormone that directly activates the VDR (36). The molecule 1,25(OH)2D3 has three hydroxyl groups, each of which is specifically contacted by a pair of polar amino acids within VDR's ligand-binding pocket (37, 38). In contrast, 25(OH)D lacks the hydroxyl group at carbon 1 and therefore binds with lower affinity to the receptor, but takes the same agonistic conformation within the ligand-binding pocket (39).
In this study, we analyzed the transcriptome-wide effects of 25(OH)D3 and 25(OH)D2 in comparison to that of 1,25(OH)2D3 in freshly isolated human PBMCs. We will demonstrate that 25(OH)D3 and 25(OH)D2 are equally potent in modulating the transcriptome of PBMCs and cannot exclude the possibility that both vitamin D metabolites directly activate the VDR.
Materials and Methods
Sample Collection
Peripheral blood was collected from a single healthy individual (male, age 57 years) the vitamin D responsiveness of whom had already been characterized in the VitDbol trial (NCT02063334) (32, 40). The ethics committee of the Northern Savo Hospital District had approved the study protocol (#9/2014). The individual gave written informed consent to participate in the study and the experiments were performed in accordance with relevant guidelines and regulations.
PBMC Isolation and Stimulation
PBMCs were isolated immediately after collecting 100 ml peripheral blood using Vacutainer CPT Cell Preparation Tubes with sodium citrate (Becton Dickinson) according to manufacturer's instructions. After washing with phosphate-buffered saline, the cells were grown at a density of 0.5 million/ml in 5 ml RPMI1640 medium supplemented with 10% charcoal-depleted fetal calf serum, 2 mM L-glutamine, 0.1 mg/ml streptomycin and 100 U/ml penicillin at 37 °C in a humidified 95% air/5% CO2 incubator and treated for 4, 8 or 24 h either with 1,25(OH)2D3 (0.1, 1 and 10 nM) 25(OH)D3 (100, 250, 500, 750, and 1,000 nM), 25(OH)D2 (100, 250, 500, 750, and 1,000 nM) or solvent (0.1% EtOH). All experiments were performed in three repeats. Deconvolution of RNA-seq data using the algorithm CIBERSORTx (41) and its LM6 signature matrix estimated the relative amounts of B cells (7%), CD8+ T cells (32%), CD4+ T cells (20%), natural killer cells (21%) and monocytes/macrophages (20%) within the PBMC pool.
RNA-Seq Data Generation and Processing
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche) according to manufacturer's instructions. RNA quality was assessed on an Agilent 2100 Bioanalyzer system (RNA integrity number ≥ 8). rRNA depletion and cDNA library preparation were performed using the New England Biolabs kits NEBNext rRNA Depletion, NEBNext Ultra II Directional RNA Library Prep for Illumina and NEBNext Multiplex Oligos for Illumina (Index Primers Sets 1 and 2) according to manufacturer's protocols. RNA-seq libraries went through quality control on an Agilent 2100 Bioanalyzer and were sequenced on a NextSeq 500 system (Illumina) at 75 bp read length using standard protocols at the Gene Core facility of the EMBL (Heidelberg, Germany). The libraries were sequenced as four batches. Fastq files of the 66 libraries can be found at Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) with accession number GSE199273.
Single-end, reverse-stranded cDNA sequence reads were aligned to the reference genome (version GRCh38) with Ensembl annotation (version 103) by using default settings of the nf-core/rnaseq STAR-Salmon pipeline (version 3.0) (http://doi.org/10.5281/zenodo.4323183) (42). The number of nucleotide sequence reads are shown in Supplementary Table 1. Ensembl gene identifiers were annotated with gene symbol, description, genomic location and biotype by accessing the Ensembl database (version 104) via the R package BiomaRt (version 2.46.0) (43). Ensembl gene identifiers missing HGNC gene symbol annotation, Entrez ID, genomic location information or being mitochondrially encoded were removed from the datasets. When a gene name appeared more than once, the entry with the highest average gene counts was kept.
Transcriptome Data Analysis
Differential gene expression analysis was computed in R (version 4.1.2) in the Rocky Linux 8.5 operating system using the tool EdgeR (version 3.36.0) (44). The analysis focused on the 19,142 protein-coding genes, in order to reduce transcriptional noise expected by non-coding genes. Read counts were normalized for differences in library size to counts per million (CPM). Genes with very low expression were filtered out by applying the function FilterByExpr(), in order to mitigate the multiple testing problem and to not interfere with the statistical approximations of the EdgeR pipeline. This requirement was fulfilled by 12,939 genes. After filtering, library sizes were recomputed and trimmed mean of M-value normalization was applied. The transcriptome data structure was explored via the dimensionality reduction method multidimensional scaling (MDS) (Supplementary Figure 1). MDS was computed via EdgeR's function plotMDS(), in which distances approximate the typical log2 fold change (FC) between the samples. This distance was calculated as the root mean square deviation (Euclidean distance) of the largest 500 log2FCs between a given pair of samples, i.e., for each pair a different set of top genes was selected. The inspection of the MDS plot showed that (i) the time-dependent divergence from the native transcriptome state and (ii) the concentration-dependent modulation by treatment with 1,25(OH)2D3, 25(OH)D3 or 25(OH)D2 are the two principal factors driving differences in the gene expression profiles of PBMCs. The gene-wise statistical test for differential expression was computed using the generalized linear model quasi-likelihood pipeline (45). The empirical Bayes shrinkage was robustified against outlier dispersions as recommended (45). The glmTreat approach was used to test for differential expression relative to FC > 1.5 at the early time points (4 and 8 h) and FC > 2 at 24 h. Taking all treatments together, 553 genes with a Benjamini-Hochberg corrected p-value [i.e., false discovery rate (FDR)] < 0.05 and a total trimmed mean of M-value normalized CPM count > 10 (i.e., the sum of the average gene expression level of the reference 10 nM 1,25(OH)2D3 and EtOH-treated samples at each time point) were considered as vitamin D targets (Supplementary Table 2). Mean-Difference (MA) plots (Figures 1A, 3A; Supplementary Figure 2) for the 19 different treatment conditions were generated with vizzy (version 1.0.0, https://github.com/ATpoint/vizzy).
