Abstract

A DNA molecule is highly electrically charged in solution. The electrical potential at the molecular surface is known to vary strongly with the local geometry of the double helix and plays a pivotal role in DNA–protein interactions. Further out from the molecular surface, the electrical field propagating into the surrounding electrolyte bears fingerprints of the three-dimensional arrangement of the charged atoms in the molecule. However, precise extraction of the structural information encoded in the electrostatic “far field” has remained experimentally challenging. Here, we report an optical microscopy-based approach that detects the field distribution surrounding a charged molecule in solution, revealing geometric features such as the radius and the average rise per basepair of the double helix with up to sub-Angstrom precision, comparable with traditional molecular structure determination techniques like X-ray crystallography and nuclear magnetic resonance. Moreover, measurement of the helical radius furnishes an unprecedented view of both hydration and the arrangement of cations at the molecule–solvent interface. We demonstrate that a probe in the electrostatic far field delivers structural and chemical information on macromolecules, opening up a new dimension in the study of charged molecules and interfaces in solution.
Nucleic acids play a central role in biological function. Investigation of the structure of nucleic acids has had a long and compelling history and continues to have far-reaching impact in fields ranging from molecular biology, genetics and disease, to nanotechnology. A range of powerful techniques such as X-ray crystallography, nuclear magnetic resonance (NMR), atomic force microscopy (AFM), small-angle X-ray scattering (SAXS), Forster resonance energy transfer, and optical trapping have generated an unprecedented structural view of DNA, covering all length scales from the atomic to the macroscopic polymer contour level.1−8 The structural properties and function of this biopolymer in solution are strongly governed not only by steric and mechanical aspects but also by electrostatic considerations, as it is among the most highly charged linear polymers known.9,10 Indeed, electrical mobility measurements provided an early demonstration of the link between nucleic acid electrostatics and double helix geometry and molecular topology.11 More recently, magnetic tweezers and SAXS have been used to infer molecular properties of nucleic acids via the measurement of an intra- or intermolecular interaction potential.12,13 Furthermore, anomalous SAXS and atomic emission spectroscopy (AES) have probed the properties of the counterion atmosphere enveloping nucleic acid molecules,14,15 while X-ray photoelectron spectroscopy (XPS) has shed light on the interface between a charged nanoparticle and the surrounding electrolyte.16 To our knowledge, the ability to glean structural information on a diffusing macromolecule and its interface with the electrolyte through precise measurement of the electrical repulsion due to the molecule has not been demonstrated.
Experimental Approach
We optically visualize and measure the strength
of electrostatic
repulsions between a charged molecule and like-charged probe surfaces
in solution using wide-field fluorescence microscopy and the recently
developed escape time electrometry (ETe) approach.17 In contrast to scanning probe techniques where
a nanoscale entity is placed in near contact with a stationary object
of interest, our experiment involves a pair of flat, featureless probe
surfaces placed in the “far field” of a diffusing charged
molecular species in solution. We qualitatively define the electrostatic
“far field” as the region in the electrolyte at a distance
greater than a Debye length,  , from
the object. Here,
, from
the object. Here,  nm is a length
scale governing the decay
of electrostatic interactions in aqueous solution at temperature T = 298 K, where the salt concentration in solution, c ≈ 1–1.5 mM in this work, implies
nm is a length
scale governing the decay
of electrostatic interactions in aqueous solution at temperature T = 298 K, where the salt concentration in solution, c ≈ 1–1.5 mM in this work, implies  8 nm. ETe measures the
reduction in system free energy associated with transferring a charged
molecule from a gap between like-charged parallel plates into a nanostructured
“trap” region of very weak confinement where the molecule–plate
repulsion is negligible18 (Figure 1a,b). The system is at thermodynamic
equilibrium, and there are no externally applied fields. We create
an array of such electrostatic fluidic traps using periodic nanostructured
indentations in one surface of a parallel plate slit composed of silica
surfaces separated by a gap of typical height, 2h = 75 nm. We introduce nucleic acid molecules at a concentration
of 50–100 pM labeled with exactly two fluorescent dye molecules
of ATTO 532, suspended in 1 mM Tris buffer and ≈1.2 mM monovalent
salt solution, pH 9, into a system with multiple parallel lattices
of traps (Figure 1a).
Alkaline pH in the experiment ensures that the weakly acidic SiO2 walls of our nanoslit system are strongly charged.19 A low (mM) concentration of monovalent salt,
in turn, ensures that the electrostatic interactions between a charged
molecule and the walls of the slit are sufficiently strong and long-ranged,
yielding long-lived trap states of ≈50–200 ms duration.
Analytical characterization of the molecular species in the study
using, for example, circular dichroism spectroscopy verifies that
the solution conditions in our measurement conditions do not perturb
the molecules’ structural integrity (see Supporting Information Figure S2).
8 nm. ETe measures the
reduction in system free energy associated with transferring a charged
molecule from a gap between like-charged parallel plates into a nanostructured
“trap” region of very weak confinement where the molecule–plate
repulsion is negligible18 (Figure 1a,b). The system is at thermodynamic
equilibrium, and there are no externally applied fields. We create
an array of such electrostatic fluidic traps using periodic nanostructured
indentations in one surface of a parallel plate slit composed of silica
surfaces separated by a gap of typical height, 2h = 75 nm. We introduce nucleic acid molecules at a concentration
of 50–100 pM labeled with exactly two fluorescent dye molecules
of ATTO 532, suspended in 1 mM Tris buffer and ≈1.2 mM monovalent
salt solution, pH 9, into a system with multiple parallel lattices
of traps (Figure 1a).
Alkaline pH in the experiment ensures that the weakly acidic SiO2 walls of our nanoslit system are strongly charged.19 A low (mM) concentration of monovalent salt,
in turn, ensures that the electrostatic interactions between a charged
molecule and the walls of the slit are sufficiently strong and long-ranged,
yielding long-lived trap states of ≈50–200 ms duration.
Analytical characterization of the molecular species in the study
using, for example, circular dichroism spectroscopy verifies that
the solution conditions in our measurement conditions do not perturb
the molecules’ structural integrity (see Supporting Information Figure S2).
Figure 1.
High-precision ETe measurements on nucleic acid
fragments. (a) Schematic representation (not-to-scale) of fluorescently
labeled nucleic acid molecules confined in an array of electrostatic
fluidic traps and imaged using wide-field optical microscopy (top).
Maximum intensity projection of 500 fluorescence images of parallel
arrays of ≈700 traps imaged for 20 s (bottom). (b) Calculated
spatial distribution of minimum axial electrostatic free energy,  , in a representative trap (top). Labels
“1” and “2” denote locations of the molecule
outside and inside the potential well, respectively, and refer to
spatial locations in the trapping nanostructure depicted in the device
schematic in (a). A time course of optical images in a single trap
(bottom) displays the duration of a single recorded residence event
of duration, Δt. (c) Probability density distributions, P(Δt), of escape times, Δt, for N = 104 escape events
for measurements on double-stranded B-DNA (solid lines) and A-RNA
(dashed lines) in 1.23 mM LiCl for fragment length nb = 30 (red), 40 (blue), and 60 (green) basepairs fitted
to the expression
, in a representative trap (top). Labels
“1” and “2” denote locations of the molecule
outside and inside the potential well, respectively, and refer to
spatial locations in the trapping nanostructure depicted in the device
schematic in (a). A time course of optical images in a single trap
(bottom) displays the duration of a single recorded residence event
of duration, Δt. (c) Probability density distributions, P(Δt), of escape times, Δt, for N = 104 escape events
for measurements on double-stranded B-DNA (solid lines) and A-RNA
(dashed lines) in 1.23 mM LiCl for fragment length nb = 30 (red), 40 (blue), and 60 (green) basepairs fitted
to the expression  . In order to enable comparison across different
molecular species, P(Δt) data
series are rescaled such that the maximum value is 1. Average escape
times, tesc, and measured effective charge
values, qm, are as follows: tesc,30B = 52.2 ± 0.3 ms (qm,30B = −25.28 ± 0.07e), tesc,40B 93.9 ± 0.4 ms (
. In order to enable comparison across different
molecular species, P(Δt) data
series are rescaled such that the maximum value is 1. Average escape
times, tesc, and measured effective charge
values, qm, are as follows: tesc,30B = 52.2 ± 0.3 ms (qm,30B = −25.28 ± 0.07e), tesc,40B 93.9 ± 0.4 ms ( 30.46 ± 0.06e), and
30.46 ± 0.06e), and  242.5 ± 1.1 ms (
242.5 ± 1.1 ms ( 40.71 ± 0.07e) for
B-DNA and
40.71 ± 0.07e) for
B-DNA and  46.3 ± 0.2 ms (−23.86
±
0.04e),
46.3 ± 0.2 ms (−23.86
±
0.04e),  70.4 ± 0.8 ms (−28.35
±
0.13e), and
70.4 ± 0.8 ms (−28.35
±
0.13e), and  192.5 ± 0.6 ms (−37.26
±
0.04e) for A-RNA. B-DNA systematically displays 10–20%
longer escape times and higher magnitudes of effective charge than
A-RNA. Space filling structures of B-DNA and A-RNA reproduced with
permission from ref (3) (right).
192.5 ± 0.6 ms (−37.26
±
0.04e) for A-RNA. B-DNA systematically displays 10–20%
longer escape times and higher magnitudes of effective charge than
A-RNA. Space filling structures of B-DNA and A-RNA reproduced with
permission from ref (3) (right).
