Abstract
Cyclin-dependent kinase 6 (CDK6) is linked with a cyclin partner and plays a crucial role in the early stages of cancer development. It is currently a potential drug target for developing therapeutic molecules targeting cancer therapy. Here, we have identified taurine as an inhibitor of CDK6 using combined in silico and experimental studies. We performed various experiments to find the binding affinity of taurine with CDK6. Molecular docking analysis revealed critical residues of CDK6 that are involved in taurine binding. Fluorescence measurement studies showed that taurine binds to CDK6 with a significant binding affinity, with a binding constant of K = 0.7 × 107 M–1 for the CDK6–taurine complex. Enzyme inhibition assay suggested taurine as a good inhibitor of CDK6 possessing an IC50 value of 4.44 μM. Isothermal titration calorimetry analysis further confirmed a spontaneous binding of taurine with CDK6 and delineated the thermodynamic parameters for the CDK6–taurine system. Altogether, this study established taurine as a CDK6 inhibitor, providing a base for using taurine and its derivatives in CDK6-associated cancer and other diseases.
Introduction
Deregulation and reprogramming in fundamental cellular processes are known as cancer cell hallmarks.1 Unidirectional cell proliferation and metabolic pathway modifications are cancer’s main events.2 Protein kinases control most cellular pathways, such as cell growth, cell development, DNA damage, transcription, translation, intercellular interaction, cell metabolism, and apoptosis,3,4 and are becoming crucial players in cancer therapeutics.5 Phosphorylation is a vital post-translational modification carried out by several protein kinases in eukaryotes. Cyclin-dependent kinases (CDKs), serine/threonine-protein kinases,6 regulate the cell cycle (G1-S phase) through the phosphorylation process.7,8 Every cell cycle step is a well-regulated process controlled by CDK and its associated cyclin partners. CDK inhibitors and cyclins significantly regulate its catalytic activity, which ultimately participates in the precise progression of cell cycle events.9
CDK6 belongs to CDK’s family protein, and its gene is present on a small arm of chromosome 7 (∼1000bp) and encodes a ∼37 kDa protein.10 During the G1 stage of the cell cycle, CDK6 regulates cell growth and differentiation through the Rb-E2F pathway.11 Rb is an essential signaling player inhibiting E2F transcription factors, which control cell proliferation,12 via regulating transcription of crucial genes for DNA replication and cell cycle progression.13 Cyclin D1 and CDKs regulate the G1/S transition of the cell cycle via phosphorylation of Rb.11,14 Upregulation of CDK6 activates the Rb-E2F signaling pathway observed during the subsequent progression of cancer.10
CDK6 participates in the metabolic switching of the glycolysis pathway through inhibition of phosphofructokinase (PFK) and the pyruvate kinase (PK) activity.15−17 Biomolecules are essential for the proper growth and development of the human body.8,18,19 They participate in various signaling pathways to maintain the biochemical and physiological properties of the body. When these biomolecules decrease below threshold values, we must obtain them by outsourcing, leading to diseased conditions.20 Biological processes such as DNA replication transcription, translation, glucose, lipid metabolism, and other pathways are continuous in the human body. These cellular pathways are disturbed during disease conditions and produce reactive oxygen species (ROS) that cause oxidative damage.21
Biomolecules show several biological activities, including antioxidant activity, protecting the cell from oxidative damage. Hence, they can be used in cancer, diabetes, inflammation, and microbial and protozoan therapeutics.22−24 Natural products interact with several cellular components and regulate the ROS production that prevents the development of diseases, such as AP-1-associated signaling pathways, MAP kinase pathway, RB-E2F pathway, PI3K/Akt/mTOR pathway, and the NFκB-signaling pathway.25−27 Several biomolecules such as protein, lipids, carbohydrates, peptides, and other amino acids have various biological properties.28,29 The low level of these biomolecules is responsible for the development of several diseases.30,31
Plants, animals, microbes, and marine plants are rich sources of natural products.32−34 Natural products are used in several diseases, such as cancer, diabetes, inflammation, and microbial and protozoan infections.35−41 Most natural compounds are bioactive products that bind to different biological molecules and regulate their activity.42−44 Due to their chemical and structural diversity, natural compounds are used in treating various diseases.45−49 Natural products target several signaling pathways, mainly associated with cancer, such asAP-1-associated signaling pathways, RB-E2F pathway, PI3K/Akt/mTOR pathway, NFκB-signaling pathway, MAP kinase pathways, and CCR5 signaling.
Taurine is a sulfur-containing nonessential amino acid known as 2-amino-ethane-sulfonic acid50 that is broadly distributed in different parts of our body. Taurine is an essential nutrient in some species but is considered semi-essential in man, although cells that lack taurine show a major pathology.51 A serum level of taurine of 46–70 μmol/l is found in the human body. It plays a role in several essential functions such as body homeostasis, cell growth, development, and cell death.52−55 It maintains the phosphorylation process, calcium concentration, membrane stability, neuromodulation, detoxification of xenobiotics, regulation, and inflammation process.56−58 Previous studies have reported that serum taurine level is significantly reduced in cancer patients, directly responsible for the reduced expression level of apoptotic marker genes and the development of angiogenesis, highlighting taurine’s importance in cancer therapeutics.59
Taurine is effectively used to treat mitochondrial diseases. In addition, it has also been used in diabetes and arthritis therapeutics.51 Taurine is an effective antioxidant that prevents ROS accumulation in tumor cells and controls the progression of cancer.60 Taurine improves drug efficacy by minimizing its toxicity.61 In addition, taurine activates tumor suppressors PTEN and p53 and is considered an anticancer agent.62 These findings suggest the importance of taurine in various diseases.
Cancer cells use different mechanisms to show resistance against multiple drugs.63 Previous studies demonstrated that nonselective synthetic drugs are used for cancer therapy after cancer regains resistance against drugs.64,65 Non-selective synthetic compounds show cytotoxicity against normal cells. Thus, selectively targeting CDK6 through natural inhibitors may be successful as a therapeutic target for cancer cell growth and alterations. Taurine shows antioxidant properties to treat tumors and associated diseases.66,67 This study shows that taurine binds to the CDK6 protein and controls its expression/activity.
Upregulation of the CDK6 protein expression and activity is associated with various cancers and thus used as a potential drug design and development target. Natural compounds with antioxidant activity and low cytotoxicity give rise to a new era of research where they can cure and treat cancer and other associated diseases. Previous studies have reported that taurine can be used to treat cancer,68,69 but the inhibition of expression and activity of CDK6 by taurine provides a novel avenue for cancer therapy.
This study determined the kinase inhibitory potential of taurine against CDK6. Molecular docking was done to find the binding pattern of taurine to CDK6. Fluorescence binding assay and isothermal titration calorimetry (ITC) measurements ascertained the actual binding affinity and mechanism. Enzyme assay revealed the CDK6 inhibitory potential of taurine.
