Abstract
The Xtra Amp tube, Isocode paper, Instagene matrix, and PrepMan matrix methods were evaluated for their ability to rapidly extract PCR-quality DNAs from Escherichia coli O157:H7 and Cryptosporidium parvum. All methods provided satisfactory DNA from E. coli, and the Xtra Amp and Instagene reagents provided satisfactory DNA from C. parvum.
Traditional methods of assaying water samples for the presence of pathogens rely on the culture of enteric bacteria (enterococci or coliforms) in conjunction with biochemical tests. These methods have several advantages: they are inexpensive, easy to perform, reproducible, and acceptable by those government agencies responsible for setting water quality standards. Molecular biology-based techniques such as PCR may offer distinct advantages in terms of sensitivity and specificity; however, one of the more critical factors in the use of PCR is the ability to provide a quality nucleic acid template free of any inhibitory substances (11). Investigators have evaluated inexpensive sample preparation methods for a variety of pathogens. Orlandi and Lampel (9) used the FTA filter paper-based method to rapidly extract PCR-ready DNAs from Cyclospora spp., Cryptosporidium parvum, and microsporidia, while Lampel et al. (8) used FTA paper to extract DNAs from bacteria (Shigella, Salmonella, and Listeria spp.). Carnevale et al. (2) used Whatman filter paper to obtain DNA from stool samples containing the microsporidian Enterocytozoon bieneusi. Another type of rapid sample preparation matrix is Chelex resin by Bio-Rad (Hercules, Calif.); Hallier-Soulier and Guillot (5) used this to extract DNA from Cryptosporidium parvum oocysts for PCR. A newer rapid extraction reagent is the Xtra Bind matrix which Kozwich et al. (7) used to detect C. parvum viral symbiont RNA via a reverse transcriptase PCR assay.
Here, we report on our use of Instagene matrix (Bio-Rad), Prepman matrix (PE Biosystems, Foster City, Calif.), Xtra Amp tubes (Ansys Diagnostics, Lake Forest, Calif.), and Isocode paper (Schleicher and Schuell, Keene, N.H.) to extract PCR-ready DNAs from pure cultures of E. coli O157:H7 and C. parvum oocysts.
Sample collection and DNA extraction.
Overnight cultures of bacterial stocks were done in Luria-Bertani broth, or minimal lactose broth, at 37°C on a rocking platform. The tubes containing the cultures were centrifuged at a medium speed (1,000 × g) on a tabletop centrifuge to pellet the cells. All but 100 μl of the supernatant was discarded, and the pellet was resuspended. Aliquots were removed and processed as described below. Colonies of E. coli O157:H7 on blood agar plates were removed with a sterile plastic loop and vortexed in 100-μl volumes of sterile water in a microcentrifuge tube to dislodge the bacteria. The sample was then centrifuged, the supernatant was discarded, and the pellet was resuspended in 30 μl of sterile water and processed as described below.
C. parvum oocysts (2 to 8 weeks of age) of the Beltsville strain were recovered from the diarrhea and manure of experimentally infected calves and enumerated by immunofluorescent-antibody microscopy using Merifluor reagent (Meridian Diagnostics, Cincinnati, Ohio) according to the procedure of Fayer et al. (4).
Prior to DNA extraction, oocysts were lysed by freezing and thawing them in a methanol dry-ice bath and heat block. Initially we performed 5 to 10 freeze-thaw cycles, but this number was later reduced to 2 when microscopic examination of oocysts indicated that the majority were lysed by 2 cycles (M. Jenkins, unpublished data).
DNAs were extracted from samples using the Instagene matrix according to the manufacturer's protocol. Briefly, a 30-μl sample was suspended in 200 μl of Instagene matrix and vortexed, followed by heating at 56°C for 15 min. The samples were vortexed again and heated at 100°C for 8 min and then centrifuged to pellet the matrix. Aliquots of 10 and 20 μl (the recommended amount) were used as templates for PCR.
Extraction with the Isocode paper was done according to the manufacturer's instructions. Briefly, 10-μl aliquots of bacterial cultures were spotted directly onto 8-mm-diameter disks of the paper and DNA was eluted in a 100-μl volume of sterile water, with 10 to 20 μl used as the template for PCR.
Sample processing with the PrepMan reagent involved adding up to 30 μl of the sample to 200 μl of the PrepMan reagent, vortexing the mixture, and then heating it at 100°C for 10 min. The preparation was centrifuged to pellet the matrix, and 1 to 5 μl of the supernatant was used as the template for PCR.