Imaging the escape dynamics of
trapped single molecules permits
us to identify individual molecular residence events of duration Δt in each trap. Photobleaching of the fluorescent dyes and
any potential impact thereof on the measurement have been carefully
explored in previous work.17 Because molecular
residence times in the trap are much shorter than dye photobleaching
times, we expect dye photophysics and photochemistry not to influence
the accuracy of our measured escape times. Overdamped escape of an
object from a potential well can be treated as a Poisson process with
residence times that are exponentially distributed.20 Fits of the measured probability density function of residence
times, P(Δt), to an exponential
function of the form  permit us to extract precise measurements
of the molecular species’ average time to escape,
permit us to extract precise measurements
of the molecular species’ average time to escape,  (Figure 1b,c). The average
escape time, in turn, is expected
to depend exponentially on well depth, according to the relation
(Figure 1b,c). The average
escape time, in turn, is expected
to depend exponentially on well depth, according to the relation  ,20 permitting
us to relate measured
,20 permitting
us to relate measured  values
to the depth of the trap, W, in the regime of W > 4kBT. In practice, we use Brownian dynamics
(BD) simulations of the escape process in order to accurately convert
measurements of
values
to the depth of the trap, W, in the regime of W > 4kBT. In practice, we use Brownian dynamics
(BD) simulations of the escape process in order to accurately convert
measurements of  to
the well depth, W,
as described previously17,21,22 (Supporting Information Section S2).
to
the well depth, W,
as described previously17,21,22 (Supporting Information Section S2).
In our BD simulations, we treat molecules as effective spheres
of a radius equal to the measured hydrodynamic radius of the molecule.
The hydrodynamic radius, rH, of each molecular
species was measured using fluorescence correlation spectroscopy as
described in the Supporting Information (Supplementary Methods). The use of an effective hydrodynamic radius,
which ignores the anisotropic diffusive behavior of non-spherical
objects, is valid when the translational diffusive length scale of
interest, ls, is much larger than the
length of the molecule, l, or in other words, when
the ratio of rotational and translational diffusive timescales  . For a rigid cylinder of length l, this ratio is approximately l2/ls2 (see ref (23)). The relevant length scale for translational diffusion, ls, in an ETe measurement corresponds
to the radius of a nanostructured pocket which is typically 250–300
nm. Given the contour length of a 60 bp B-DNA (l ≈
20 nm), which is the longest fragment considered here, we have l2/ls ≈ 0.01 ≪ 1, which ensures
that the translational diffusion of an anisometric object may be treated
as equivalent to that of an effective sphere for large displacements.
It is worth noting that we ignore inertial effects in our BD simulations
on the grounds that the momentum relaxation time of the molecule is
very small.24 Although inertial BD simulations
of large supercoiled DNA plasmids (∼1000 bp) have shown that
mass can have some effects on conformation transition rates in equilibrium,
they do also demonstrate that the translational diffusion coefficient
of these molecules is accurately captured by conventional BD simulations.24,25 Thus, for short nucleic acid fragments, which are expected to behave
like rigid rods, BD simulations in the overdamped regime are expected
to provide an accurate description of our escape time problem.
. For a rigid cylinder of length l, this ratio is approximately l2/ls2 (see ref (23)). The relevant length scale for translational diffusion, ls, in an ETe measurement corresponds
to the radius of a nanostructured pocket which is typically 250–300
nm. Given the contour length of a 60 bp B-DNA (l ≈
20 nm), which is the longest fragment considered here, we have l2/ls ≈ 0.01 ≪ 1, which ensures
that the translational diffusion of an anisometric object may be treated
as equivalent to that of an effective sphere for large displacements.
It is worth noting that we ignore inertial effects in our BD simulations
on the grounds that the momentum relaxation time of the molecule is
very small.24 Although inertial BD simulations
of large supercoiled DNA plasmids (∼1000 bp) have shown that
mass can have some effects on conformation transition rates in equilibrium,
they do also demonstrate that the translational diffusion coefficient
of these molecules is accurately captured by conventional BD simulations.24,25 Thus, for short nucleic acid fragments, which are expected to behave
like rigid rods, BD simulations in the overdamped regime are expected
to provide an accurate description of our escape time problem.
The highly non-linear dependence of the measurand (escape time,  ) on
the measurable (well depth, W) facilitates precise
interaction energy measurements.
Observation of a large number of escape events, N ≈ 104, reduces the fractional statistical uncertainty
in the determination of W to about 0.1%.22 Importantly, the dominant contribution to the
trap depth, W, is the electrostatic free energy of
interaction,
) on
the measurable (well depth, W) facilitates precise
interaction energy measurements.
Observation of a large number of escape events, N ≈ 104, reduces the fractional statistical uncertainty
in the determination of W to about 0.1%.22 Importantly, the dominant contribution to the
trap depth, W, is the electrostatic free energy of
interaction,  , which
has robust theoretical underpinnings
in the Poisson–Boltzmann (PB) framework for solution phase
electrostatics as discussed further later.26−28 Correction
of a contribution from axial spatial fluctuations to the total free
energy, W, permits us to determine
, which
has robust theoretical underpinnings
in the Poisson–Boltzmann (PB) framework for solution phase
electrostatics as discussed further later.26−28 Correction
of a contribution from axial spatial fluctuations to the total free
energy, W, permits us to determine  with
high precision, as described further
in Supporting Information, Section S2.
We have previously shown that
with
high precision, as described further
in Supporting Information, Section S2.
We have previously shown that  may
be regarded in terms of the product
of the effective charge of the molecule in solution,
may
be regarded in terms of the product
of the effective charge of the molecule in solution,  , and
the electrical potential, ϕm, at the midplane of
the slit, such that
, and
the electrical potential, ϕm, at the midplane of
the slit, such that  =
=  .29,30 If ϕm is accurately known,
the measurable in our experiment is the effective
charge,
.29,30 If ϕm is accurately known,
the measurable in our experiment is the effective
charge,  , of
the molecular species under the experimental
conditions. Note that our values of
, of
the molecular species under the experimental
conditions. Note that our values of  for
charged spheres and cylinders are comparable
to those encountered in other charge renormalization theories.30−32 Furthermore, our interaction-energy-based definition of
for
charged spheres and cylinders are comparable
to those encountered in other charge renormalization theories.30−32 Furthermore, our interaction-energy-based definition of  (i.e.,
(i.e.,  =
=  ) is identical to that in Kjellander’s
dressed ion theory.33−36
) is identical to that in Kjellander’s
dressed ion theory.33−36
The principle behind the present study may be summarized as
follows:
Accurate measurements of the electrostatic free energy,  permit us to measure the effective
charge,
permit us to measure the effective
charge,  , of
three different lengths of a nucleic
acid species (e.g., A- or B-form helix in this work). Theoretically
expected effective charge values may also be calculated using the
PB theoretical framework for each length of the fragment, as described
previously (see Supporting Information Section
S7).30 As described further below, calculations
show that
, of
three different lengths of a nucleic
acid species (e.g., A- or B-form helix in this work). Theoretically
expected effective charge values may also be calculated using the
PB theoretical framework for each length of the fragment, as described
previously (see Supporting Information Section
S7).30 As described further below, calculations
show that  depends
strongly on geometrical dimensions
of the molecular species of interest, for example, the rise per basepair, b, and helical radius, r. The precise functional
form of this dependence is itself a function of the length of each
fragment, as shown in Figures 3a and S4a. Thus,
we have three independent theoretical relationships relating effective
charge with molecular geometry for the fragment lengths under consideration.
Since the effective charge of the molecular conformation under study
(e.g., either the A-form or the B-form helix) may be described by
a common pair of underlying geometric parameters
(e.g., rise per basepair, b, and helical radius, r), a comparison of the measured effective charge values
with the theoretically expected values for the three lengths of the
double helix permits us to extract estimates of the two geometric
properties of interest (described in detail in Supporting Information, Section S4). The third parameter we
extract from the analysis characterizes the measurement device. We
find that measurements of the helical radius in electrolytes containing
cations of different radii further permit us to make inferences on
the structure of the molecular interface with the electrolyte.
depends
strongly on geometrical dimensions
of the molecular species of interest, for example, the rise per basepair, b, and helical radius, r. The precise functional
form of this dependence is itself a function of the length of each
fragment, as shown in Figures 3a and S4a. Thus,
we have three independent theoretical relationships relating effective
charge with molecular geometry for the fragment lengths under consideration.
Since the effective charge of the molecular conformation under study
(e.g., either the A-form or the B-form helix) may be described by
a common pair of underlying geometric parameters
(e.g., rise per basepair, b, and helical radius, r), a comparison of the measured effective charge values
with the theoretically expected values for the three lengths of the
double helix permits us to extract estimates of the two geometric
properties of interest (described in detail in Supporting Information, Section S4). The third parameter we
extract from the analysis characterizes the measurement device. We
find that measurements of the helical radius in electrolytes containing
cations of different radii further permit us to make inferences on
the structure of the molecular interface with the electrolyte.
Figure 3.
Measuring the helical rise per basepair and radius of
the double
helix. (a) Principle behind the measurement of the helical rise per
basepair, b, and radius, r, of the
double helix, for an ideal experiment, free of systematic measurement
uncertainty (i.e., fM = 1). Schematic
representations of three lengths of a double-stranded nucleic acid
species surrounded by a cloud of screening counterions (left). A measured
value  for each molecular species of length n bp, in conjunction
with the corresponding calculated 2D
function (colored surface) for the effective charge,
for each molecular species of length n bp, in conjunction
with the corresponding calculated 2D
function (colored surface) for the effective charge,  , generates a curve of possible solutions
in b and r. Intersection of three
such curves for n = 30, 40, and 60 bp yields a probability-weighted
manifold of solutions from which measured values, bm and rm, for the rise and
radius, respectively, of each helix form can be obtained. (b) Measured b–r probability manifolds for B-DNA
(top) and A-RNA (bottom) for an experiment performed in 1.2 mM CsCl.
Since fM ≠ 1 in experiments, measured b–r manifolds are broader than those
in the ideal case depicted in (a) yielding
, generates a curve of possible solutions
in b and r. Intersection of three
such curves for n = 30, 40, and 60 bp yields a probability-weighted
manifold of solutions from which measured values, bm and rm, for the rise and
radius, respectively, of each helix form can be obtained. (b) Measured b–r probability manifolds for B-DNA
(top) and A-RNA (bottom) for an experiment performed in 1.2 mM CsCl.