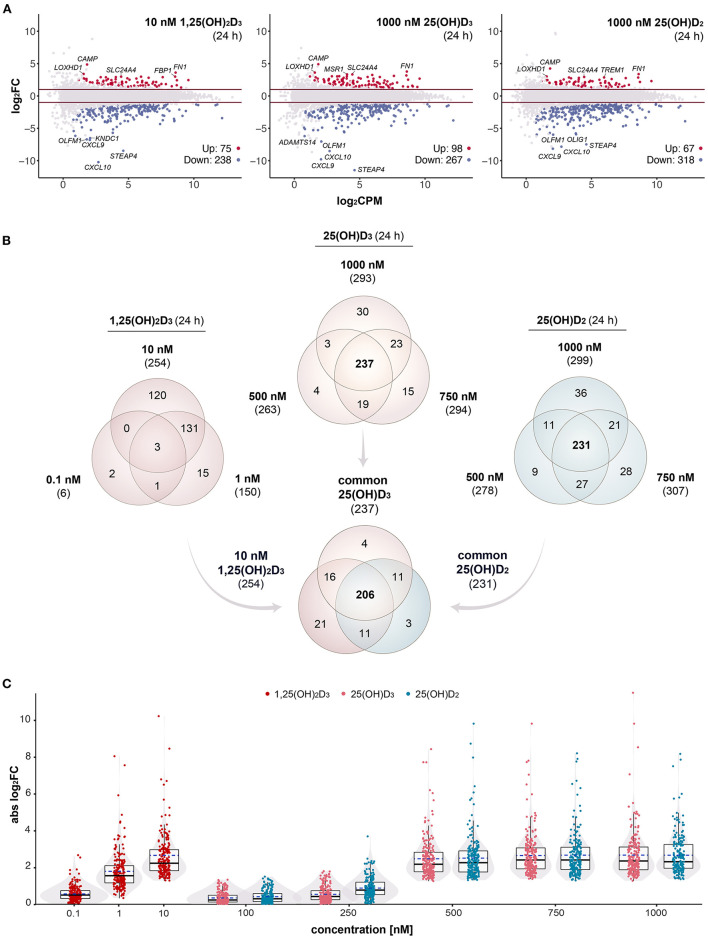
Figure 1.
Gene-regulatory potential of vitamin D metabolites. MA plots display the genome-wide transcriptional response of PBMCs treated for 24 h with 10 nM 1,25(OH)2D3, 1000 nM 25(OH)D3 or 1000 nM 25(OH)D2 in comparison to solvent (0.1% EtOH) (A). For each gene, the change of expression (log2FC) between treated and control samples is shown in relation to its mean expression level (log2CPM). Differential expression analysis was performed as a pairwise comparison per each concentration by using glmTreat test. Significantly (FDR < 0.05) up- and down-regulated genes are highlighted in red and blue, respectively. Horizonal lines (red) indicate the applied statistical testing threshold (absolute FC > 2). The top 5 responsive genes (up- and down-regulated) are labeled. Venn diagrams show the overlap of vitamin D target genes per metabolite (B). The relations between all treatments and concentrations (at 24 h) are provided in Supplementary Figure 3. Box and violin plots summarize the distribution of the magnitude of expression change (absolute log2FC) of the 206 common genes for each vitamin D metabolite and concentration (C). Solid lines within the boxes indicate medians, while dashed lines mark the mean.
Dose Response Analysis
The effect of vitamin D metabolites on the change in mRNA levels (absolute log2FC) was modeled with the three-parameter log-logistic function LL.3 from the R package drc (46) having lower limit fixed at 0. The estimated relative EC50 values and their standard errors were retrieved from the curve fits via the summary() function. The quality of fitting was checked by manual inspection. EC50 values were reported only for those 206 genes, for which the value could be estimated for all three vitamin D metabolites. Pairwise comparisons of EC50 values between different metabolites as well as groups of target genes were performed using Tukey's test implemented in the R package multcomp (version 1.4.18) and family-wise error rate (FWER)-adjusted p-values retrieved. Comparisons with a FWER < 0.05 were considered as significant. The code of the analysis can be found at https://github.com/andreahanel/2022_Doseresponse.
Results
Transcriptome-Wide Responses to 1,25(OH)2D3, 25(OH)D3, and 25(OH)D2
In order to obtain maximal responses of the transcriptome, PBMCs from an healthy individual were treated immediately after isolation for 24 h with either 10 nM 1,25(OH)2D3, 1,000 nM 25(OH)D3, 1,000 nM 25(OH)D2 or solvent (0.1% EtOH). In three repeats RNA-seq was performed on the basis of total RNA. As in comparable studies (31, 47, 48), strict statistical thresholds of FDR < 0.05 and FC > 2 were applied. This resulted in 313 genes (75 up-regulated, 238 down-regulated) responding to 1,25(OH)2D3, 365 target genes of 25(OH)D3 (98 up-regulated, 267 down-regulated) and 385 genes modulated by 25(OH)D2 (67 up-regulated, 318 down-regulated) (Figure 1A). For all three vitamin D metabolites the genes CAMP (cathelicidin antimicrobial peptide), FN1 (fibronectin 1), LOXHD1 (lipoxygenase homology PLAT domains 1) and SLC24A4 (solute carrier family 24 member 4) were the top up-regulated and the genes CXCL10 (C-X-C motif chemokine ligand 10), STEAP4 (STEAP4 metalloreductase), CXCL9 and OLFM1 (olfactomedin 1) the most down-regulated.
