Abstract
Genes responsible for the synthesis of poly(3-hydroxybutyrate) (PHB) in Azotobacter sp. FA8 were cloned and analyzed. A PHB polymerase gene (phbC) was found downstream from genes coding for β-ketothiolase (phbA) and acetoacetyl-coenzyme A reductase (phbB). A PHB synthase mutant was obtained by gene inactivation and used for genetic studies. The phbC gene from this strain was introduced into Ralstonia eutropha PHB-4 (phbC-negative mutant), and the recombinant accumulated PHB when either glucose or octanoate was used as a source of carbon, indicating that this PHB synthase cannot incorporate medium-chain-length hydroxyalkanoates into PHB.
Polyhydroxyalkanoates (PHAs) are a group of polyesters produced by a large number of bacteria, which accumulate them in intracellular granules as a response to environmental stress and nutrient imbalance (2, 7). These thermoplastic polymers have drawn great interest since their discovery due to their degradability and the potential to produce them from renewable carbon sources. Azotobacter sp. FA8 is an aerobic, nitrogen-fixing bacterium that accumulates poly(3-hydroxybutyrate) (PHB) when cultivated on several carbon sources, including sucrose (13). Although the capacity of Azotobacter strains to accumulate PHAs is well known (2, 9), except for a recently described β-ketothiolase from Azotobacter vinelandii (18), the genes responsible for their synthesis have not yet been identified. In this paper we report the identification, cloning, and molecular analysis of the phb gene cluster of Azotobacter sp. FA8.
Cloning and molecular analysis of the PHB synthase gene from Azotobacter sp. FA8.
Genomic DNA of Azotobacter sp. FA8 was partially digested with XhoI and ligated to the mobilizable cosmid pVK102 (5) digested with the same enzyme and dephosphorylated. The resulting ligation mixture was packaged using an in vitro packaging system (Stratagene, La Jolla, Calif.) and used to transfect Escherichia coli S17-1, a strain that contains the tra genes of plasmid RP4 integrated into the chromosome (19). Transductants were selected on Luria-Bertani plates containing 10 μg of tetracycline/ml. The library was screened for the presence of the PHB synthase (phbC) gene by complementation analysis. Ralstonia eutropha PHB-4 (17), a phbC mutant, was used as recipient. The complementation was carried out on a mineral salts medium (16) containing 1.5% (wt/vol) agar, 0.05% (wt/vol) NH4Cl, 0.5% (wt/vol) fructose, and 5 μg of tetracycline/ml. One of the E. coli clones, C1, gave rise to opaque, PHB-producing transconjugants. The polymer produced by the recombinant was extracted from lyophilized cells with hot chloroform and ethanol precipitated, and the methyl ester derivatives were analyzed by gas chromatography (1) using a Gow Mac Series 580 gas chromatograph (Bridgewater, N.J.) equipped with a flame ionization detector and a packing column of Carbowax 20 M-TPA–Chromosorb W-AW (SUPELCO, Bellefonte, Pa.). The polymer obtained was found to be a homopolymer of 3-hydroxybutyrate.
The recombinant cosmid from clone C1, pC1 (Table 1), was purified using the Concert High Purity system (BRL, Rockville, Md.) and subjected to restriction analysis with several enzymes, giving an estimated insert size of around 20 kb. Digestion with BamHI originated two bands of 6 and 3 kb. The cosmid cut with this enzyme was diluted, religated, and used to transform E. coli S17-1, giving rise to cosmid pC1B, containing a 9-kb deletion, which was not able to complement the R. eutropha phbC mutation. The two BamHI restriction fragments were then subcloned in pBluescript SK (Stratagene) to obtain the recombinant plasmids pAC3 and pAC6, containing the 3- and 6-kb BamHI fragments, respectively (Table 1), and the ends of the inserts from both plasmids were sequenced using M13 forward and reverse universal sequencing primers. An ABI 373 A automatic sequencer was used for sequencing. The sequences obtained were compared with sequences available in the databanks using the Blast program (Bioinformatics Center, Institute for Genomic Research, Kyoto, Japan [http://www.blast.genome.ad.jp]), and a region highly homologous to the amino-terminal end of PHA synthase genes occurred 41 bp downstream from one end of the 6-kb insert. Sequencing of the complete corresponding 1,700-bp open reading frame (ORF) was obtained from this clone by primer walking. The deduced amino acid sequence from this ORF (ORF1) showed a striking homology with the PhbC gene product of Pseudomonas sp. 61-3 (GenBank accession no. AB014757) (67% identity; 82% similarity). ORF1 also showed great homology with other PHB synthases, such as PhbC from R. eutropha (GenBank accession no. J05003) (55% identity; 73% similarity) and PhaC from Burkholderia sp. (GenBank accession no. AF153086) (52% identity; 71% similarity), and was consequently designated phbC.