Since fM ≠ 1 in experiments, measured b–r manifolds are broader than those
in the ideal case depicted in (a) yielding  3.2 Å and
3.2 Å and  10.4 Å and
10.4 Å and  2.6 Å and
2.6 Å and  12.5 Å for B-DNA and A-RNA, respectively.
12.5 Å for B-DNA and A-RNA, respectively.
For a highly charged molecule in solution, it has
been shown that  , where η is a molecular
geometry-dependent
charge renormalization factor.30−32,37,38
, where η is a molecular
geometry-dependent
charge renormalization factor.30−32,37,38 denotes
the net electrical charge in the
molecular structure and stems from the sum of charge carried by the
ionized structural groups and bound ions from the electrolyte. A highly
acidic molecule like DNA, n basepairs in length and
carrying a chemical modification at both 5′-end phosphates,
has a structural charge
denotes
the net electrical charge in the
molecular structure and stems from the sum of charge carried by the
ionized structural groups and bound ions from the electrolyte. A highly
acidic molecule like DNA, n basepairs in length and
carrying a chemical modification at both 5′-end phosphates,
has a structural charge  at pH 7 and higher (see Supporting Information, Section S3.2). Here, e is the elementary charge
and
at pH 7 and higher (see Supporting Information, Section S3.2). Here, e is the elementary charge
and  is the amount
of charge due to the backbone
phosphate groups on the molecule which are all fully ionized in our
experiments. However, if a number of positively charged counterions,
δ, associate with the molecule, for example, via energetic interactions
beyond the purely Coulombic that are already accounted for within
the PB model, then
is the amount
of charge due to the backbone
phosphate groups on the molecule which are all fully ionized in our
experiments. However, if a number of positively charged counterions,
δ, associate with the molecule, for example, via energetic interactions
beyond the purely Coulombic that are already accounted for within
the PB model, then  , where
, where  is an inverse ion affinity parameter which
tends to zero as
is an inverse ion affinity parameter which
tends to zero as  .
.
To a first approximation, a periodic
linear charged structure such
as a short fragment of a double-stranded nucleic acid may be viewed
as a smooth, charged cylinder of finite length.39,40 Here, η depends on the charge density of the polyelectrolyte
and therefore on the axial base spacing, b, and the
radius of the polyelectrolyte backbone, r. Considering
a short stretch of a nucleic acid whose contour length, l = nb, is of the order of the Debye length, η
further depends on l.17,41,42 Upon approximating a short stretch of DNA (≤60
bp) by a rigid cylinder of radius r and length l, we thus have  which can be calculated for a range of b and r values using the PB framework (Figures 2 and 3).29,30 Finally, for a given molecular
geometry and structural charge, η is essentially independent
of ion affinity for
which can be calculated for a range of b and r values using the PB framework (Figures 2 and 3).29,30 Finally, for a given molecular
geometry and structural charge, η is essentially independent
of ion affinity for  0.7. Although η does exhibit
some
dependence on the salt concentration, c, this variation
is negligible over the small range in experimental uncertainty in c in a given measurement.30,31
0.7. Although η does exhibit
some
dependence on the salt concentration, c, this variation
is negligible over the small range in experimental uncertainty in c in a given measurement.30,31
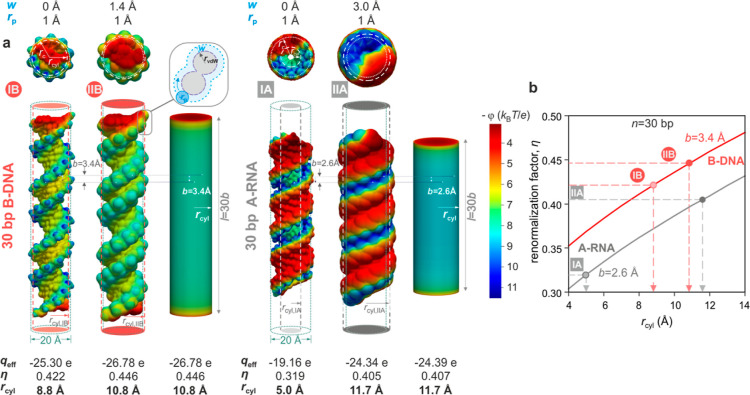
Figure 2.
Modeling the
double helix as a smooth charged cylinder of finite
length. (a) Distributions of surface electrostatic potential, ϕ,
for two molecular models of a 30 bp fragment of B-DNA (IB and IIB—left)
and A-RNA (IA and IIA—right) generated based on atomic coordinates
with rolling probe radius (rp = 1 Å)
and solvent accessible surface (w) parameter values
as listed and pictured (inset) alongside axial projections of the
molecular models (top panel). Surface potential distributions for
corresponding smooth charged cylinders equivalent to models IIB and
IIA carrying a total charge  60e with radii,
60e with radii,  10.8 Å and
10.8 Å and  11.7 Å, respectively, and length 30b Å in each case. The radius of the equivalent cylinder,
11.7 Å, respectively, and length 30b Å in each case. The radius of the equivalent cylinder,  (dashed
lines), may be compared with a
nominal double-helical radius rc = 10
Å (dotted lines). (b) Calculated trends for the renormalization
factor,
(dashed
lines), may be compared with a
nominal double-helical radius rc = 10
Å (dotted lines). (b) Calculated trends for the renormalization
factor,  , for cylinders of radius
, for cylinders of radius  and length 30b Å,
with nominal values of b = 3.4 Å for B-DNA (red
line) and 2.6 Å for A-RNA (gray line). η values for the
four molecular models can be related to those for smooth cylinders
and correspond to
and length 30b Å,
with nominal values of b = 3.4 Å for B-DNA (red
line) and 2.6 Å for A-RNA (gray line). η values for the
four molecular models can be related to those for smooth cylinders
and correspond to  8.8 Å(effective
vdW surface),
8.8 Å(effective
vdW surface),  10.8 Å (effective SAS),
10.8 Å (effective SAS),  5 Å (vdWS),
and
5 Å (vdWS),
and  11.7 Å (SAS), two of which are depicted
in (a). Panels are reproduced from ref (48), with the permission of AIP Publishing.
11.7 Å (SAS), two of which are depicted
in (a). Panels are reproduced from ref (48), with the permission of AIP Publishing.
In view
of the grooved molecular surface of double-stranded nucleic
acids and the helicoid distribution of charge on the molecular backbone,
we first test the quality of the smooth cylinder electrostatic model
for DNA in the context of our experiment (Figure 2). We calculate  and
therefore determine
and
therefore determine  values
for molecular models of the full
3D structure of 30 bp B-DNA and A-RNA molecules constructed using
the 3DNA platform (Supporting Information Section S7).43 We then determine
values
for molecular models of the full
3D structure of 30 bp B-DNA and A-RNA molecules constructed using
the 3DNA platform (Supporting Information Section S7).43 We then determine  values
for smooth cylinders of variable
radii, r, and the same axial rise per basepair, b, as the molecular helices. Cylinders of radius
values
for smooth cylinders of variable
radii, r, and the same axial rise per basepair, b, as the molecular helices. Cylinders of radius  whose
whose  values
are identical to those of the molecular
helices within computational error (estimated at <0.1%) are termed
equivalent cylinders. Physically, this means that the computed electrostatic
free energy difference between the "free" and "trapped"
states (states1
and 2 in Fig. 1b respectively)of
the molecular helix,
values
are identical to those of the molecular
helices within computational error (estimated at <0.1%) are termed
equivalent cylinders. Physically, this means that the computed electrostatic
free energy difference between the "free" and "trapped"
states (states1
and 2 in Fig. 1b respectively)of
the molecular helix,  , is indistinguishable from that due to
a smooth cylinder of radius
, is indistinguishable from that due to
a smooth cylinder of radius  . Importantly,
a domain decomposition of
the free energy in the system demonstrates that the electrostatic
well depth of the trap,
. Importantly,
a domain decomposition of
the free energy in the system demonstrates that the electrostatic
well depth of the trap,  , stems in nearly equal proportions from
the “near field” (the region within about 2 nm) of both
the molecule and the slit surfaces48. Note
that high-resolution structural studies have shown that the double
helix can have local structural variability, for example, sequence-dependent
and thermally induced variation of the rise per basepair along the
molecular contour, which is not captured in the uniformly charged
cylinder model.44−47 Our approach measures an averaged interaction response from the
molecule. Whereas thermal variations are expected to average out in
the measurement, local sequence-dependent variations will be interpreted
in terms of an average rise per basepair parameter
characterizing the molecule. Therefore, for the current work, we assume
that a coarse-grained model that treats the double helix as a uniformly
charged cylinder provides a sufficient description of the measurement.
Although mapping of the molecular problem on to that of a uniform
cylinder can be highly informative, future work could directly compare
electrometry measurements with expectations for molecular structural
models.
, stems in nearly equal proportions from
the “near field” (the region within about 2 nm) of both
the molecule and the slit surfaces48. Note
that high-resolution structural studies have shown that the double
helix can have local structural variability, for example, sequence-dependent
and thermally induced variation of the rise per basepair along the
molecular contour, which is not captured in the uniformly charged
cylinder model.44−47 Our approach measures an averaged interaction response from the
molecule. Whereas thermal variations are expected to average out in
the measurement, local sequence-dependent variations will be interpreted
in terms of an average rise per basepair parameter
characterizing the molecule. Therefore, for the current work, we assume
that a coarse-grained model that treats the double helix as a uniformly
charged cylinder provides a sufficient description of the measurement.
Although mapping of the molecular problem on to that of a uniform
cylinder can be highly informative, future work could directly compare
electrometry measurements with expectations for molecular structural
models.