In addition to saturating ligand concentrations, PBMCs were treated for 24 h with 0.1 and 1 nM 1,25(OH)2D3, with 100, 250, 500, and 750 nM 25(OH)D3 as well as with 100, 250, 500, and 750 nM 25(OH)D2. This resulted in 7 and 179 target genes for 0.1 and 1 nM 1,25(OH)2D3, no targets for 100 and 250 nM 25(OH)D3, 322 and 392 responding genes for 500 and 750 nM 25(OH)D3, no targets for 100 nM 25(OH)D2 as well as 30, 342, and 397 genes that reacted on a stimulation with 250, 500, and 750 nM 25(OH)D2 (Supplementary Figure 3A). These in total 526 different genes were filtered with a list of 662 genes, which were detected by time course analysis of treatment of PBMCs with 1,25(OH)2D3 (31). This led to 389 confirmed vitamin D target genes, of which 6, 150, and 254 responded to 0.1, 1 and 10 nM 1,25(OH)2D3, 263, 294, and 293 to 500, 750, and 1,000 nM 25(OH)D3 as well as 25, 278, 307, and 299 to 250, 500, 750, and 1,000 nM 25(OH)D2 (Supplementary Figure 3B). Venn diagrams indicated that there are 237 common targets of 500, 750, and 1,000 nM 25(OH)D3 as well as 231 common targets of 500, 750, and 1,000 nM 25(OH)D2 (Figure 1B). From both lists 206 genes are identical with the 254 targets responding to 10 nM 1,25(OH)2D3.
The 206 common targets represent a stable set of genes responding to variant concentrations of all three tested vitamin D metabolites (Figure 1C). Combined box plots and violin plots monitored the overall change in the expression of these 206 genes with raising metabolite concentrations. Interestingly, 250 nM of both 25(OH)D3 and 25(OH)D2 were insufficient for general gene regulation, while 500 nM of both vitamin D metabolites was clearly above this threshold.
In summary, both by number as well as by most responsive target genes the transcriptome-wide responses to saturating concentrations of 1,25(OH)2D3, 25(OH)D3 and 25(OH)D2 are very comparable. At concentrations of 250 nM and below, 25(OH)D3 and 25(OH)D2 are largely insufficient to significantly modulate the expression of vitamin D target genes.
Gene-Specific Sensitivity to Vitamin D Metabolites
Plotting the FC of the 206 common vitamin D target genes over vitamin D metabolite concentration and using the three-parameter log-logistic model, determined the EC50 values 0.48 nM for 1,25(OH)2D3, 322 nM for 25(OH)D3 and 295 nM for 25(OH)D2 (Figure 2A). There is statistically no significant difference (FWER > 0.05, Tukey's test) between the potencies of 25(OH)D3 and 25(OH)D2, but their average EC50 is some 640-fold higher than that of 1,25(OH)2D3.
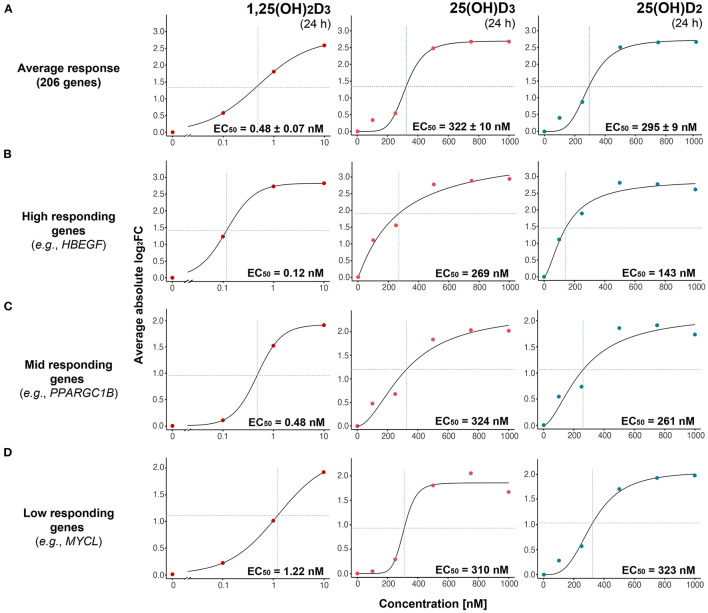
Figure 2.
Gene-specific response to vitamin D metabolites. The magnitude of expression change (absolute log2FC) as a function of vitamin D metabolite concentration is modeled with a three-parameter log-logistic model based on the average of 206 common targets (A) or indicated individual genes (B–D). Genes are segregated into high-, mid- and low-responding based on their estimated EC50 values in response to 1,25(OH)2D3. Standard errors of the EC50 estimates are indicated.
Based on the reference dataset (31), the 206 common targets were subdivided either into 85 primary targets (25 up- and 60 down-regulated) and 121 secondary targets (17 up- and 104 down-regulated) or into 94 direct targets (31 up- and 63 down-regulated) and 112 indirect targets (11 up- and 101 down-regulated). For 1,25(OH)2D3 the EC50-values of the eight different categories varied from 0.29 to 0.73 nM but did not differ significantly (FWER > 0.05, Tukey's test) between each other (Supplementary Figure 4). Similarly, for 25(OH)D3 the range was 318 to 389 nM and for 25(OH)D2 192 to 313 nM, but the difference was not statistically significant (FWER > 0.05, Tukey's test).
Since neither gene categories nor up- or down-regulation allowed a distinction of the sensitivity of vitamin D target genes to vitamin D metabolites, the dose responses of the 206 genes were investigated on an individual gene level. Manual inspection of the dose response curves provided for 130 genes convincing fits for all three vitamin D metabolites. Interestingly, for 1,25(OH)2D3 the EC50-values ranged from 0.10 nM [ENPP2 (ectonucleotide pyrophosphatase/phosphodiesterase 2)] to 2.39 nM [LMNA (lamin A/C)], for 25(OH)D3 from 121 nM [NXPH4 (neurexophilin 4)] to 461 nM [ENTPD7 (ectonucleoside triphosphate diphosphohydrolase 7)] and for 25(OH)D2 from 132 nM (SLC11A1) to 421 nM [STAB1 (stabilin 1)] (Supplementary Table 2). The wide range of gene-specific sensitivity to 1,25(OH)2D3 allowed the categorization of the representative 130 vitamin target genes into 59 high responders (EC50 value range from 0.10 to 0.39 nM), 59 mid responders (0.41 to 1.06 nM) and 12 low responders (1.20 to 2.39 nM). Representative genes for the three categories are HBEGF (heparin binding EGF like growth factor) as high responding gene (Figure 2B), PPARGC1B (PPARG coactivator 1 beta) as mid responding gene (Figure 2C) and MYCL (MYCL proto-oncogene, BHLH transcription factor) as low responding gene (Figure 2D). Importantly, the far smaller range of the gene-specific EC50 values for 25(OH)D3 and 25(OH)D2 did not allow a categorization of the vitamin D target genes by their response to these metabolites.
Taken together, the average EC50 values of the response of vitamin D target genes to 25(OH)D3 and 25(OH)D2 are not significantly different and are in the order of 300 nM, i.e., some 600-fold higher as those for 1,25(OH)2D3. Categorization of the target genes into primary/secondary, direct/indirect or up-regulated/down-regulated does not allow any significant distinction in their response to the three vitamin D metabolites. However, individual target genes can be classified by their response to 1,25(OH)2D3 (but not to 25(OH)D3 and 25(OH)D2) as high, mid and low responding genes.
Time Course Response of Vitamin D Target Genes
In order to investigate whether 25(OH)D3 and 25(OH)D2 may activate vitamin D signaling without enzymatic conversion by CYP27B1 to 1,25(OH)2D3 and 1,25(OH)2D2, respectively, the transcriptome-wide response to saturating concentrations of all three vitamin D metabolites was assessed by RNA-seq at earlier time points. After 4 h stimulation with 10 nM 1,25(OH)2D3, 16 genes (13 up- and 3 down-regulated) passed the statistical threshold (FDR < 0.05, FC > 1.5), with 1,000 nM 25(OH)D3 32 genes (27 up- and 5 down-regulated) and with 1,000 nM 25(OH)D2 20 genes (19 up- and 1 down-regulated) (Figure 3A). Common top up-regulated genes were HBEGF and G0S2 (G0/G1 switch 2). After filtering with the reference dataset of vitamin D target genes in PBMCs (Supplementary Figure 5) (31), Venn diagrams indicated that there are 13 common genes (out of 31 in total) of the three vitamin D metabolites responding already after 4 h, 93 (out of 159 in total) after 8 h and 229 (out of 337 in total) after 24 h (Figure 3B). As observed in previous time course studies (29, 31), at earlier time points there are more up- than down-regulated genes, since the process of up-regulation is more straightforward and quicker than that of down-regulation. When comparing the response of the 206 common target genes to the three vitamin D metabolites, there was no significant difference in the response to 10 nM 1,25(OH)2D3, 1,000 nM 25(OH)D3 or 1,000 nM 25(OH)D2 at 4, 8 and 24 h (Figure 3C).
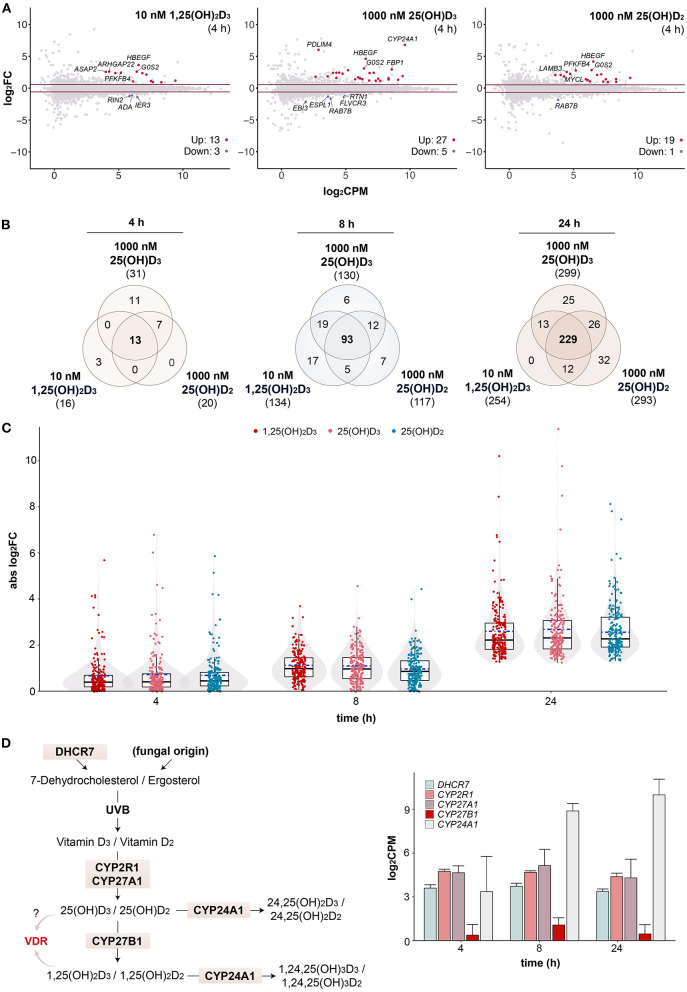
Figure 3.