For the Xtra Amp tubes, up to 30 μl of sample was added to an Xtra Amp tube containing 75 μl of lysis buffer and the total volume was brought up to 150 μl with sterile water. The mixture was incubated for 10 min at room temperature and then discarded, and the tube was washed twice with 175 μl of wash buffer. The wash buffer was discarded, and laboratory tissues were used to dry the interior of the tube. The PCR master mix (50 μl, described below), including 5 μl of Xtra Amp Enhance reagent, was pipetted directly into the tube, and cycling was increased by three cycles as per the manufacturer's instructions.
DNA amplification protocols.
The E. coli lacZ TaqMan probe was 5′ (6-carboxyfluorescein) CGC CTT ACT GCC TGT TTT GAC 6-carboxy-N,N′,N,N′-tetramethylrhodamine, and the primers were lacZ forward (5′ GTC CCG CAG CGC AGA C; nucleotides 5889 to 5904) and lacZ reverse (5′ GCA GCG TTG TTG CAG TGC; nucleotides 6236 to 6253), which amplify a 364-bp region of the E. coli lacZ gene (GenBank accession no. AE000141.1 [1]). A variety of C. parvum genes were used as targets for PCR, namely, Cp11 (3), 18S rRNA (12), and Cp41 (GenBank accession no. AF144621 [6]). Primers for this gene were outer forward primer Cp41OF, 5′ GAG GAG ATG GAC TAT TCT AGG; outer reverse primer Cp41OR, 5′ GCA ACA GTA GTA AGA GTG GTA; inner forward primer Cp41 IF, 5′ TGT ATG AAT TGG ATA TAT TAT TA; and inner reverse primer Cp41 IR, 5′ GTA AAA GCA ACA CCA TTA CTA. To confirm that the extracted DNA was of sufficient quality for use in PCR, 50 ng of an internal positive control consisting of a cloned fragment of a 15-kDa C. parvum gene was spiked into PCR mixtures and amplified with the Cp11 primer set.
PCRs were done in 50-μl volumes containing 1 U of Taq polymerase (Life Technologies, Gaithersburg, Md.), 10 mM deoxynucleoside triphosphate mix, 1.5 mM MgCl2, 5 μl of 10× PCR buffer, 50 pmol of each primer, 10 pmol of the TaqMan probe, and 5 μl of 10× PCR buffer. For E. coli PCR, cycling parameters were 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 1 min. For the C. parvum Cp41 gene PCR, cycling conditions were 95°C for 1 min, followed by 35 cycles of 95°C for 15 s, 50°C for 30 s, and 60°C for 1 min. The PCR cycling conditions for the Cp11 and 18S rRNA assays were according to published protocols (3, 12).
lacZ amplification from E. coli cultures.
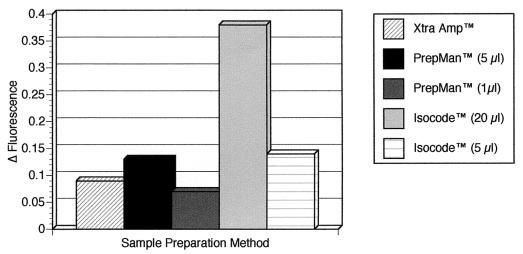
Results from 10-ml overnight cultures of E. coli O157:H7 of one of three replicate experiments evaluating three different DNA extraction methods are shown in Fig. 1. Based on the fluorescence intensities of the lacZ TaqMan probe reactions, the strongest positive reaction was obtained with 20 μl of the Isocode-extracted DNA. Results with 5 μl of PrepMan-extracted DNA and with 5 μl of Isocode-extracted DNA were very similar, with greater fluorescence emissions than those observed for Xtra Amp-extracted DNA and 1 μl of PrepMan-extracted DNA. This pattern was consistent for all three replicates.
FIG. 1.
Results of E. coli lacZ gene TaqMan assay performed on DNAs extracted from broth cultures of the E. coli O157:H7 Odwalla strain using the Xtra Amp tube, Isocode paper, and PrepMan matrix reagents. Thirty microliters (out of 100 μl) of an overnight culture pellet was extracted with the Xtra Amp and PrepMan reagents, and 10 μl was extracted with the Isocode paper. Fifty microliters of the PCR master mix plus 5 μl of Amp Enhance solution were used in the Xtra Amp tube. For the other two methods, 5- and 1-μl aliquots of the PrepMan-extracted DNA and 20- and 5-μl aliquots of Isocode paper-extracted DNA were used as the templates. Fluorescence readings for the 6-carboxyfluorescein reporter dye were obtained after 43 cycles of PCR, were corrected for background, and are plotted on the y axis. Δ Fluorescence, change in fluorescence.