We considered two molecular models each for B-DNA and
A-RNA, with
all molecular surfaces generated using rolling probe radii, rp = 1 Å. Models-IA and -IB were generated
using reference van der Waals (vdW) values for all atoms, while models-II
A and -IIB entail atomic radii that are all w = 3
Å and w = 1.4 Å larger than the vdW values,
respectively (Figure 2a). While model-I is expected to capture the vdW surface (vdWS) of
the molecule, a larger atomic radius in model-II is expected to mimic
a “solvent accessible surface” (SAS) which defines the
distance of closest approach of the center of a water molecule to
the macromolecular structure. For B-DNA, we find that models-I and
-II yield equivalent electrostatic cylinder radii,  9 Å
and
9 Å
and  11 Å, respectively,
which are in remarkable
agreement with the nominal outer helical radius, rc ≈ 10 Å, inferred from molecular crystal
structures (Figure 2b).28 Interestingly, for A-RNA, the
11 Å, respectively,
which are in remarkable
agreement with the nominal outer helical radius, rc ≈ 10 Å, inferred from molecular crystal
structures (Figure 2b).28 Interestingly, for A-RNA, the  values
for the two structures considered
are rather different:
values
for the two structures considered
are rather different:  5 Å
and
5 Å
and  12 Å, suggesting that an experimental
measurement with sufficient accuracy may be able to distinguish between
the two models, shedding light on molecular interfacial structural
detail in an electrolyte (Supporting Information Section S7). The modeling procedure has been described in detail
previously48 and is summarized in Supporting Information Section S7.
12 Å, suggesting that an experimental
measurement with sufficient accuracy may be able to distinguish between
the two models, shedding light on molecular interfacial structural
detail in an electrolyte (Supporting Information Section S7). The modeling procedure has been described in detail
previously48 and is summarized in Supporting Information Section S7.
Precise
measurements (uncertainty<1%) of  on
three nucleic acid fragments of different
lengths may be compared with calculated
on
three nucleic acid fragments of different
lengths may be compared with calculated  values
for charged cylinders in order to
extract measures of three unknown quantities of interest (Figure 3a). Two of these
three unknowns describe geometric properties of the underlying molecular
structure, namely, the radius of the helix, r, and
the axial helical rise per basepair, b. The third
unknown relates to experimental measurement conditions and the associated
uncertainty. Experiments generally contain parameters that need to
be well controlled, or accounted for, in order to foster accurate
measurements. We account for uncertainties in various experimental
quantities through the use of two correction terms: one is a multiplicative
factor, fM, and the other is an additive
quantity, fA, such that the measured effective
charge for each fragment of size n bases is given
by
values
for charged cylinders in order to
extract measures of three unknown quantities of interest (Figure 3a). Two of these
three unknowns describe geometric properties of the underlying molecular
structure, namely, the radius of the helix, r, and
the axial helical rise per basepair, b. The third
unknown relates to experimental measurement conditions and the associated
uncertainty. Experiments generally contain parameters that need to
be well controlled, or accounted for, in order to foster accurate
measurements. We account for uncertainties in various experimental
quantities through the use of two correction terms: one is a multiplicative
factor, fM, and the other is an additive
quantity, fA, such that the measured effective
charge for each fragment of size n bases is given
by  . The correction factor, fM = fionRfϕ, accounts
for effects that influence the measured effective charge in a multiplicative
fashion and is, in turn, composed of two terms. fϕ reflects a property of the measurement apparatus
and involves the overall uncertainty in the midplane electrical potential,
ϕm, in the slit. ϕm directly relates
to the effective surface potential of the silica surfaces, ϕs, via the relation ϕm = 2ϕs exp(−
. The correction factor, fM = fionRfϕ, accounts
for effects that influence the measured effective charge in a multiplicative
fashion and is, in turn, composed of two terms. fϕ reflects a property of the measurement apparatus
and involves the overall uncertainty in the midplane electrical potential,
ϕm, in the slit. ϕm directly relates
to the effective surface potential of the silica surfaces, ϕs, via the relation ϕm = 2ϕs exp(− h), and we use
a nominal
value of ϕs = −2.8
h), and we use
a nominal
value of ϕs = −2.8 for our experimental conditions as noted
in previous work.21 Examples of factors
that contribute to variations in fϕ include the finite accuracy of the order of he ≈ 1 nm in the height of the slit, the particular value
of the surface charge density on the confining walls, the salt concentration,
and possible ionic species effects on ϕs. fion, in turn, represents a relative "inverse affinity" of
cations for
the nucleic acid molecule, measured with respect to Na+ ions, such that fNaR = 1. Finally, fA is an additive term, the main contribution to which is
for our experimental conditions as noted
in previous work.21 Examples of factors
that contribute to variations in fϕ include the finite accuracy of the order of he ≈ 1 nm in the height of the slit, the particular value
of the surface charge density on the confining walls, the salt concentration,
and possible ionic species effects on ϕs. fion, in turn, represents a relative "inverse affinity" of
cations for
the nucleic acid molecule, measured with respect to Na+ ions, such that fNaR = 1. Finally, fA is an additive term, the main contribution to which is  0.5e, the effective charge
of the fluorescent label covalently coupled to both 5′-phosphates
of the double helix, which is determined by measurement (see Supporting Information, Section S3).
0.5e, the effective charge
of the fluorescent label covalently coupled to both 5′-phosphates
of the double helix, which is determined by measurement (see Supporting Information, Section S3).
We
constructed 30, 60, and 40 bp fragments of dsDNA and dsRNA and
measured the effective charge for each molecular species. We then
compared the measured effective charge values, qm, with the corresponding calculated values,  , for
cylinders with linear charge spacing
corresponding to rise per basepair values, b, ranging
from 2 to 5 Å and the radius, r, in the range
of 6–30 Å. In principle, simultaneously solving the three
known relationships for
, for
cylinders with linear charge spacing
corresponding to rise per basepair values, b, ranging
from 2 to 5 Å and the radius, r, in the range
of 6–30 Å. In principle, simultaneously solving the three
known relationships for  with
with  for the three fragments should yield values
for the unknowns b and r when fM = 1 (Figure 3a). However, in general, fM ≠ 1, and the measurement data, which are of the form
for the three fragments should yield values
for the unknowns b and r when fM = 1 (Figure 3a). However, in general, fM ≠ 1, and the measurement data, which are of the form  , are not single-valued but rather carry
Gaussian-distributed uncertainties of width
, are not single-valued but rather carry
Gaussian-distributed uncertainties of width  about the mean value, qm. Thus, we have three functions of the form
about the mean value, qm. Thus, we have three functions of the form  Pairwise division of these three
equations
eliminates fM and results in two functions
that may be numerically solved to yield a probability-weighted manifold
of solutions in b and r (Figure 3b). We determine
the most probable measured values bm and rm using an algorithm developed based on simulated
input data. fM is then determined self-consistently
by substitution into one of the three equations for
Pairwise division of these three
equations
eliminates fM and results in two functions
that may be numerically solved to yield a probability-weighted manifold
of solutions in b and r (Figure 3b). We determine
the most probable measured values bm and rm using an algorithm developed based on simulated
input data. fM is then determined self-consistently
by substitution into one of the three equations for  (Supporting Information Figure S4 and Section
S4).
(Supporting Information Figure S4 and Section
S4).
Results
We measured the radius, r, and
axial rise per
basepair, b, for dsDNA and dsRNA in solution containing
alkali metal chlorides LiCl, NaCl, RbCl, and CsCl. Although the bare
cationic radius decreases in the order Cs → Li, in an electrolyte,
hydrated ionic radii increase with decreasing ionic radius due to
favorable interactions between the ionic core and the surrounding
polarizable water molecules (Figure 4). We found that our measured rise per basepair values
for B-DNA and A-RNA are essentially insensitive to the nature of the
cation in solution, and we obtained rise values averaged over all
measurements of  3.1 ± 0.1 Å and
3.1 ± 0.1 Å and  2.5 ± 0.1 Å for B-form and A-form
helices, respectively (Figure 4a, top). These measurements compare well with values from
crystallography and NMR.2,3,5,45,49
2.5 ± 0.1 Å for B-form and A-form
helices, respectively (Figure 4a, top). These measurements compare well with values from
crystallography and NMR.2,3,5,45,49
Figure 4.
Inferring
the structure of the molecule–electrolyte interface.
(a) Measured helical rise per basepair, bm (top), and radius, rm (bottom), as a
function of the hydrated cation radius, aH. Error bars denote s.e.m. Rise per basepair values show no significant
variation with aH and yield average values
of  3.1 ± 0.1 Å and
3.1 ± 0.1 Å and  2.5 ± 0.1 Å.
Helical radius data
were fit with a function of the form
2.5 ± 0.1 Å.
Helical radius data
were fit with a function of the form  , yielding
, yielding  10.5 ± 0.6 Å
and
10.5 ± 0.6 Å
and  11.8 ± 0.6 Å. The slope, k = 0.8 ±
0.2, is a shared fit parameter in both relationships.
(b) Cylinder of radius
11.8 ± 0.6 Å. The slope, k = 0.8 ±
0.2, is a shared fit parameter in both relationships.
(b) Cylinder of radius  10.5
10.5  (blue
dashed cylinder) depicting that the
effective cylinder in model-IIB of B-DNA is superimposed for comparison
on the vdW molecular surface in model-IB (gray dashed cylinder). k = 0.8 ± 0.2 suggests that the distance of the closest
approach of screening cations to the molecular surface is directly
related to the radius of the hydrated cation species, aH. The resulting effective “ion accessible surface”
(IAS) is the distance from the molecular axis beyond which the point-ion
description of the electrolyte may be invoked (red, green, and blue
dotted lines). The molecular structure may carry bound ions (yellow
spheres) whose charge is included in
(blue
dashed cylinder) depicting that the
effective cylinder in model-IIB of B-DNA is superimposed for comparison
on the vdW molecular surface in model-IB (gray dashed cylinder). k = 0.8 ± 0.2 suggests that the distance of the closest
approach of screening cations to the molecular surface is directly
related to the radius of the hydrated cation species, aH. The resulting effective “ion accessible surface”
(IAS) is the distance from the molecular axis beyond which the point-ion
description of the electrolyte may be invoked (red, green, and blue
dotted lines). The molecular structure may carry bound ions (yellow
spheres) whose charge is included in  . (c)
For A-RNA, model-IIA which includes
a SAS of thickness w = 3 Å meets the condition
. (c)
For A-RNA, model-IIA which includes
a SAS of thickness w = 3 Å meets the condition  12 Å
(blue dashed cylinder). (d) Extrapolating
the inferred structure of the molecule–electrolyte interface
in (b) to a view of a macroscopic interface in solution where w < 3 Å.