Changes in transcriptional profiles over time. MA plots show early gene expression changes in PBMCs treated for 4 h with 10 nM 1,25(OH)2D3, 1000 nM 25(OH)D3 or 1000 nM 25(OH)D2 in comparison to solvent (0.1% EtOH, i.e., time-matched control) (A). For each gene, difference in expression (log2FC) between treated and control samples is shown in relation to its mean expression level (log2CPM). Genes were tested for differential expression relative to an absolute FC > 1.5 using glmTreat method. Horizonal lines (red) indicate the applied statistical testing threshold. The top 5 most responsive up- and down regulated genes (if any; FDR < 0.05) are highlighted. Venn diagrams show the overlap of vitamin D target genes per time point (B). The relations between all treatments are provided in Supplementary Figure 5. Box and violin plots summarize the distribution of the magnitude of expression change (absolute log2FC) of the 206 common genes for each vitamin D metabolite and time point (C). Solid lines within the boxes indicate medians, while dashed lines mark the mean. Please note that the data of the 24 h time point serve as a reference and are identical to those presented in Figure 1C. A map of the human vitamin D metabolism pathway marks key enzymes [(D), left]. The mean of 1,25(OH)2D3-treated and untreated mRNA expression of the five indicated genes is displayed in log2-scale as columns for all three time points [(D), right]. Error bars indicate standard deviation.
The enzymes DHCR7 (7-dehydrocholesterol reductase), CYP2R1, CYP27A1, CYP27B1 and CYP24A1 are critical nodes in the vitamin D metabolism pathway (Figure 3D, left). Therefore, the relative mRNA expression of the genes encoding for these enzymes were extracted from the transcriptome datasets and compared at the time points 4, 8 and 24 h (Figure 3D, right). DHCR7, CYP2R1 and CYP27A1 show comparable mid-range expression, while the average expression of CYP27B1 is 10- to 32-fold lower. In this way, CYP27B1 belongs to the 5% lowest expressed genes in PBMCs. Since the CYP24A1 gene is a well-known vitamin D target gene (49), its expression is even 671-fold higher than that of CYP27B1. For comparison, the relative expression values of all five genes in PBMCs of 12 participants of our VitDHiD study (50) are displayed (Supplementary Figure 6). The individuals were ranked by increasing CYP27B1 expression, which varied by a factor of 12.5, but being in average some 200-fold lower as the CYP24A1 expression. Thus, in vitamin D-triggered PBMCs the synthesis of 1,25(OH)2D on the basis of 25(OH)D is far less prominently covered by enzyme expression as the degradation of 25(OH)D and 1,25(OH)2D to 24,25(OH)D and 1,24,25(OH)3D, respectively.
In summary, already at early time points (4 and 8 h) the PBMC transcriptome responds to a stimulation with saturating concentrations of 25(OH)D3 and 25(OH)D2 as prominently as to 1,25(OH)2D3. The expression of CYP27B1 protein in PBMCs is very likely too low for an efficient conversion of 25(OH)D3 and 25(OH)D2 into 1,25(OH)2D3 and 1,25(OH)2D2, respectively, in particular in the presence of highly expressed CYP24A1 protein. However, it cannot be excluded that metabolic formation of 1,25(OH)2D is partly contributing to the results obtained.
Discussion
The focus of this study was to compare the gene regulatory potential of the vitamin D metabolites 25(OH)D3 and 25(OH)D2 to each other and in reference to 1,25(OH)2D3. At supra-physiological concentrations of 10 nM 1,25(OH)2D3 (100-times the natural serum concentration) and 1,000 nM 25(OH)D (10-times the recommended serum level) the response of the PBMC transcriptome is very comparable, i.e., the expression of some 300 genes is significantly modulated showing after 24 h stimulation some 3-times more down-regulation than up-regulation. Moreover, the target gene lists of 25(OH)D3, 25(OH)D2 and 1,25(OH)2D3 are to 85% identical, i.e., our experimental series had relatively low transcriptional noise. These observations suggest that all three vitamin D metabolites use identical mechanisms in the modulation of vitamin D target gene expression. Form a structural point of view this is obvious, since 25(OH)D will bind in the same agonistic VDR conformation as 1,25(OH)2D (51).
The 206 common vitamin D target genes, on which we focus in this study, represent the majority of the vitamin D sensitive part of the PBMC transcriptome, although (in order to further reduce transcriptional noise) they had been filtered by a reference dataset from a recent 1,25(OH)2D3 time course study in PBMCs (31). The estimation of average EC50 values of 322 nM for 25(OH)D3 and 295 nM for 25(OH)D2 compared to 0.48 nM for 1,25(OH)2D3 is the first report on the sensitivity of the transcriptome to vitamin D metabolites. These results provide an additional argument that there is no difference in the response of the transcriptome to 25(OH)D3 and 25(OH)D2. Moreover, our findings indicate with the factor of ~600 a good estimation of the relative gene regulatory potential of 25(OH)D compared to 1,25(OH)2D. Since serum levels of 25(OH)D are even 1,000-times higher than that of 1,25(OH)2D, this supports the option that 25(OH)D can directly modulate the expression of vitamin D target genes. However, 25(OH)D concentrations in the order of 300 nM are far higher than the recommended 100 nM. Therefore, irrespective of the mechanism of action, for persons with a normal vitamin D status the transcriptome as a whole may not be regulated by 25(OH)D. However, there are a few very sensitive genes, such as NXPH4, SLC11A1, ADGR3 (adhesion G protein-coupled receptor G3), G0S2, HBEGF, and PMEPA1 (prostate transmembrane protein, androgen induced 1), which showed EC50 values for 25(OH)D below 150 nM. Thus, in healthy persons with a very high vitamin D status, a few genes may be directly affected by elevated 25(OH)D serum levels.
