Abstract
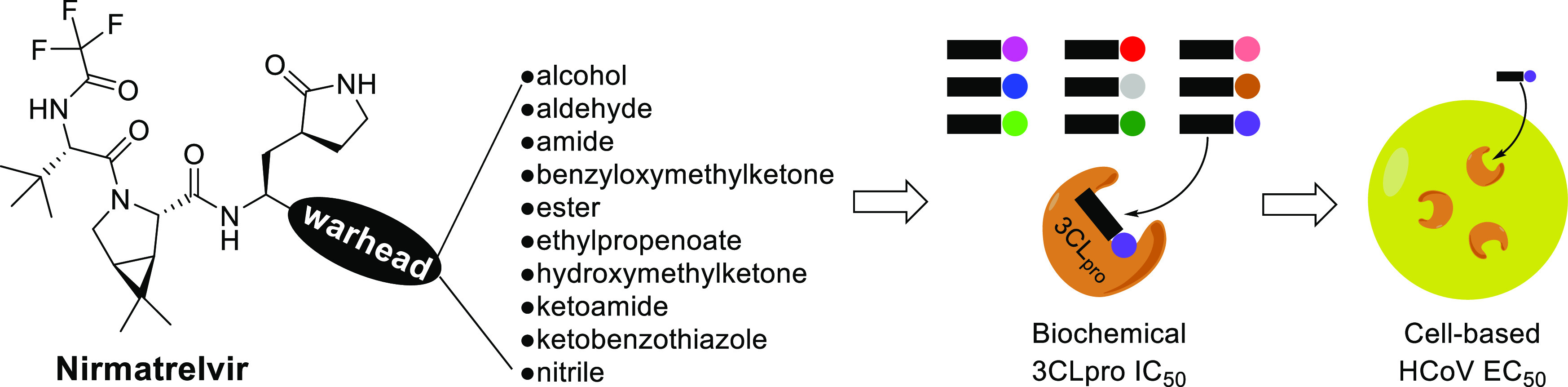
The SARS-CoV-2 pandemic is currently causing an unprecedented global health emergency since its emergence in December 2019. In December 2021, the FDA granted emergency use authorization to nirmatrelvir, a SARS-CoV-2 main protease inhibitor, for treating infected patients. This peptidomimetic is designed with a nitrile warhead, which forms a covalent bond to the viral protease. Herein, we investigate nirmatrelvir analogs with different warheads and their inhibitory activities. In addition, antiviral activities against human alphacoronavirus 229E was also investigated along with a cell-based assay. We discovered that the hydroxymethylketone and ketobenzothiazole warheads were equipotent to the nitrile warhead, suggesting that these analogs can also be used for treating coronavirus infections.
Keywords: SARS-CoV-2, COVID-19, 3CLpro, Mpro, nirmatrelvir, Paxlovid
The current severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic is causing an unprecedented global health emergency. Infection symptoms include sore throat, dry cough, pyrexia, lethargy, body aches, headaches, anosmia and ageusia. More serious symptoms include pneumonia and dyspnea, which can lead to death. First reported in Wuhan, China, in December 2019, it was named “coronavirus disease 2019” (COVID-19) by the World Health Organization (WHO) in 2020.1−3 It has since spread worldwide, and by 10 July 2022, more than 550 million cases and 6 million fatalities have been reported,4 highlighting the urgent need for antiviral drugs.