TABLE 1.
Recombinant plasmids used in this work
| Plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| pC1 | pVK102 containing a 20-kb chromosomal fragment from Azotobacter sp. FA8 inserted at the XhoI site | This work |
| pC1B | a deletion derivative of pC1, obtained by self-ligation after restriction with BamHI | This work |
| pAC3 | pBluescript containing a 3-kb BamHI fragment from pC1 | This work |
| pAC6 | pBluescript containing a 6-kb BamHI fragment from pC1 | This work |
| pATpoliK | pAT18 (21) containing a 578-bp amplification fragment from the Azotobacter sp. FA8 phbC gene, interrupted by a kanamycin cassette | This work |
| pRKpolC1 | pRK404 (3) containing a 2.3-kb fragment from pAC6 containing the phbC coding region downstream of lac promoter | This work |
| pHP1014::2000 | Tcr CmrphaC1PaphaDPaphaC2Pa ORF1 ORF2 ORF3 ORF4′ | 21 |
No genes related to PHB synthesis were found downstream from the phbC gene. Sequencing of the upstream region from cosmid pC1 revealed the presence of the carboxy-terminal region of an ORF (ORF2) with great (>80%) homology with previously described β-ketoacyl-coenzyme A (CoA) thiolase genes.
Cloning of the β-ketoacyl-CoA thiolase and acetoacetyl-CoA reductase genes from Azotobacter sp. strain FA8.
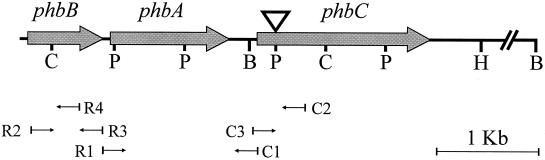
In order to clone the rest of ORF2, presumably the Azotobacter sp. FA8 β-ketoacyl-CoA thiolase (phbA) gene, primers were designed from the conserved sequences corresponding to the amino-terminal region of several β-ketoacyl-CoA thiolase genes available in the databanks and used together with C1 (5′ GAC ATT GAT CCT GAA AAG CG 3′), a primer corresponding to the region immediately upstream from the phbC gene from Azotobacter sp. FA8 (Fig. 1), but no amplification fragment was obtained. pha genes are normally organized in clusters (7). phb genes were not found downstream of phbC, and we hypothesized that the acetoacetyl-CoA reductase gene (phbB) might be upstream from ORF2. Based on this hypothesis, several primers were designed from conserved regions of phbB genes. The following primers were used to obtain a 1,500-bp amplification fragment: R1 [5′ GCN GA(C/T) TT(G/T) (A/T)(G/C)N (G/T/C)TN AA(C/T) GGN GG 3′], a degenerate primer corresponding to the conserved carboxy-terminal region of available phbB genes, and C1, described above. The 1,500-bp amplification fragment was cloned into vector pGemT-Easy (Promega, Madison, Wis.) and sequenced as indicated previously. The analysis of its sequence revealed the presence of a 1,176-bp ORF (ORF2) highly homologous to previously described phbA genes. The deduced amino acid sequence of ORF2, designated phbA, comprised 392 residues and showed similarity with other β-ketoacyl-CoA thiolases, such as the biosynthetic ketothiolase (PhbA) of A. vinelandii (GenBank accession no. AF267243) (94% identity; 96% similarity), the acetyl-CoA acetyltransferase of Pseudomonas aeruginosa PAO1 (AtoB) (GenBank accession no. C83396) (83% identity; 91% similarity), and the β-ketoacyl-CoA thiolases (PhbA) of Pseudomonas sp. strain 61-3 (GenBank accession no. T44362) (80% identity; 89% similarity) and R. eutropha (GenBank accession no. J05003) (67% identity; 79% similarity).