12 Å
(blue dashed cylinder). (d) Extrapolating
the inferred structure of the molecule–electrolyte interface
in (b) to a view of a macroscopic interface in solution where w < 3 Å.
In contrast to the response of the helical rise to the cationic
species in solution, we found that the inferred helical radii tended
to increase in the order Cs → Li (Figure 4a, bottom). We further systematically found
that  with an average
difference in helical radii
between A and B forms of about 1–2 Å. Using values for
hydrated ionic radii, aH, determined from
ionic mobilities and slip hydrodynamic boundary conditions, and plotting
measured helical radii, rm, against aH, revealed a linear relationship between the
two quantities.50 Extrapolating the measured rm values to aH =
0 yielded values for r0 that may be thought
to represent the measured radii of equivalent cylinders in a hypothetical
electrolyte containing point ions (Figure 4a). We obtained
with an average
difference in helical radii
between A and B forms of about 1–2 Å. Using values for
hydrated ionic radii, aH, determined from
ionic mobilities and slip hydrodynamic boundary conditions, and plotting
measured helical radii, rm, against aH, revealed a linear relationship between the
two quantities.50 Extrapolating the measured rm values to aH =
0 yielded values for r0 that may be thought
to represent the measured radii of equivalent cylinders in a hypothetical
electrolyte containing point ions (Figure 4a). We obtained  10.5 ± 0.6
Å and
10.5 ± 0.6
Å and  11.8 ± 0.6
Å for B-DNA and A-RNA,
respectively (Figure 4a). Atomic models of B-DNA and A-RNA display axial radii of gyration
of ≈6.7 and ≈7.8 Å and have helical radii of ≈8
and ≈9.5 Å based on the main backbone carbon atoms, respectively
(Supporting Information Figure S10a). Thus,
in addition to the average axial charge separation, our measurement
is sensitive to the radial arrangement of atoms in the double helix.
The latter appears to contribute to an effective electrical molecular
surface topography, the geometry of which can be sensed even by a
probe in the electrostatic far field, according to our measurements
(Figure 2a and Supporting Information Figure S10b).10Figure S9 further
examines the influence of various literature estimates of hydrated
cationic radii on the inferred trends in rm.
11.8 ± 0.6
Å for B-DNA and A-RNA,
respectively (Figure 4a). Atomic models of B-DNA and A-RNA display axial radii of gyration
of ≈6.7 and ≈7.8 Å and have helical radii of ≈8
and ≈9.5 Å based on the main backbone carbon atoms, respectively
(Supporting Information Figure S10a). Thus,
in addition to the average axial charge separation, our measurement
is sensitive to the radial arrangement of atoms in the double helix.
The latter appears to contribute to an effective electrical molecular
surface topography, the geometry of which can be sensed even by a
probe in the electrostatic far field, according to our measurements
(Figure 2a and Supporting Information Figure S10b).10Figure S9 further
examines the influence of various literature estimates of hydrated
cationic radii on the inferred trends in rm.
Our measured r0 values may be
thought
to reflect a hypothetical experimental scenario involving point ions
in solution (Supporting Information Section
S7.5). We therefore expect these values to be amenable to direct comparison
with the quantity  computed
for the molecular models. We find
that
computed
for the molecular models. We find
that  10.5 ± 0.6
Å is comparable to
10.5 ± 0.6
Å is comparable to  10.8 Å obtained for model-II of B-DNA
that incorporates a SAS region of width w = 1.4 Å
(Figures 4b and 2a). For A-RNA, we obtain agreement between the measured
value of
10.8 Å obtained for model-II of B-DNA
that incorporates a SAS region of width w = 1.4 Å
(Figures 4b and 2a). For A-RNA, we obtain agreement between the measured
value of  11.8 ± 0.6
Å and a molecular
model constructed using w = 3 Å, yielding
11.8 ± 0.6
Å and a molecular
model constructed using w = 3 Å, yielding  11.7 Å, as reflected in model II-A.
Taken together, the measurements and the molecular electrostatic models
for both B-DNA and A-RNA would point to the presence of a hydration
layer of thickness 1 ≲ w ≲ 3 Å.
This is in general agreement with the value of 1.4 ± 0.6 Å
reported in a study using XPS of the Stern layer at the silica–water
interface.16,51 Furthermore, the large disparity
between the measured rm value for A-RNA
and the
11.7 Å, as reflected in model II-A.
Taken together, the measurements and the molecular electrostatic models
for both B-DNA and A-RNA would point to the presence of a hydration
layer of thickness 1 ≲ w ≲ 3 Å.
This is in general agreement with the value of 1.4 ± 0.6 Å
reported in a study using XPS of the Stern layer at the silica–water
interface.16,51 Furthermore, the large disparity
between the measured rm value for A-RNA
and the  value
calculated for model-IA would appear
to strongly preclude a molecular electrostatic model that neglects
hydration at the molecular interface. A combination of the “hollow
spine” along the A-RNA molecular axis, the deep and narrow
major groove, and the closer packing of charged atoms in general would
appear to render a measurement of the electrostatic free energy of
A-RNA a more sensitive probe of interfacial structural detail and
the finite size of ions in solution compared to B-DNA.52,53 Finally, our inferred slope for the rm versus aH relationship, k = 0.8 ± 0.2 ≈ 1, suggests that the radius of the effective
cylindrical molecular surface contour in solution is enlarged by an
amount that correlates with the radius of the hydrated cation (Figure 4c). Thus, in our
picture, the thickness of the “Stern layer” at the molecular
interface has a strong contribution from the size of the counterion
in the electrolyte (Figure 4c,d).
value
calculated for model-IA would appear
to strongly preclude a molecular electrostatic model that neglects
hydration at the molecular interface. A combination of the “hollow
spine” along the A-RNA molecular axis, the deep and narrow
major groove, and the closer packing of charged atoms in general would
appear to render a measurement of the electrostatic free energy of
A-RNA a more sensitive probe of interfacial structural detail and
the finite size of ions in solution compared to B-DNA.52,53 Finally, our inferred slope for the rm versus aH relationship, k = 0.8 ± 0.2 ≈ 1, suggests that the radius of the effective
cylindrical molecular surface contour in solution is enlarged by an
amount that correlates with the radius of the hydrated cation (Figure 4c). Thus, in our
picture, the thickness of the “Stern layer” at the molecular
interface has a strong contribution from the size of the counterion
in the electrolyte (Figure 4c,d).
Importantly, we find that a PB model of the electrostatics in conjunction with a geometric modification of the object—a slight inflation of the cylindrical radius in this case—is sufficient to model measured free energies in an experimental system with finite-sized ions.42 A comparison between all-atom molecular dynamics (MD) simulations and a PB model of nucleic acids reveals that the latter is capable of capturing many features evident in MD simulations, for example, integrated spatial free energy density profiles which are central to our work. However, it has also been pointed out that detailed agreement between a PB model and MD simulations, for example, at the level of spatial ionic densities in the major groove of A-RNA, will likely require a suitably modified PB theory.53,54 In future, a modified PB model for a charged cylinder of a fixed radius, which self-consistently accounts for hydration and finite ion-size effects, is likely to provide a common underlying framework to explain the results for both A- and B-form helices.55 Such a model will likely furnish more refined estimates of the interfacial parameters of interest, for example, w, k, and aH.
To conclude the study, we focus on fM, a parameter describing the experimental apparatus, determined
in
the measurement alongside bm and rm. Like rm, we found
that fM displayed a systematic dependence
on the cationic species in solution. For measurements that hold fixed
all other experimental parameters, such as the slit height and salt
concentration, any cation species-dependent variation in fM = fionRfϕ is expected
to stem from either, or both, of the two interfacial sources: (1)
cation-specific surface potential dependence of the silica surfaces,
reflected in fϕ and/or (2) non-electrostatic
cation interactions with the double helix captured by a relative ion
binding affinity factor, given by fion. Our measured fM values for various cationic species relative to those
for the Na+ ion yielded on average  1.1,
1.1,  0.9, and
0.9, and  0.9 (where
0.9 (where  ), and the affinity factors lie in the order
Li > Na > Rb ≈ Cs (Supporting Information Figure S8). These values prove to be close to the “Hofmeister
series”-dependent zeta (ζ) potentials reported for silica
surfaces in alkali metal chloride solutions of concentration 10–3 M to 1 M, where
), and the affinity factors lie in the order
Li > Na > Rb ≈ Cs (Supporting Information Figure S8). These values prove to be close to the “Hofmeister
series”-dependent zeta (ζ) potentials reported for silica
surfaces in alkali metal chloride solutions of concentration 10–3 M to 1 M, where  1.1 ±
0.1 and
1.1 ±
0.1 and  0.8 ±
0.1 (Supporting Information Figure S8b).56,57 Assuming that the reported
trend for the ζ-potential reflects the behavior of the effective
surface electrical potential, ϕs, in our experiments,
our measured trends for fM would suggest
that most of the observed ion-dependent variation in qm stems from the variation of surface potential of silica,
captured by fϕ. Our estimate of
0.9 ≲ fLiR ≲ 1 would therefore point to a 10%
reduction in
0.8 ±
0.1 (Supporting Information Figure S8b).56,57 Assuming that the reported
trend for the ζ-potential reflects the behavior of the effective
surface electrical potential, ϕs, in our experiments,
our measured trends for fM would suggest
that most of the observed ion-dependent variation in qm stems from the variation of surface potential of silica,
captured by fϕ. Our estimate of
0.9 ≲ fLiR ≲ 1 would therefore point to a 10%
reduction in  , at the most, due to binding of
Li+ cations to the molecule, that is,
, at the most, due to binding of
Li+ cations to the molecule, that is,  0.1
0.1 . Therefore, at present, we do
not obtain
evidence of relative cation affinity values, fion, that depart
substantially from 1. To compare these observations with other techniques,
Na23 NMR reports little significant sodium
binding to DNA, with dissociation constants on the order of several
molar.58 MD studies find that while monovalent
cations do reside in the major and minor grooves of DNA, there is
little preferential long-lived binding of monovalent cations (e.g.,
Li+ compared to Na+).59 However, AES reports weak affinities for Li+ cations
corresponding to an amount of bound charge of ≈5–10%
of
. Therefore, at present, we do
not obtain
evidence of relative cation affinity values, fion, that depart
substantially from 1. To compare these observations with other techniques,
Na23 NMR reports little significant sodium
binding to DNA, with dissociation constants on the order of several
molar.58 MD studies find that while monovalent
cations do reside in the major and minor grooves of DNA, there is
little preferential long-lived binding of monovalent cations (e.g.,
Li+ compared to Na+).59 However, AES reports weak affinities for Li+ cations
corresponding to an amount of bound charge of ≈5–10%
of  , and transport measurements
report decreased
electrical mobility of DNA in the presence of Li+ cations.14,60,61 Our observation of an absence
of substantial variation in relative affinity of alkali metal cations,
and a possible weak affinity of Li+ for the double helix,
is thus in broad agreement with previous observations.