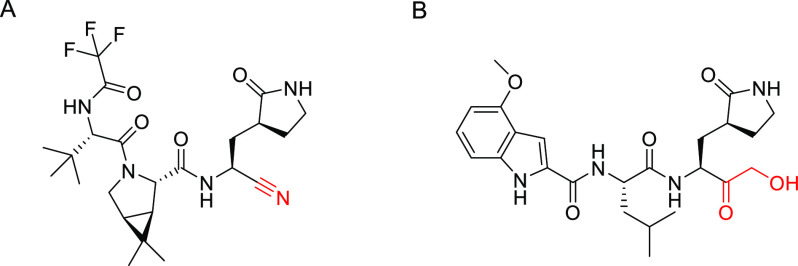
On 22 December 2021, the US Food and Drug Administration (FDA) granted Pfizer’s nirmatrelvir (Figure 1A) Emergency Use Authorization (EUA) for treating infected patients.5−8 The drug inhibits the SARS-CoV-2 main protease, also known as 3C-like protease (3CLpro), which plays a crucial role in cleaving the coronavirus polyprotein to form smaller essential proteins required for virus replication and pathogenesis.6,9−11 Indeed, 3CLpro is deemed an attractive drug target as it has no known human homologues and hence inhibitor drugs should lack off-target side effects.6,9 Nirmatrelvir is a peptidomimetic resembling 3CLpro’s natural polypeptide substrate leucine–glutamine recognition sequence.6 Its P1 residue is a 5-membered lactam resembling glutamine, while its P2 residue is a rigidified leucine analog (Figure 1A). Upon binding to 3CLpro’s active site, its C-terminal nitrile reacts with the Cys145–SH moiety to form a covalent thioimidate bond, thereby inhibiting 3CLpro.6
Figure 1.
(A) Nirmatrelvir. (B) PF-00835231. Warheads depicted in red.
We were surprised by the choice of the nitrile warhead found on nirmatrelvir as a past report revealed peptidomimetics with nitrile warheads possessed only moderate 3CLpro inhibitory activities (IC50 values of 5–49 μM).12 In addition, peptide nitriles were also found to be inactive toward enterovirus 3C proteases.13 Intrigued, we conducted a structure–activity relationship study on 10 nirmatrelvir analogs with various warheads to investigate how different warheads affected inhibitory activities. In addition, their inhibitory activities against human coronavirus (HCoV) 229E 3CLpro was also investigated with a cell-based assay to gauge their potential to be used as pan-coronavirus inhibitors for future coronavirus pandemics.
Nirmatrelvir and the test compounds were synthesized based on reported methods and procedures can be found in the Supporting Information. The biochemical 3CLpro inhibition assay is based on a published procedure11 and details can be found in our recent paper.14 The 3CLpro peptide substrate (DABCYL)-KTSAVLQSGFRKM-(Glu)(EDANS) was synthesized by Genscript. The cell-based HCoV 229E inhibition assay protocol can be found in the Supporting Information.
Nirmatrelvir and its 10 analogs with various electrophilic warheads were synthesized, and their inhibitory activities against SARS-CoV-2 and HCoV 229E 3CLpro were determined using a FRET-based biochemical assay, collated in Table 1. HCoV 229E was first reported in Europe in 1966 and is now globally distributed.15,16 It causes a range of respiratory symptoms with varying severity, from mild upper respiratory tract irritation to potentially life-threatening bronchiolitis and pneumonia.16 As its 3CLpro shares only 40% sequence identity to that of SARS-CoV-2 based on a BLAST search,17 a 3CLpro inhibitor with inhibitory activities against both viral proteases can potentially be used as a pan-coronavirus antiviral for future coronavirus pandemics.
Table 1. Inhibitory Activity of Nirmatrelvir and Its Analogs against SARS-CoV-2 and HCoV 229E Arranged Based on Increasing Molecular Weightsa.