FIG. 1.
Organization of the Azotobacter sp. FA8 genomic region containing the genes phbA, phbB, and phbC. Relevant restriction sites are indicated with capital letters (B, BamHI; C, ClaI; H, HindIII; P, PstI). Thin black arrows indicate the positions and directions of primers used in this work. The restriction site used for mutagenesis by gene disruption is indicated by an inverted triangle.
A similar approach was used for the cloning of the acetoacetyl-CoA reductase gene (phbB) from Azotobacter sp. FA8. A degenerate primer, R2 (5′ TNA CNG GNG GNA TGG GNG G 3′), was designed from a conserved region situated approximately 30 bp downstream from the amino-terminal end of available acetoacetyl-CoA reductase genes and used together with primer R3 (5′GCA TGT TCA GAC CAC CGT TG 3′), corresponding to the already sequenced carboxy-terminal end of the gene (Fig. 1), to obtain a 714-bp amplification fragment, which was cloned and sequenced as indicated above. The analysis of its sequence revealed the presence of an ORF (ORF3) highly homologous to previously cloned phbB genes. The inverse PCR technique was used to obtain the first 30 bases from the nonconserved amino-terminal end of ORF3. BglI-digested genomic DNA from Azotobacter sp. FA8 was treated with T4 DNA ligase (New England Biolabs, Beverly, Mass.) at a final concentration of approximately 10 ng/μl. Two microliters of the ligation mixture was used for PCR amplification with primers R1 and R4 (5′CGG TCA CGC CCT TGC TGG 3′), corresponding to the already sequenced part of ORF3 (Fig. 1). The 1,724-bp amplification fragment that was obtained (which was cloned and sequenced as previously described) contained an incomplete ORF corresponding to the amino-terminal end of ORF3. The deduced amino acid sequence of ORF3 showed great homology with previously described phbB genes, such as the phbB genes from Pseudomonas sp. strain 61-3 (GenBank accession no. T44362) (76% identity; 87% similarity), Burkholderia sp. (GenBank accession no. AF153086) (67% identity; 82% similarity), and R. eutropha (GenBank accession no. J05003) (64% identity; 81% similarity), and was consequently designated phbB. Several consensus sequences of ς70-dependent promoters were found in the region upstream from phbB.
Construction of a PHB synthase mutant by gene inactivation.
A 578-bp fragment from the Azotobacter sp. strain FA8 phbC gene containing a PstI internal restriction site was obtained by PCR amplification of genomic DNA using primers C2 (5′CGC AAT CCC GTT GAT AAG 3′) and C3 (5′CGC TTT TCA GGA TCA ATG TC 3′) (Fig. 1). The amplification fragment was cloned in the pGEM-T Easy cloning vector (Promega), digested with PstI, and ligated with a kanamycin cassette obtained from plasmid pUC4K (Pharmacia, San Francisco, Calif.) cut with PstI. A 1,750-bp EcoRI fragment carrying the whole insert (phbC gene fragment with kanamycin cassette) was cloned into the mobilizable Emr plasmid pAT18 (22), which cannot replicate in Azotobacter, and used to transform E. coli S17-1. Kmr, Emr transformants were selected on Luria-Bertani plates containing 50 μg of kanamycin/ml and 100 μg of erythromycin/ml, and their recombinant plasmids were purified using standard techniques and checked by restriction analysis. One of these transformants was used as a conjugation donor in order to introduce the recombinant plasmid, designated pATpoliK (Table 1), into Azotobacter sp. FA8. Transconjugants were selected on Burk's medium (9) containing 1.5% (wt/vol) agar, 0.5% (wt/vol) sucrose, and 5 μg of kanamycin/ml. Their PHB phenotype was verified by gas chromatography of the methyl ester derivatives as previously described (Table 2). One PHB-negative mutant, Azotobacter sp. UBA 60-3, was chosen for further studies. In order to characterize the mutant by complementation, we cloned a 2.3-kb HindIII fragment from pAC6, containing the complete phbC gene, into the mobilizable Tetr plasmid pRK404 (3), downstream from the lac promoter, giving rise to plasmid pRKpolC1 (Table 1). This construction was introduced in E. coli S17-1 by transformation and transferred to mutant Azotobacter sp. UBA 60-3 by conjugation. All the Tetr transconjugants were able to accumulate PHB, indicating that the mutation in Azotobacter sp. UBA 60-3 was complemented by the phbC gene (Table 2). The capacity of the transconjugants to accumulate PHB was analyzed by gas chromatography as indicated above.