Subsequently, the PrepMan, Instagene matrix, and Isocode methods were evaluated on E. coli O157:H7 colonies removed from agar plates. DNAs extracted using these techniques were subjected to PCR for the virulence gene eae. Results from three replicate experiments indicated that the use of either 1 or 5 μl of PrepMan reagent, 20 μl of Instagene matrix, and 5-μl and 20-μl aliquots of Isocode paper-extracted DNA all yielded a positive reaction for the E. coli O157:H7 colonies (data not shown).
PCR on DNA extracted from oocysts.
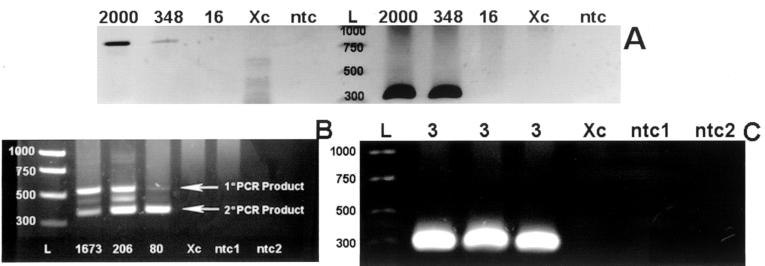
Results from over 30 replicate samples indicated that larger numbers of oocysts (>200) provided positive PCR results with one round of amplification but that obtaining strong positives from 100 or fewer oocysts required a nested PCR. As an example, results from Cp11, Cp41, and 18S rRNA PCR assays done on Instagene-extracted DNAs are shown in Fig. 2. Figure 2A shows positive reactions for aliquots of 2,000 and 348 oocysts, but not 16 oocysts, amplified by one round of PCR with either Cp11 or 18S rRNA. When Cp41 gene PCR was performed on another dilution series of 1,673, 206, and 80 oocysts, samples with 1,673 and 206 oocysts were positive by primary PCR and samples with all three amounts were positive by nested PCR (Fig. 2B). In another experiment, nested Cp11 gene PCR was done on very small quantities of oocysts (n = 3) and positive reactions were observed for three replicate samples (Fig. 2C). If we assume uniform distribution of oocyst DNA in the Instagene matrix and we use 20 μl (of the total 200 μl) of Instagene-derived DNA, the theoretical number of oocysts detected by the nested Cp11 gene PCR under these idealized conditions is less than 1. However, in contrast to a report describing the use of a Chelex-based DNA extraction procedure in conjunction with one round of 18S rRNA PCR to detect 1 oocyst in 20 liters of source water (5), we found that nested PCR was necessary to detect even 80 or fewer oocysts in a 50-μl PCR mixture. A similar requirement for nested 18S rRNA PCR to detect small numbers of oocysts enumerated by the use of micromanipulators was reported by Sturbaum et al. (10).
FIG. 2.
Results of PCR performed on DNA extracted from purified C. parvum oocysts using the Instagene matrix. The quantity of oocysts assayed is indicated above each lane. Lanes L contain the DNA ladder (rung sizes are labeled), lanes Xc contain the extraction control, and lanes ntc, ntc1, and ntc2 contain the no-template controls. (A) 18S rRNA gene (left side of the DNA ladder) and Cp11 gene (right side of the DNA ladder) PCR results with DNAs from 2,000, 348, and 16 oocysts. (B) Results of nested Cp41 gene PCR on 1,673, 206, and 80 oocysts. (The band located between the primary (1°) and secondary (2°) PCR products is an artifact of the gel electrophoresis.) (C) Results of nested Cp11 gene PCR on three replicate samples containing three oocysts.