, and transport measurements
report decreased
electrical mobility of DNA in the presence of Li+ cations.14,60,61 Our observation of an absence
of substantial variation in relative affinity of alkali metal cations,
and a possible weak affinity of Li+ for the double helix,
is thus in broad agreement with previous observations.
Discussion
It is important to note that although it may in principle be possible
to evaluate electrostatic interaction free energies,  , using
molecular simulations such as Monte
Carlo (MC) or MD methods,53,62,63 these techniques are computationally resource-intensive as the system
size increases. Statistical simulation approaches such as MC require
an exhaustive sampling of the configuration space in order to provide
reliable results with acceptable accuracy.64 On the other hand, PB theory ignores correlations between ions but
is nonetheless expected to provide satisfactory theoretical description
of experiments involving monovalent salts in solution which is typical
for ETe measurements. The PB approximation relies
on the basic assumption that the potential of mean force for each
ion type is equivalent to the mean electrostatic potential.42,65 This assumption neglects all higher-order ion correlations which
manifest both through a long-range coulombic interaction and a short-range
volume-excluded effect.42 These correlations
are particularly important at high concentrations and in the presence
of multivalent ions in solution.66−68 Nevertheless, comparison
of PB ion densities with MC simulations involving finite-sized ions69,70 reveals unexpectedly good agreement, despite the fact that PB theory
is typically thought of as a “point-ion” description
of the problem.42 Although the reason for
this behavior has not been fully understood, it might be attributed
to fortuitous error cancellation within the PB approximation.42
, using
molecular simulations such as Monte
Carlo (MC) or MD methods,53,62,63 these techniques are computationally resource-intensive as the system
size increases. Statistical simulation approaches such as MC require
an exhaustive sampling of the configuration space in order to provide
reliable results with acceptable accuracy.64 On the other hand, PB theory ignores correlations between ions but
is nonetheless expected to provide satisfactory theoretical description
of experiments involving monovalent salts in solution which is typical
for ETe measurements. The PB approximation relies
on the basic assumption that the potential of mean force for each
ion type is equivalent to the mean electrostatic potential.42,65 This assumption neglects all higher-order ion correlations which
manifest both through a long-range coulombic interaction and a short-range
volume-excluded effect.42 These correlations
are particularly important at high concentrations and in the presence
of multivalent ions in solution.66−68 Nevertheless, comparison
of PB ion densities with MC simulations involving finite-sized ions69,70 reveals unexpectedly good agreement, despite the fact that PB theory
is typically thought of as a “point-ion” description
of the problem.42 Although the reason for
this behavior has not been fully understood, it might be attributed
to fortuitous error cancellation within the PB approximation.42
Furthermore, in recent years, several attempts have been made to incorporate missing additional physics into the standard PB model by introducing different forms of modified PB equation.68,71−73 Although these modified PB models provide results that compare well with MC/MD simulations, their application to situations involving monovalent ions and dilute solutions does not lead to results which are significantly different from those of standard PB theory. Thus, in general, the PB approximation gives a satisfactory description for long-range electrostatic interactions of DNA molecules in monovalent electrolytes, which is also in agreement with MC simulations and hypernetted chain approximations.67 The fact that our measurements of the rise per basepair and radius of two classes of the double helix are so close to values known from high-resolution structural biology techniques, such as X-ray crystallography and NMR, may be viewed as evidence of the validity and applicability of the combined experimental and modeling approach described here.
In conclusion, although molecular simulations are gaining dramatically in sophistication and power, field theoretical descriptions of these systems remain important due to the high computational cost of problems involving explicit atoms in a many-body problem. We demonstrate that precise measurements of interaction free energies readily distinguish between structurally or conformationally distinct states of a molecular species. Viewed through the lens of the standing theoretical model for electrostatics, such measurements also provide information on molecular and interfacial structure. Although the approach does not furnish single-atom locations, it is capable of delivering more coarse-grained molecular structural information at high resolution, which could prove useful in analyzing molecular species that are challenging to crystallize or to isotope-label for NMR. Our findings further provide estimates of geometric parameters that describe the far-field properties and interactions of a polyelectrolyte in solution, for example, the effective molecular radius, the SAS, and the radii of ions at an interface. With the surface electrical characteristics of the system (given by fM) determined with high accuracy, we expect that in future, molecular electrometry measurements will be capable of yielding similar information on a molecular species using fewer independent measurements. For example, it may be possible to use our approach to directly measure sequence-dependent differences in the rise per basepair between different oligonucleotide species.74 Furthermore, given the sensitivity of the method to small differences in 3D conformation, for example, in helical geometry as shown in this work, it is likely that molecular electrometry will provide sensitive detection of more complex 3D conformational states and structural features such as loops and bubbles in molecules. Besides, the method is not limited to the study of rod-like molecules but can be readily extended to longer nucleic acids, as long as the measurements are then compared with free energies calculated for relevant molecular structural models.53 Although the present work relies on optical observation of about 1 zmol of a species, label-free optical detection could foster such measurements at the level of one molecule in solution, enabling analysis of biomolecular conformational or structural heterogeneity at the highest sensitivity.75 Finally, since ions and water tend to be disordered, they generally evade detection by high-resolution structural methods. Thus, beyond the structural properties of the molecule, our study furnishes a parameter-free, atomic-level view of the contact region between a molecule and the electrolyte phase (Figure 4b,c), reporting directly on the structure of the “Stern layer” at the liquid–solid interface in solution (Figure 4d).
Materials and Methods
ETe Experimental Procedure: The Measurement of Molecular Escape Time, tesc
Devices for ETe measurements were fabricated using silicon/silicon dioxide and glass substrates as previously described.18 Nanofabricated fluidic slits and nanostructured pocket regions were extensively characterized by scanning electron microscopy (SEM), AFM, and profilometry. We used nanoslits of height 2h = 71–77 nm and a width of about 5 μm and pockets of depth d = 140–160 nm and radii of either 250 or 300 nm. Nanoslits were loaded with a suspension of the molecular species of interest at a concentration of 50–70 pM using pressure-driven flow for about 1 min. The flow was then stopped, and the inlet and outlet reservoirs were filled with the same suspension and sealed to prevent evaporation. The system was allowed to equilibrate for 5–10 min and maintained in an argon atmosphere during the whole measurement.
The salt concentration in the electrolyte was monitored before and after the measurement by measuring solution conductivity with a microconductivity meter (Laquatwin, Horiba Scientific, Japan). The conductivity meter was calibrated for each salt species: LiCl, NaCl, RbCl, and CsCl (Supporting Information Figure S2d). Solution pH was measured before and after the measurement using a micro-pH electrode (InLab, Mettler Toledo, UK) and pH meter (Orion Star A215, Thermo Scientific, UK).
Optical measurements
were performed using wide-field fluorescence
imaging. Fluorescence excitation was achieved by illuminating the
labeled molecules with a 532 nm DPSS laser (MGL_III-532_100 mW, PhotonTec,
Berlin) that was focused at the back aperture of a 60×, NA =
1.35 oil immersion objective (Olympus, UK). Images were acquired using
an sCMOS camera (Prime95B, Photometrics). Time-lapse videos were recorded
using an exposure time  5 ms and a variable lag time between exposures,
5 ms and a variable lag time between exposures,  . The
sampling frequency is the inverse
of
. The
sampling frequency is the inverse
of  , where
, where  =
=  +
+  is
a factor 2–4 smaller than the
average escape time,
is
a factor 2–4 smaller than the
average escape time,  , for
the molecular species of interest.
Typical cycle times were in the range of 40–65 ms for 60 bp
DNA/RNA, 25–40 ms for 40 bp DNA/RNA, and 15–25 ms for
30 bp DNA/RNA. Therefore, typical imaging frequencies were around
15–25 Hz for 60 bp DNA/RNA, 25–40 Hz for 40 bp DNA/RNA,
and 40–67 Hz for 30 bp DNA/RNA.
, for
the molecular species of interest.
Typical cycle times were in the range of 40–65 ms for 60 bp
DNA/RNA, 25–40 ms for 40 bp DNA/RNA, and 15–25 ms for
30 bp DNA/RNA. Therefore, typical imaging frequencies were around
15–25 Hz for 60 bp DNA/RNA, 25–40 Hz for 40 bp DNA/RNA,
and 40–67 Hz for 30 bp DNA/RNA.
Fluorescence images of
molecular trapping were analyzed as described
previously.17 Briefly, regions of interest
(ROIs) centered on the locations of the individual traps were identified
in an automated fashion. Intensity time traces for ROIs were analyzed
using threshold intensity values to identify durations of trapping
events, and the extracted residence times were pooled to construct
escape time histograms (Supporting Information Figure S1a). Operating in the rapid escape regime, corresponding
to average molecular residence times of Δt ≈20–350
ms, we were able to acquire ≈104 escape events within
a total imaging time of 10–20 min for each molecular species
of interest. Fitting the probability density of Δt values with an exponential function of the form  yields the
value of average escape time,
yields the
value of average escape time,  , in any given measurement with an uncertainty
of ≈1% (Supporting Information Figure
S1a).
, in any given measurement with an uncertainty
of ≈1% (Supporting Information Figure
S1a).
Purification and Characterization of DNA and RNA Samples
All nucleic acid fragments were purchased from IBA Lifesciences (Germany) with a single ATTO 532 dye molecule coupled to either one 5′ end or both 5′ termini (Supporting Information Figure S2a). The oligomers were purified with reversed-phase high-performance liquid chromatography using a Reprosil-Pur 200 C18 AQ column (Dr. Maisch, Germany) and elution with a gradient of acetonitrile in an aqueous 0.1 M triethylammonium acetate solution at a flow rate of 5 mL/min. The integrity of DNA and RNA fragments was examined with 20% polyacrylamide native gel electrophoresis (Supporting Information Figure S2c), and the helical structures (A-form for dsRNA and B-form for dsDNA) were confirmed by acquiring circular dichroism (CD) spectra using a CD spectrometer (Chirascan, Applied Photophysics, UK). Nucleic acid samples in CD spectrometry measurements contained 1 mM NaCl and 1–1.3 mM Tris, similar to the electrometry measurements. CD spectra with a data resolution of 0.5 nm per point were recorded three times for each fragment and averaged (Supporting Information Figure S2b).
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 724180) and from the Alexander von Humboldt Foundation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.macromol.2c00657.
Considerations influencing the choice of NA fragments for the study; Converting measured escape times, tesc, to measured molecular effective charge, qm; Accounting for experimental inaccuracies; Inferring values of r, b and fM from measurements of effective charge; Dependence of the inferred rm vs. aH relationship on the assumed values of aH; Comparing the inferred radii of two forms of the double helix; Electrostatic modeling and free energy calculations for B-DNA and A-RNA (PDF)
Author Contributions
M.B. and N.K. performed experiments and analyzed the data. A.B. and R.W.-G. performed molecular modeling. M.B., A.B., and R.W.-G. participated in manuscript preparation. M.K. designed and supervised the project and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Watson J. D.; Crick F. H. C. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wing R.; Drew H.; Takano T.; Broka C.; Tanaka S.; Itakura K.; Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E.; Drew H. R.; Conner B. N.; Wing R. M.; Fratini A. V.; Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science 1982, 216, 475–485. 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Bustamante C.; Marko J. F.; Siggia E. D.; Smith S. Entropic elasticity of lambda-phage DNA. Science 1994, 265, 1599–1600. 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Tjandra N.; Tate S.-i.; Ono A.; Kainosho M.; Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000, 122, 6190–6200. 10.1021/ja000324n. [DOI] [Google Scholar]
- Woźniak A. K.; Schröder G. F.; Grubmüller H.; Seidel C. A. M.; Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 18337–18342. 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Meisburger S. P.; Pabit S. A.; Sutton J. L.; Webb W. W.; Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 799–804. 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heenan P. R.; Perkins T. T. Imaging DNA Equilibrated onto Mica in Liquid Using Biochemically Relevant Deposition Conditions. ACS Nano 2019, 13, 4220–4229. 10.1021/acsnano.8b09234. [DOI] [PubMed] [Google Scholar]
- Rybenkov V. V.; Cozzarelli N. R.; Vologodskii A. V. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 5307–5311. 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; West S. M.; Sosinsky A.; Liu P.; Mann R. S.; Honig B. The role of DNA shape in protein-DNA recognition. Nature 2009, 461, 1248–1253. 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc. Natl. Acad. Sci. U.S.A. 1979, 76, 200–203. 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffeo C.; Schöpflin R.; Brutzer H.; Stehr R.; Aksimentiev A.; Wedemann G.; Seidel R. DNA-DNA interactions in tight supercoils are described by a small effective charge density. Phys. Rev. Lett. 2010, 105, 158101. 10.1103/PhysRevLett.105.158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.; Kwok L. W.; Park H. Y.; Lamb J. S.; Andresen K.; Pollack L. Measuring inter-DNA potentials in solution. Phys. Rev. Lett. 2006, 96, 138101. 10.1103/PhysRevLett.96.138101. [DOI] [PubMed] [Google Scholar]
- Bai Y.; Greenfeld M.; Travers K. J.; Chu V. B.; Lipfert J.; Doniach S.; Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007, 129, 14981–14988. 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen K.; Das R.; Park H. Y.; Smith H.; Kwok L. W.; Lamb J. S.; Kirkland E. J.; Herschlag D.; Finkelstein K. D.; Pollack L. Spatial distribution of competing ions around DNA in solution. Phys. Rev. Lett. 2004, 93, 248103. 10.1103/PhysRevLett.93.248103. [DOI] [PubMed] [Google Scholar]
- Brown M. A.; Abbas Z.; Kleibert A.; Green R. G.; Goel A.; May S.; Squires T. M. Determination of Surface Potential and Electrical Double-Layer Structure at the Aqueous Electrolyte-Nanoparticle Interface. Phys. Rev. X 2016, 6, 011007. 10.1103/PhysRevX.6.011007. [DOI] [Google Scholar]
- Ruggeri F.; Zosel F.; Mutter N.; Różycka M.; Wojtas M.; Ożyhar A.; Schuler B.; Krishnan M. Single-molecule electrometry. Nat. Nanotechnol. 2017, 12, 488–495. 10.1038/nnano.2017.26. [DOI] [PubMed] [Google Scholar]
- Krishnan M.; Mojarad N.; Kukura P.; Sandoghdar V. Geometry-induced electrostatic trapping of nanometric objects in a fluid. Nature 2010, 467, 692–695. 10.1038/nature09404. [DOI] [PubMed] [Google Scholar]
- Behrens S. H.; Grier D. G. The charge of glass and silica surfaces. J. Chem. Phys. 2001, 115, 6716–6721. 10.1063/1.1404988. [DOI] [Google Scholar]
- Kramers H. A. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica 1940, 7, 284–304. 10.1016/s0031-8914(40)90098-2. [DOI] [Google Scholar]
- Ruggeri F.; Krishnan M. Lattice diffusion of a single molecule in solution. Phys. Rev. E 2017, 96, 062406. 10.1103/PhysRevE.96.062406. [DOI] [PubMed] [Google Scholar]
- Ruggeri F.; Krishnan M. Entropic Trapping of a Singly Charged Molecule in Solution. Nano Lett. 2018, 18, 3773–3779. 10.1021/acs.nanolett.8b01011. [DOI] [PubMed] [Google Scholar]
- Doi M.; Edwards S. F.. The Theory of Polymer Dynamics; Clarendon Press, 1986. [Google Scholar]
- Schlick T.Molecular Dynamics: Further Topics. Molecular Modeling and Simulation: an Interdisciplinary Guide, 2nd ed.; Springer, 2010; Vol. 21, pp 463–517. [Google Scholar]
- Beard D. A.; Schlick T. Inertial stochastic dynamics. II. Influence of inertia on slow kinetic processes of supercoiled DNA. J. Chem. Phys. 2000, 112, 7323–7338. 10.1063/1.481371. [DOI] [Google Scholar]
- Theodoor J.; Overbeek G. The role of energy and entropy in the electrical double layer. Colloids Surf. 1990, 51, 61–75. 10.1016/0166-6622(90)80132-N. [DOI] [Google Scholar]
- Sharp K. A.; Honig B. Calculating total electrostatic energies with the nonlinear Poisson-Boltzmann equation. J. Phys. Chem. 1990, 94, 7684–7692. 10.1021/j100382a068. [DOI] [Google Scholar]
- Jayaram B.; Sharp K. A.; Honig B. The electrostatic potential of B-DNA. Biopolymers 1989, 28, 975–993. 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- Krishnan M. Electrostatic free energy for a confined nanoscale object in a fluid. J. Chem. Phys. 2013, 138, 114906. 10.1063/1.4795087. [DOI] [PubMed] [Google Scholar]
- Krishnan M. A simple model for electrical charge in globular macromolecules and linear polyelectrolytes in solution. J. Chem. Phys. 2017, 146, 205101. 10.1063/1.4983485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz R. R.; Orland H. Variational charge renormalization in charged systems. Eur. Phys. J. E: Soft Matter Biol. Phys. 2003, 11, 301–311. 10.1140/epje/i2002-10159-0. [DOI] [PubMed] [Google Scholar]
- Aubouy M.; Trizac E.; Bocquet L. r. Effective charge versus bare charge: an analytical estimate for colloids in the infinite dilution limit. J. Phys. A: Math. Gen. 2003, 36, 5835–5840. 10.1088/0305-4470/36/22/302. [DOI] [Google Scholar]
- Kjellander R.; Ulander J. Effective ionic charges, permittivity and screening length: dressed ion theory applied to 1 : 2 electrolyte solutions. Mol. Phys. 1998, 95, 495–505. 10.1080/00268979809483184. [DOI] [Google Scholar]
- Kjellander R. A multiple decay-length extension of the Debye-Huckel theory: to achieve high accuracy also for concentrated solutions and explain under-screening in dilute symmetric electrolytes. Phys. Chem. Chem. Phys. 2020, 22, 23952–23985. 10.1039/d0cp02742a. [DOI] [PubMed] [Google Scholar]
- Ramirez R.; Kjellander R. Dressed molecule theory for liquids and solutions: An exact charge renormalization formalism for molecules with arbitrary charge distributions. J. Chem. Phys. 2003, 119, 11380–11395. 10.1063/1.1623179. [DOI] [Google Scholar]
- Ramirez R.; Kjellander R. Effective multipoles and Yukawa electrostatics in dressed molecule theory. J. Chem. Phys. 2006, 125, 144110. 10.1063/1.2355486. [DOI] [PubMed] [Google Scholar]
- Manning G. S. Limiting laws and counterion condensation in polyelectrolyte solutions. I. Colligative properties. J. Chem. Phys. 1969, 51, 924–933. 10.1063/1.1672157. [DOI] [Google Scholar]
- Manning G. S. Electrostatic free energies of spheres, cylinders, and planes in counterion condensation theory with some applications. Macromolecules 2007, 40, 8071–8081. 10.1021/ma071457x. [DOI] [Google Scholar]
- Schellman J. A.; Stigter D. Electrical double layer, zeta potential, and electrophoretic charge of double-stranded DNA. Biopolymers 1977, 16, 1415–1434. 10.1002/bip.1977.360160704. [DOI] [PubMed] [Google Scholar]
- Vologodskii A.; Cozzarelli N. Modeling of long-range electrostatic interactions in DNA. Biopolymers 1995, 35, 289–296. 10.1002/bip.360350304. [DOI] [PubMed] [Google Scholar]
- Manning G. S.; Mohanty U. Counterion condensation on ionic oligomers. Physica A 1997, 247, 196–204. 10.1016/s0378-4371(97)00413-5. [DOI] [Google Scholar]
- Anderson C. F.; Record M. T. Ion Distributions Around DNA and Other Cylindrical Polyions: Theoretical Descriptions and Physical Implications. Annu. Rev. Biophys. Biophys. Chem. 1990, 19, 423–463. 10.1146/annurev.bb.19.060190.002231. [DOI] [PubMed] [Google Scholar]
- Lu X.-J.; Olson W. K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornyshev A. A.; Lee D. J.; Leikin S.; Wynveen A. Structure and interactions of biological helices. Rev. Mod. Phys. 2007, 79, 943–996. 10.1103/RevModPhys.79.943. [DOI] [Google Scholar]
- Wynveen A.; Lee D. J.; Kornyshev A. A.; Leikin S. Helical coherence of DNA in crystals and solution. Nucleic Acids Res. 2008, 36, 5540–5551. 10.1093/nar/gkn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornyshev A. A.; Lee D. J.; Wynveen A.; Leikin S. Signatures of DNA flexibility, interactions and sequence-related structural variations in classical X-ray diffraction patterns. Nucleic Acids Res. 2011, 39, 7289–7299. 10.1093/nar/gkr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E.; Klug A. Base sequence and helical structure variation byin B-DNA and A-DNA. J. Mol. Biol. 1983, 166, 419–441. 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Behjatian A.; Krishnan M. Electrostatic free energies carry structural information on nucleic acid molecules in solution. J. Chem. Phys. 2022, 156, 134201. 10.1063/5.0080008. [DOI] [PubMed] [Google Scholar]
- Verdaguer N.; Aymamí J.; Fernández-Forner D.; Fita I.; Coll M.; Huynh-Dinh T.; Igolen J.; Subirana J. A. Molecular structure of a complete turn of A-DNA. J. Mol. Biol. 1991, 221, 623–635. 10.1016/0022-2836(91)80077-8. [DOI] [PubMed] [Google Scholar]
- Pau P. C. F.; Berg J. O.; McMillan W. G. Application of Stokes’ law to ions in aqueous solution. J. Phys. Chem. 1990, 94, 2671–2679. 10.1021/j100369a080. [DOI] [Google Scholar]
- Stern O. The theory of the electrolytic double layer. Z. Elektrochem. Angew. Phys. Chem. 1924, 30, 508–516. [Google Scholar]
- Pabit S. A.; Qiu X.; Lamb J. S.; Li L.; Meisburger S. P.; Pollack L. Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA. Nucleic Acids Res. 2009, 37, 3887–3896. 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. A.; Marucho M.; Baker N. A.; Pappu R. V. Simulations of RNA interactions with monovalent ions. Methods Enzymol. 2009, 469, 411–432. 10.1016/S0076-6879(09)69020-0. [DOI] [PubMed] [Google Scholar]
- Borukhov I.; Andelman D.; Orland H. Steric effects in electrolytes: A modified Poisson-Boltzmann equation. Phys. Rev. Lett. 1997, 79, 435–438. 10.1103/PhysRevLett.79.435. [DOI] [Google Scholar]
- Brown M. A.; Bossa G. V.; May S. Emergence of a Stern Layer from the Incorporation of Hydration Interactions into the Gouy-Chapman Model of the Electrical Double Layer. Langmuir 2015, 31, 11477–11483. 10.1021/acs.langmuir.5b02389. [DOI] [PubMed] [Google Scholar]
- Franks G. V. Zeta potentials and yield stresses of silica suspensions in concentrated monovalent electrolytes: Isoelectric point shift and additional attraction. J. Colloid Interface Sci. 2002, 249, 44–51. 10.1006/jcis.2002.8250. [DOI] [PubMed] [Google Scholar]
- Beloqui Redondo A.; Jordan I.; Ziazadeh I.; Kleibert A.; Giorgi J. B.; Wörner H. J.; May S.; Abbas Z.; Brown M. A. Nanoparticle-Induced Charge Redistribution of the Air-Water Interface. J. Phys. Chem. C 2015, 119, 2661–2668. 10.1021/jp511915b. [DOI] [Google Scholar]
- Denisov V. P.; Halle B. Sequence-specific binding of counterions to B-DNA. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 629–633. 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.; Aksimentiev A. Competitive binding of cations to duplex DNA revealed through molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 12946–12954. 10.1021/jp306598y. [DOI] [PubMed] [Google Scholar]
- Stellwagen E.; Dong Q.; Stellwagen N. C. Monovalent cations affect the free solution mobility of DNA by perturbing the hydrogen-bonded structure of water. Biopolymers 2005, 78, 62–68. 10.1002/bip.20260. [DOI] [PubMed] [Google Scholar]
- Kowalczyk S. W.; Wells D. B.; Aksimentiev A.; Dekker C. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012, 12, 1038–1044. 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills P.; Anderson C. F.; Record M. T. Grand Canonical Monte-Carlo Calculations of Thermodynamic Coefficients for a Primitive Model of DNA Salt-Solutions. J. Phys. Chem. 1986, 90, 6541–6548. 10.1021/j100282a025. [DOI] [Google Scholar]
- Olmsted M. C.; Anderson C. F.; Record M. T. Monte Carlo description of oligoelectrolyte properties of DNA oligomers: range of the end effect and the approach of molecular and thermodynamic properties to the polyelectrolyte limits. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 7766–7770. 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T.Monte Carlo Techniques. Molecular Modeling and Simulation: An Interdisciplinary Guide, 2nd ed.; Springer, 2010; Vol. 21, pp 385–423. [Google Scholar]
- Kjellander R.Statistical Mechanics of Liquids and Solutions: Intermolecular Forces, Structure and Surface Interactions; CRC Press, 2020; Vol. I. [Google Scholar]
- Gavryushov S.; Zielenkiewicz P. Electrostatics of a DNA-like polyelectrolyte: Effects of solvent dielectric saturation and polarization of ion hydration shells. J. Phys. Chem. B 1999, 103, 5860–5868. 10.1021/jp983081i. [DOI] [Google Scholar]
- Gavryushov S. Electrostatics of B-DNA in NaCl and CaCl2 solutions: Ion size, interionic correlation, and solvent dielectric saturation effects. J. Phys. Chem. B 2008, 112, 8955–8965. 10.1021/jp711339d. [DOI] [PubMed] [Google Scholar]
- Gavryushov S. Dielectric saturation of the ion hydration shell and interaction between two double helices of DNA in mono- and multivalent electrolyte solutions: Foundations of the epsilon-modified Poisson-Boltzmann theory. J. Phys. Chem. B 2007, 111, 5264–5276. 10.1021/jp067120z. [DOI] [PubMed] [Google Scholar]
- Bret M. L.; Zimm B. H. Monte-Carlo determination of the distribution of ions about a cylindrical poly-electrolyte. Biopolymers 1984, 23, 271–285. 10.1002/bip.360230208. [DOI] [PubMed] [Google Scholar]
- Mills P.; Anderson C. F.; Record M. T. Monte-Carlo studies of counterion DNA interactions - comparison of the radial-distribution of counterions with predictions of other poly-electrolyte theories. J. Phys. Chem. 1985, 89, 3984–3994. 10.1021/j100265a012. [DOI] [Google Scholar]
- Antypov D.; Barbosa M. C.; Holm C. Incorporation of excluded-volume correlations into Poisson-Boltzmann theory. Phys. Rev. E 2005, 71, 061106. 10.1103/PhysRevE.71.061106. [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov D.; Andelman D.; Podgornik R.; Harries D. Ion-specific hydration effects: Extending the Poisson-Boltzmann theory. Curr. Opin. Colloid Interface Sci. 2011, 16, 542–550. 10.1016/j.cocis.2011.04.012. [DOI] [Google Scholar]
- Gongadze E.; van Rienen U.; Kralj-Iglič V.; Iglič A. Spatial variation of permittivity of an electrolyte solution in contact with a charged metal surface: a mini review. Comput. Methods Biomech. Biomed. Eng. 2013, 16, 463–480. 10.1080/10255842.2011.624769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J.; Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature 1981, 292, 375–378. 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Li Y.; Struwe W. B.; Kukura P. Single molecule mass photometry of nucleic acids. Nucleic Acids Res. 2020, 48, e97 10.1093/nar/gkaa632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.