TABLE 2.
Analysis of polymer production from different carbon sources in Azotobacter sp. FA8 and several recombinant strainsa
| Strain | Plasmid | Carbon source(s) (% concn) | PHA produced | PHB content (% [wt/wt] of CDW)b |
|---|---|---|---|---|
| Azotobacter sp. FA8 | None | Glucose (3) | PHB | 38 |
| Glucose (3) + octanoate (0.125) | PHB | 26 | ||
| Glucose (0.5) | PHB | 4.5 | ||
| Glucose (0.5) + octanoate (0.125) | PHB | 3.5 | ||
| Azotobacter sp. UBA 60-3 | None | Glucose (3) | NDc | ND |
| Azotobacter sp. UBA 60-3 | pC1 | Glucose (3) | PHB | 25 |
| Glucose (3) + octanoate (0.125) | PHB | 14 | ||
| Azotobacter sp. UBA 60-3 | pHP1014::2000 | Glucose (3) | ND | ND |
| Glucose (3) + octanoate (0.125) | ND | ND | ||
| Azotobacter sp. UBA 60-3 | pRK404 | Glucose (3) | ND | ND |
| Azotobacter sp. UBA 60-3 | pRKpolC1 | Glucose (3) | PHB | 28.5 |
| R. eutropha PHB-4 | None | Glucose (1.5) | ND | ND |
| R. eutropha PHB-4 | pRKpolC1 | Glucose (1.5) | PHB | 4 |
| Octanoate (0.1) | PHB | 1.9 | ||
| Octanoate (0.25) | PHB | 3.9 | ||
| Octanoate (0.5) | PHB | 5 |
Azotobacter strains were grown for 24 h in Burk's medium supplemented with carbon sources as indicated. Ralstonia eutropha recombinants were grown for 48 h in mineral salts medium supplemented with carbon sources as indicated.
CDW = cell dry weight.
ND, not detected (≤1% [wt/wt] of CDW).
Production of polyhydroxyalkanoates from different carbon sources in Azotobacter sp. strain FA8.
Bacterial PHA synthases can be classified in three classes, depending on the type of hydroxyalkanoates they can use as substrates and their subunit composition (14). PHA synthases that prefer three to five carbon substrates and are composed of one type of subunit are called short-chain-length (SCL) PHA synthases and belong to class I. PHA synthases that use substrates with 6 to 14 carbons are also composed of one type of subunit, are called medium-chain-length (MCL) PHA synthases, and belong to class II. The best known example of class I PHASCL synthases is the R. eutropha PHB synthase. Most pseudomonads have PHA synthases belonging to the second group, PHAMCL synthases. Class III PHA synthases prefer three to five carbon substrates and are composed of two different subunits: PhaC (PHA synthase homologue) and PhaE (20). An exception of the class III enzymes is the Thiocapsa pfennigii PHA synthase, which also accepts six to eight carbon substrates (6). Class II PHAMCL synthase genes expressed in E. coli facilitated the accumulation of PHAMCL from fatty acids when fatty acid β-oxidation was truncated (10, 11, 12).