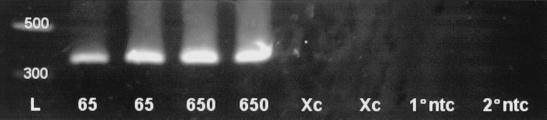
In separate experiments, the PrepMan reagent allowed Cp11 gene amplification of the 2,000-oocyst sample but not the 348- or 16-oocyst samples (data not shown); increasing the amount of PrepMan used as the template from 5 to 20 μl (the same amount used by the Instagene matrix) resulted in no amplification at all, indicating that higher volumes of PrepMan-derived DNA may contain PCR inhibitors. We also evaluated the use of Xtra Amp tubes for DNA extraction from C. parvum oocysts. An example of one assay is shown in Fig. 3; here, nested Cp41 gene PCR was done on quantities of 65 and 650 oocysts. While no bands were observed for the primary PCR, secondary PCR yielded strongly positive bands. This was observed in replicate experiments using Cp11 and 18S rRNA gene PCRs (data not shown).
FIG. 3.
Results of nested Cp41 gene PCR performed on two replicate samples of DNA extracted from 650 and 65 oocysts of C. parvum using Xtra Amp tubes. Lane L, DNA ladder with rung sizes indicated; lanes Xc, extraction control; lanes 1° ntc and 2° ntc, primary and secondary no-template controls, respectively.
In summary, the PrepMan, Instagene, Xtra Amp, and Isocode paper methods all yielded PCR-quality DNA from E. coli colonies and broth culture and the Xtra Amp and Instagene methods yielded satisfactory templates from C. parvum oocysts. All the methods cost under $1.80 per sample, and up to 10 samples can be extracted in 45 min or less. These methods also require less ancillary equipment and reagents than many standard nucleic acid extraction protocols.
Acknowledgments
We thank Rob Palmer, Christina Hohn, Kristy Ludwig, and James Trout for providing technical assistance.
Funding was provided by Water Environment Research Foundation project no. 00-HHE-2a (J. Higgins) and American Water Works Association Research Foundation project no. 2502 (M. Jenkins).
REFERENCES
- 1.Blattner F R, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Carnevale S, Velasquez J N, Labbe J H, Chertcoff A, Cabrera M G, Rodriguez M I. Diagnosis of Enterocytozoon bieneusi by PCR in stool samples eluted from filter paper disks. Clin Diagn Lab Immunol. 2000;7:504–506. doi: 10.1128/cdli.7.3.504-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fayer R, Lewis E J, Trout J M, Graczyk T K, Jenkins M C, Higgins J, Xiao L, Lal A A. Cryptosporidium parvum detection in oysters from commercial harvesting sites in the Chesapeake Bay. Emerg Infect Dis. 1999;5:706–710. doi: 10.3201/eid0505.990513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fayer R, Trout J M, Graczyk T K, Lewis E J. Prevalence of Cryptosporidium, Giardia and Eimeria infections in post-weaned and adult cattle on three Maryland farms. Vet Parasitol. 2000;93:103–112. doi: 10.1016/s0304-4017(00)00356-3. [DOI] [PubMed] [Google Scholar]
- 5.Hallier-Soulier S, Guillot E. Detection of cryptosporidia and Cryptosporidium parvum oocysts in environmental water samples by immunomagnetic separation-polymerase chain reaction. J Appl Microbiol. 2000;89:5–10. doi: 10.1046/j.1365-2672.2000.01029.x. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins M C, Trout J M, Murphy C, Harp J A, Higgins J, Wergin W, Fayer R. Cloning and expression of a DNA sequence encoding a 41-kilodalton Cryptosporidium parvum oocyst wall protein. Clin Diagn Lab Immunol. 1999;6:912–920. doi: 10.1128/cdli.6.6.912-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozwich D, Johansen K A, Landau K, Roehl C A, Woronoff S, Roehl P A. Development of a novel, rapid integrated Cryptosporidium parvum detection assay. Appl Environ Microbiol. 2000;66:2711–2717. doi: 10.1128/aem.66.7.2711-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampel K A, Orlandi P A, Komegay L. Improved template preparation for PCR-based assays for detection of food-borne bacterial pathogens. Appl Environ Microbiol. 2000;66:4539–4542. doi: 10.1128/aem.66.10.4539-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlandi P A, Lampel K A. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J Clin Microbiol. 2000;38:2271–2277. doi: 10.1128/jcm.38.6.2271-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturbaum G D, Reed C, Hoover P J, Jost B H, Marshall M M, Sterling C R. Species-specific, nested PCR-restriction fragment length polymorphism detection of single Cryptosporidium parvum oocysts. Appl Environ Microbiol. 2001;67:2665–2668. doi: 10.1128/AEM.67.6.2665-2668.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]