Results and Discussion
Expression and Purification of CDK6
CDK6 gene was successfully cloned into bacterial expression vector pET28a+and purified using Ni-NTA affinity chromatography. The CDK6 gene was amplified from pCDNA using specific primers. The amplified product was inserted into the pET28a+plasmed vector containing 6-histidine through restriction and ligation enzymes. The colony PCR and sequencing method confirmed the cloning of the cdk6 gene. Overexpressed CDK6 protein was purified using Ni-NTA affinity column chromatography.
Molecular Docking
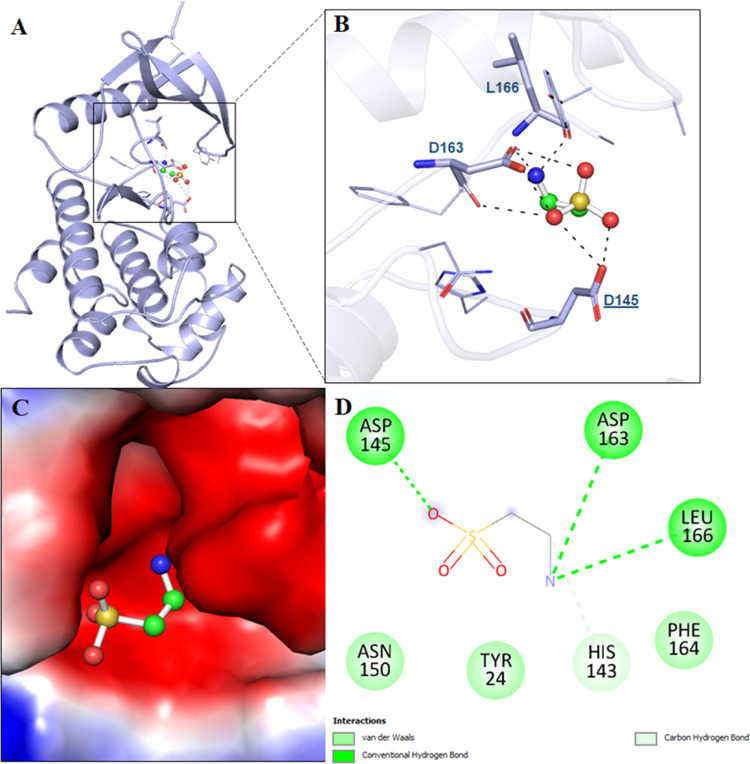
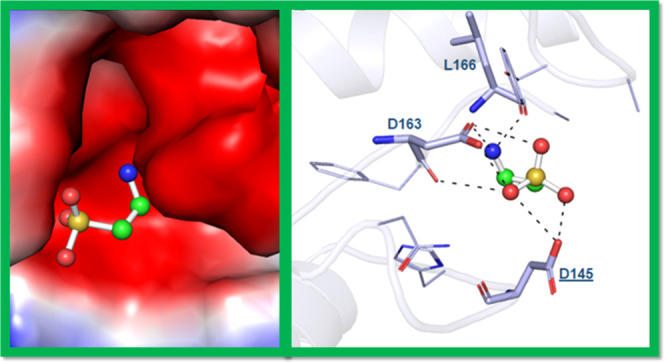
Molecular docking tools are used in estimating the binding affinity and mode of ligand interactions with the target protein.70−73 Taurine is a pharmacologically significant natural compound, promising in anticancer therapy. Molecular docking studies showed that taurine interacts with the ATP-binding pocket of CDK6. We estimated the binding affinity of taurine to CDK6 with a docking score of −3.3 kcal/mol, indicating an appreciable binding. It shows the ligand efficiency of 0.46 kcal/mol/non-H atom with CDK6, which is appreciable. Docking results were analyzed to determine the taurine binding site of CDK6. The selected conformation was based on its preferable interaction and the highest affinity toward the active site of CDK6. Here, we found that taurine occupied the binding pocket of CDK6 (Figure 1A) and not the ATP-binding site. It directly interacts with the active site residue of CDK6, i.e., Asp145, and forms a conventional hydrogen bond (Figure 1B). Asp145 is the most critical residue of the active site of CDK6, and taurine is found to form H-bonds with this critical residue, highlighting the importance of this interaction. Taurine interacts with CDK6 at multiple alternative binding sites (Figure S1). Other CDK6 inhibitors such as palbociclib are known to bind to the ATP-binding pocket of CDK6, interacting with the vital Lys43 of this ATP-binding site. Taurine resides in the deep cavity of CDK6 and forms a significant number of noncovalent interactions with the pocket residues (Figure 1C).
Figure 1.
Molecular interactions of CDK6 with taurine. (A) Cartoon model representation, (B) interacting residues, and (C) potential surface cavity of CDK6 in complex with taurine (white element) and taurine (cyan element). (D) Two-dimensional (2D) structural representation of CDK6 residues interacting with taurine.
The detailed interaction analysis of the docked complex shows that taurine interacts with many functionally important residues of the binding pocket of CDK6. It was observed that residues of CDK6 such as Asp145, Asp163, and Leu166 offer significant interactions as conventional hydrogen bonding to taurine (Figure 1D). At the same time, His143 forms a carbon–hydrogen bond, and Tyr24, Asn150, and Phe164 are involved in van der Waals interactions with taurine (Figure 1D). A significant number of interactions between functionally essential residues of CDK6 and taurine suggest an excellent binding affinity.
Fluorescence Binding Assay
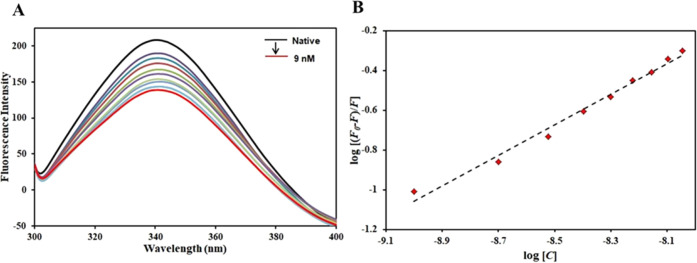
Fluorescence spectroscopy is a technique routinely deployed to study the binding pattern with the protein.74 In the present work, the fluorescence quenching of CDK6 was observed with decreasing concentrations of taurine (0–9 nM), and it has been used to obtain detailed binding information. This experiment aimed to investigate taurine’s actual binding affinity to CDK6. The CDK6 protein was excited at 280 nm, with emission spectra recorded at 300–400 nm. Based on the fluorescence emission spectra, an increase in taurine concentration (0–9 nM) in the CDK6–taurine system leads to a gradual decrease in the fluorescence intensity of the system. No shift was recorded, coupled with a gradual decrease in the fluorescence (Figure 2A). This visible decrease in the CDK6 fluorescence with increasing taurine concentrations is known as fluorescence quenching.
Figure 2.
(A) Fluorescence emission spectra of CDK6 in the absence and presence of taurine (0–9 nM). (B) Modified Stern–Volmer equation of CDK6–taurine.
The quenching data provide essential information regarding the actual binding of the ligand with the protein. Fluorescence quenching data were fitted into the modified Stern–Volmer (MSV) equation, with the intercept giving the binding constant (K) (Figure 2B). Taurine was found to bind with CDK6 with a very high binding affinity (K = 0.7 × 107 M–1). The binding constant of this order is in line with other well-known inhibitors of CDK6 implying the strength of this interaction. A recent study reported quercetin as a potent CDK6 inhibitor with a binding constant of 107 M–1.75 Another study in the literature reported that vanillin binds to CDK6 with a binding constant of 4.1 × 107 M–1.76 Another natural compound having a significant role in cancer therapeutics, ellagic acid, binds to CDK6 with K of the order of 107 M–1, inhibiting the ATPase activity of CDK6.77 All of the above studies highlight the importance of natural compounds in anticancer therapeutics targeting inhibition of CDK6 to manage CDK6-directed diseases. The present study demonstrates that taurine binds to CDK6 with an appreciable affinity affirming molecular docking observations. Thus, the fluorescence binding assay reveals the strong binding of taurine to CDK6, resulting in stable CDK6–taurine complex formation. Further, the binding is ascertained using another sophisticated technique, namely, isothermal titration calorimetry, and the CDK6 inhibitory potential of taurine is estimated using ATPase assay.
Isothermal Titration Calorimetry
ITC is a sensitive technique used to study the protein–drug interaction pattern and provide information about the system’s different binding parameters giving a complete description of the binding energetics of the protein–ligand complex.74 The result of titration of 200 μM taurine (present in syringe) into 20 μM CDK6 present in the sample cell is depicted in Figure 3 and is corrected by deduction of dilution heat. It is evident from the obtained isotherm that taurine binds spontaneously to CDK6. The upper panel indicates the raw data obtained from consecutive taurine injections into CDK6.
Figure 3.
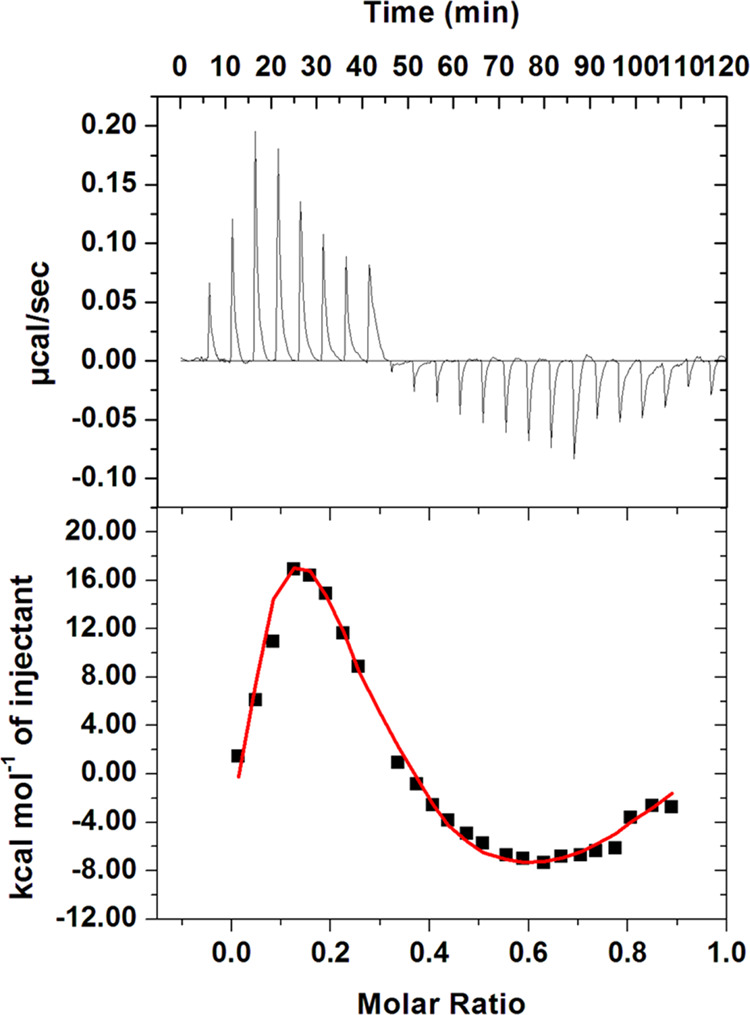
ITC isotherm was obtained upon the titration of 200 μM taurine into 20 μM CDK6. The data are plotted as a four-site model.
In contrast, the lower panel represents the binding curves obtained after subtracting heat dilution from both ligand and protein. There might be variations in thermodynamic parameters obtained from fluorescence spectroscopy and ITC. This variation is attributed to the fact that ITC measures a global change in the thermodynamic property. In contrast, fluorescence spectroscopy considers only the local changes around the fluorophore (Trp-214).78 The obtained isotherm implies that taurine binds to CDK6 spontaneously. MicroCal 8.0 was used to obtain the final figure and the associated thermodynamic parameters for the CDK6–taurine system (Table 1). Fluorescence binding assay coupled with molecular docking analysis and ITC establishes that taurine binds to CDK6 with a significant affinity resulting in the formation of a stable CDK6–taurine complex.
Table 1. Thermodynamic Parameters Obtained for the CDK6–Taurine Complex from ITC.
| Ka (association constant) M–1 | ΔH (enthalpy change) cal/mol | ΔS (cal/mol/deg) |
|---|---|---|
| Ka1 = 1.93 × 104 ± 1.1 × 104 | ΔH1 = −1.895 × 104 ± 1.54 × 104 | ΔS1 = −43.9 |
| Ka2 = 1.54 × 105 ± 4.1 × 104 | ΔH2 = 1.04 × 106 ± 3.18 × 105 | ΔS2 = 3.52 × 103 |
| Ka3 = 2.29 × 105 ± 5.2 × 104 | ΔH3 = −2.88 × 106 ± 6.05 × 105 | ΔS3 = −9.64 × 103 |
| Ka4 = 2.29 × 105 ± 5.2 × 104 | ΔH4 = 3.519 × 106 ± 5.45 × 105 | ΔS4 = 1.18 × 104 |
Kinase Assay
Kinase assay was performed in the presence of a fixed concentration of the CDK6 protein and subsequent increasing concentration of taurine (0–20 μM), and the activity of purified CDK6 was taken as 100%. Figure 4 shows the kinase inhibition profile of CDK6 with varying taurine concentrations. From the obtained data, it is clear that a concomitant decrease in the kinase activity of CDK6 protein was observed with increasing taurine concentrations. The concentration of ligand at which its 50% inhibitory effect is observed is IC50 and was found to be 4.44 μM for taurine. This obtained value is in the range of those of other well-reported CDK6 inhibitors,77 suggesting that taurine is a potent CDK6 inhibitor. Recently, many studies have targeted identifying potential kinase inhibitors because these kinases govern various essential steps of cellular pathways.79 Thus, any aberrant changes in the expression/activity of these kinases are directly associated with the pathology of lethal diseases ranging from neurodegenerative diseases to different cancers.80,81 Thus, these data suggest that taurine binds to CDK6 and inhibits its kinase activity. This inhibition will be used in diseases in which CDK6 is overexpressed. Recently published literature has established other compounds as strong inhibitors of kinases, and the obtained IC50 for taurine against CDK6 lies in the same range establishing it as a strong CDK6 inhibitor. Other CDK6 inhibitors in the clinical trial have much higher IC50 values, highlighting the strength of taurine as a CDK6 inhibitor. Additionally, minimal side effects associated with natural compounds are their additional advantage signifying the importance of establishing taurine as a potent CDK6 inhibitor. Altogether, the above data suggest that taurine binds to CDK6 with a very good affinity, inhibiting its kinase activity significantly.
Figure 4.
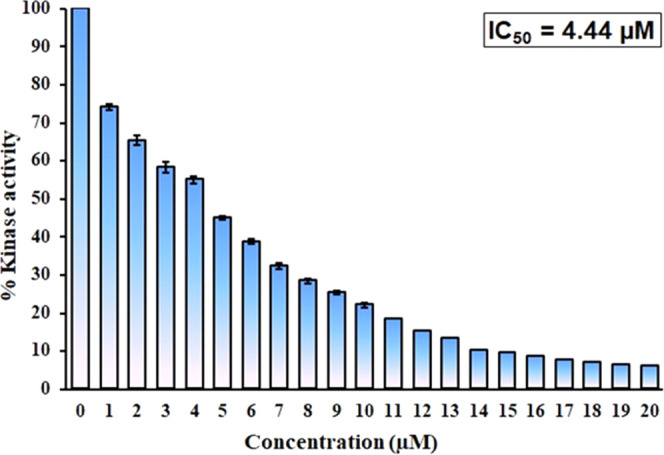
Kinase inhibition assay of CDK6 with increasing taurine concentrations (0–20 μM).
Conclusions
Protein kinase inhibitors are rapidly emerged as vital players in cancer therapeutics and hold an important place in the drug design and discovery field. Kinases govern various critical steps of important cellular pathways; thus, any changes in their activity are directly related to the progression of a disease. Overexpression of CDK6 is directly linked to various cancers, signifying the importance of identifying CDK6 inhibitors that can be used in managing CDK6-directed cancers. The present study establishes taurine as a potent inhibitor of CDK6, employing in-silico and in vitro approaches. With the aid of molecular docking analysis, functionally essential residues of CDK6 are involved in the CDK6–taurine complex formation was discovered. Further, the fluorescence-based binding assay demonstrated the actual binding of taurine with CDK6. Additionally, the ITC experiment validated the spontaneous binding and formation of a stable CDK6–taurine complex. The kinase assay further delineated the CDK6 inhibitory activity of taurine with an admirable IC50 value. Overall, the present study suggests that taurine inhibits CDK6, and can be used to treat CDK6-directed cancers. For future prospects, this study provides a platform to identify and synthesize derivatives of taurine that can interact with CDK6 inhibiting its activity more efficaciously.
Materials and Methods
Materials
Bacterial growth culture medium (Difco LB broth Miller) was purchased from Becton, Dickinson. Kanamycin, isopropyl β-d-1-thiogalactopyranoside (IPTG), lysozyme, and Tris-Hcl were procured from Sisco Research Laboratories (India). Phenylmethylsulfonyl fluoride (PMSF) catalog #P-470 was obtained from Gold Biotechnology St. Louis, MO. Taurine was purchased from Sigma-Aldrich. The Kinase assay kit was procured from Enzo (New York). All reagents and cloning enzymes used were analytical grade. pCDNA of the CDK6 gene was obtained from the Harvard Medical School repository unit. DH5α were procured for cloning purposes from Invitrogen. Taq polymerase, dNTPs, MgCl2, fast digestion restriction endonuclease, and ligation enzyme were purchased from Thermo Fisher Scientific.
Cloning, Expression, and Purification
Using our standard published protocol, we have expressed CDK6 in BL21 codon+ cells.75 The bacterial primary culture cells were grown overnight at 37 °C, induced by IPTG, and further incubated for 16 h at 18 °C. The culture was centrifuged the next day at 10,000 rpm for 10 min to separate the inclusion body. The CDK6 protein was purified from the inclusion body using Ni-NTA affinity chromatography as described earlier.77 The purity of CDK6 protein was determined on 12% SDS PAGE.
Molecular Docking Analysis
A molecular docking study was performed to obtain insights into the structural basis of taurine interactions with CDK6.82,83 The atomic coordinates of CDK6 were taken from the RCSB Protein Data Bank (PDB ID: 6OQO), and heteroatoms, including co-crystallized water molecules, were removed through SwissPDB-Viewer and the MGL AutoDock tools.84 The three-dimensional structure of taurine was retrieved from the PubChem database (PubChem ID: 1123). The docking was performed using InstaDock GUI software,85 which uses AutoDockVina86 for docking calculation. The blind search space for the ligands was set to a grid size of 78, 55, and 64 Å, centralized at 14.25, 33.66, and 0.18 for X, Y, and Z coordinates, respectively, with a grid spacing of 1.00 Å.87,88
Fluorescence Measurements
Fluorescence binding studies were performed at 25 ± 0.1 °C using a 5 mm quartz cuvette on a Jasco spectrofluorimeter (FP-6200, Jasco, Japan) containing a thermostat Peltier device. The excitation wavelength was fixed to 280 nm for the CDK6 protein. The emission spectra were recorded in the 300–400 nm range with a slit width of 10 nm.89 The stock solution of taurine was prepared and subsequently diluted to working concentration. The concentration of CDK6 was fixed at 4 μM, while the taurine concentration was varied from 0 to 9 nM. The obtained fluorescence quenching data were evaluated using earlier published protocols with the aid of the modified Stern–Volmer equation.90
Isothermal Titration Calorimetry
We used a VP-ITC microcalorimeter from MicroCal, Inc (GE, MicroCal) to further confirm the binding of CDK6 with taurine. The sample cell and syringe were filled with the purified CDK6 protein (20 μM) and taurine solution (200 μM). An equal amount of DMSO (1.0% v/v) was added to the CDK6 protein sample and ligand solution to avoid signal hindrance. We used a standard protocol in which the first injection of 2 μL was false, followed by successive injections of 10 μL of taurine solutions into the sample cell containing CDK6. These injections were at a span of 260 s intervals with a stirring speed set at 320 rpm. The spacing between two injections was set up at 280 s with the stirring speed and reference power set before the loading. The data obtained from titration were analyzed. The stoichiometry of binding (n), association constant (Ka), and enthalpy change (ΔH) were estimated using Origin 8.0 software.
Enzyme Inhibition Assay
An enzyme inhibition assay was performed to determine the effect of taurine on the CDK6 ATPase activity as per a previously published protocol.77 In brief, an increasing concentration of taurine (0–20 μM) was added to the fixed amount of purified CDK6 protein (2 μM) and incubated at 25 °C for 1 h. Freshly prepared ATP solution (200 μM) and MgCl2 (10 mM) were added to the reaction mixture and incubated for 30 min at 25 °C. Malachite green was added to this reaction mixture to terminate the reaction and incubated for 20–30 min until the development of green color. CDK6 utilized the ATP and released the free inorganic phosphate measured on a multiplate ELISA reader at 620 nm. If the ligand molecule binds to the CDK6 and inhibits the kinase activity, the amount of free inorganic phosphate released from the reaction is depleted. All of the experiments were performed in triplicate.
Acknowledgments
The Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia has funded this project, under grant no. (KEP-19-130-42). M.I.H. acknowledges the Council of Scientific and Industrial Research for financial support [Project No. 27(0368)/20/EMR-II]. M.Y. thanks the Indian Council of Medical Research for the award of the Senior Research Fellowship (F. No.45/54/2018-PHA/ BMS/OL).
Glossary
List of Abbreviations
- CDKs
cyclin-dependent kinases
- PFK
phosphofructokinase
- PK
pyruvate kinase
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- ITC
isothermal titration calorimetry
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03479.
Figure S1: Multiple binding sites of taurine on CDK6 (PDF)
Author Contributions
All authors have read and given approval to the final version of the manuscript. Conceptualization: M.Y., A.S., A.I., and M.I.H.; methodology: M.Y., T.M., A.S., and N.A.; software: M.Y., A.S., A.M.E., and S.Y.M.A.; validation: M.Y., A.S., Q.M.R.H., A.M.A., and M.I.H.; formal analysis: M.Y., A.S., and M.I.H.; investigation: M.Y., A.M.E., T.M., Q.M.R.H., and M.I.H.; resources and data curation: M.I.H.; writing—original draft preparation: M.Y., A.S., A.I., and T.M. and writing—review and editing: Q.M.R.H.; visualization: M.Y., T.M., A.I., and A.S.; supervision: M.I.H. and Q.M.R.H.; and project administration: M.I.H. and Q.M.R.H.
The authors declare no competing financial interest.
Notes
All data generated or analyzed during this study are included in this manuscript and supplementary materials attached to this article.
Supplementary Material
References
- Hanahan D.; Weinberg R. A. The hallmarks of cancer. Cell 2000, 100, 57–70. 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Xia Y.; Lu Z. Metabolic features of cancer cells. Cancer Commun. 2018, 38, 1–65. 10.1186/s40880-018-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D. H.; Kim N. R.; Kim T.; Ahn J.; Lee S.; Lee Y. D.; Cho H. Y. Malignant glomus tumor of the thyroid gland where is heretofore an unreported organ: a case report and literature review. Endocr. Pathol. 2015, 26, 37–44. 10.1007/s12022-014-9352-5. [DOI] [PubMed] [Google Scholar]
- Kim Y. J.; Kim E. H.; Hahm K. B. Oxidative stress in inflammation-based gastrointestinal tract diseases: challenges and opportunities. J. Gastroenterol. Hepatol. 2012, 27, 1004–1010. 10.1111/j.1440-1746.2012.07108.x. [DOI] [PubMed] [Google Scholar]
- Bhullar K. S.; Lagarón N. O.; McGowan E. M.; Parmar I.; Jha A.; Hubbard B. P.; Rupasinghe H. V. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol. cancer 2018, 17, 48 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L.; Fisher D. Non-cell cycle functions of the CDK Network in ciliogenesis: recycling the cell cycle oscillator. BioEssays 2018, 40, 1800016 10.1002/bies.201800016. [DOI] [PubMed] [Google Scholar]
- Son K. H.; Park C. H.; Park K. Y.; Choi C. H. Which Variables Should be Considered as Confounders of p38-Mitogen Activated Protein Kinase Activation Measurements?. Ann. Thorac. Surg. 2016, 102, 1764–1765. 10.1016/j.athoracsur.2016.03.118. [DOI] [PubMed] [Google Scholar]
- Tran N. N. Q.; Chun K. H. ROCK2-Specific Inhibitor KD025 Suppresses Adipocyte Differentiation by Inhibiting Casein Kinase 2. Molecules 2021, 26, 4747 10.3390/molecules26164747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar U.; Witkiewicz A. K.; Turner N. C.; Knudsen E. S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discovery 2015, 14, 130–146. 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. A.; Tong L.; Lee J.-O.; Jeffrey P. D.; Pavletich N. P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16 INK4a. Nature 1998, 395, 237–243. 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- Matsushime H.; Quelle D.; Shurtleff S.; Shibuya M.; Sherr C.; Kato J. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 1994, 14, 2066–2076. 10.1128/mcb.14.3.2066-2076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R. C.; Nevins J. R. Signaling networks that link cell proliferation and cell fate. J. Biol. Chem. 2002, 277, 11617–11620. 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- Ghanem N.; Andrusiak M. G.; Svoboda D.; Al Lafi S. M.; Julian L. M.; McClellan K. A.; De Repentigny Y.; Kothary R.; Ekker M.; et al. The Rb/E2F pathway modulates neurogenesis through direct regulation of the Dlx1/Dlx2 bigene cluster. J. Neurosci. 2012, 32, 8219–8230. 10.1523/JNEUROSCI.1344-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J.-Y.; Matsushime H.; Hiebert S.; Ewen M.; Sherr C. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993, 7, 331–342. 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Wang H.; Nicolay B. N.; Chick J. M.; Gao X.; Geng Y.; Ren H.; Gao H.; Yang G.; Williams J. A.; et al. The metabolic function of cyclin D3–CDK6 kinase in cancer cell survival. Nature 2017, 546, 426–430. 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W.; Clark P. M.; Mason D. E.; Keenan M. C.; Hill C.; Goddard W. A.; Peters E. C.; Driggers E. M.; Hsieh-Wilson L. C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D.; Poulogiannis G.; Asara J. M.; Boxer M. B.; Jiang J.-k.; Shen M.; Bellinger G.; Sasaki A. T.; Locasale J. W.; et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 334, 1278–1283. 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teli M. K.; Kumar S.; Yadav D. K.; Kim M. H. In silico identification of hydantoin derivatives: a novel natural prolyl hydroxylase inhibitor. J. Biomol. Struct. Dyn. 2021, 39, 703–717. 10.1080/07391102.2020.1714480. [DOI] [PubMed] [Google Scholar]
- Yadav D. K.; Kumar S.; Choi E. H.; Chaudhary S.; Kim M. H. Computational Modeling on Aquaporin-3 as Skin Cancer Target: A Virtual Screening Study. Front. Chem. 2020, 8, 250 10.3389/fchem.2020.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa A. K.; Fatima T.; Mattoo A. K. Polyamines: bio-molecules with diverse functions in plant and human health and disease. Front. Chem. 2018, 6, 10 10.3389/fchem.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmanoglu E.; Kurutas E. B. Induction of oxidative/nitrosative stress following Tc-99m pertechnetate thyroid scintigraphy in human. Cumhuriyet Med. J. 2019, 41, 592–598. 10.7197/cmj.vi.486246. [DOI] [Google Scholar]
- Bouarab Chibane L.; Degraeve P.; Ferhout H.; Bouajila J.; Oulahal N. Plant antimicrobial polyphenols as potential natural food preservatives. J. Sci. Food Agric. 2019, 99, 1457–1474. 10.1002/jsfa.9357. [DOI] [PubMed] [Google Scholar]
- Iranshahy M.; Iranshahi M.; Abtahi S. R.; Karimi G. The role of nuclear factor erythroid 2-related factor 2 in hepatoprotective activity of natural products: a review. Food Chem. Toxicol. 2018, 120, 261–276. 10.1016/j.fct.2018.07.024. [DOI] [PubMed] [Google Scholar]
- Jamshidi-Kia F.; Wibowo J. P.; Elachouri M.; Masumi R.; Salehifard-Jouneghani A.; Abolhasanzadeh Z.; Lorigooini Z. Battle between plants as antioxidants with free radicals in human body. J. HerbMed Pharmacol. 2020, 9, 191–199. 10.34172/jhp.2020.25. [DOI] [Google Scholar]
- Kim C.; Kim B. Anti-cancer natural products and their bioactive compounds inducing ER stress-mediated apoptosis: A review. Nutrients 2018, 10, 1021 10.3390/nu10081021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X.; Nawab O.; Patel T.; Kossenkov A. V.; Halama N.; Jaeger D.; Pestell R. G. Recent advances targeting CCR5 for cancer and its role in immuno-oncology. Cancer Res. 2019, 79, 4801–4807. 10.1158/0008-5472.CAN-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamse H. Photobiomodulation and Oxidative/Nitrosative Stress. Photobiomodulation, Photomed., Laser Surg. 2021, 39, 685–686. 10.1089/photob.2021.0084. [DOI] [PubMed] [Google Scholar]
- Ahsan H. The biomolecules of beauty: Biochemical pharmacology and immunotoxicology of cosmeceuticals. J. Immunoassay Immunochem. 2019, 40, 91–108. 10.1080/15321819.2018.1555766. [DOI] [PubMed] [Google Scholar]
- Davies M. J.; Hawkins C. L. The role of myeloperoxidase in biomolecule modification, chronic inflammation, and disease. Antioxid. Redox Signaling 2020, 32, 957–981. 10.1089/ars.2020.8030. [DOI] [PubMed] [Google Scholar]
- Mirończuk-Chodakowska I.; Witkowska A. M.; Zujko M. E. Endogenous non-enzymatic antioxidants in the human body. Adv. Med. Sci. 2018, 63, 68–78. 10.1016/j.advms.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Verma M. K.; Verma P. Role of Oxidant Alteration of Biomolecules in Diabetes and Other Associated Diseases. Int. J. Life-Sci. S. Res. 2018, 4, 1542–1549. 10.21276/ijlssr.2018.4.1.6. [DOI] [Google Scholar]
- Alam M.; Hasan G. M.; Ansari M. M.; Sharma R.; Yadav D. K.; Hassan M. I. Therapeutic implications and clinical manifestations of thymoquinone. Phytochemistry 2022, 200, 113213 10.1016/j.phytochem.2022.113213. [DOI] [PubMed] [Google Scholar]
- Alam M.; Ahmed S.; Elasbali A. M.; Adnan M.; Alam S.; Hassan M. I.; Pasupuleti V. R. Therapeutic Implications of Caffeic Acid in Cancer and Neurological Diseases. Front. Oncol. 2022, 12, 860508 10.3389/fonc.2022.860508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M.; Ali S.; Ashraf G. M.; Bilgrami A. L.; Yadav D. K.; Hassan M. I. Epigallocatechin 3-gallate: From green tea to cancer therapeutics. Food Chem. 2022, 379, 132135 10.1016/j.foodchem.2022.132135. [DOI] [PubMed] [Google Scholar]
- Alam M.; Ali S.; Ahmed S.; Elasbali A. M.; Adnan M.; Islam A.; Hassan M. I.; Yadav D. K. Therapeutic Potential of Ursolic Acid in Cancer and Diabetic Neuropathy Diseases. Int. J. Mol. Sci. 2021, 22, 12162 10.3390/ijms222212162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R.; Mohammad T.; Roy S.; Anwar S.; Gupta P.; Haque A.; Khan P.; Kazim S. N.; Islam A.; et al. Investigation of inhibitory potential of quercetin to the pyruvate dehydrogenase kinase 3: Towards implications in anticancer therapy. Int. J. Biol. Macromol. 2019, 136, 1076–1085. 10.1016/j.ijbiomac.2019.06.158. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Mohammad T.; Dahiya R.; Roy S.; Noman O. M. A.; Alajmi M. F.; Hussain A.; Hassan M. I. Evaluation of binding and inhibition mechanism of dietary phytochemicals with sphingosine kinase 1: Towards targeted anticancer therapy. Sci Rep 2019, 9, 18727 10.1038/s41598-019-55199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Mohammad T.; Khan P.; Alajmi M. F.; Hussain A.; Rehman M. T.; Hassan M. I. Evaluation of ellagic acid as an inhibitor of sphingosine kinase 1: A targeted approach towards anticancer therapy. Biomed. Pharm. 2019, 118, 109245 10.1016/j.biopha.2019.109245. [DOI] [PubMed] [Google Scholar]
- Khan P.; Queen A.; Mohammad T.; Smita; Khan N. S.; Hafeez Z. B.; Hassan M. I.; Ali S. Identification of α-Mangostin as a Potential Inhibitor of Microtubule Affinity Regulating Kinase 4. J. Nat. Prod. 2019, 82, 2252–2261. 10.1021/acs.jnatprod.9b00372. [DOI] [PubMed] [Google Scholar]
- Mohammad T.; Arif K.; Alajmi M. F.; Hussain A.; Islam A.; Rehman M. T.; Hassan I. Identification of high-affinity inhibitors of pyruvate dehydrogenase kinase-3: towards therapeutic management of cancer. J. Biomol. Struct. Dyn. 2021, 39, 586–594. 10.1080/07391102.2020.1711810. [DOI] [PubMed] [Google Scholar]
- Mohammad T.; Batra S.; Dahiya R.; Baig M. H.; Rather I. A.; Dong J. J.; Hassan I. Identification of high-affinity inhibitors of cyclin-dependent kinase 2 towards anticancer therapy. Molecules 2019, 24, 4589 10.3390/molecules24244589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T.; Khan F. I.; Lobb K. A.; Islam A.; Ahmad F.; Hassan M. I. Identification and evaluation of bioactive natural products as potential inhibitors of human microtubule affinity-regulating kinase 4 (MARK4). J. Biomol. Struct. Dyn. 2019, 37, 1813–1829. 10.1080/07391102.2018.1468282. [DOI] [PubMed] [Google Scholar]
- Mohammad T.; Siddiqui S.; Shamsi A.; Alajmi M. F.; Hussain A.; Islam A.; Ahmad F.; Hassan M. I. Virtual Screening Approach to Identify High-Affinity Inhibitors of Serum and Glucocorticoid-Regulated Kinase 1 among Bioactive Natural Products: Combined Molecular Docking and Simulation Studies. Molecules 2020, 25, 823 10.3390/molecules25040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A.; Anwar S.; Mohammad T.; Alajmi M. F.; Hussain A.; Rehman M. T.; Hasan G. M.; Islam A.; Hassan M. I. MARK4 inhibited by AChE inhibitors, donepezil and rivastigmine tartrate: Insights into alzheimer’s disease therapy. Biomolecules 2020, 10, 789 10.3390/biom10050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan P.; Rahman S.; Queen A.; Manzoor S.; Naz F.; Hasan G. M.; Luqman S.; Kim J.; Islam A.; et al. Elucidation of Dietary Polyphenolics as Potential Inhibitor of Microtubule Affinity Regulating Kinase 4: In silico and in vitro Studies. Sci. Rep. 2017, 7, 9470 10.1038/s41598-017-09941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.; Khan F. I.; Mohammad T.; Khan P.; Hasan G. M.; Lobb K. A.; Islam A.; Ahmad F.; Imtaiyaz Hassan M. Exploring molecular insights into the interaction mechanism of cholesterol derivatives with the Mce4A: A combined spectroscopic and molecular dynamic simulation studies. Int. J. Biol. Macromol. 2018, 111, 548–560. 10.1016/j.ijbiomac.2017.12.160. [DOI] [PubMed] [Google Scholar]
- Naz F.; Khan F. I.; Mohammad T.; Khan P.; Manzoor S.; Hasan G. M.; Lobb K. A.; Luqman S.; Islam A.; et al. Investigation of molecular mechanism of recognition between citral and MARK4: A newer therapeutic approach to attenuate cancer cell progression. Int. J. Biol. Macromol. 2018, 107, 2580–2589. 10.1016/j.ijbiomac.2017.10.143. [DOI] [PubMed] [Google Scholar]
- Naz H.; Khan P.; Tarique M.; Rahman S.; Meena A.; Ahamad S.; Luqman S.; Islam A.; Ahmad F.; et al. Binding studies and biological evaluation of β-carotene as a potential inhibitor of human calcium/calmodulin-dependent protein kinase IV. Int. J. Biol. Macromol. 2017, 96, 161–170. 10.1016/j.ijbiomac.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Naz H.; Tarique M.; Khan P.; Luqman S.; Ahamad S.; Islam A.; Ahmad F.; Hassan M. I. Evidence of vanillin binding to CAMKIV explains the anti-cancer mechanism in human hepatic carcinoma and neuroblastoma cells. Mol. Cell. Biochem. 2018, 438, 35–45. 10.1007/s11010-017-3111-0. [DOI] [PubMed] [Google Scholar]
- Lourenco R.; Camilo M. Taurine: a conditionally essential amino acid in humans? An overview in health and disease. Nutr. Hosp 2002, 17, 262–270. [PubMed] [Google Scholar]
- Schaffer S.; Kim H. W. Effects and mechanisms of taurine as a therapeutic agent. Biomol. Ther. 2018, 26, 225–241. 10.4062/biomolther.2017.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E. K.; Lambert I. H.; Pedersen S. F. Physiology of cell volume regulation in vertebrates. Physiol. Rev. 2009, 89, 193–277. 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- Sturman J. A. Taurine in development. J. Nutr. 1988, 118, 1169–1176. 10.1093/jn/118.10.1169. [DOI] [PubMed] [Google Scholar]
- Li X.-W.; Gao H.-Y.; Liu J. The role of taurine in improving neural stem cells proliferation and differentiation. Nutr. Neurosci. 2017, 20, 409–415. 10.1080/1028415X.2016.1152004. [DOI] [PubMed] [Google Scholar]
- Seidel U.; Huebbe P.; Rimbach G. Taurine: a regulator of cellular redox homeostasis and skeletal muscle function. Mol. Nutr. Food Res. 2019, 63, 1800569 10.1002/mnfr.201800569. [DOI] [PubMed] [Google Scholar]
- Kilb W.; Fukuda A. Taurine as an essential neuromodulator during perinatal cortical development. Front. Cell. Neurosci. 2017, 11, 328 10.3389/fncel.2017.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Borges R. H.; González M. J.; Gupta R. C.; Ayeotan O. Taurine as Anticancer and Antiviral: Case Report and Prospective Update. Global J. Cancer Case Rep. 2020, 01, 1–14. 10.47733/GJCCR.2020.1205. [DOI] [Google Scholar]
- Qaradakhi T.; Gadanec L. K.; McSweeney K. R.; Abraham J. R.; Apostolopoulos V.; Zulli A. The anti-inflammatory effect of taurine on cardiovascular disease. Nutrients 2020, 12, 2847. 10.3390/nu12092847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agouza I. M.; Eissa S.; El Houseini M.; El-Nashar D. E.; Abd El Hameed O. Taurine: a novel tumor marker for enhanced detection of breast cancer among female patients. Angiogenesis 2011, 14, 321–330. 10.1007/s10456-011-9215-3. [DOI] [PubMed] [Google Scholar]
- Mates J. M.; Segura J. A.; Alonso F. J.; Marquez J. Sulphur-containing non enzymatic antioxidants: therapeutic tools against cancer. Front. Biosci. 2012, S4, 722–748. 10.2741/s296. [DOI] [PubMed] [Google Scholar]
- Okamoto K.; Sugie S.; Ohnishi M.; Makita H.; Kawamori T.; Watanabe T.; Tanaka T.; Mori H. Chemopreventive Effects of Taurine on Diethylnitrosamine and Phenobarbitalinduced Hepatocarcinogenesis in Male F344 Rats. Jpn. J. Cancer Res. 1996, 87, 30–36. 10.1111/j.1349-7006.1996.tb00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F.; Ma N.; Midorikawa K.; Hiraku Y.; Oikawa S.; Mo Y.; Zhang Z.; Takeuchi K.; Murata M.. Anti-cancer mechanisms of taurine in human nasopharyngeal carcinoma cells. In Taurine 11; Springer, 2019; pp 533–541, 10.1007/978-981-13-8023-5_49. [DOI] [PubMed] [Google Scholar]
- Szakács G.; Paterson J. K.; Ludwig J. A.; Booth-Genthe C.; Gottesman M. M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discovery 2006, 5, 219–234. 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Creighton C. J.; Li X.; Landis M.; Dixon J. M.; Neumeister V. M.; Sjolund A.; Rimm D. L.; Wong H.; Rodriguez A.; et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Nat. Acad. Sci. 2009, 106, 13820–13825. 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniak D.; Sabri S.; Xu Y.; Graham K.; Bhatnagar P.; Suresh M.; Abdulkarim B. Spontaneous epithelial-mesenchymal transition and resistance to HER-2-targeted therapies in HER-2-positive luminal breast cancer. PLoS One 2013, 8, e71987 10.1371/journal.pone.0071987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliou S.; Kyriakopoulos A. M.; Spandidos D. A.; Zoumpourlis V. Role of taurine, its haloamines and its lncRNA TUG1 in both inflammation and cancer progression. On the road to therapeutics?. Int. J. Oncol. 2020, 57, 631–664. 10.3892/ijo.2020.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Xia Y.; Zhang X.; Liu L.; Tu S.; Zhu W.; Yu L.; Wan H.; Yu B.; Wan F. Roles of the MST1-JNK signaling pathway in apoptosis of colorectal cancer cells induced by Taurine. Libyan J. Med. 2018, 13, 1500346 10.1080/19932820.2018.1500346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.; Li H.; Tang Y. The Effect of Suppression Taurine on Relocation and Epithelial-Mesenchymal Transition in Mankind Lung Cancer Cells. J. Healthcare Eng. 2021, 2021, 1–11. 10.1155/2021/6656080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lu B.; Yi M.; Hu S.; Wu D.; Zhu Z.; Wu C.; Wang Z.; Li Y.; Zhang J. Taurine-Based Ionic Liquids for Transdermal Protein Delivery and Enhanced Anticancer Activity. ACS Sustainable Chem. Eng. 2021, 9, 5991–6000. 10.1021/acssuschemeng.1c01064. [DOI] [Google Scholar]
- Beg A.; Khan F. I.; Lobb K. A.; Islam A.; Ahmad F.; Hassan M. I. High throughput screening, docking, and molecular dynamics studies to identify potential inhibitors of human calcium/calmodulin-dependent protein kinase IV. J. Biomol. Struct. Dyn. 2019, 37, 2179–2192. 10.1080/07391102.2018.1479310. [DOI] [PubMed] [Google Scholar]
- Gulzar M.; Ali S.; Khan F. I.; Khan P.; Taneja P.; Hassan M. I. Binding mechanism of caffeic acid and simvastatin to the integrin linked kinase for therapeutic implications: a comparative docking and MD simulation studies. J. Biomol. Struct. Dyn. 2019, 37, 4327–4337. 10.1080/07391102.2018.1546621. [DOI] [PubMed] [Google Scholar]
- Jairajpuri D. S.; Hussain A.; Nasreen K.; Mohammad T.; Anjum F.; Tabish Rehman M.; Mustafa Hasan G.; Alajmi M. F.; Imtaiyaz Hassan M. Identification of natural compounds as potent inhibitors of SARS-CoV-2 main protease using combined docking and molecular dynamics simulations. Saudi J. Biol. Sci. 2021, 28, 2423–2431. 10.1016/j.sjbs.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A. A. T.; Mohammad T.; Hasan G. M.; Hassan M. I. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 2018, 18, 1755–1768. 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- Anwar S.; Shamsi A.; Shahbaaz M.; Queen A.; Khan P.; Hasan G. M.; Islam A.; Alajmi M. F.; Hussain A.; et al. Rosmarinic acid exhibits anticancer effects via MARK4 inhibition. Sci. Rep. 2020, 10, 10300 10.1038/s41598-020-65648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf M.; Khan P.; Shamsi A.; Shahbaaz M.; Hasan G. M.; Haque Q. M. R.; Christoffels A.; Islam A.; Hassan M. I. Inhibiting CDK6 activity by quercetin is an attractive strategy for cancer therapy. ACS omega 2020, 5, 27480–27491. 10.1021/acsomega.0c03975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf M.; Shamsi A.; Queen A.; Shahbaaz M.; Khan P.; Hussain A.; Alajmi M. F.; Rizwanul Haque Q. M.; Imtaiyaz Hassan M. Targeting cyclin-dependent kinase 6 by vanillin inhibits proliferation of breast and lung cancer cells: Combined computational and biochemical studies. J. Cell. Biochem. 2021, 122, 897–910. 10.1002/jcb.29921. [DOI] [PubMed] [Google Scholar]
- Yousuf M.; Shamsi A.; Khan P.; Shahbaaz M.; AlAjmi M. F.; Hussain A.; Hassan G. M.; Islam A.; Rizwanul Haque Q. M.; Hassan M. I. Ellagic acid controls cell proliferation and induces apoptosis in breast cancer cells via inhibition of cyclin-dependent kinase 6. Int. J. Mol. Sci. 2020, 21, 3526 10.3390/ijms21103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman M. T.; Shamsi H.; Khan A. U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharmaceutics 2014, 11, 1785–1797. 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Yang P. L.; Gray N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- Khan P.; Rahman S.; Queen A.; Manzoor S.; Naz F.; Hasan G. M.; Luqman S.; Kim J.; Islam A.; et al. Elucidation of dietary polyphenolics as potential inhibitor of microtubule affinity regulating kinase 4: in silico and in vitro studies. Sci. Rep. 2017, 7, 9470 10.1038/s41598-017-09941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martinez C.; Gelbert L. M.; Lallena M. J.; de Dios A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorg. Med. Chem. Lett. 2015, 25, 3420–3435. 10.1016/j.bmcl.2015.05.100. [DOI] [PubMed] [Google Scholar]
- Yousuf M.; Shamsi A.; Khan S.; Khan P.; Shahwan M.; Elasbali A. M.; Haque Q. M. R.; Hassan M. I. Naringenin as a potential inhibitor of human cyclin-dependent kinase 6: Molecular and structural insights into anti-cancer therapeutics. Int. J. Biol. Macromol. 2022, 213, 01207. 10.1016/j.ijbiomac.2022.06.013. [DOI] [PubMed] [Google Scholar]
- Yang C.; Alam A.; Alhumaydhi F. A.; Khan M. S.; Alsagaby S. A.; Al Abdulmonem W.; Hassan M. I.; Shamsi A.; Bano B.; Yadav D. K. Bioactive Phytoconstituents as Potent Inhibitors of Tyrosine-Protein Kinase Yes (YES1): Implications in Anticancer Therapeutics. Molecules 2022, 27, 3060 10.3390/molecules27103060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T.; Mathur Y.; Hassan M. I. InstaDock: A single-click graphical user interface for molecular docking-based virtual high-throughput screening. Briefings Bioinf. 2020, bbaa279 10.1093/bib/bbaa279. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A. A. T.; Jairajpuri D. S.; Noman O. M. A.; Hussain A.; Islam A.; Ahmad F.; Alajmi M. F.; Hassan M. I. Evaluation of pyrazolopyrimidine derivatives as microtubule affinity regulating kinase 4 inhibitors: Towards therapeutic management of Alzheimer’s disease. J. Biomol. Struct. Dyn. 2020, 38, 3892–3907. 10.1080/07391102.2019.1666745. [DOI] [PubMed] [Google Scholar]
- Naqvi A. A. T.; Mohammad T.; Hasan G. M.; Hassan M. I. Advancements in Docking and Molecular Dynamics Simulations Towards Ligand-receptor Interactions and Structure-function Relationships. Curr. Top. Med. Chem. 2018, 18, 1755–1768. 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- Anwar S.; Shamsi A.; Kar R. K.; Queen A.; Islam A.; Ahmad F.; Hassan M. I. Structural and biochemical investigation of MARK4 inhibitory potential of cholic acid: Towards therapeutic implications in neurodegenerative diseases. Int. J. Biol. Macromol. 2020, 161, 596–604. 10.1016/j.ijbiomac.2020.06.078. [DOI] [PubMed] [Google Scholar]
- Anwar S.; Mohammad T.; Shamsi A.; Queen A.; Parveen S.; Luqman S.; Hasan G. M.; Alamry K. A.; Azum N.; et al. Discovery of Hordenine as a potential inhibitor of pyruvate dehydrogenase kinase 3: implication in lung Cancer therapy. Biomedicines 2020, 8, 119 10.3390/biomedicines8050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.