To determine if Azotobacter sp. FA8 could produce MCL polyhydroxyalkanoates, polymer formation in cultures grown in octanoate-supplemented medium was evaluated. Growth of this strain was not observed in Burk's medium containing this fatty acid or hexanoic acid as sole carbon sources. When Azotobacter sp. strain FA8 was grown in Burk's medium containing glucose and sodium octanoate, PHB was the only polymer detected (Table 2). This result could be due to (i) the inability of the cells to metabolize MCL fatty acids to intermediates that could be used by the PHA polymerase, or (ii) the inability of the PHA polymerase to incorporate MCL monomers into the polyhydroxyalkanoate. As an approach to studying these possibilities, plasmid pHP1014::2000 (21), containing genes encoding PHAMCL synthases PhaC1 and PhaC2 from P. aeruginosa, was introduced in Azotobacter sp. UBA 60-3 by conjugation. The recombinants were grown in Burk's medium with the addition of octanoate, and their polymer contents were determined (Table 2). No polymer was detected in mutant Azotobacter sp. UBA 60-3 containing pHP1014::2000. We cannot rule out the possibility that the Pseudomonas genes could not be expressed in this strain. R. eutropha is able to use octanoate as a carbon source, and, when grown in a medium supplemented with this fatty acid, recombinants containing a PHAMCL synthase are able to synthesize PHAs containing 3-hydroxyhexanoic acid and 3-hydroxyoctanoic acid (21). Plasmid pRKpolC1, containing the Azotobacter sp. strain FA8 phbC gene downstream from the lac promoter (Table 1) was introduced into R. eutropha PHB-4 to determine if the Azotobacter sp. FA8 synthase could incorporate MCL monomers into the polymer. The only polymer produced by the recombinant was PHB (Table 2).
These collective findings indicate that the product of the phbC gene from Azotobacter sp. FA8, situated downstream of the genes for a β-ketoacyl-CoA thiolase and acetoacetyl-CoA reductase (Fig 1), is a class I PHA synthase. This conclusion is supported by the fact that (i) this enzyme cannot incorporate MCL hydroxyalkanoates into PHA, and (ii) the product of the phbC gene reestablishes the capacity of R. eutropha PHB-4 to accumulate PHB. The Azotobacter sp. FA8 phb genes showed great homology to the phb genes of a PHB-producing Pseudomonas strain, Pseudomonas sp. 61-3 (8). Similarities in the genes between members of these genera have been previously described (4) and could be due to a common phylogenetic origin or horizontal gene transfer.
The importance of PHAs to the fitness and survival of bacteria during periods of starvation has been proposed (2, 7). Recent work has established a correlation between polymer utilization and nucleotide accumulation and provides insight into the mechanism by which PHAs enhance the survival capabilities of bacteria (15). The isolation of the genes described in the present study will facilitate both applied and basic research on polyhydroxyalkanoates in nitrogen-fixing soil bacteria.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the EMBL database under the accession numbers AJ319748 (corresponding to the Azotobacter sp. FA8 phbC and phbA genes) and AJ311166 (corresponding to the Azotobacter sp. FA8 phbB gene).
While the manuscript of this report was in the process of being evaluated, the sequences corresponding to the phb gene cluster of A. vinelandii were released. These sequences, annotated under the GenBank accession number AF267243, include the phbC and phbB genes, which exhibit high amino acid homology (91% identity and 95% similarity for phbC; 94% identity and 97% similarity for phbB) with the corresponding Azotobacter sp. FA8 genes described in this study.
Acknowledgments
We thank Patrice Courvalin for his kind gift of plasmid pAT18 and Luciano Chaneton for help in some of the cloning experiments.
This work was funded by the European Commission under contract ERB5514IC18-CT97-0201 and the Agencia Nacional de Promoción Científica y Tecnológica. G.J.V. was the recipient of a graduate student fellowship from the University of Buenos Aires. B.S.M is a researcher from CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas).