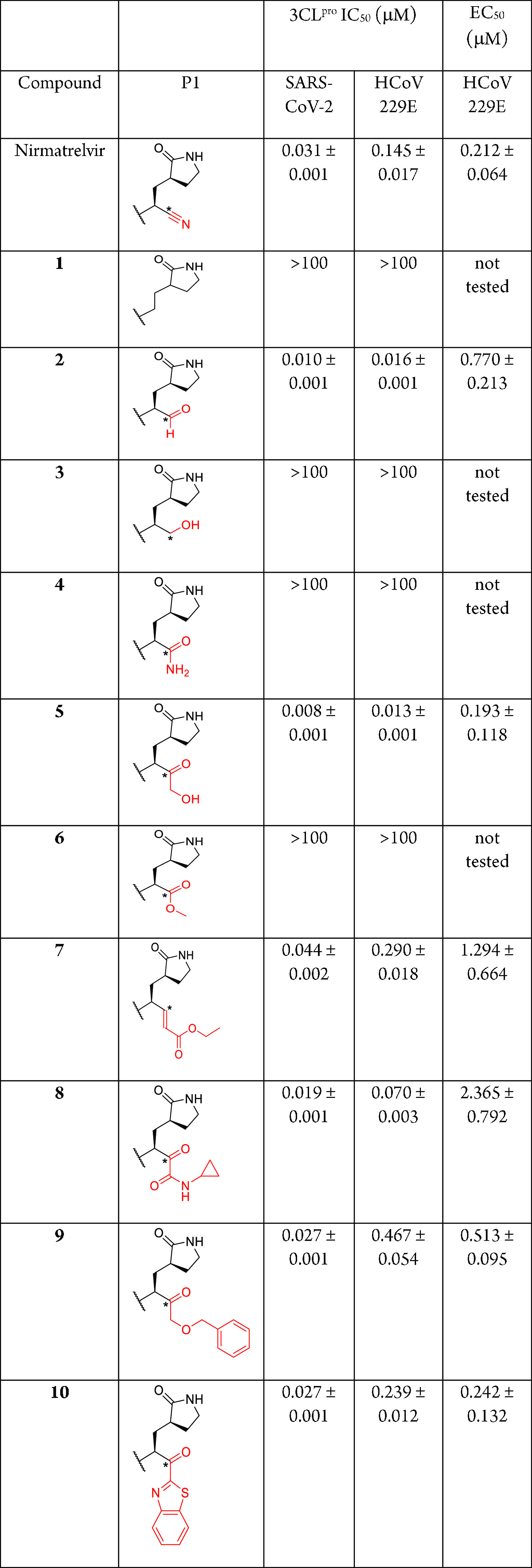
Warheads are depicted in red. Asterisks represent potentially reactive electrophilic carbons.
Our biochemical 3CLpro inhibition assay data revealed nirmatrelvir to be highly potent against SARS-CoV-2 and HCoV 229E 3CLpro with IC50 values of 0.031 and 0.145 μM, respectively (Table 1). In addition, the cell-based HCoV 229E inhibition assay using MRC5 human lung fibroblast cells revealed an EC50 of 0.212 μM, 3-fold less potent than the reported 0.075 μM against SARS-CoV-2 using human bronchial epithelial cells.6 These sub-micromolar EC50 values suggest that nirmatrelvir can potentially be utilized as a pan-coronavirus antiviral drug for future coronavirus pandemics.
Our first test candidate (1) is a nirmatrelvir analog without the nitrile warhead. “Warheadless” peptidomimetic inhibitors have been shown to inhibit the West Nile virus and Murray Valley encephalitis virus proteases with single-digit micromolar IC50 values,18,19 suggesting that a reactive electrophilic warhead may not be needed for protease inhibition. However, our biochemical 3CLpro inhibition assay data revealed 1 to be devoid of any inhibitory activity against SARS-CoV-2 and HCoV 229E 3CLpro (IC50 > 100 μM; Table 1), suggesting that an electrophilic warhead is essential for inhibiting coronavirus 3CLpro.
Our second candidate (2) replaces nirmatrelvir’s nitrile with an aldehyde. Aldehyde peptides are well-known protease inhibitors and have been reported extensively in the literature.20,21 In the context of SARS-CoV-2 3CLpro inhibition, reported IC50 values range from 9 nM to 11 μM, depending on the residues used in the peptide aldehyde inhibitor.11,14,22,23 Expectedly, our biochemical 3CLpro inhibition assay data revealed 2 to potently inhibit SARS-CoV-2 and HCoV 229E 3CLpro with IC50 values of 0.010 and 0.016 μM, respectively (Table 1). It is noteworthy that 2 exhibited a 9-fold inhibitory activity improvement for HCoV 229E 3CLpro compared to nirmatrelvir (0.016 vs 0.145 μM, respectively), suggesting that 2 may be more efficacious than nirmatrelvir. However, our cell-based HCoV 229E inhibition assay revealed its EC50 to be 0.770 μM, almost 4 times less potent than nirmatrelvir (EC50 0.212 μM), suggesting that the aldehyde moiety hindered cell penetration. In addition, aldehydes are also known to be metabolically unstable due to their susceptibility to oxidation and reduction by human liver enzymes, making them unsuitable for drug development.24,25 Hence, we believe peptide aldehydes will be hampered by pharmacological liabilities and opine that peptide aldehyde 2 lacks potential for further drug development.
The third candidate (3) replaces nirmatrelvir’s nitrile with a primary alcohol. A peptide alcohol has been reported by Pfizer with mild inhibitory activity (IC50 = 68 μM) against SARS-CoV-1 3CLpro in their 2005 patent application.26 Interestingly, our biochemical 3CLpro inhibition assay data revealed 3 to be devoid of any inhibitory activity against SARS-CoV-2 and HCoV 229E 3CLpro (IC50 > 100 μM; Table 1). This indicates that for a warhead to react with 3CLpro’s active site Cys145, the warhead’s electrophilic carbon may need to be linear or planar (sp or sp2 hybridized) and not tetrahedral (sp3 hybridized).
Our fourth candidate (4) replaces nirmatrelvir’s nitrile with a primary amide. Peptide amides have been reported to inhibit the dengue and Zika virus nonstructural proteases with single- to double-digit micromolar potencies.27,28 However, our biochemical 3CLpro inhibition assays revealed 4 to be impotent against SARS-CoV-2 and HCoV 229E 3CLpro (IC50 > 100 μM; Table 1), suggesting that primary amides, although planar, are unsuitable warheads for inhibiting coronavirus 3CLpro. The same observation was also reported in a recent publication involving a linear tetrapeptide amide with a P1 glutamine and P2 leucine against SARS-CoV-2 3CLpro.29
The fifth candidate (5) replaces nirmatrelvir’s nitrile with a hydroxymethylketone moiety. Peptidomimetics with this warhead have been reported to be highly potent against SARS-CoV-1 3CLpro with single- to double-digit nanomolar IC50 values.30 As the SARS-CoV-1 3CLpro share 96% sequence identity to SARS-CoV-2,31,32 it was thus unsurprising to observe 5 exhibiting IC50 values of 0.008 and 0.013 μM against SARS-CoV-2 and HCoV 229E 3CLpro, respectively (Table 1). It is noteworthy that these are 4–11-fold improvements over nirmatrelvir (IC50 of 0.031 and 0.145 μM respectively; Table 1), suggesting that a hydroxymethylketone warhead is more reactive than a nitrile. Similarly, 6–8 nM IC50 values were reported recently for Pfizer’s indole dipeptide hydroxymethylketone, PF-00835231 (Figure 1B), against SARS-CoV-2.14,33 Despite this, our cell-based HCoV 229E inhibition assay revealed inhibitor 5 and nirmatrelvir to exhibit very similar EC50 values (0.193 vs 0.212 μM; Table 1), suggesting that the hydroxymethylketone moiety was less efficient than a nitrile for cell membrane penetration. Despite this, the phosphate prodrug of PF-00835231 entered clinical trials in September 2020 for treating hospitalized COVID-19 patients (clinical trial identifier NCT04535167).34 Hence, we opine that inhibitor 5 has the potential for further development as a pan-coronavirus antiviral drug.
Our sixth candidate (6) replaces nirmatrelvir’s nitrile with a methyl ester. A peptide methyl ester has been shown to moderately inhibit the hepatitis C virus nonstructural protease by Boehringer lngelheim researchers (IC50 = 17 μM).35 However, our biochemical 3CLpro inhibition assays revealed 6 to be impotent against SARS-CoV-2 and HCoV 229E 3CLpro (IC50 > 100 μM; Table 1), suggesting that methyl esters are unsuitable warheads for inhibiting coronavirus 3CLpro.
The seventh candidate (7) replaces nirmatrelvir’s nitrile with an ethyl propenoate moiety. Peptidomimetics with this warhead were first reported by Agouron Pharmaceuticals to inhibit human rhinovirus 3C protease in 1998 and were later shown to inhibit the enterovirus 3C protease with single-digit micromolar IC50 values.36−39 Our recent investigation showed that a 3-residue ethyl propenoate peptidomimetic designed by TaiGen Biotechnology was able to inhibit SARS-CoV-2 3CLpro with an IC50 of 286 nM.14,40 Interestingly, when the warhead was incorporated into nirmatrelvir, peptidomimetic 7 exhibited comparable IC50 values against SARS-CoV-2 and HCoV 229E 3CLpro compared to nirmatrelvir (0.044 vs 0.031 μM and 0.290 vs 0.145 μM respectively; Table 1), initially highlighting 7’s drug development potential. However, our cell-based HCoV 229E inhibition assay revealed its EC50 to be 1.294 μM, 6-fold less potent than nirmatrelvir (EC50 0.212 μM), suggesting that the ethyl propenoate moiety hindered cell penetration. It is also noteworthy that rupintrivir (formerly AG7088), another peptidomimetic armed with an ethyl propenoate warhead, was abandoned by Pfizer after phase 2 clinical trials due to a lack of efficacy against rhinovirus infection.41 Hence, we opine that inhibitor 7 lacks drug development potential.
Our eighth candidate (8) replaces nirmatrelvir’s nitrile with a ketoamide warhead. Peptide ketoamides have been approved for the treatment of hepatitis C virus infection, targeting the hepatitis C virus nonstructural protease, and are exemplified by the approved antivirals boceprevir and telaprevir.42 Peptide ketoamides have also been reported to inhibit SARS-CoV-2 3CLpro with IC50 values ranging from 65 nM to 5.7 μM, depending on the residues in the peptide ketoamide.11,14,32 In our biochemical 3CLpro inhibition assays, inhibitor 8 exhibited IC50 values of 0.019 and 0.070 μM against SARS-CoV-2 and HCoV 229E 3CLpro, respectively, approximately 2-fold more potent than nirmatrelvir (Table 1). However, our cell-based HCoV 229E inhibition assay revealed its EC50 to be 2.365 μM, 11-fold less potent than nirmatrelvir (EC50 0.212 μM; Table 1), indicating that the ketoamide moiety hindered cell penetration. Hence, we believe peptide ketoamide 8 lacks further drug development potential.
The ninth candidate (9) replaces nirmatrelvir’s nitrile with a benzyloxymethylketone warhead. Such peptidomimetics were first reported by SmithKline Beecham Pharmaceuticals as potent sub-micromolar cathepsin K cysteine protease inhibitors in 1999.43 In our biochemical 3CLpro inhibition assays, 9 exhibited IC50 values of 0.027 and 0.467 μM against SARS-CoV-2 and HCoV 229E 3CLpro respectively, on par with nirmatrelvir for SARS-CoV-2 3CLpro (IC50 0.031 μM) but 3-fold less potent against HCoV 229E 3CLpro (IC50 0.145 μM). A plausible reason could be due to steric effect caused by the warhead’s benzyl moiety, hindering inhibitor 9 from binding to the HCoV 229E 3CLpro active site. The cell-based HCoV 229E inhibition assay revealed its EC50 to be 0.513 μM, approximately 2-fold less potent than nirmatrelvir (EC50 = 0.212 μM), suggesting peptidomimetic 9 to be an inferior drug candidate compared to nirmatrelvir.
Our last inhibitor (10) replaces nirmatrelvir’s nitrile with a ketobenzothiazole warhead and was first published by Pfizer in 2021.6 Such peptidomimetics were first reported in 2000 by Agouron Pharmaceuticals and could bind to rhinovirus 3C protease with nanomolar binding affinities.44 Binding affinities toward SARS-CoV-2 3CLpro were recently reported to be between 8 and 230 nM,6,45,46 while IC50 values ranged between 94 nM and >10 μM, depending on the residues used in the inhibitors.14 In our biochemical 3CLpro inhibition assays, 10 exhibited IC50 values of 0.027 and 0.239 μM against SARS-CoV-2 and HCoV 229E 3CLpro, respectively, similar to nirmatrelvir (IC50 of 0.031 and 0.145 μM respectively; Table 1). The cell-based HCoV 229E inhibition assay revealed its EC50 value to be on par with that of nirmatrelvir (EC50 0.212 vs 0.242 μM, respectively; Table 1), indicating inhibitor 10 to be a plausible candidate for further development as a pan-coronavirus antiviral for future pandemics.
In conclusion, 10 nirmatrelvir analogs with varying warheads were synthesized and their coronavirus 3CLpro inhibitory activities were determined (Table 1) to gauge their potential for antiviral drug development as pan-coronavirus inhibitors. Inhibitors 2 (aldehyde), 5 (hydroxymethylketone), 8 (ketoamide) and 10 (ketobenzothiazole) were found to be more or equipotent to nirmatrelvir against SARS-CoV-2 and HCoV 229E 3CLpro based on biochemical inhibition (IC50) assays. A subsequent HCoV 229E cell-based inhibition (EC50) assay revealed inhibitors 5 and 10 to be equipotent to nirmatrelvir, indicating both are plausible candidates for further drug development as pan-coronavirus inhibitors for future coronavirus pandemics. We recommend subjecting these candidates to further biochemical IC50 studies using 3CL proteases from other coronaviruses followed by animal coronavirus infection models.
Glossary
Abbreviations
- 3CLpro
3C-like protease
- BLAST
basic local alignment search tool
- CoV
coronavirus
- COVID-19
coronavirus disease 2019
- DMSO
dimethyl sulfoxide
- EC50
half-maximal effective concentration
- EMEM
Earles’s minimum essential medium
- EUA
emergency use authorization
- FBS
fetal bovine serum
- FDA
Food and Drug Administration
- FRET
fluorescence resonance energy transfer
- HCoV
human coronavirus
- HCV
hepatitis C virus
- HPLC
high performance liquid chromatography
- IC50
half-maximal inhibitory concentration
- MOI
multiplicity of infection
- Mpro
main protease
- SARS
severe acute respiratory syndrome
- WHO
World Health Organization
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00260.
Synthetic procedures, NMR characterization, HRMS spectra, protease production, biochemical and cell-based assay protocols (PDF)
Author Contributions
‡ M.D.D., J.Y.F., and Q.Y.O. contributed equally.
This work was funded by Agency for Science, Technology and Research (A*STAR), Singapore.
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Li Q.; Guan X.; Wu P.; Wang X.; Zhou L.; Tong Y.; Ren R.; Leung K. S. M.; Lau E. H. Y.; Wong J. Y.; Xing X.; Xiang N.; Wu Y.; Li C.; Chen Q.; Li D.; Liu T.; Zhao J.; Liu M.; Tu W.; Chen C.; Jin L.; Yang R.; Wang Q.; Zhou S.; Wang R.; Liu H.; Luo Y.; Liu Y.; Shao G.; Li H.; Tao Z.; Yang Y.; Deng Z.; Liu B.; Ma Z.; Zhang Y.; Shi G.; Lam T. T. Y.; Wu J. T.; Gao G. F.; Cowling B. J.; Yang B.; Leung G. M.; Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H.; Zhu Y.; Li B.; Huang C.; Chen H.; Chen J.; Luo Y.; Guo H.; Jiang R.; Liu M.; Chen Y.; Shen X.; Wang X.; Zheng X.; Zhao K.; Chen Q.; Deng F.; Liu L.; Yan B.; Zhan F.; Wang Y.; Xiao G.; Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020, 579, 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int, 2022. (accessed 10 July 2022).
- FDA authorizes first oral antiviral for treatment of COVID-19. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19, 2021. (accessed 23 December 2021).
- Owen D. R.; Allerton C. M. N.; Anderson A. S.; Aschenbrenner L.; Avery M.; Berritt S.; Boras B.; Cardin R. D.; Carlo A.; Coffman K. J.; Dantonio A.; Di L.; Eng H.; Ferre R. A.; Gajiwala K. S.; Gibson S. A.; Greasley S. E.; