Abstract
Plastic has become established over the world as an essential basic need for our daily life. Current global plastic production exceeds 300 million tons annually. Plastics have many characteristics such as low production costs, inertness, relatively low weight, and durability. The primary disadvantage of plastics is their extremely slow natural degradation. The latter results in an accumulation of plastic waste in nature. The amount of plastic waste as of 2015 was 6300 million tons worldwide, and 79% of this was placed in landfills or left in the natural environment. Moreover, recent estimates report that 12,000 million tons of plastic waste will have been accumulated on the earth by 2050. Therefore, it is necessary to develop an effective plastic biodegradation process to accelerate the natural degradation rate of plastics. More than 400 microbes have been identified as capable of plastic degradation. This is the first paper of the series on plastic-degrading fungi. This paper provides a summary of the current global production of plastic and plastic waste accumulation in nature. A list is given of all the plastic-degrading fungi recorded thus far, based on the available literature, and comments are made relating to the major fungal groups. In addition, the phylogenetic relationships of plastic-degrading fungi were analyzed using a combined ITS, LSU, SSU, TEF, RPB1, and RPB2 dataset consisting of 395 strains. Our results confirm that plastic-degrading fungi are found in eleven classes in the fungal phyla Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes, Saccharomycetes, and Sordariomycetes), Basidiomycota (Agaricomycetes, Microbotryomycetes, Tremellomycetes, Tritirachiomycetes, and Ustilaginomy-cetes), and Mucoromycota (Mucoromycetes). The taxonomic placement of plastic-degrading fungal taxa is briefly discussed. The Eurotiomycetes include the largest number of plastic degraders in the kingdom Fungi. The results presented herein are expected to influence the direction of future research on similar topics in order to find effective plastic-degrading fungi that can eliminate plastic wastes. The next publication of the series on plastic-degrading fungi will be focused on major metabolites, degradation pathways, and enzyme production in plastic degradation by fungi.
Keywords: fungi, global plastic production, plastic waste accumulation, synthetic polymers, multi-gene phylogeny
1. Introduction
Plastic is one of the most abundant human-produced, versatile materials on the earth. Its high stability and long durability facilitate its integral role in our day-to-day lives, from the kitchen to an industrial level [1,2]. Plastic is a polymer and consists of the elements carbon, hydrogen, silicon, oxygen, chlorine, and nitrogen [1]. Plastic production can be bio-based or synthetic. Bio-based plastics are made from natural compounds such as lignin, cellulose, hemicellulose, terpenes, vegetable oils, carbohydrates, and food waste [3,4,5]. In contrast, crude oil is the main component of synthetic plastics [6,7], which are generally referred to as non-biodegradable [8].
The present study is focused on synthetic plastics. Seven major types of synthetic plastic are used around the world. These are polyethylene terephthalate (PET), high-density polyethylene (HDPE), polyvinyl chloride (PVC), low-density polyethylene (LDPE), polypropylene (PP), polystyrene (PS), and various other plastics that include acrylic, polycarbonates, polylactic acid (PLA), fibers, and nylon [3,9].
Global plastic production is rising year by year. In 2018, the annual global plastic production was 359 million tons, while it was 368 million tons in 2019 [10]. China is the world leader in plastic production, accounting for 31% of world plastic production in 2019 [10]. Although the production of new plastic increases annually, the rate of plastic waste management and plastic recycling rate has not reached precise levels [10]. In Europe, total plastic production in 2018 was 61.8 million tons. However, the amount of plastic post-consumer waste collected in Europe in 2018 was only 9.4 million tons [10]. The estimated global total for virgin plastic production was 8300 million tons in 2017 [11]. The amount of plastic waste accumulated as of 2015 was 6300 million tons; from that, only 9% were recycled, 12% were incinerated, and 79% were placed in landfills or introduced to the natural environment [11]. Every year 25 million tons of synthetic plastics are accumulated along seacoasts and in terrestrial environments [12]. In 2019, the amount of plastic waste released into the aquatic environments was 6.1 million tonnes (Mt), and 1.7 Mt flowed into oceans. Currently, there has been a dramatic increase. At least eight million tons of plastic get into oceans, which have the potential of breaking down into tiny microplastics that possibly make their way into our food chains and result in unknown effects [13]. Current plastic accumulation in reverse is 109 Mt [14]. The build-up of plastics in rivers implies that leakage into the ocean will continue for decades, even if mismanaged plastic waste could be significantly reduced [14]. Current estimations reveal that 80% of marine litter accumulation is due to plastic debris [4]. This marine plastic waste accumulation in amounts is around 30 Mt. The total amount of plastic waste only in the ocean is expected to grow from 50 million tons in 2015 to 150 million tons by 2025 [15]. If current production and waste management trends continue, roughly 12,000 million tons of plastic waste will be in landfills or the natural environment by 2050, with an annual accumulation of ~339 million tons [11,16].
Moreover, when considering the plastic accumulation based on major plastic producing regions globally, in just Europe, 75,000 to 300,000 tons of microplastics accumulate every year [4]. In the United States, the landfill rate for discarded plastics exceeds 75% [17].
Therefore, these various types of plastics, after being used and then discarded, not only pervade in every nook and cranny of the planet but also cause major negative impacts at the ecosystem level [18]. At present, plastic pollution has become one of the major global environmental issues [4], and it is the leading cause of biodiversity reduction. Polyethylene causes blockages in the intestines of birds, fish, and marine mammals. Entanglement in or ingesting this waste has endangered hundreds of different species [19]. Fragmented plastic debris, referred to as microplastics, were reported entrapping in subtropical gyres that run over a million square kilometers. These microplastics ingested by a wide range of organisms subsequently cause substantial negative impacts on their existence [20]. Moreover, plastics can cause health effects; for instance, airborne microplastics can be inhaled, thus representing a direct risk to human health [21].
The natural degradation rate of plastic is extremely slow, and that causes plastics wastes to accumulate in all components of the environment [22,23]. The long chain polymer structure, high molecular weight, and hydrophobicity cause plastics to be resistant to biodegradation [24]. In fact, some plastics take up to 1000 years to degrade [25]. These facts are the reason for the rapid plastic accumulation in natural environments. Hence, developing an efficient process to accelerate the plastic degradation rate is essential to avoid this annual accumulation. Several solutions have been provided in the scientific community and have been experimentally proved to some extent. Those methods include photo-degradation (degraded by light), chemical degradation, thermal degradation (degraded by heat), irradiation using gamma rays, and biodegradation (degraded by biological additives or microorganisms) [25,26,27]. However, a method with minimum harmful effects to nature but without or at most minimum toxic by-products is required [27]. The processes such as photo-degradation, chemical degradation, thermal degradation, and irradiation using gamma rays cause many negative impacts on nature, such as accumulation of heavy metals in ecosystems and disturbance in natural ecosystem functioning. Moreover, those methods require high costs and energy levels to perform. Therefore, scientists across the globe have tended to investigate better biodegradation methods that do not result in harmful effects and represent an eco-friendly approach for managing plastics [27]. Furthermore, biodegradation is a proper solution since it is cost-effective and does not require much energy. More than 400 microbes have been recorded to be capable of plastic degradation [28]. The present study was focused only on plastic-degrading fungi among plastic-degrading microbes.
Several studies have been carried out on plastic-degrading fungi. For example, Lacerda et al. [29] investigated the fungi from the plastisphere within the aquatic environments of the western South Atlantic and Antarctic Peninsula. Microorganisms and enzymes that are able to degrade a variety of generally used synthetic plastics were comprehensively summarized. The microbial metabolic pathways for plastic depolymerization products and the current attempts toward utilization of such products as feedstocks for microbial production of chemicals with high value were highlighted by Ru et al. [30]. Sánchez [31] described the natural and unique ability of fungi to invade macro- and microplastic substrates by using enzymes that have the capacity to detoxify pollutants. Sáenz et al. [32] experimented with biodegradation of low-density polyethylene (LDPE) by Aspergillus niger and A. terreus to increase the degradation rate without any co-substrate or photothermal treatment. Iram et al. [33] reviewed some of the most common strategies for the degradation of various types of polymers, along with a list of potential microbes capable of feeding on them. The role of marine fungi was also recently reviewed by Zeghal et al. [34]. Microbial degradation of plastics was reviewed by Kale et al. [22]. However, a recent complete study on plastic-degrading fungi in all environments, including both aquatic and terrestrial environments, which addresses their detailed phylogenetic relationships, is lacking. As such, the information presented herein reviews plastic-degrading fungi reported thus far and their phylogenetic relationships.
The present study attempted to summarize current plastic production and plastic waste accumulation in natural environments worldwide. Moreover, a list of all taxa of fungi known to be capable of degrading plastic-degrading fungi was developed from the available literature, and notes were compiled on the major fungal groups involved. In addition, the phylogenetic relationships of plastic-degrading fungi were analyzed based on multi-gene analyses (ITS, LSU, SSU, TEF, RPB1, and RPB2). As such, the overall objective of the study was to summarize all currently known basic information on plastic-degrading fungi to influence future research on similar topics.
2. Materials and Methods
2.1. Literature Review
The data presented herein was acquired from several different sources—the published literature, online databases, and personal communication. We selected seven major synthetic plastic types used around the world based on recent literature [3,9] and the species of fungi reported to degrade those plastic types. Plastic-degrading fungal species data were collected primarily from “Google Scholar”, “Research Gate”, “PubMed”, and “Web of Science”. The main phrases used in the online literature search for the plastic types mentioned above and the taxa of fungi involved in their biodegradation were “plastic types”, “global plastic production”, “global plastic waste management”, “fungal bio-degradation of plastics”, and “bio-degradable plastics”. A list of major types of synthetic plastic with their major uses and annual production was prepared (Table 1). Taxa of fungi reported to degrade plastics are listed in Table 2.
Table 1.
| Plastic Type and Structure [9] | Main Uses [9] | Annual Production/ Million Metric Tons [35] |
Specific Surface Degradation Rate (Min−Max; μm Tear−1) [17] |
|
|---|---|---|---|---|
| Land | Marine | |||
| Polyethylene Terephthalate (PET or PETE) [36] | Beverage bottles, Food bottles/jars and polyester clothing or rope | 30.5 | 0 | - |
| High-Density Polyethylene (HDPE) | Milk cartons, detergent bottles, cereal box liners, toys, buckets, park benches, and rigid pipes | 66.96 | 1.0 (0.91−1.1) |
4.3 (0−11) |
| Polyvinyl Chloride (PVC or Vinyl) | Plumbing pipes, credit cards, human and pet toys, rain gutters, teething rings, IV fluid bags and medical tubing, and oxygen masks | 44.3 | 0 | - |
| Low-Density Polyethylene (>LDPE) | Plastic wrap, sandwich and bread bags, bubble wrap, garbage bags, grocery bags and beverage cups | >1500 | 11 | 15 (0−37) |
| Polypropylene (PP) | Straws, bottle caps, prescription bottles, hot food containers, packaging tape, disposable diapers, and DVD/CD boxes | 56 | - | 7.5 |
| Polystyrene (PS or Styrofoam) | Cups, takeout food containers, shipping and product packaging, egg cartons, cutlery and building insulation | 15.61 | 0 | - |
| Other | Eyeglasses, baby and sports bottles, electronics, CD/DVDs, lighting fixtures, and clear plastic cutlery | - | 270 (20−1400) |
16 (7.5−29) |
Table 2.
Recorded taxa of fungi that are known to degrade plastics.
| Fungus | Polymer Hydrolysed | Class | Family | Environment | References |
|---|---|---|---|---|---|
| Ascomycota | |||||
| Acremonium kiliense | PE | Sordariomycetes | Bionectriaceae | Soils | [37] |
| Acremonium sp. | PHB, Poly[3 HB-co-(10 mol%) 3HV] | Sordariomycetes | Bionectriaceae | Soils | [38] |
| Alternaria alternata | PE, LDPE | Dothideomycetes | Pleosporaceae | Dumpsites, Mangrove stands | [19,39] |
| Alternaria brassicicola | - | Dothideomycetes | Pleosporaceae | Not mentioned | [40] |
| Alternaria dauci | PUR | Dothideomycetes | Pleosporaceae | Rainforest | [41] |
| Alternaria solani | PS-PUR | Dothideomycetes | Pleosporaceae | Soils, Wall paints (Latex), Pieces of plastic debris | [42] |
| Alternaria sp. | PUR | Dothideomycetes | Pleosporaceae | Rainforest | [41] |
| Arxula adeninivorans | - | Saccharomycetes | Trichomonascaceae | Not mentioned | [43] |
| Aspergillus caespitosus | LDPE | Eurotiomycetes | Aspergillaceae | Mangrove stands | [19] |
| Aspergillus fischeri | PCL | Eurotiomycetes | Aspergillaceae | Soils | [44,45] |
| Aspergillus flavus | PE, HDPE, LDPE, PVC, PCL, PS-PUR, PEA, PPA, PBA | Eurotiomycetes | Aspergillaceae | Soils | [12,42,44,46,47,48] |
| Aspergillus fumigatus | PHB, Poly[3HB-co-(10 mol%) 3HV], HDPE, LDPE, PS-PUR, Sky-Green, Poly[3HB-co-(7–77 mol%) 3HV], PHV, Poly[3HB-co-(13–61 mol%) 4HB], PES, PEA, PBA, PCL, PBS | Eurotiomycetes | Aspergillaceae | Soils | [12,38,42,49,50,51,52,53,54] |
| Aspergillus glaucus | PE | Eurotiomycetes | Aspergillaceae | Mangrove Soils | [55] |
| Aspergillus japonicus | LDPE | Eurotiomycetes | Aspergillaceae | Polythene polluted sites | [56] |
| Aspergillus nidulans | LDPE | Eurotiomycetes | Aspergillaceae | Dumpsite | [54,57] |
| Aspergillus Niger | PE, HDPE, LDPE, PVC, Sky-Green, PEA, PPA, PBA | Eurotiomycetes | Aspergillaceae | Soils | [12,47,48,50,56,58,59,60,61,62,63,64] |
| Aspergillus nomius | LDPE | Eurotiomycetes | Aspergillaceae | Landfill soils | [65] |
| Aspergillus ochraceus | LDPE | Eurotiomycetes | Aspergillaceae | Pleurotus ostreatus (Oyster mushroom) baglog | [63] |
| Aspergillus oryzae | LDPE | Eurotiomycetes | Aspergillaceae | Not mentioned | [54,66,67] |
| Aspergillus penicilloides | PHB | Eurotiomycetes | Aspergillaceae | Biological products | [68] |
| Aspergillus sp. | PE | Eurotiomycetes | Aspergillaceae | Sea water | [69] |
| Aspergillus sydowii | PE | Eurotiomycetes | Aspergillaceae | Dumping sites, Mangrove rhizosphere soils | [2] |
| Aspergillus terreus | LDPE, HDPE, PS-PUR, PE | Eurotiomycetes | Aspergillaceae | Soils | [2,12,19,42,45,49] |
| Aspergillus tubingensis | PU | Eurotiomycetes | Aspergillaceae | Soils | [70] |
| Aspergillus ustus | Sky-Green, PHB | Eurotiomycetes | Aspergillaceae | Soils, Deep Sea | [50,71] |
| Aspergillus versicolor | HDPE, LDPE, PVC, PEA, PPA, PBA | Eurotiomycetes | Aspergillaceae | Soils, Degraded polyimides, Marine water | [47,48,71,72] |
| Aureobasidium pullulans | PCL, PEA, PPA, PBA | Dothideomycetes | Saccotheciaceae | Not mentioned | [48,73] |
| Bionectria sp. | PUR | Sordariomycetes | Bionectriaceae | Rainforest | [41] |
| Candida guilliermondii | PHB | Saccharomycetes | Candidaceae | Deep sea | [71] |
| Cephalosporium sp. | PHB | Sordariomycetes | Incertae sedis | Not mentioned | [74] |
| Chaetomium globosum | HDPE, LDPE, PVC, PCL, PEA, PPA, PBA | Sordariomycetes | Chaetomiaceae | Soils | [44,47,48] |
| Chaetomium sp. | PE | Sordariomycetes | Chaetomiaceae | Groundnut | [46] |
| Chrysonilia setophila | HDPE, LDPE, PVC | Sordariomycetes | Sordariaceae | Soils | [47] |
| Cladosporium cladosporioides | PU | Sordariomycetes | Chaetomiaceae | Plastic debris in a shoreline of a lake | [6] |
| Cladosporium sp. | PHB | Sordariomycetes | Chaetomiaceae | Not mentioned | [74] |
| Colletotrichum fructicola | LDPE | Sordariomycetes | Glomerellaceae | Not mentioned | [64] |
| Curvularia lunata | PE | Dothideomycetes | Pleosporaceae | Dumpsites | [39] |
| Curvularia protuberata | Sky-Green | Dothideomycetes | Pleosporaceae | Soils | [50] |
| Debaryomyces hansenii | PHB | Saccharomycetes | Debaryomycetaceae | Deep sea | [71] |
| Diaporthe italiana | LDPE | Sordariomycetes | Diaporthaceae | Not mentioned | [64] |
| Edenia gomezpompae | PUR | Dothideomycetes | Phaeosphaeriaceae | Rainforest | [41] |
| Emericellopsis minima | PHB, Poly[3HB-co-(30 mol%) 3HV] | Sordariomycetes | Incertae sedis | Not mentioned | [75] |
| Eupenicillium hirayamae | - | Eurotiomycetes | Aspergillaceae | Mangrove stand | [19] |
| Eupenicillium rubidurum | - | Eurotiomycetes | Aspergillaceae | Not mentioned | [45] |
| Eupenicillium sp. | PHB | Eurotiomycetes | Aspergillaceae | Soils | [76] |
| Exophiala jeanselmei | Polyether | Eurotiomycetes | Herpotrichiellaceae | Soils | [77] |
| Fusarium moniiforme | PCL | Sordariomycetes | Nectriaceae | Not mentioned | [78] |
| Fusarium oxysporium | Poly[3HB-co-(12 mol%) 3HV], HDPE, LDPE, PVC, PET | Sordariomycetes | Nectriaceae | Soils | [47,79,80,81] |
| Fusarium solani | LDPE, HDPE, PVC, PCL, PS-PUR, PHB, PET | Sordariomycetes | Nectriaceae | Soils | [12,42,47,49,50,78,81,82,83,84] |
| Fusarium sp. | PE, PCL | Sordariomycetes | Nectriaceae | Soils, Dumpsites | [44,85] |
| Gliocladium roseum | PS-PUR | Sordariomycetes | Hypocreaceae | Not mentioned | [86] |
| Gliocladium virens | LDPE | Sordariomycetes | Hypocreaceae | Not mentioned | [60] |
| Glomerella cingulata | - | Sordariomycetes | Glomerellaceae | Not mentioned | [87,88] |
| Guignardia mangiferae | PUR | Dothideomycetes | Phyllostictaceae | Rainforest | [41] |
| Humicola insolens | - | Sordariomycetes | Chaetomiaceae | Not mentioned | [89] |
| Lasiodiplodia sp. | PUR | Dothideomycetes | Botryosphaeriaceae | Rainforest | [41] |
| Leptosphaeria sp. | PU | Dothideomycetes | Botryosphaeriaceae | Plastic debris in a shoreline of a lake | [6] |
| Malbranchea cinnamomea | - | Eurotiomycetes | Onygenaceae | Not mentioned | [90] |
| Monascus sp. | PU | Eurotiomycetes | Monascaceae | Plastic contaminated soils | [91] |
| Monilinia fructicola | - | Leotiomycetes | Sclerotiniaceae | Not mentioned | [92] |
| Myceliophthora thermophila | - | Sordariomycetes | Chaetomiaceae | Not mentioned | [93] |
| Nectria sp. | PUR | Sordariomycetes | Nectriaceae | Rainforest | [41] |
| Paecilomyces farinosus | Poly[3HB-co-(12 mol%) 3HV], PHB, Sky-Green | Eurotiomycetes | Thermoascaceae | Soils | [50,79] |
| Paecilomyces lilacinus | PHB, PCL, Poly[3HB-co-(12 mol%) 3HV] | Eurotiomycetes | Thermoascaceae | Soils | [79,94] |
| Paecilomyces marquandii | PHB | Eurotiomycetes | Thermoascaceae | Biological products | [67] |
| Paecilomyces variotii | - | Eurotiomycetes | Thermoascaceae | Mangrove stands | [19] |
| Paraphoma-like | - | Dothideomycetes | Phaeosphaeriaceae | Barley phylloplane | [95,96] |
| Penicillium adametzii | PHB | Eurotiomycetes | Aspergillaceae | Biological products | [68] |
| Penicillium argillaceum | PCL | Eurotiomycetes | Aspergillaceae | Not mentioned | [53] |
| Penicillium chermisinum | PHB | Eurotiomycetes | Aspergillaceae | Freshwater | [97] |
| Penicillium crysosporium | Poly[3HB-co-(7 mol%) 3HV] | Eurotiomycetes | Aspergillaceae | Not mentioned | [98] |
| Penicillium daleae | PHB | Eurotiomycetes | Aspergillaceae | Biological products | [68] |
| Penicillium dupontii | PCL | Eurotiomycetes | Aspergillaceae | Not mentioned | [53] |
| Penicillium funiculosum | PCL, PHB, PHV, Poly[3HB-co-(7, 14%) 4HB], Poly[3HB-co-(7, 27, 45, 71%) 3HV], PEA, PPA, PBA | Eurotiomycetes | Aspergillaceae | Soils | [44,48,50,94,99,100] |
| Penicillium griseofulvum | PU | Eurotiomycetes | Aspergillaceae | Plastic debris in a shoreline of a lake | [6] |
| Penicillium janthinellum | PHB | Eurotiomycetes | Aspergillaceae | Freshwater | [97] |
| Penicillium minioluteum | PHB | Eurotiomycetes | Aspergillaceae | Soils | [50] |
| Penicillium orchrochloron | PHB | Eurotiomycetes | Aspergillaceae | Biological products | [68] |
| Penicillium oxalicum | HDPE, LDPE | Eurotiomycetes | Aspergillaceae | Soils of a plastic dumping site | [26] |
| Penicillium pinophilium | PHB, LDPE | Eurotiomycetes | Aspergillaceae | Soils | [50,60,101] |
| Penicillium restricum | PHB | Eurotiomycetes | Aspergillaceae | Biological products | [68] |
| Penicillium roqueforti | PLA | Eurotiomycetes | Aspergillaceae | Not mentioned | [102] |
| Penicillium simplicissimum | PE, PHB, Poly[3HB-co-(7 mol%) 3HV], Sky-Green | Eurotiomycetes | Aspergillaceae | Soils | [50,76,85,97,98,103] |
| Penicillium sp. | PHB, HDPE, LDPE, PVC, PEA, PCL, polyalkylene dicarboxylic acids | Eurotiomycetes | Aspergillaceae | Soils | [47,104,105,106] |
| Penicillium verruculosum | Mater-Bi | Eurotiomycetes | Aspergillaceae | Soils | [50] |
| Pestalotiopsis microspora | PUR | Sordariomycetes | Sporocadaceae | Rainforest | [41] |
| Pestalotiopsis sp. | PUR | Sordariomycetes | Sporocadaceae | Rainforest | [41] |
| Phaeosphaeria sp. | PUR | Dothideomycetes | Phaeosphaeriaceae | Rainforest | [41] |
| Phialophora alba | - | Eurotiomycetes | Herpotrichiellaceae | Mangrove stands | [19] |
| Phoma sp. | HDPE, LDPE, PVC | Dothideomycetes | Didymellaceae | Soils | [45,47] |
| Plectosphaerella sp. | PUR | Sordariomycetes | Plectosphaerellaceae | Rainforest | [41] |
| Pleosporales sp. | PUR | Dothideomycetes | Rainforest | [41] | |
| Pullularia pullulans | PEA, PPA, PBA | Dothideomycetes | Saccotheciaceae | Not mentioned | [48] |
| Sirococcus conigenus | - | Sordariomycetes | Gnomoniaceae | Not mentioned | [107] |
| Spicaria spp. | PS-PUR | Incertae sedis | Incertae sedis | Soils, Wall paints (Latex), Plastic debris | [42] |
| Stagonosporopsis citrulli | LDPE | Dothideomycetes | Didymellaceae | Not mentioned | [64] |
| Talaromyces islandicus | - | Eurotiomycetes | Aspergillaceae | Not mentioned | [45] |
| Thermoascus aurantiacus | PHB, PCL, PBS | Eurotiomycetes | Thermoascaceae | Not mentioned | [53] |
| Thielavia terrestris | - | Sordariomycetes | Chaetomiaceae | Soils | [108,109] |
| Thyrostroma jaczewskii | LDPE | Dothideomycetes | Botryosphaeriaceae | Not mentioned | [64] |
| Trichoderma hamatum | LDPE, PS, PVC | Sordariomycetes | Hypocreaceae | Plastic waste material | [110,111] |
| Trichoderma reesei | - | Sordariomycetes | Hypocreaceae | Not mentioned | [112] |
| Trichoderma viride | - | Sordariomycetes | Hypocreaceae | Landfill soils | [65] |
| Verticillium Lecanii | PE | Sordariomycetes | Plectosphaerellaceae | Soils | [37] |
| Verticillium leptobactrum | PHB | Sordariomycetes | Plectosphaerellaceae | Soils | [38] |
| Xepiculopsis gramineae | PU | Sordariomycetes | Incertae sedis | Plastic debris in a shoreline of a lake | [6] |
| Zopfiella karachiensis | PUR | Sordariomycetes | Lasiosphaeriaceae | Rainforest | [41] |
| Basidiomycota | |||||
| Cryptococcus laurentii | PCL | Tremellomycetes | Tremellaceae | Soils | [44] |
| Cryptococcus magnus | - | Tremellomycetes | Tremellaceae | Barley Phylloplane | [95] |
| Cryptococcus sp. | PBS, PBSA | Tremellomycetes | Tremellaceae | - | [113] |
| Papiliotrema laurentii | PBS, PBSA | Tremellomycetes | Rhynchogastremaceae | Part of a microbiome analysis of an aircraft | [114] |
| Phanerochaete chrysosporium | LDPE, Poly[3HB-co-(7 mol%) 3HV], PVC | Agaricomycetes | Phanerochaetaceae | Soils | [60,108,115,116,117] |
| Pleurotus ostreatus | PE, LDPE | Agaricomycetes | Pleurotaceae | - | [16] |
| Polyporus circinatus | PHB | Agaricomycetes | Hymenochaetaceae | - | [74] |
| Pseudozyma antarctica | - | Ustilaginomycetes | Ustilaginaceae | Obtained from the culture collection of the Japan Collection of Microorganisms (JCM) of the Riken BioResource Center in Wako, Japan. | [118] |
| Rhodosporidium sphaerocarpum | PHB | Microbotryomycetes | Sporidiobolaceae | Deep sea | [71] |
| Tritirachium album | PLA | Tritirachiomycetes | Tritirachiaceae | - | [119] |
| Mucoromycota | |||||
| Mucor sp. | PHB, PVC | Mucoromycetes | Mucoraceae | - | [74,106] |
| Mucor hiemalis | HDPE, LDPE, PVC | Mucoromycetes | Mucoraceae | Soils | [47] |
| Rhizopus arrhizus | PCL, polyalkylene dicarboxylic acids | Mucoromycetes | Rhizopodaceae | - | [120] |
| Rhizopus delemar | PPA, PET copolymers with dicarboxylic acids | Mucoromycetes | Rhizopodaceae | - | [121,122] |
PHB: Polyhydroxybutyrate, PUR: Polyurethane, PCL: Polycaprolactone, PPA: Polyphthalamide, PBA: Polybutanamide, PHV: Poly(3-hydroxybutyrate-co-3-hydroxyvalerate), PES: Polyethersulfone, PEA: Polyesteracetals, PBS: Polybutylene succinate, PBSA: Poly(butylene succinate-co-butylene adipate), PET: Polyethylene terephthalate, HDPE: High-density polyethylene, PVC: Polyvinyl chloride, LDPE: Low-density polyethylene, PS-PUR: polyester-polyurethane, PP: Polypropylene, PS: Polystyrene, PE: Polyethylene, PU: Polyurethane. The “-” is used to show the data is not available in literature.
2.2. Phylogenetic Analyses
2.2.1. Taxon Sampling
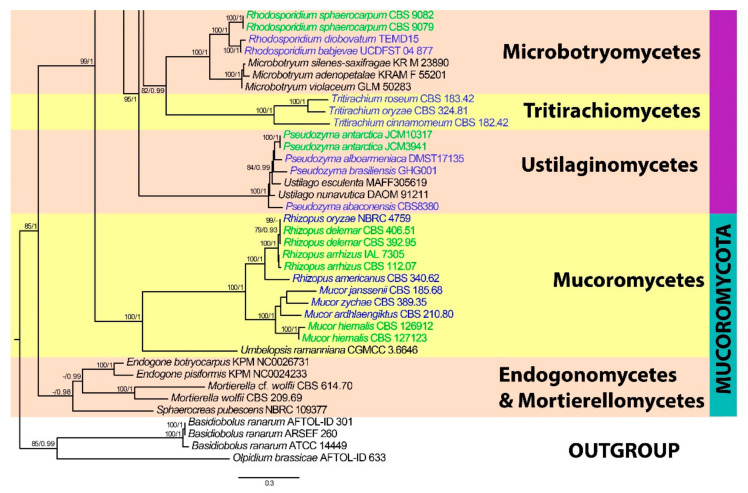
A data matrix containing 395 taxa, including four out-group taxa (Basidiobolus ranarum AFTOL-ID 301, Basidiobolus ranarum ARSEF 260, Basidiobolus ranarum ATCC 14449, and Olpidium brassicae AFTOL-ID 633) were generated in the present study. ITS, LSU, SSU, TEF, RPB1, and RPB2 sequence data and their GenBank accession numbers are provided in Table S1.
2.2.2. Initial Phylogenetic Analyses
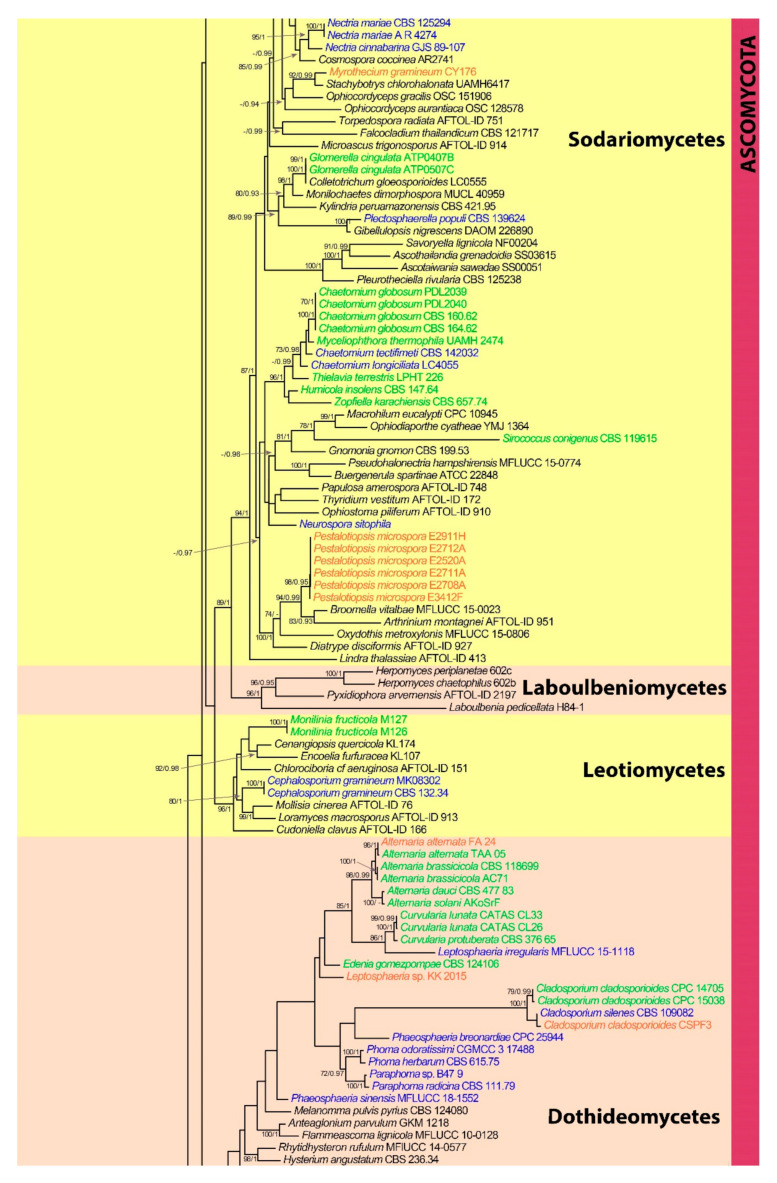
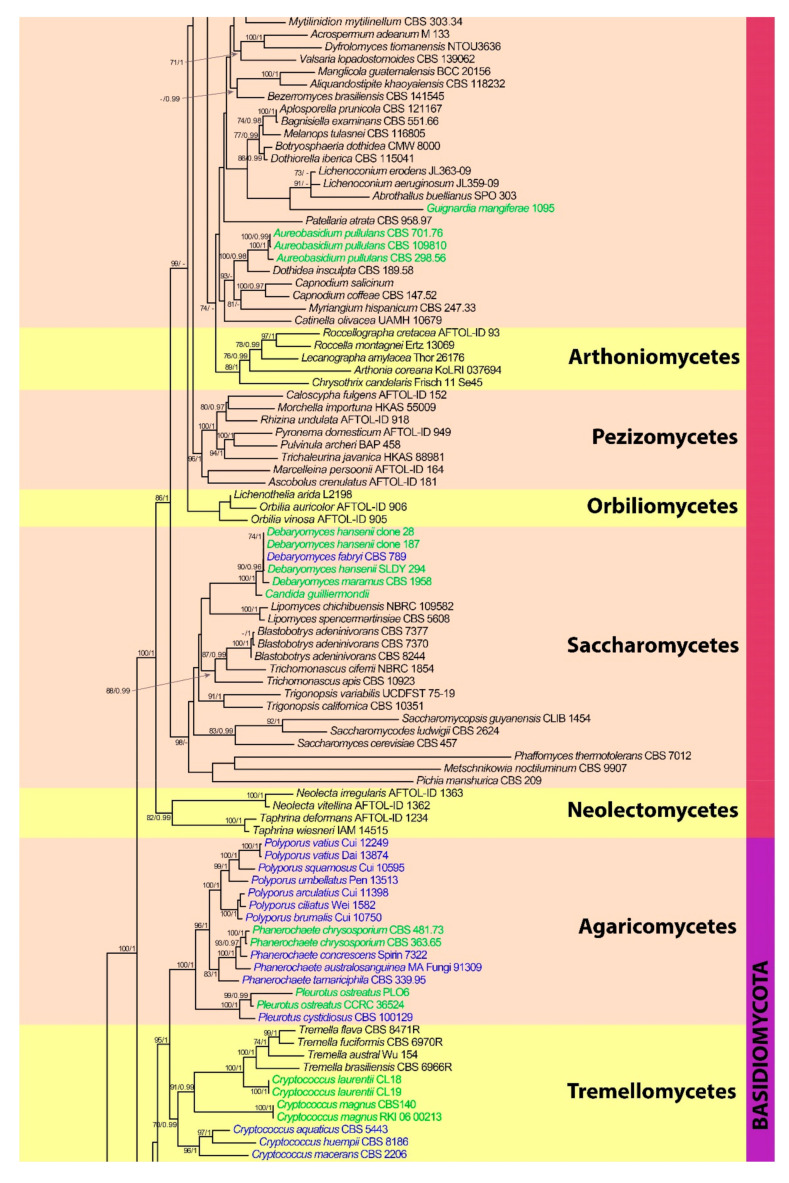
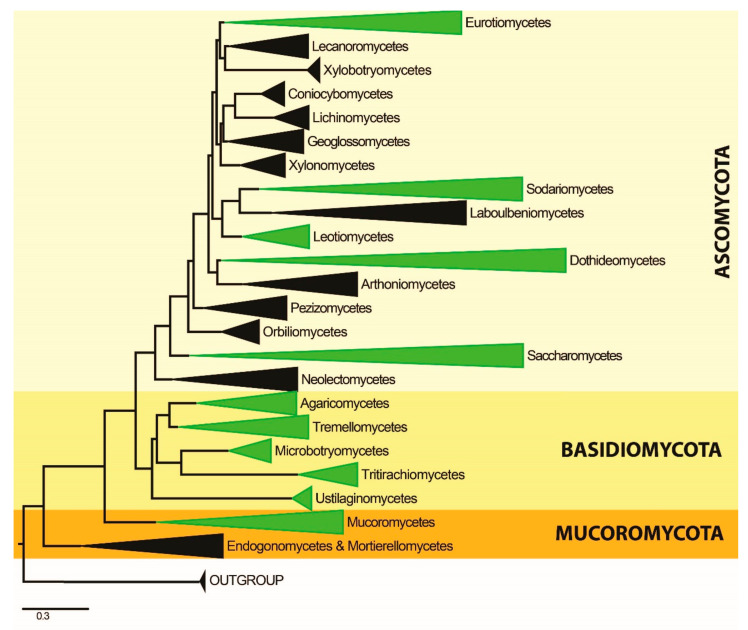
Multiple sequence alignments for each gene (ITS, LSU, SSU, TEF, RPB1, and RPB2) were generated with MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/, accessed on 22 April 2022). Long alignment gaps were removed using trimAl [123] available at Phylemon2 web server [124] and manually adjusted in BioEdit v. 7.0.4 [125] where necessary. The individual datasets were concatenated into a combined dataset using FaBox (1.41) [126]. Ambiguously aligned regions were excluded, and gaps were treated as missing data. Maximum likelihood phylogenetic analyses were performed in the CIPRES web portal [127] using RAxML-HPC2 on XSEDE (8.2.12) tool [128]. The bootstrap analysis for each ML tree was performed with 1000 thorough bootstrap replicates with the same parameter settings using the GTR+G+I substitution model selected by MrModel Test 2.2 [129]. Posterior probabilities (PP) [130,131] were determined by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.0b4 [132]. Four simultaneous Markov chains were run for 20,000,000 generations, and the trees were sampled every 1000th generation. MCMC heated chain was set with a “temperature” value of 0.2. The distribution of loglikelihood scores was examined to determine the stationary phase for each search and to decide if extra runs were required to achieve convergence using the program Tracer 1.5 [133]. All sampled topologies beneath the asymptote (40%) were discarded as part of a burn-in procedure, while the remaining trees (12,000) were used for calculating posterior probabilities in the majority rule consensus tree. Bayesian Posterior Probabilities (BYPP) equal to or greater than 0.90 are given below or above each node (Figure 1). The resulting trees were viewed with FigTree v.1.4.0 [134]. The compressed overview of the phylogram resulting from the phylogenetic analysis is presented in Figure 2, where classes including plastic-degrading fungi are colored green.
Figure 1.
Phylogenetic relationships of plastic-degrading fungi. Phylogram generated from a maximum likelihood analysis of ITS, LSU, SSU, TEF, RPB1 and RPB2 fungal sequence data. MLBP values ≥ 70% and BYPP ≥ 0.90 values are given as the first and the second set of numbers near the nodes. Strain/culture numbers are given after the taxon names. The tree is rooted with Basidiobolus ranarum AFTOL-ID 301, Basidiobolus ranarum ARSEF 260, Basidiobolus ranarum ATCC 14449, and Olpidium brassicae AFTOL-ID 633. Strains in Red: Strains directly reported as plastic degraders. Green: Other strain of the same species reported as plastic degraders. Blue: Other species in the same genus reported as plastic degraders.
Figure 2.
Compressed overview of the phylogram for the phylogenetic relationships of plastic-degrading fungi. Class level taxonomic ranks in Green include fungal species that were reported as plastic degraders.
3. Results
3.1. Phylogenetic Analyses
The present study investigated the phylogenetic relationships of plastic-degrading fungi based on an analysis of ITS, LSU, SSU, TEF, RPB1, and RPB2 sequence data. The alignment of combined genes included 6289 bp (ITS-1–388, LSU-389–1434, SSU-1435–2502, TEF-2503–3382, RPB1-3383–4569, and RPB2-4570–6289). The topology of the tree from maximum likelihood analysis was similar to the tree from Bayesian analysis. The best scoring RAxML tree with a final likelihood value of −275,048.218811 is presented. The matrix had 5830 distinct alignment patterns, with 67.51% undetermined characters or gaps.
The phylogenetic tree (Figure 1 and Figure 2) representing those taxa of fungi known to be able to degrade fungi comprises three major clades, with these representing the three major phyla Ascomycota, Basidiomycota, and Mucoromycota. The basal clade Mucoromycota includes the class Mucoromycetes. Consequently, these taxa are the basal groups of plastic degraders in the fungi kingdom. The phylum Basidiomycota also can be recognized as a clade, and it is comprised of the Agaricomycetes, Microbotryomycetes, Tremellomycetes, Tritirachiomycetes, and Ustilaginomycetes. For Clade 3, the upper clade is the phylum Ascomycota and includes the Dothideomycetes, Eurotiomycetes, Leotiomycetes, Saccharomycetes, and Sordariomycetes. Based on the phylogeny of the present study, the Eurotiomycetes are the most evolved group of plastic-degrading fungi. The highest numbers of plastic-degrading fungi are included in Clade 3.
3.2. Phylum Ascomycota
3.2.1. Class Dothideomycetes
The Dothideomycetes is the largest class within the phylum Ascomycota. Most of the taxa within this class are recorded as saprobes in various habitats and substrates [135]. Our investigations revealed that some members of Dothideomycetes are capable of plastic degradation. Most of the recorded species of the Dothideomycetes belong to the order Pleosporales, but a few taxa are recorded from the Dothideales and Botryosphaeriales (Table 2, Figure 1 and Figure 2). Plastic degraders in the Dothideomycetes have the ability to degrade LDPE, PUR, PS, PCL, PEA, PPA, PBA, HDPE, PVC, PE, PU, and Sky-Green plastics (Table 2). Recent studies on plastic-degrading members of the Dothideomycetes are those of Khruengsai et al. [64] and Brunner et al. [6]. In the phylogenetic tree, the Dothideomycetes formed a well-supported clade sister to the Arthoniomycetes. Most of the plastic-degrading Dothideomycetes were placed in the upper subclade of the main Dothideomycetes clade, and the rest were grouped in a basal subclade (Figure 1 and Figure 2).
3.2.2. Class Eurotiomycetes
The Eurotiomycetes are extremely common saprobes in diverse habitats and substrates [136]. Based on our results, most plastic-degrading fungal records belong to the Eurotiomycetes. Many plastic-degrading members of the Eurotiomycetes are taxonomically placed under the Eurotiales, and the most common plastic-degrading fungal genera are Aspergillus and Penicillium (Table 2, Figure 1 and Figure 2). The plastic types they are reported to degrade are HDPE, LDPE, PCL, PE, PVC, PS-PUR, PEA, PPA, PBA, PHB, Poly[3HB-co-(10 mol%) 3HV], Sky-Green, PHV, PBS, PLA and PVC (Table 2). Recent studies on plastic-degrading Eurotiomycetes are those of Rani & Singh [12], Ndahebwa Muhonja et al. [54], Bermúdez-García et al. [57], Laila [63], Khruengsai et al. [64], Munir et al. [65], Alshehrei [69], Sangale et al. [2], Duan et al. [90], El-Morsy et al. [92], Brunner et al. [6] and Ojha et al. [26]. The Eurotiomycetes is the uppermost clade within our phylogram (Figure 1 and Figure 2). The genera Aspergillus and Penicillium contain a large number of species with a worldwide distribution and a huge range of ecological habitats [137]. They are mostly widespread saprobes and can be found in both indoor and outdoor environments, including in both the air and soil. In addition, some species of Aspergillus and Penicillium have the ability to grow under extreme conditions [137]. Hence, further research on these genera would provide better solutions for the environmental accumulation of plastics.
3.2.3. Class Leotiomycetes
Most of the Leotiomycetes are saprobes on a wide variety of substrates. However, this class also includes many important plant pathogens [138]. Our results showed that the few records of plastic-degrading fungi are phylogenetically related to the Leotiomycetes (Table 2, Figure 1 and Figure 2). In the phylogenetic tree of this study, the Leotiomycetes formed a well-supported clade sister to the Laboulbeniomycetes (Figure 1 and Figure 2). However, there is a single record found that clearly belongs to the Leotiomycetes and possibly has the ability of plastic degradation [92]. The other records that grouped within Leotiomycetes in our phylogeny were Cephalosporium gramineum, which is currently placed under the Sordariomycetes. Therefore, studies with a wide range of taxon sampling are required to resolve the phylogenetic position of Cephalosporium gramineum. Furthermore, a recent study on the biodegradation of bio-based and biodegradable plastic, polybutylene succinate-co-adipate (PBSA), identified a species (Tetracladium furcatum) within the Leotiomycetes that has the ability to degrade PBSA [139]. However, recent studies on synthetic plastic-degrading members of the Leotiomycetes are very few. As a result, additional studies on the Leotiomycetes are required to assess their ability to degrade plastics.
3.2.4. Class Saccharomycetes
The Saccharomycetes is a small class of yeasts with a single order of about 1000 known species, which are classified under the Ascomycota [140]. Most of the Saccharomycetes are saprobes, and few are recorded as human and plant pathogens [140]. The present study discovered five records of five members of the Saccharomycetes capable of degrading plastics (Table 2, Figure 1 and Figure 2). The Saccharomycetes is the second basal clade within the phylum Ascomycota. All Saccharomycetes members that are plastic degraders are grouped in the upper subclade of the main Saccharomycetes clade (Figure 1 and Figure 2). Arxula, Candida, and Debaryomyces are the genera to which those plastic-degrading members of the Saccharomycetes belong [43,71]. Some members of the Saccharomycetes are widely used in industrial and biotechnological processes. Species such as Saccharomyces cerevisiae are model organisms in many types of research [140]. A recent study used Genetic engineering techniques on two strains of Saccharomyces cerevisiae to produce the heterologous protein Polyethylene Terephthalate (PET) hydrolase enzyme, which has been shown to have the capability of degrading PET into its subsequent monomers [141]. We believe future research on the Saccharomycetes would find a better solution for plastic accumulation in nature.
3.2.5. Class Sordariomycetes
The Sordariomycetes is the second largest class within the phylum Ascomycota. The majority of the Sordariomycetes are saprobes, but the group also includes some important plant pathogens [142]. They have a wide ecological distribution in both terrestrial and aquatic habitats [142]. Our research recorded many members of the Sordariomycetes with the ability to degrade plastics (Table 2, Figure 1 and Figure 2). The plastic types they are reported to degrade are PE, PS, PHB, Poly[3HB-co-(10 mol%) 3HV], PUR, PS-PUR, HDPE, LDPE, PVC, PCL, PEA, PPA, and PBA (Table 2). Recent studies of plastic-degrading Sordariomycetes are those of Munir et al. [65], (Yang et al. [93], Brunner et al. [6], and Khruengsai et al. [64]. The Sordariomycetes formed a middle clade within the main Ascomycota clade that is sister to the Laboulbeniomycetes (Figure 1 and Figure 2). Many plastic degraders in the Sordariomycetes are classified under the Hypocreales, while others belong to the Amphisphaeriales, Glomerellales, Phyllachorales, and Sordariales. Even though the Eurotiomycetes include the highest number of records of plastic-degrading fungi, the Sordariomycetes contain the highest number of genera with the capability to degrade plastics.
3.3. Phylum Basidiomycota
3.3.1. Class Agaricomycetes
The Agaricomycetes is a morphologically diverse class of macrofungi within the Basidiomycota, containing around 36,000 described species. They are ecologically diverse and include saprobes, mycorrhizal symbionts, and pathogens [143]. Moreover, the Agaricomycetes encompasses several important commercially growing edible mushrooms [144]. The present study found several records of Agaricomycetes with the ability to degrade plastics (Table 2, Figure 1 and Figure 2). They are reported to degrade Poly[3HB-co-(7 mol%) 3HV], LDPE, PVC, polyethylene, and PHB (Table 2). Additionally, da Luz et al. [16] performed a study on plastic-degrading Agaricomycetes. They investigated the degradation of oxo-biodegradable plastic bags and green polyethylene by Pleurotus ostreatus. The Agaricomycetes is the upper clade within phylum Basidiomycota, and it is highly statistically supported within the present phylogeny (Figure 1 and Figure 2). Further studies of edible mushrooms belonging to the Agaricomycetes and their ability to degrade plastics would increase world food production and reduce the plastic accumulation in nature.
3.3.2. Class Microbotryomycetes
The Microbotryomycetes include mainly mycoparasites, saprobic yeasts, and plant pathogens [145]. A single record of plastic-degrading fungi was found in the order Sporidiobolales of the Microbotryomycetes in the present study (Table 2, Figure 1 and Figure 2). In our phylogenetic tree, the Microbotryomycetes formed a well-supported clade sister to Tritirachiomycetes (Figure 1 and Figure 2).
3.3.3. Class Tremellomycetes
The Tremellomycetes are classified under the Basidiomycota and consist of saprobic yeasts, dimorphic taxa, and species that form hyphae and/or complex fruiting bodies [146]. The present study identified a few records from the Tremellomycetes that are capable of degrading plastics (Table 2, Figure 1 and Figure 2). All the records belong to the genera Cryptococcus and Papiliotrema (Tremellales), andthey are capable of degrading PCL, PBS, and PBSA (Table 2). The Tremellomycetes formed a well-supported clade sister to Agaricomycetes in the present phylogenetic tree (Figure 1 and Figure 2). A recent study on synthetic plastic-degrading Tremellomycetes was published by Hung et al. [114]. A recent study on biodegradable plastic mulch films (BDMs) and their associated soil microbial communities found that the Tremellomycetes are capable of degrading agriculturally-weathered BDMs [147].
3.3.4. Class Tritirachiomycetes
The Tritirachiomycetes is a small class within the Basidiomycota that is made up of filamentous fungi. They are mainly saprobes, but some have been recorded as human pathogens [148]. A single record of a fungus from this class that is capable of degrading plastics was found in the present study (Table 2, Figure 1 and Figure 2). The Tritirachiomycetes formed a well-supported clade sister to the Microbotryomycetes in our phylogenetic tree (Figure 1 and Figure 2).
3.3.5. Class Ustilaginomycetes
The majority of the Ustilaginomycetes are economically important plant pathogens. They are usually unicellular yeasts (sporidia), but some are simple multicellular forms, such as a pseudomycelium, multicellular cluster, or mycelium [149]. Moreover, the Ustilaginomycetes have a comparatively short life cycle, which makes them easy to handle under laboratory conditions. As a result, the Ustilaginomycetes can be considered model organisms for studying fungi [149]. The present study found a single record from the Ustilaginomycetes of a fungus capable of degrading plastics (Table 2, Figure 1 and Figure 2). The Ustilaginomycetes is the basal clade within the main Basidiomycota clade, and it is statistically highly supported (Figure 1 and Figure 2).
3.4. Phylum Mucoromycota
Class Mucoromycetes
The Mucoromycetes are a class within the phylum Mucoromycota and consist of mainly filamentous fungi with a saprobic lifestyle. Several species are also life-threatening human pathogens, plant parasites, and food spoilage organisms. Moreover, members of the Mucoromycetes are used as a traditional fermenting agent for Asian and African foods, such as soybean products and several varieties of European cheese. Fungi belonging to the Mucoromycetes are common in the environment and able to colonize all kinds of wet, organic substrates [150]. The present study found several Mucoromycetes fungi with the ability to degrade plastics. All the recorded plastic-degrading members of the Mucoromycetes belong to the genera Mucor and Rhizopus. These fungi are capable of degrading PHB, HDPE, LDPE, PVC, PCL, polyalkylene dicarboxylic acids, PPA, and PET copolymers with dicarboxylic acids (Table 2). A recent study of plastic-degrading Mucoromycetes was that of Pardo-Rodríguez & Zorro-Mateus [106]. The Mucoromycota formed the basal clade within our phylogenetic tree (Figure 1 and Figure 2). However, the phylum presented a polyphyletic nature in the present phylogenetic tree and was separated into two basal clades.
4. Discussion
As global plastic production and plastic waste accumulation increase rapidly, we need to look for a quick and efficient solution to save nature and, indeed, the entire planet [4,7]. The problem becomes even more acute as the natural degradation rate of all types of plastic is very slow [23]. Hence, it is essential to look at ways to accelerate plastic degradation methods. Several solutions that have been proposed are photo-degradation (degrade by light), chemical degradation, thermal degradation (degrade by heat), irradiation using gamma rays, and biodegradation (degrade by biological additives or microorganisms) [25,26,27]. Biodegradation has been suggested as the best solution as it is an eco-friendly approach [27]. However, based on the results obtained in previous studies, people select biodegradation since the other options are not cost-effective [26]. In this study, we reviewed the literature on plastic-degrading fungi from different sources and summarized a list of records on plastic degraders in the fungi kingdom. Moreover, we analyzed phylogenetic relationships of plastic-degrading fungi based on a combined ITS, LSU, SSU, TEF, RPB1, and RPB2 dataset from 395 strains and provided brief taxonomy at the level of class.
The present study identified more than 200 records of fungi capable of degrading different plastic types under a wide range of conditions. These are listed in Table 2. Most plastic-degrading fungal records are found in the class Eurotiomycetes of the phylum Ascomycota. However, a wide range of generic level diversity in plastic-degrading fungi was found in the class Sordariomycetes of the phylum Ascomycota. Considering phylum level diversity in relation to the ability to degrade plastic, most of the plastic degraders belong to the phylum Ascomycota. The second group with the highest number of recorded plastic degraders was in the phylum Basidiomycota, with a few records also found in the phylum Mucoromycota. However, very few sequences are available directly from the plastic-degrading fungi. Hence, we used sequence data from another strain of the same species, which the species was recorded as a plastic degrader. Furthermore, to establish a detailed and accurate phylogenetic tree for plastic-degrading fungi, sequence data need to be obtained exactly from the strains which were recorded as plastic-degrading fungi. Moreover, future studies on whole-genome sequencing or sequencing of more gene regions and evolutionary relationships analyses of plastic-degrading fungi concerning the specific plastic type the fungi can degrade would be beneficial to develop commercially available plastic degrading fungal strains.
Recent studies have reported some common saprobic fungi and plant pathogenic fungi that can degrade plastic [6]. Our study agrees with this statement as we found many plastic degraders that belong to the Eurotiomycetes and Dothideomycetes when most of the saprobic fungi are taxonomically members of the Sordariomycetes, where many important plant pathogens are classified. Moreover, recent research has reported that fungi that can degrade complex C-polymers, such as lignin and protein, can also degrade plastics [6]. Therefore, future studies of the Dothideomycetes, Eurotiomycetes, and Sordariomycetes are essential. Since these classes include plant pathogens, saprobes, and degraders of complex C-polymers, it is possible that they can degrade plastics. Moreover, these findings also proved that enzymes produced by common saprobes and plant pathogens can degrade or even transform plastics. Hence further studies on the metabolic pathways of those fungi and the discovery of the essential enzymes involved in plastic degradation can significantly contribute to reducing plastic accumulation in nature.
Plastics accumulating in the marine environment are an increasingly important environmental issue [34]. Most of the plastic waste is accumulated in the sea, and there will be more plastics in the sea than fish by 2050. Recently, plastic-fungi interactions in marine environments were reviewed by Zeghal et al. [34]. The same study listed more than 60 records of fungi identified to the species level and belonging to the Ascomycota and Basidiomycota as having the ability to degrade plastic debris in marine environments [34]. Our study listed plastic-degrading fungi from all environmental conditions on the earth. When comparing the number of records, only about twenty-five percent of records are from marine environments, even though the highest environmental pollution from plastics is recorded from marine environments. This clearly shows the requirement for more research on plastic-degrading fungi in marine environments. Moreover, Zeghal et al. [34] concluded that future studies are required to identify species of fungi that can degrade plastics in the marine environment. They also concluded that these studies should address their enzymatic potential, which might then serve biotechnological applications for plastic waste bioremediation [34]. Furthermore, the effect of physicochemical parameters in marine environments (such as salinity and pH) on plastic-degrading fungi and the differences between the metabolic pathways of marine plastic degraders from terrestrial plastic degraders need to be focused on in future research fields to save the sea from plastic accumulation.
Plastic use in the agricultural sector is growing rapidly. Some of the main examples of LDPE use are in greenhouse and mulch film as well as in structures to provide crop protection against environmental conditions and insects and pests. HDPE is used in containers for pesticides and pesticides as well as in containers for nurseries or slips for irrigation, and PP is used in containers for nurseries or floating covers and polystyrene in trays for a plant nursery [151]. Recent research by Khan & Stevenson [151] investigated the plastic-degrading ability of soil fungi and found that Aspergillus tubingensis can successfully colonize plastic surfaces. Additionally, they found that the enzymes this fungus produces can break the chemical bonds between the plastic molecules. Moreover, the genus Aspergillus is very important in plastic degradation. Several species from different habitats have been reported that degrade various plastic types (Table 2). As such, further research on the members of the genus Aspergillus and their ability to degrade plastics needs more attention. Not only Aspergillus, but many other fungi have also been reported (Table 2), including the ability to degrade multiple plastic types by the same fungi. Some examples are Chaetomium globosum that can degrade HDPE, LDPE, PVC, PCL, PEA, PPA, and PBA, Fusarium solani that can degrade LDPE, HDPE, PVC, PCL, PS-PUR, PHB, and PET, and Penicillium funiculosum that can degrade PCL, PHB, PHV, Poly[3HB-co-(7, 14%) 4HB], Poly[3HB-co-(7, 27, 45, 71%) 3HV], PEA, PPA, and PBA. Those fungi that can degrade multiple plastics have also been recorded from different habitats. Hence, they are the perfect candidates for strain improvements for commercial applications. Molecular studies of those plastic-degrading fungi, specifically on genes related to the metabolic pathways of those fungi involved in plastic degradation, are critical. Simultaneously, even though there are commercially developed strains for plastic degradation in agricultural lands, there must be experimental confirmation that those fungi do not have pathogenic effects on crop plants and the beneficial flora and fauna associated with the farmlands.
Low-Density Polyethylene (LDPE) is the most common type of plastic, generally known as polythene [9,152]. There are more than 15 species of fungi, including several members of Aspergillus recorded as capable of degrading LDPE (i.e., Aspergillus caespitosus, Aspergillus flavus, Aspergillus fumigatus, Aspergillus nidulans, Aspergillus niger, Aspergillus nomius, Aspergillus ochraceus, Aspergillus oryzae, Aspergillus terreus, Chaetomium globosum, Chrysonilia setophila, Diaporthe italiana, Fusarium solani, Gliocladium virens, Phoma sp., Thyrostroma jaczewskii, Trichoderma hamatum, and Mucor hiemalis) (Table 2). Therefore, we suggest that the genus Aspergillus would be a perfect fungal genus for developing commercial levels to degrade plastics. For the basement of such a lengthy biotechnological application, comprehensive molecular studies related to the ecology of the genus Aspergillus are very important. Moreover, investigations on enzyme production and metabolic pathways of the genus Aspergillus are helpful research areas.
The slow degradation rate of synthetic plastics and the growing demand for increasingly more limited oil reserves drive the effort to produce bio-based plastics [3,16]. Bio-based plastics originate from renewable resources such as plant and animal waste products from industry [3]. However, the global markets for bio-based plastics are still small. The annual global production of bio-based plastics in 2020 was approximately 2 Mt [153]. Moreover, bio-based plastics do not represent a better solution for the slow degradation rate of synthetic plastics or plastic accumulation in nature. Recent records indicate that only half of bio-based plastics are biodegradable, while the other half is non-biodegradable [3,4,5]. Bio-based plastics naturally degrade only to some extent. For the remainder, we once again need to look for suitable microorganisms that are capable of their biodegradation.
Although there are more than 400 records of plastic-degrading microbes, including over 200 records of species of plastic-degrading fungi, many studies have failed to differentiate losses caused by the leaching or degradation of polymers [28]. Moreover, many studies have used more highly crystalline polymers with species of fungi under laboratory conditions [28]. Hence, even the species of fungi involved provided few clues as to their ability to degrade those crystallized polymers. There was no confirmation of their biodegradation rate in a natural environment where large-sized plastic wastes have accumulated. Herein we propose a more detailed in situ research approach that delivers a clearer picture of plastic-fungi interactions, including changes in the polymer structure, mass loss, and confirmation of fungal strains and their enzymatic activity with related genes to degrade large-sized plastic polymers. Furthermore, genetic and biotechnology research is required to identify enzymes and their related genes for plastic degradation from currently identified plastic-degrading fungi. We expect to continue our following review on enzyme production and metabolic pathways of plastic-degrading fungi and their evolutionary relationships. This information can then be applied to develop more powerful hybrid fungal species to degrade plastics. Moreover, we expect this study to influence the research and applications of plastic-degrading fungi to save Earth’s planet.
5. Conclusions
Even though plastic is currently an important material in the global environment, it is becoming a huge threat to nature. Because current global plastic production is increasing rapidly (300 million tons annually) and plastics have a very low natural degradation rate, they accumulate in natural environments and cause considerable damage to biodiversity and natural ecosystems. At present, scientists and researchers are assessing the usefulness of microorganisms in accelerating plastic degradation. In this study, we reviewed plastic degradation using fungi. Herein we list more than 200 records of fungi capable of degrading fungi based on the available literature. Their phylogenetic relationships were analyzed using a combined ITS, LSU, SSU, TEF, RPB1, and RPB2 dataset generated from 395 strains. Our results confirm that plastic-degrading fungi are taxonomically diverse and belong to three major fungal phyla—the Ascomycota, Basidiomycota, and Mucoromycota. The Ascomycota plastic degraders belong to five major classes: Dothideomycetes, Eurotiomycetes, Leotiomycetes, Saccharomycetes, and Sordariomycetes. Plastic-degrading Basidiomycota fall within the Agaricomycetes, Microbotryomycetes, Tremellomycetes, Tritirachiomycetes, and Ustilaginomycetes. Mucoromycota fungi capable of degrading plastics were found under the Mucoromycetes. The Eurotiomycetes include the highest number of recorded plastic degraders in the fungi kingdom. However, a wide range of plastic-degrading fungal genera was found within the class Sordariomycetes. Moreover, there is an acute need for future research on similar topics to resolve the global problem of plastic accumulation in nature.
Acknowledgments
Qujing Normal University is thanked for the facilities provided for research work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8080772/s1, Table S1: Taxa table for phylogenetic analysis.
Author Contributions
Conceptualization, S.C.K. and A.H.E.; methodology, S.C.K. and A.H.E.; formal analysis, A.H.E.; writing—original draft preparation, A.H.E. and S.C.K.; writing—review and editing, A.H.E., S.C.K., S.T., C.D., R.X. and S.L.S.; visualization, D.D., N.S. and S.T.; supervision, S.C.K., D.D., N.S. and S.T.; project administration, S.C.K.; funding acquisition, S.C.K. and D.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
Authors declare that they have no competing interest.
Funding Statement
The research was supported by the National Natural Science Foundation of China (No. NSFC 31760013, 31950410558), High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program) and Chiang Mai University.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ogunbayo A., Olanipekun O., Adamu I. Preliminary Studies on the Microbial Degradation of Plastic Waste Using Aspergillus niger and Pseudomonas sp. J. Environ. Prot. 2019;10:625–631. doi: 10.4236/jep.2019.105037. [DOI] [Google Scholar]
- 2.Sangale M.K., Shahnawaz M., Ade A.B. Potential of Fungi Isolated from the Dumping Sites Mangrove Rhizosphere Soil to Degrade Polythene. Sci. Rep. 2019;9:5390. doi: 10.1038/s41598-019-41448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baheti P. How Is Plastic Made? [(accessed on 15 September 2021)]. Available online: https://www.bpf.co.uk/plastipedia/how-is-plastic-made.aspx.
- 4.Di Bartolo A., Infurna G., Dintcheva N.T. A Review of Bioplastics and Their Adoption in the Circular Economy. Polymers. 2021;13:1229. doi: 10.3390/polym13081229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstein J.E., Dekle J.L., Leads R.R., Hunter R.A. Degradation of Bio-Based and Biodegradable Plastics in a Salt Marsh Habitat: Another Potential Source of Microplastics in Coastal Waters. Mar. Pollut. Bull. 2020;160:111518. doi: 10.1016/j.marpolbul.2020.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner I., Fischer M., Rüthi J., Stierli B., Frey B. Ability of Fungi Isolated from Plastic Debris Floating in the Shoreline of a Lake to Degrade Plastics. PLoS ONE. 2018;13:e0202047. doi: 10.1371/journal.pone.0202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plastics Europe How Plastics Are Made. [(accessed on 13 September 2021)]. Available online: https://www.plasticseurope.org/en/about-plastics/what-are-plastics/how-plastics-are-made.
- 8.Wayman C., Niemann H. The Fate of Plastic in the Ocean Environment—A Minireview. Environ. Sci. Process. Impacts. 2021;23:198–212. doi: 10.1039/D0EM00446D. [DOI] [PubMed] [Google Scholar]
- 9.Hardin T. Plastic: It’s Not All the Same; The Basics on 7 Common Types of Plastic. [(accessed on 4 September 2021)]. Available online: https://plasticoceans.org/7-types-of-plastic.
- 10.Plastics Europe Plastics—The Facts 2020, an Analysis of European Plastics Production, Demand and Waste Data. [(accessed on 18 September 2021)]. Available online: https://www.plasticseurope.org/en/resources/publications/4312-plastics-facts-2020.
- 11.Geyer R., Jambeck J.R., Law K.L. Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rani A., Singh P. Screening of Polyethylene Degrading Fungi from Polyethylene Dump Site. Int. J. Chem. Tech. Res. 2017;10:699–704. [Google Scholar]
- 13.United Nations Environment Programme Fungi Research Lifts Lid on Shy Organisms That Break down Plastic. [(accessed on 12 September 2021)]. Available online: https://www.unep.org/news-and-stories/story/fungi-research-lifts-lid-shy-organisms-break-down-plastic.
- 14.Organisation for Economic Co-Operation and Development . Plastic Pollution Is Growing Relentlessly as Waste Management and Recycling Fall Short, Says OECD. Organisation for Economic Co-Operation and Development; Paris, France: 2022. [Google Scholar]
- 15.Jambeck J.R., Geyer R., Wilcox C., Siegler T.R., Perryman M., Andrady A., Narayan R., Law K.L. Plastic Waste Inputs from Land into the Ocean. Science. 2015;347:768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 16.da Luz J.M.R., da Silva M.d.C.S., dos Santos L.F., Kasuya M.C.M. Microorganisms. IntechOpen; Vienna, Austria: 2019. Plastics Polymers Degradation by Fungi. [Google Scholar]
- 17.Chamas A., Moon H., Zheng J., Qiu Y., Tabassum T., Jang J.H., Abu-Omar M., Scott S.L., Suh S. Degradation Rates of Plastics in the Environment. ACS Sustain. Chem. Eng. 2020;8:3494–3511. doi: 10.1021/acssuschemeng.9b06635. [DOI] [Google Scholar]
- 18.Oberbeckmann S., Osborn A.M., Duhaime M.B. Microbes on a Bottle: Substrate, Season and Geography Influence Community Composition of Microbes Colonizing Marine Plastic Debris. PLoS ONE. 2016;11:e0159289. doi: 10.1371/journal.pone.0159289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameen F., Moslem M., Hadi S., Al-Sabri A.E. Biodegradation of Low Density Polyethylene (LDPE) by Mangrove Fungi from the Red Sea Coast. Prog. Rubber Plast. Recycl. Technol. 2015;31:125–143. doi: 10.1177/147776061503100204. [DOI] [Google Scholar]
- 20.Debroas D., Mone A., Ter Halle A. Plastics in the North Atlantic Garbage Patch: A Boat-Microbe for Hitchhikers and Plastic Degraders. Sci. Total Environ. 2017;599:1222–1232. doi: 10.1016/j.scitotenv.2017.05.059. [DOI] [PubMed] [Google Scholar]
- 21.Gascueña D. Nature’s Allies Against Plastic: Algae, Bacteria, and Fungi. [(accessed on 12 September 2021)]. Available online: https://www.bbvaopenmind.com/en/science/environment/natures-allies-against-plastic-algae-bacteria-and-fungi.
- 22.Kale S.K., Deshmukh A.G., Dudhare M.S., Patil V.B. Microbial Degradation of Plastic: A Review. J. Biochem. Technol. 2015;6:952–961. [Google Scholar]
- 23.Webb H.K., Arnott J., Crawford R.J., Ivanova E.P. Plastic Degradation and Its Environmental Implications with Special Reference to Poly (Ethylene terephthalate) Polymers. 2013;5:1–18. doi: 10.3390/polym5010001. [DOI] [Google Scholar]
- 24.Wilkes R.A., Aristilde L. Degradation and Metabolism of Synthetic Plastics and Associated Products by Pseudomonas sp.: Capabilities and Challenges. J. Appl. Microbiol. 2017;123:582–593. doi: 10.1111/jam.13472. [DOI] [PubMed] [Google Scholar]
- 25.Pramila R., Ramesh K.V. Biodegradation of Low Density Polyethylene (LDPE) by Fungi Isolated from Municipal Landfill Area. J. Microbiol. Biotechnol. Res. 2011;1:e136. [Google Scholar]
- 26.Ojha N., Pradhan N., Singh S., Barla A., Shrivastava A., Khatua P., Rai V., Bose S. Evaluation of HDPE and LDPE Degradation by Fungus, Implemented by Statistical Optimization. Sci. Rep. 2017;7:39515. doi: 10.1038/srep39515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soundararajan N. Pune Researchers Discover Fungi That Can Break up Polythene. [(accessed on 11 September 2021)]. Available online: https://researchmatters.in/news/pune-researchers-discover-fungi-can-break-polythene.
- 28.Lear G., Kingsbury J., Franchini S., Gambarini V., Maday S., Wallbank J., Weaver L., Pantos O. Plastics and the Microbiome: Impacts and Solutions. Environ. Microbiome. 2021;16:2. doi: 10.1186/s40793-020-00371-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacerda A.L.d.F., Proietti M.C., Secchi E.R., Taylor J.D. Diverse Groups of Fungi Are Associated with Plastics in the Surface Waters of the Western South Atlantic and the Antarctic Peninsula. Mol. Ecol. 2020;29:1903–1918. doi: 10.1111/mec.15444. [DOI] [PubMed] [Google Scholar]
- 30.Ru J., Huo Y., Yang Y. Microbial Degradation and Valorization of Plastic Wastes. Front. Microbiol. 2020;11:442. doi: 10.3389/fmicb.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez C. Fungal Potential for the Degradation of Petroleum-Based Polymers: An Overview of Macro-and Microplastics Biodegradation. Biotechnol. Adv. 2020;40:107501. doi: 10.1016/j.biotechadv.2019.107501. [DOI] [PubMed] [Google Scholar]
- 32.Sáenz M., Borodulina T., Diaz L., Banchon C. Minimal Conditions to Degrade Low Density Polyethylene by Aspergillus terreus and niger. J. Ecol. Eng. 2019;20:40–51. doi: 10.12911/22998993/108699. [DOI] [Google Scholar]
- 33.Iram D., Riaz R., Iqbal R.K. Usage of Potential Micro-Organisms for Degradation of Plastics. Open J. Environ. Biol. 2019;4:7–15. [Google Scholar]
- 34.Zeghal E., Vaksmaa A., Vielfaure H., Boekhout T., Niemann H. The Potential Role of Marine Fungi in Plastic Degradation—A Review. Front. Mar. Sci. 2021;8:738877. doi: 10.3389/fmars.2021.738877. [DOI] [Google Scholar]
- 35.Statista Statista: Global No.1 Business Data Platform. 2022. [(accessed on 22 April 2022)]. Available online: https://www.statista.com/
- 36.Alyamaç-Seydibeyoğlu E. Master’s Thesis. Middle East Technical University; Ankara, Turkey: 2004. Impact Modified Poly(Ethylene Terephthalate)-Organoclay Nanocomposites. [Google Scholar]
- 37.Karlsson S., Ljungquist O., Albertsson A.-C. Biodegradation of Polyethylene and the Influence of Surfactants. Polym. Degrad. Stab. 1988;21:237–250. doi: 10.1016/0141-3910(88)90030-4. [DOI] [Google Scholar]
- 38.Mergaert J., Webb A., Anderson C., Wouters A., Swings J. Microbial Degradation of Poly (3-Hydroxybutyrate) and Poly (3-Hydroxybutyrate-Co-3-Hydroxyvalerate) in Soils. Appl. Environ. Microbiol. 1993;59:3233–3238. doi: 10.1128/aem.59.10.3233-3238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sowmya H., Ramalingappa B., Nayanashree G., Thippeswamy B., Krishnappa M. Polyethylene Degradation by Fungal Consortium. Int. J. Environ. Res. 2015;9:823–830. [Google Scholar]
- 40.Koschorreck K., Liu D., Kazenwadel C., Schmid R.D., Hauer B. Heterologous Expression, Characterization and Site-Directed Mutagenesis of Cutinase CUTAB1 from Alternaria brassicicola. Appl. Microbiol. Biotechnol. 2010;87:991–997. doi: 10.1007/s00253-010-2533-3. [DOI] [PubMed] [Google Scholar]
- 41.Russell J.R., Huang J., Anand P., Kucera K., Sandoval A.G., Dantzler K.W., Hickman D., Jee J., Kimovec F.M., Koppstein D. Biodegradation of Polyester Polyurethane by Endophytic Fungi. Appl. Environ. Microbiol. 2011;77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibrahim I.N., Maraqa A., Hameed K.M., Saadoun I.M., Maswadeh H.M. Assessment of Potential Plastic-Degrading Fungi in Jordanian Habitats. Turk. J. Biol. 2011;35:551–557. doi: 10.3906/biy-0901-9. [DOI] [Google Scholar]
- 43.Bischoff F., Litwińska K., Cordes A., Baronian K., Bode R., Schauer F., Kunze G. Three New Cutinases from the Yeast Arxula adeninivorans That Are Suitable for Biotechnological Applications. Appl. Environ. Microbiol. 2015;81:5497–5510. doi: 10.1128/AEM.00894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benedict C.V., Cook W.J., Jarrett P., Cameron J., Huang S.J., Bell J.P. Fungal Degradation of Polycaprolactones. J. Appl. Polym. Sci. 1983;28:327–334. doi: 10.1002/app.1983.070280128. [DOI] [Google Scholar]
- 45.Nakei M.D. Ph.D. Thesis. Sokoine University of Agriculture; Morogoro, Tanzania: 2015. Isolation and Identification of Plastics—Degrading Microorganisms from Soils of Morogoro, Tanzania. [Google Scholar]
- 46.Soumya S., Nair B.R. Antifungal Efficacy of Capsicum Frutescens L. Extracts against Some Prevalent Fungal Strains Associated with Groundnut Storage. J. Agric. Technol. 2012;8:739–750. [Google Scholar]
- 47.Sakhalkar S., Mishra R. Screening and Identification of Soil Fungi with Potential of Plastic Degrading Ability. Indian J. Appl. Res. 2013;3:3. doi: 10.15373/2249555X/DEC2013/16. [DOI] [Google Scholar]
- 48.Darby R.T., Kaplan A.M. Fungal Susceptibility of Polyurethanes. Appl. Microbiol. 1968;16:900–905. doi: 10.1128/am.16.6.900-905.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zahra S., Abbas S.S., Mahsa M.-T., Mohsen N. Biodegradation of Low-Density Polyethylene (LDPE) by Isolated Fungi in Solid Waste Medium. Waste Manag. 2010;30:396–401. doi: 10.1016/j.wasman.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 50.Kim M.-N., Lee A.-R., Yoon J.-S., Chin I.-J. Biodegradation of Poly (3-Hydroxybutyrate), Sky-Green® and Mater-Bi® by Fungi Isolated from Soils. Eur. Polym. J. 2000;36:1677–1685. doi: 10.1016/S0014-3057(99)00219-0. [DOI] [Google Scholar]
- 51.Scherer T.M., Fuller R.C., Lenz R.W., Goodwin S. Production, Purification and Activity of an Extracellular Depolymerase from Aspergillus fumigatus. J. Environ. Polym. Degrad. 1999;7:117–125. doi: 10.1023/A:1022881204565. [DOI] [Google Scholar]
- 52.Iyer S., Shah R., Sharma A., Jendrossek D., Desai A. Purification of Aspergillus fumigatus (Pdf1) Poly (β-Hydroxybutyrate)(PHB) Depolymerase Using a New, Single-Step Substrate Affinity Chromatography Method: Characterization of the PHB Depolymerase Exhibiting Novel Self-Aggregation Behavior. J. Polym. Environ. 2000;8:197–203. doi: 10.1023/A:1015249811314. [DOI] [Google Scholar]
- 53.Sanchez J.G., Tsuchii A., Tokiwa Y. Degradation of Polycaprolactone at 50 °C by a Thermotolerant Aspergillus sp. Biotechnol. Lett. 2000;22:849–853. doi: 10.1023/A:1005603112688. [DOI] [Google Scholar]
- 54.Ndahebwa Muhonja C., Magoma G., Imbuga M., Makonde H.M. Molecular Characterization of Low-Density Polyethene (LDPE) Degrading Bacteria and Fungi from Dandora Dumpsite, Nairobi, Kenya. Int. J. Microbiol. 2018;2018:4167845. doi: 10.1155/2018/4167845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kathiresan K. Polythene and Plastics-Degrading Microbes from the Mangrove Soil. Rev. Biol. Trop. 2003;51:629–633. [PubMed] [Google Scholar]
- 56.Raaman N., Rajitha N., Jayshree A., Jegadeesh R. Biodegradation of Plastic by Aspergillus spp. Isolated from Polythene Polluted Sites around Chennai. J. Acad. Indus. Res. 2012;1:313–316. [Google Scholar]
- 57.Bermúdez-García E., Peña-Montes C., Castro-Rodríguez J.A., González-Canto A., Navarro-Ocaña A., Farrés A. ANCUT2, a Thermo-Alkaline Cutinase from Aspergillus nidulans and Its Potential Applications. Appl. Biochem. Biotechnol. 2017;182:1014–1036. doi: 10.1007/s12010-016-2378-z. [DOI] [PubMed] [Google Scholar]
- 58.Raghavan D., Torma A. DSC and FTIR Characterization of Biodegradation of Polyethylene. Polym. Eng. Sci. 1992;32:438–442. doi: 10.1002/pen.760320609. [DOI] [Google Scholar]
- 59.Shabani F., Kumar L., Esmaeili A. A Modelling Implementation of Climate Change on Biodegradation of Low-Density Polyethylene (LDPE) by Aspergillus niger in Soil. Glob. Ecol. Conserv. 2015;4:388–398. doi: 10.1016/j.gecco.2015.08.003. [DOI] [Google Scholar]
- 60.Volke-Sepúlveda T., Saucedo-Castañeda G., Gutiérrez-Rojas M., Manzur A., Favela-Torres E. Thermally Treated Low Density Polyethylene Biodegradation by Penicillium pinophilum and Aspergillus niger. J. Appl. Polym. Sci. 2002;83:305–314. doi: 10.1002/app.2245. [DOI] [Google Scholar]
- 61.Esmaeili A., Pourbabaee A.A., Alikhani H.A., Shabani F., Esmaeili E. Biodegradation of Low-Density Polyethylene (LDPE) by Mixed Culture of Lysinibacillus xylanilyticus and Aspergillus niger in Soil. PLoS ONE. 2013;8:e71720. doi: 10.1371/journal.pone.0071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nyyssölä A., Pihlajaniemi V., Järvinen R., Mikander S., Kontkanen H., Kruus K., Kallio H., Buchert J. Screening of Microbes for Novel Acidic Cutinases and Cloning and Expression of an Acidic Cutinase from Aspergillus niger CBS 513.88. Enzym. Microb. Technol. 2013;52:272–278. doi: 10.1016/j.enzmictec.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Laila M.D. Ph.D Thesis. Universitas Andalas; Padang, Indonesia: 2021. The Potential of Fungi Isolated from Pleurotus ostreatus (Oyster Mushroom) Baglog to Degrade Plastics. [Google Scholar]
- 64.Khruengsai S., Sripahco T., Pripdeevech P. Low-Density Polyethylene Film Biodegradation Potential by Fungal Species from Thailand. J. Fungi. 2021;7:594. doi: 10.3390/jof7080594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munir E., Harefa R., Priyani N., Suryanto D. Plastic Degrading Fungi Trichoderma viride and Aspergillus nomius Isolated from Local Landfill Soil in Medan; IOP Publishing: Bristol, UK, 2018; Volume 126, p. 01. :2145. [Google Scholar]
- 66.Maeda H., Yamagata Y., Abe K., Hasegawa F., Machida M., Ishioka R., Gomi K., Nakajima T. Purification and Characterization of a Biodegradable Plastic-Degrading Enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005;67:778–788. doi: 10.1007/s00253-004-1853-6. [DOI] [PubMed] [Google Scholar]
- 67.Liu Z., Gosser Y., Baker P.J., Ravee Y., Lu Z., Alemu G., Li H., Butterfoss G.L., Kong X.-P., Gross R. Structural and Functional Studies of Aspergillus oryzae Cutinase: Enhanced Thermostability and Hydrolytic Activity of Synthetic Ester and Polyester Degradation. J. Am. Chem. Soc. 2009;131:15711–15716. doi: 10.1021/ja9046697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mergaert J., Anderson C., Wouters A., Swings J., Kersters K. Biodegradation of Polyhydroxyalkanoates. FEMS Microbiol. Rev. 1992;9:317–321. doi: 10.1111/j.1574-6968.1992.tb05853.x. [DOI] [PubMed] [Google Scholar]
- 69.Alshehrei F. Biodegradation of Low Density Polyethylene by Fungi Isolated from Red Sea Water. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:1703–1709. doi: 10.20546/ijcmas.2017.608.204. [DOI] [Google Scholar]
- 70.Khan S., Nadir S., Shah Z.U., Shah A.A., Karunarathna S.C., Xu J., Khan A., Munir S., Hasan F. Biodegradation of Polyester Polyurethane by Aspergillus tubingensis. Environ. Pollut. 2017;225:469–480. doi: 10.1016/j.envpol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 71.Gonda K., Jendrossek D., Molitoris H.-P. Life at Interfaces and Under Extreme Conditions. Springer; Berlin/Heidelberg, Germany: 2000. Fungal Degradation of the Thermoplastic Polymer Poly-β-Hydroxybutyric Acid (PHB) under Simulated Deep Sea Pressure; pp. 173–183. [Google Scholar]
- 72.Gu J.-D., Mitton D., Ford T., Mitchell R. Microbial Degradation of Polymeric Coatings Measured by Electrochemical Impedance Spectroscopy. Biodegradation. 1998;9:39–45. doi: 10.1023/A:1008252301377. [DOI] [PubMed] [Google Scholar]
- 73.Fields R., Rodriguez F., Finn R. Microbial Degradation of Polyesters: Polycaprolactone Degraded by P. Pullulans. J. Appl. Polym. Sci. 1974;18:3571–3579. doi: 10.1002/app.1974.070181207. [DOI] [Google Scholar]
- 74.Matavulj M., Molitoris H.-P. Fungal Degradation of Polyhydroxyalkanoates and a Semiquantitative Assay for Screening Their Degradation by Terrestrial Fungi. FEMS Microbiol. Rev. 1992;9:323–331. doi: 10.1111/j.1574-6968.1992.tb05854.x. [DOI] [PubMed] [Google Scholar]
- 75.Kim H., Lee J.W. Effect of Ultrasonic Wave on the Degradation of Polypropylene Melt and Morphology of Its Blend with Polystyrene. Polymer. 2002;43:2585–2589. doi: 10.1016/S0032-3861(02)00017-4. [DOI] [Google Scholar]
- 76.McLellan D.W., Halling P.J. Acid-Tolerant Poly (3-Hydroxybutyrate) Hydrolases from Moulds. FEMS Microbiol. Lett. 1988;52:215–218. doi: 10.1111/j.1574-6968.1988.tb02598.x. [DOI] [Google Scholar]
- 77.Owen S., Otani T., Masaoka S., Ohe T. The Biodegradation of Low-Molecular-Weight Urethane Compounds by a Strain of Exophiala jeanselmei. Biosci. Biotechnol. Biochem. 1996;60:244–248. doi: 10.1271/bbb.60.244. [DOI] [PubMed] [Google Scholar]
- 78.Murphy C.A., Cameron J., Huang S.J., Vinopal R.T. Fusarium Polycaprolactone Depolymerase Is Cutinase. Appl. Environ. Microbiol. 1996;62:456–460. doi: 10.1128/aem.62.2.456-460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sang B.-I., Hori K., Tanji Y., Unno H. Fungal Contribution to in Situ Biodegradation of Poly (3-Hydroxybutyrate-Co-3-Hydroxyvalerate) Film in Soil. Appl. Microbiol. Biotechnol. 2002;58:241–247. doi: 10.1007/s00253-001-0884-5. [DOI] [PubMed] [Google Scholar]
- 80.Dimarogona M., Nikolaivits E., Kanelli M., Christakopoulos P., Sandgren M., Topakas E. Structural and Functional Studies of a Fusarium oxysporum Cutinase with Polyethylene Terephthalate Modification Potential. Biochim. Biophys. Acta Gen. Subj. 2015;1850:2308–2317. doi: 10.1016/j.bbagen.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 81.Nimchua T., Punnapayak H., Zimmermann W. Comparison of the Hydrolysis of Polyethylene Terephthalate Fibers by a Hydrolase from Fusarium oxysporum LCH I and Fusarium solani f. sp. pisi. Biotechnol. J. Healthc. Nutr. Technol. 2007;2:361–364. doi: 10.1002/biot.200600095. [DOI] [PubMed] [Google Scholar]
- 82.Kwon M.-A., Kim H.S., Yang T.H., Song B.K., Song J.K. High-Level Expression and Characterization of Fusarium solani Cutinase in Pichia Pastoris. Protein Expr. Purif. 2009;68:104–109. doi: 10.1016/j.pep.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 83.Longhi S., Czjzek M., Lamzin V., Nicolas A., Cambillau C. Atomic Resolution (1.0 Å) Crystal Structure of Fusarium solani Cutinase: Stereochemical Analysis. J. Mol. Biol. 1997;268:779–799. doi: 10.1006/jmbi.1997.1000. [DOI] [PubMed] [Google Scholar]
- 84.Prompers J.J., Hilbers C.W., Pepermans H.A. Tryptophan Mediated Photoreduction of Disulfide Bond Causes Unusual Fluorescence Behaviour of Fusarium solani pisi Cutinase. FEBS Lett. 1999;456:409–416. doi: 10.1016/S0014-5793(99)00990-4. [DOI] [PubMed] [Google Scholar]
- 85.Sowmya H., Krishnappa M., Thippeswamy B. Degradation of Polyethylene by Penicillium simplicissimum Isolated from Local Dumpsite of Shivamogga District. Environ. Dev. Sustain. 2015;17:731–745. doi: 10.1007/s10668-014-9571-4. [DOI] [Google Scholar]
- 86.Pathirana R., Seal K. Gliocladium Roseum (Bainier), a Potential Biodeteriogen of Polyester Polyurethane Elastomers. John and Wiley and Sons; Hoboken, NJ, USA: 1983. [Google Scholar]
- 87.Seman W.W., Bakar S., Bukhari N., Gaspar S., Othman R., Nathan S., Mahadi N., Jahim J., Murad A., Bakar F.A. High Level Expression of Glomerella cingulata Cutinase in Dense Cultures of Pichia Pastoris Grown under Fed-Batch Conditions. J. Biotechnol. 2014;184:219–228. doi: 10.1016/j.jbiotec.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 88.Nyon M.P., Rice D.W., Berrisford J.M., Hounslow A.M., Moir A.J., Huang H., Nathan S., Mahadi N.M., Bakar F.D.A., Craven C.J. Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad. J. Mol. Biol. 2009;385:226–235. doi: 10.1016/j.jmb.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 89.Kazenwadel C., Eiben S., Maurer S., Beuttler H., Wetzl D., Hauer B., Koschorreck K. Thiol-Functionalization of Acrylic Ester Monomers Catalyzed by Immobilized Humicola Insolens Cutinase. Enzym. Microb. Technol. 2012;51:9–15. doi: 10.1016/j.enzmictec.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Duan X., Liu Y., You X., Jiang Z., Yang S., Yang S. High-Level Expression and Characterization of a Novel Cutinase from Malbranchea Cinnamomea Suitable for Butyl Butyrate Production. Biotechnol. Biofuels. 2017;10:223. doi: 10.1186/s13068-017-0912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.El-Morsy E., Hassan H., Ahmed E. Biodegradative Activities of Fungal Isolates from Plastic Contaminated Soils. Mycosphere. 2017;8:1071–1087. doi: 10.5943/mycosphere/8/8/13. [DOI] [Google Scholar]
- 92.Wang G.Y., Michailides T.J., Hammock B.D., Lee Y.-M., Bostock R.M. Molecular Cloning, Characterization, and Expression of a Redox-Responsive Cutinase from Monilinia fructicola (Wint.) Honey. Fungal Genet. Biol. 2002;35:261–276. doi: 10.1006/fgbi.2001.1320. [DOI] [PubMed] [Google Scholar]
- 93.Yang S., Liu M., Long L., Zhang R., Ding S. Characterization of a Cutinase from Myceliophthora thermophila and Its Application in Polyester Hydrolysis and Deinking Process. Process Biochem. 2018;66:106–112. doi: 10.1016/j.procbio.2017.11.021. [DOI] [Google Scholar]
- 94.Oda Y., Asari H., Urakami T., Tonomura K. Microbial Degradation of Poly (3-Hydroxybutyrate) and Polycaprolactone by Filamentous Fungi. J. Ferment. Bioeng. 1995;80:265–269. doi: 10.1016/0922-338X(95)90827-M. [DOI] [Google Scholar]
- 95.Suzuki K., Noguchi M.T., Shinozaki Y., Koitabashi M., Sameshima-Yamashita Y., Yoshida S., Fujii T., Kitamoto H.K. Purification, Characterization, and Cloning of the Gene for a Biodegradable Plastic-Degrading Enzyme from Paraphoma-Related Fungal Strain B47-9. Appl. Microbiol. Biotechnol. 2014;98:4457–4465. doi: 10.1007/s00253-013-5454-0. [DOI] [PubMed] [Google Scholar]
- 96.Sameshima-Yamashita Y., Koitabashi M., Tsuchiya W., Suzuki K., Watanabe T., Shinozaki Y., Yamamoto-Tamura K., Yamazaki T., Kitamoto H. Enhancement of Biodegradable Plastic-Degrading Enzyme Production from Paraphoma-like Fungus, Strain B47-9. J. Oleo Sci. 2016;65:ess15207. doi: 10.5650/jos.ess15207. [DOI] [PubMed] [Google Scholar]
- 97.Mergaert J., Wouters A., Swings J., Anderson C. In Situ Biodegradation of Poly (3-Hydroxybutyrate) and Poly (3-Hydroxybutyrate-Co-3-Hydroxyvalerate) in Natural Waters. Can. J. Microbiol. 1995;41:154–159. doi: 10.1139/m95-182. [DOI] [PubMed] [Google Scholar]
- 98.Renstad R., Karlsson S., Albertsson A.-C. The Influence of Processing Induced Differences in Molecular Structure on the Biological and Non-Biological Degradation of Poly (3-Hydroxybutyrate-Co-3-Hydroxyvalerate), P (3-HB-Co-3-HV) Polym. Degrad. Stab. 1999;63:201–211. doi: 10.1016/S0141-3910(98)00092-5. [DOI] [Google Scholar]
- 99.Brucato C.L., Wong S.S. Extracellular Poly (3-Hydroxybutyrate) Depolymerase from Penicillium funiculosum: General Characteristics and Active Site Studies. Arch. Biochem. Biophys. 1991;290:497–502. doi: 10.1016/0003-9861(91)90572-Z. [DOI] [PubMed] [Google Scholar]
- 100.Miyazaki S., Takahashi K., Shiraki M., Saito T., Tezuka Y., Kasuya K. Properties of a Poly (3-Hydroxybutyrate) Depolymerase from Penicillium funiculosum. J. Polym. Environ. 2000;8:175–182. doi: 10.1023/A:1015245710406. [DOI] [Google Scholar]
- 101.Han J.-S., Son Y.-J., Chang C.-S., Kim M.-N. Purification and Properties of Extracellular Poly (3-Hydroxybutyrate) Depolymerase Produced by Penicillium pinophilum. J. Microbiol. 1998;36:67–73. [Google Scholar]
- 102.Torres A., Li S., Roussos S., Vert M. Screening of Microorganisms for Biodegradation of Poly (Lactic-Acid) and Lactic Acid-Containing Polymers. Appl. Environ. Microbiol. 1996;62:2393–2397. doi: 10.1128/aem.62.7.2393-2397.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamada-Onodera K., Mukumoto H., Katsuyaya Y., Saiganji A., Tani Y. Degradation of Polyethylene by a Fungus, Penicillium simplicissimum YK. Polym. Degrad. Stab. 2001;72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- 104.Tokiwa Y., Suzuki T. Purification and Some Properties of Polyethylene Adipate-Degrading Enzyme Produced by Penicillium sp. Strain 14–3. Agric. Biol. Chem. 1977;41:265–274. doi: 10.1271/bbb1961.41.265. [DOI] [Google Scholar]
- 105.Tokiwa Y. Degradation of Polycaprolactone by a Fungus. J. Appl. Polym. Sci. 1976;28:327–334. [Google Scholar]
- 106.Pardo-Rodríguez M.L., Zorro-Mateus P.J.P. Biodegradation of Polyvinyl Chloride by Mucor sp. and Penicillium sp. Isolated from Soil. Rev. Investig. Desarro. Innovación. 2021;11:387–400. doi: 10.19053/20278306.v11.n2.2021.12763. [DOI] [Google Scholar]
- 107.Nyyssölä A., Pihlajaniemi V., Häkkinen M., Kontkanen H., Saloheimo M., Nakari-Setälä T. Cloning and Characterization of a Novel Acidic Cutinase from Sirococcus conigenus. Appl. Microbiol. Biotechnol. 2014;98:3639–3650. doi: 10.1007/s00253-013-5293-z. [DOI] [PubMed] [Google Scholar]
- 108.Yang S., Xu H., Yan Q., Liu Y., Zhou P., Jiang Z. A Low Molecular Mass Cutinase of Thielavia terrestris Efficiently Hydrolyzes Poly (Esters) J. Ind. Microbiol. Biotechnol. 2013;40:217–226. doi: 10.1007/s10295-012-1222-x. [DOI] [PubMed] [Google Scholar]
- 109.Xu H., Yan Q., Duan X., Yang S., Jiang Z. Characterization of an Acidic Cold-Adapted Cutinase from Thielavia terrestris and Its Application in Flavor Ester Synthesis. Food Chem. 2015;188:439–445. doi: 10.1016/j.foodchem.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 110.Rubio M.B., Cardoza R.E., Hermosa R., Gutiérrez S., Monte E. Cloning and Characterization of the Thcut1 Gene Encoding a Cutinase of Trichoderma harzianum T34. Curr. Genet. 2008;54:301–312. doi: 10.1007/s00294-008-0218-6. [DOI] [PubMed] [Google Scholar]
- 111.Malachová K., Novotný Č., Adamus G., Lotti N., Rybková Z., Soccio M., Šlosarčíková P., Verney V., Fava F. Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films. Processes. 2020;8:467. doi: 10.3390/pr8040467. [DOI] [Google Scholar]
- 112.Roussel A., Amara S., Nyyssölä A., Mateos-Diaz E., Blangy S., Kontkanen H., Westerholm-Parvinen A., Carrière F., Cambillau C. A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases. J. Mol. Biol. 2014;426:3757–3772. doi: 10.1016/j.jmb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 113.Thirunavukarasu K., Purushothaman S., Sridevi J., Aarthy M., Gowthaman M.K., Nakajima-Kambe T., Kamini N.R. Degradation of Poly (Butylene Succinate) and Poly (Butylene Succinate-Co-Butylene Adipate) by a Lipase from Yeast Cryptococcus sp. Grown on Agro-Industrial Residues. Int. Biodeterior. Biodegrad. 2016;110:99–107. doi: 10.1016/j.ibiod.2016.03.005. [DOI] [Google Scholar]
- 114.Hung C.-S., Barlow D.E., Varaljay V.A., Drake C.A., Crouch A.L., Russell J.N., Jr., Nadeau L.J., Crookes-Goodson W.J., Biffinger J.C. The Biodegradation of Polyester and Polyester Polyurethane Coatings Using Papiliotrema laurentii. Int. Biodeterior. Biodegrad. 2019;139:34–43. doi: 10.1016/j.ibiod.2019.02.002. [DOI] [Google Scholar]
- 115.Glenn J.K., Gold M.H. Decolorization of Several Polymeric Dyes by the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1983;45:1741–1747. doi: 10.1128/aem.45.6.1741-1747.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tien M., Kirk T.K. Lignin-Degrading Enzyme from the Hymenomycete Phanerochaete chrysosporium Burds. Science. 1983;221:661–663. doi: 10.1126/science.221.4611.661. [DOI] [PubMed] [Google Scholar]
- 117.Ali M., Ahmed S., Javed I., Ali N., Atiq N., Hameed A., Robson G. Biodegradation of Starch Blended Polyvinyl Chloride Films by Isolated Phanerochaete chrysosporium PV1. Int. J. Environ. Sci. Technol. 2014;11:339–348. doi: 10.1007/s13762-013-0220-5. [DOI] [Google Scholar]
- 118.Shinozaki Y., Morita T., Cao X., Yoshida S., Koitabashi M., Watanabe T., Suzuki K., Sameshima-Yamashita Y., Nakajima-Kambe T., Fujii T. Biodegradable Plastic-Degrading Enzyme from Pseudozyma antarctica: Cloning, Sequencing, and Characterization. Appl. Microbiol. Biotechnol. 2013;97:2951–2959. doi: 10.1007/s00253-012-4188-8. [DOI] [PubMed] [Google Scholar]
- 119.Jarerat A., Tokiwa Y. Degradation of Poly (L-lactide) by a Fungus. Macromol. Biosci. 2001;1:136–140. doi: 10.1002/1616-5195(20010601)1:4<136::AID-MABI136>3.0.CO;2-3. [DOI] [Google Scholar]
- 120.Tokiwa H., Ohnishi Y., Rosenkranz H.S. Mutagenicity and Carcinogenicity of Nitroarenes and Their Sources in the Environment. CRC Crit. Rev. Toxicol. 1986;17:23–58. doi: 10.3109/10408448609037070. [DOI] [PubMed] [Google Scholar]
- 121.Walter T., Augusta J., Müller R.-J., Widdecke H., Klein J. Enzymatic Degradation of a Model Polyester by Lipase from Rhizopus delemar. Enzym. Microb. Technol. 1995;17:218–224. doi: 10.1016/0141-0229(94)00007-E. [DOI] [Google Scholar]
- 122.Nagata M., Kiyotsukuri T., Takeuchi S., Tsutsumi N., Sakai W. Hydrolytic Degradation of Aliphatic Polyesters Copolymerized with Poly (Ethylene Glycol) s. Polym. Int. 1997;42:33–38. doi: 10.1002/(SICI)1097-0126(199701)42:1<33::AID-PI655>3.0.CO;2-9. [DOI] [Google Scholar]
- 123.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T. Phylemon 2.0: A Suite of Web-Tools for Molecular Evolution, Phylogenetics, Phylogenomics and Hypotheses Testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hall T. BioEdit, Version 7.0.9. Ibis Therapeutics; Carlsbad, CA, USA: 2004. [Google Scholar]
- 126.Villesen P. FaBox: An Online Toolbox for Fasta Sequences. Mol. Ecol. Notes. 2007;7:965–968. doi: 10.1111/j.1471-8286.2007.01821.x. [DOI] [Google Scholar]
- 127.Miller M.A., Schwartz T., Pfeiffer W. Embedding CIPRES Science Gateway Capabilities in Phylogenetics Software Environments. 2013. [(accessed on 22 April 2022)]. p. 1. Available online: https://dl.acm.org/doi/10.1145/2484762.2484806.
- 128.Stamatakis A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nylander J. MrModeltest, Version 2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 130.Rannala B., Yang Z. Probability Distribution of Molecular Evolutionary Trees: A New Method of Phylogenetic Inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 131.Zhaxybayeva O., Gogarten J.P. Bootstrap, Bayesian Probability and Maximum Likelihood Mapping: Exploring New Tools for Comparative Genome Analyses. BMC Genom. 2002;3:4. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huelsenbeck J., Ronquist F. MrBayes: Bayesian Inferences of Phylogeny (Software) University of Rochester; Rochester, NY, USA: 2000. [Google Scholar]
- 133.135 Rambaut A., Drummond A. Tracer. MCMC Trace Analysis Tool, Version v1.5.0. University of Oxford; Oxford, UK: 2009. [Google Scholar]
- 134.Rambaut A. FigTree. Tree Figure Drawing Tool, Version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh; Edinburgh, UK: 2006. [Google Scholar]
- 135.Hyde K.D., Jones E.G., Liu J.-K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.-Q. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 136.Geiser D.M., LoBuglio K.F., Gueidan C. Systematics and Evolution. Springer; Berlin/Heidelberg, Germany: 2015. 5 Pezizomycotina: Eurotiomycetes; pp. 121–141. [Google Scholar]
- 137.Tsang C.-C., Tang J.Y., Lau S.K., Woo P.C. Taxonomy and Evolution of Aspergillus, Penicillium and Talaromyces in the Omics Era–Past, Present and Future. Comput. Struct. Biotechnol. J. 2018;16:197–210. doi: 10.1016/j.csbj.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ekanayaka A., Hyde K., Gentekaki E., McKenzie E., Zhao Q., Bulgakov T., Camporesi E. Preliminary Classification of Leotiomycetes. Mycosphere. 2019;10:310–489. doi: 10.5943/mycosphere/10/1/7. [DOI] [Google Scholar]
- 139.Tanunchai B., Juncheed K., Wahdan S.F.M., Guliyev V., Udovenko M., Lehnert A.-S., Alves E.G., Glaser B., Noll M., Buscot F. Analysis of Microbial Populations in Plastic–Soil Systems after Exposure to High Poly (Butylene Succinate-Co-Adipate) Load Using High-Resolution Molecular Technique. Environ. Sci. Eur. 2021;33:105. doi: 10.1186/s12302-021-00528-5. [DOI] [Google Scholar]
- 140.Suh S.-O., Blackwell M., Kurtzman C.P., Lachance M.-A. Phylogenetics of Saccharomycetales, the Ascomycete Yeasts. Mycologia. 2006;98:1006–1017. doi: 10.1080/15572536.2006.11832629. [DOI] [PubMed] [Google Scholar]
- 141.Issa N. Ph.D. Thesis. University of Kent; Canterbury, UK: 2021. Towards the Biological Degradation of Plastics: Genetic Engineering of Saccharomyces cerevisiae to Secrete Ideonella Sakaiensis Derived PETase. [Google Scholar]
- 142.Maharachchikumbura S.S., Hyde K.D., Jones E.G., McKenzie E.H., Huang S.-K., Abdel-Wahab M.A., Daranagama D.A., Dayarathne M., D’souza M.J., Goonasekara I.D. Towards a Natural Classification and Backbone Tree for Sordariomycetes. Fungal Divers. 2015;72:199–301. doi: 10.1007/s13225-015-0331-z. [DOI] [Google Scholar]
- 143.Sánchez-García M., Ryberg M., Khan F.K., Varga T., Nagy L.G., Hibbett D.S. Fruiting Body Form, Not Nutritional Mode, Is the Major Driver of Diversification in Mushroom-Forming Fungi. Proc. Natl. Acad. Sci. USA. 2020;117:32528–32534. doi: 10.1073/pnas.1922539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Mattos-Shipley K.M., Ford K.L., Alberti F., Banks A., Bailey A.M., Foster G. The Good, the Bad and the Tasty: The Many Roles of Mushrooms. Stud. Mycol. 2016;85:125–157. doi: 10.1016/j.simyco.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oberwinkler F. Yeasts in Pucciniomycotina. Mycol. Prog. 2017;16:831–856. doi: 10.1007/s11557-017-1327-8. [DOI] [Google Scholar]
- 146.Liu X.-Z., Wang Q.-M., Göker M., Groenewald M., Kachalkin A., Lumbsch H.T., Millanes A., Wedin M., Yurkov A., Boekhout T. Towards an Integrated Phylogenetic Classification of the Tremellomycetes. Stud. Mycol. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bandopadhyay S., Liquet y Gonzalez J.E., Henderson K.B., Anunciado M.B., Hayes D.G., DeBruyn J.M. Soil Microbial Communities Associated with Biodegradable Plastic Mulch Films. Front. Microbiol. 2020;11:2840. doi: 10.3389/fmicb.2020.587074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Manohar C.S., Boekhout T., Müller W.H., Stoeck T. Tritirachium candoliense sp. nov., a Novel Basidiomycetous Fungus Isolated from the Anoxic Zone of the Arabian Sea. Fungal Biol. 2014;118:139–149. doi: 10.1016/j.funbio.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 149.Martínez-Soto D., Ortiz-Castellanos L., Robledo-Briones M., León-Ramírez C.G. Molecular Mechanisms Involved in the Multicellular Growth of Ustilaginomycetes. Microorganisms. 2020;8:1072. doi: 10.3390/microorganisms8071072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walther G., Wagner L., Kurzai O. Updates on the Taxonomy of Mucorales with an Emphasis on Clinically Important Taxa. J. Fungi. 2019;5:106. doi: 10.3390/jof5040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khan S., Stevenson A. Mountain Futures Inspiration and Innovation from the World’s Highlands. World Agroforestry Centre; Nairobi, Kenya: 2018. A Feast of Plastic Chewing through the World’s Waste; pp. 209–212. [Google Scholar]
- 152.Morone A. Can Tiny Fungi Solve Our Giant Plastic Problem? [(accessed on 10 September 2021)]. Available online: https://weather.com/en-IN/india/pollution/news/2019-04-29-can-tiny-fungi-be-a-solution-to-our-giant-plastic-problem.
- 153.Bioplastics Market Development Update Dynamic Growth: Global Production Capacities of Bioplastics 2020–2025; 2020. [(accessed on 10 September 2021)]. Available online: https://docs.european-bioplastics.org/conference/Report_Bioplastics_Market_Data_2020_short_version.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.