REFERENCES
- 1.Brandl H, Gross R A, Lenz R W, Fuller R C. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoate) for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawes E A, Senior P J. The role and regulation of energy reserve polymers in micro-organisms. Adv Microbial Physiol. 1973;10:135–266. doi: 10.1016/s0065-2911(08)60088-0. [DOI] [PubMed] [Google Scholar]
- 3.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X-W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 4.Durham D R, Ornston L N. Homologous structural genes and similar induction patterns in Azotobacter spp. and Pseudomonas spp. J Bacteriol. 1980;143:834–840. doi: 10.1128/jb.143.2.834-840.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 6.Liebergesell M, Rahalkar S, Steinbüchel A. Analysis of the Thiocapsa pfennigii polyhydroxyalkanoate synthase: subcloning, molecular characterization and generation of hybrid synthases with the corresponding Chromatium vinosum enzyme. Appl Microbiol Biotechnol. 2000;54:186–194. doi: 10.1007/s002530000375. [DOI] [PubMed] [Google Scholar]
- 7.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsusaki H, Manji S, Taguchi K, Kato M, Fukui T, Doi Y. Cloning and molecular analysis of the poly(3-hydroxybutyrate) biosynthesis genes in Pseudomonas sp. strain 61-3. J Bacteriol. 1998;180:6459–6467. doi: 10.1128/jb.180.24.6459-6467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page W J, Manchak J, Rudy B. Formation of poly(hydroxybutyrate-co-hydroxyvalerate) by Azotobacter vinelandii UWD. Appl Environ Microbiol. 1992;58:2866–2873. doi: 10.1128/aem.58.9.2866-2873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi Q, Rehm B H A, Steinbüchel A. Synthesis of poly(3hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 11.Qi Q, Steinbüchel A, Rehm B H A. Metabolic routing towards polyhydroxyalkanoic acid synthesis in recombinant Escherichia coli (fadR): inhibition of fatty acid β-oxidation by acrylic acid. FEMS Microbiol Lett. 1998;167:89–94. doi: 10.1111/j.1574-6968.1998.tb13212.x. [DOI] [PubMed] [Google Scholar]
- 12.Qi Q, Steinbüchel A, Rehm B H A. In vitro synthesis of poly(3-hydroxydecanoate): purification and enzymatic characterization of type II polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2000;54:37–43. doi: 10.1007/s002530000357. [DOI] [PubMed] [Google Scholar]
- 13.Quagliano J C, Alegre P, Miyazaki S S. Aislamiento y caracterización de Azotobacter sp. para la producción de poli-β -hidroxialcanoatos. Rev Argent Microbiol. 1994;26:21–27. [PubMed] [Google Scholar]
- 14.Rehm B H A, Steinbüchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol. 1999;25:3–19. doi: 10.1016/s0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz J A, López N I, Fernández R O, Méndez B S. Polyhydroxyalkanoate degradation is associated with nucleotide accumulation and enhances stress resistance and survival of Pseudomonas oleovorans in natural microcosms. Appl Environ Microbiol. 2001;67:225–230. doi: 10.1128/AEM.67.1.225-230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysyologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 17.Schlegel H G, Lafferty R, Krauss I. The isolation of mutants not accumulating poly-β-hydroxybutyric acid. Arch Mikrobiol. 1970;71:283–294. doi: 10.1007/BF00410161. [DOI] [PubMed] [Google Scholar]
- 18.Segura D, Vargas E, Espin G. Beta-ketothiolase genes in Azotobacter vinelandii. Gene. 2000;260:113–120. doi: 10.1016/s0378-1119(00)00462-5. [DOI] [PubMed] [Google Scholar]
- 19.Simon R, Priefer U U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 20.Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 21.Timm A, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxyalkanoic acid) gene locus of Pseudomonas aeruginosa PAO1. Eur J Biochem. 1992;209:15–30. doi: 10.1111/j.1432-1033.1992.tb17256.x. [DOI] [PubMed] [Google Scholar]
- 22.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. Shuttle vectors containing a multiple cloning site and a lacZ α gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene. 1991;102:99–104. doi: 10.1016/0378-1119(91)90546-n. [DOI] [PubMed] [Google Scholar]