In addition to the control of the vitamin D status of healthy individuals through careful sun exposure and vitamin D3 or D2 supplementation, there are clinical settings, where supplementation with higher doses of 25(OH)D3 or 25(OH)D2 are recommended (52). These patients may reach, at least for a limited time, far higher 25(OH)D serum levels than healthy individuals. Moreover, 25(OH)D3 is used as a food supplement in animal farming, e.g., for accelerating the growth of chicken (53). Also in these settings elevated 25(OH)D3 serum levels may be reached. Thus, there are a few scenarios, in which larger parts of the vitamin D-dependent transcriptome may be affected by 25(OH)D supplementation.
Studying the transcriptome‘s sensitivity to treatments with vitamin D metabolites led to the interesting observation that vitamin D target genes can be distinguished in high, mid and low responders. This suggests that not all vitamin D target genes respond equally to a stimulation with a given concentration of a VDR ligand. High responding genes, the best known of which are HBEGF (54) and G0S2 (55), get activated at already at 5-times lower levels than the average of all genes, while low responding genes, such as LMNA (56) and STAB1 (48), need for their response up to 5-times higher 1,25(OH)2D3 concentrations than the mean. Interestingly, high responding genes tend to be primary targets that are directly regulated by activated VDR binding to enhancers in the vicinity of the gene's transcription start sites (57), while low responding genes are preferentially secondary targets that are regulated by transcription factors encoded by primary target genes (31). This adds a new characteristic to the description of vitamin D target genes, the mechanistic basis and physiological meaning of which needs to be further explored in the future.
For the main aim of this study, the comparison of the gene regulatory potential 25(OH)D3 and 25(OH)D2, it does not matter, if the observed effects on the PBMC transcriptome are explained either (i) by the enzymatic conversion of 25(OH)D into 1,25(OH)2D during the 4–24 h duration of stimulation phase, which then activates the VDR, (ii) by a direct binding of 25(OH)D to the VDR or (iii) a combination of both. The very low expression of the CYP27B1 gene, in particular in relation to CYP24A1 expression, in PBMCs of one person used in this study is representative for other individuals. This calls into question, whether there was enough 1α-hydroxylase activity to convert within 4 h a sufficient amount of 25(OH)D into 1,25(OH)2D, which then stimulated primary vitamin D target genes. For example, in order to reach a 1,25(OH)2D level of 10 nM, 1% of the 1,000 nM 25(OH)D pool need to be handled within 4 h. However, as it is typical for in vitro cell culture stimulation experiments, supra-physiological concentrations of 1,25(OH)2D3 and 25(OH)D are compared. In fact, the tight regulation of the 1,25(OH)2D3 level in vivo via the molecule's rapid degradation by the enzyme CYP24A1 (58), indicates that concentrations used in vitro most likely never occur in vivo.
In conclusion, 25(OH)D3 and 25(OH)D2 are equally potent in modulating the transcriptome of PBMCs and regulate the same set of vitamin D target genes as the most potent VDR ligand, 1,25(OH)2D3. However, in order to observe consequences of the gene regulatory potential of 25(OH)D, concentrations of 300 nM or higher need to be available. This is three times the recommended serum level, i.e., it does not apply to healthy individuals with a regular vitamin D status.
Data Availability Statement
Fastq files of the raw data can be found at GEO with accession number GSE199273.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Northern Savo Hospital District had approved the study protocol (#9/2014). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AH performed RNA-seq data analysis. CC did RNA isolation and RNA-seq library preparation. CV synthesized the tested compounds. AH and CC wrote the manuscript, which was reviewed by CV. All authors contributed to the article and approved the submitted version.
Funding
Parts of this study were sponsored by Carbogen Amics, Ltd. CC was supported by the WELCOME2—ERA Chair European Union's Horizon2020 research and innovation program under Grant Agreement No. 952601.
Conflict of Interest
This study received funding from Carbogen Amics, Ltd. The funder had the following involvement with the study: Providing vitamin D metabolites. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Konstantin Ivanov for sharing his code as well as the UEF Bioinformatics Center for providing computing resources. Kind thanks to the Gene Core Facility at the EMBL in Heidelberg, Germany, for massively parallel sequencing services.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.910601/full#supplementary-material
References
- 1.Holick MF. Photobiology of vitamin D. Vitamin D 3rd edn. (2011), p. 13–22. 10.1016/B978-0-12-381978-9.10002-2 [DOI] [Google Scholar]
- 2.McKenzie RL, Liley JB, Bjorn LO. UV radiation: balancing risks and benefits. Photochem Photobiol. (2009) 85:88–98. 10.1111/j.1751-1097.2008.00400.x [DOI] [PubMed] [Google Scholar]
- 3.Bendik I, Friedel A, Roos FF, Weber P, Eggersdorfer M. Vitamin D: a critical and essential micronutrient for human health. Front Physiol. (2014) 5:248. 10.3389/fphys.2014.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Japelt RB, Jakobsen J. Vitamin D in plants: a review of occurrence, analysis, and biosynthesis. Front Plant Sci. (2013) 4:136. 10.3389/fpls.2013.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripkovic L, Lambert H, Hart K, Smith CP, Bucca G, Penson S, et al. Comparison of vitamin D2 and vitamin D3 supplementation in raising serum 25-hydroxyvitamin D status: a systematic review and meta-analysis. Am J Clin Nutr. (2012) 95:1357–64. 10.3945/ajcn.111.031070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamberg-Allardt C. Vitamin D in foods and as supplements. Progr Biophys Mol Biol. (2006) 92:33–8. 10.1016/j.pbiomolbio.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 7.Zhu JG, Ochalek JT, Kaufmann M, Jones G, Deluca HF. CYP2R1 is a major, but not exclusive, contributor to 25-hydroxyvitamin D production in vivo. Proc Natl Acad Sci U S A. (2013) 110:15650–5. 10.1073/pnas.1315006110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashman KD, van den Heuvel EG, Schoemaker RJ, Preveraud DP, Macdonald HM, Arcot J. 25-Hydroxyvitamin D as a biomarker of vitamin D status and its modeling to inform strategies for prevention of vitamin D deficiency within the population. Adv Nutri. (2017) 8:947–57. 10.3945/an.117.015578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollis BW. Circulating 25-hydroxyvitamin D levels indicative of vitamin D sufficiency: implications for establishing a new effective dietary intake recommendation for vitamin D. J Nutr. (2005) 135:317–22. 10.1093/jn/135.2.317 [DOI] [PubMed] [Google Scholar]
- 10.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. (2008) 29:726–76. 10.1210/er.2008-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffery LE, Raza K, Hewison M. Vitamin D in rheumatoid arthritis-towards clinical application. Nat Rev Rheumatol. (2016) 12:201–10. 10.1038/nrrheum.2015.140 [DOI] [PubMed] [Google Scholar]
- 12.Sintzel MB, Rametta M, Reder AT. Vitamin D and multiple sclerosis: a comprehensive review. Neurol Ther. (2018) 7:59–85. 10.1007/s40120-017-0086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Infante M, Ricordi C, Sanchez J, Clare-Salzler MJ, Padilla N, Fuenmayor V, et al. Influence of vitamin D on islet autoimmunity and beta-cell function in type 1 diabetes. Nutrients. (2019) 11:185. 10.3390/nu11092185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher J, Cooper SC, Ghosh S, Hewison M. The role of vitamin D in Inflammatory bowel disease: mechanism to management. Nutrients. (2019) 11:1019. 10.3390/nu11051019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang SJ, Wang XH, Liu ZD, Cao WL, Han Y, Ma AG, et al. Vitamin D deficiency and the risk of tuberculosis: a meta-analysis. Drug Des Devel Ther. (2017) 11:91–102. 10.2147/DDDT.S79870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charoenngam N, Shirvani A, Holick MF. Vitamin D and Its potential benefit for the COVID-19 pandemic. Endocr Pract Offic J Am Coll Endocrinol Am Assoc Clinic Endocrinol. (2021) 27:484–93. 10.1016/j.eprac.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maghbooli Z, Sahraian MA, Ebrahimi M, Pazoki M, Kafan S, Tabriz HM, et al. Vitamin D sufficiency, a serum 25-hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS ONE. (2020) 15:e0239799. 10.1371/journal.pone.0239799 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Pludowski P, Holick MF, Pilz S, Wagner CL, Hollis BW, Grant WB, et al. Vitamin D effects on musculoskeletal health, immunity, autoimmunity, cardiovascular disease, cancer, fertility, pregnancy, dementia and mortality-a review of recent evidence. Autoimmun Rev. (2013) 12:976–89. 10.1016/j.autrev.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 19.Tebben PJ, Singh RJ, Kumar R. Vitamin D-mediated hypercalcemia: mechanisms, diagnosis, and treatment. Endocr Rev. (2016) 37:521–47. 10.1210/er.2016-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norman AW. From vitamin D to hormone D: fundamentals of the vitamin D endocrine system essential for good health. Am J Clin Nutr. (2008) 88:491S−499S. 10.1093/ajcn/88.2.491S [DOI] [PubMed] [Google Scholar]
- 21.Bikle D, Christakos S. New aspects of vitamin D metabolism and action—addressing the skin as source and target. Nat Rev Endocrinol. (2020) 20:5. 10.1038/s41574-019-0312-5 [DOI] [PubMed] [Google Scholar]
- 22.Chun RF, Liu PT, Modlin RL, Adams JS, Hewison M. Impact of vitamin D on immune function: lessons learned from genome-wide analysis. Front Physiol. (2014) 5:151. 10.3389/fphys.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlberg C. Nutrigenomics of vitamin D. Nutrients (2019) 11:676. 10.3390/nu11030676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haussler MR, Haussler CA, Bartik L, Whitfield GK, Hsieh JC, Slater S, et al. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutri Rev. (2008) 66:S98–112. 10.1111/j.1753-4887.2008.00093.x [DOI] [PubMed] [Google Scholar]
- 25.Carlberg C. Genome-wide (over)view on the actions of vitamin D. Front Physiol. (2014) 5:167. 10.3389/fphys.2014.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell MJ. Vitamin D and the RNA transcriptome: more than mRNA regulation. Front Physiol. (2014) 5:181. 10.3389/fphys.2014.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. (2011) 39:9181–93. 10.1093/nar/gkr654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verway M, Bouttier M, Wang TT, Carrier M, Calderon M, An BS, et al. Vitamin D induces interleukin-1beta expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS pathogens. (2013) 9:e1003407. 10.1371/journal.ppat.1003407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seuter S, Neme A, Carlberg C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. (2016) 44:4090–104. 10.1093/nar/gkv1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimitrov V, Barbier C, Ismailova A, Wang Y, Dmowski K, Salehi-Tabar R, et al. Vitamin D-regulated gene expression profiles: species-specificity and cell-specific effects on metabolism and immunity. Endocrinology. (2021) 21:162. 10.1210/endocr/bqaa218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanel A, Carlberg C. Time-resolved gene expression analysis monitors the regulation of inflammatory mediators and attenuation of adaptive immune response by vitamin D. Int J Mol Sci. (2022) 23:911. 10.3390/ijms23020911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neme A, Seuter S, Malinen M, Nurmi T, Tuomainen TP, Virtanen JK, et al. In vivo transcriptome changes of human white blood cells in response to vitamin D. J Steroid Biochem Mol Biol. (2019) 188:71–6. 10.1016/j.jsbmb.2018.11.019 [DOI] [PubMed] [Google Scholar]
- 33.Wilhelm F, Mayer E, Norman AW. Biological activity assessment of the 26,23-lactones of 1,25-dihydroxyvitamin D3 and 25-hydroxyvitamin D3 and their binding properties to chick intestinal receptor and plasma vitamin D binding protein. Arch Biochem Biophys. (1984) 233:322–9. 10.1016/0003-9861(84)90452-1 [DOI] [PubMed] [Google Scholar]
- 34.Kutner A, Link RP, Schnoes HK, DeLuca HF. Photoactivable analogs for labeling 25-hydroxyvitamin D3 serum binding protein and for 1,25-dihydroxyvitamin D3 intestinal receptor protein. Bioorg Chem. (1986) 14:134–47. 10.1016/0045-2068(86)90023-4 [DOI] [PubMed] [Google Scholar]
- 35.Holick MF. Vitamin D deficiency. N Engl J Med. (2007) 357:266–81. 10.1056/NEJMra070553 [DOI] [PubMed] [Google Scholar]
- 36.Lou YR, Laaksi I, Syvala H, Blauer M, Tammela TL, Ylikomi T, et al. 25-hydroxyvitamin D3 is an active hormone in human primary prostatic stromal cells. FASEB J. (2004) 18:332–4. 10.1096/fj.03-0140fje [DOI] [PubMed] [Google Scholar]
- 37.Väisänen S, Ryhänen S, Saarela JT, Peräkylä M, Andersin T, Mäenpää PH. Structurally and functionally important amino acids of the agonistic conformation of the human vitamin D receptor. Mol. Pharmacol. (2002) 62:788–94. 10.1124/mol.62.4.788 [DOI] [PubMed] [Google Scholar]
- 38.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. Crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. (2000) 5:173–9. 10.1016/S1097-2765(00)80413-X [DOI] [PubMed] [Google Scholar]
- 39.Lou YR, Molnár F, Peräkylä M, Qiao S, Kalueff AV, St-Arnaud R, Carlberg C, Tuohimaa P. 25-Hydroxyvitamin D3 is an agonistic vitamin D receptor ligand. J Steroid Biochem Mol Biol. (2010) 118:162–70. 10.1016/j.jsbmb.2009.11.011 [DOI] [PubMed] [Google Scholar]
- 40.Vukic M, Neme A, Seuter S, Saksa N, de Mello VD, Nurmi T, et al. Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS ONE. (2015) 10:e0124339. 10.1371/journal.pone.0124339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. (2019) 37:773–782. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. (2020) 38:276–8. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- 43.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature protocols. (2009) 4:1184–91. 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Lun AT, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research. (2016) 5:1438. 10.12688/f1000research.8987.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-Response analysis Using R. PLoS ONE. (2015) 10:e0146021. 10.1371/journal.pone.0146021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanel A, Bendik I, Carlberg C. Transcriptome-wide profile of 25-hydroxyvitamin D3 in pimary immune cells from human peripheral blood. Nutrients. (2021) 13:100. 10.3390/nu13114100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malmberg HR, Hanel A, Taipale M, Heikkinen S, Carlberg C. Vitamin D treatment sequence is critical for transcriptome modulation of immune challenged primary human cells. Front Immunol. (2021) 12:754056. 10.3389/fimmu.2021.754056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlberg C, Seuter S, Heikkinen S. The first genome-wide view of vitamin D receptor locations and their mechanistic implications. Anticancer Res. (2012) 32:271–82. [PubMed] [Google Scholar]
- 50.Hanel A, Neme A, Malinen M, Hamalainen E, Malmberg HR, Etheve S, et al. Common and personal target genes of the micronutrient vitamin D in primary immune cells from human peripheral blood. Scientific reports. (2020) 10:21051. 10.1038/s41598-020-78288-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tocchini-Valentini G, Rochel N, Wurtz JM, Mitschler A, Moras D. Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands. Proc Natl Acad Sci USA. (2001) 98:5491–6. 10.1073/pnas.091018698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quesada-Gomez JM, Bouillon R. Is calcifediol better than cholecalciferol for vitamin D supplementation? Osteoporosis Int: J Establ Result Cooperat Betw Euro Foundat Osteopor Nat Osteopor Foundat USA. (2018) 29:1697–711. 10.1007/s00198-018-4520-y [DOI] [PubMed] [Google Scholar]
- 53.Vazquez JR, Gomez GV, Lopez CC, Cortes AC, Diaz AC, Fernandez SRT, et al. Effects of 25-hydroxycholecalciferol with two D3 vitamin levels on production and immunity parameters in broiler chickens. J Anim Physiol Anim Nutri. (2018) 102:e493–7. 10.1111/jpn.12715 [DOI] [PubMed] [Google Scholar]
- 54.Seuter S, Pehkonen P, Heikkinen S, Carlberg C. Dynamics of 1α,25-dihydroxyvitamin D-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim Biophys Acta. (2013) 1829:1266–75. 10.1016/j.bbagrm.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 55.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon-Cardo C, Munoz A. Genetic signatures of differentiation induced by 1α,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. (2003) 63:7799–806. [PubMed] [Google Scholar]
- 56.Kreienkamp R, Croke M, Neumann MA, Bedia-Diaz G, Graziano S, Dusso A, et al. Vitamin D receptor signaling improves Hutchinson-Gilford progeria syndrome cellular phenotypes. Oncotarget. (2016) 16:30018–31. 10.18632/oncotarget.9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nurminen V, Seuter S, Carlberg C. Primary vitamin D target genes of human monocytes. Front Physiol. (2019) 10:194. 10.3389/fphys.2019.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christakos S, Ajibade DV, Dhawan P, Fechner AJ, Mady LJ. Vitamin D: metabolism. Endocrinol Metab Clin North Am. (2010) 39:243–53. 10.1016/j.ecl.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq files of the raw data can be found at GEO with accession number GSE199273.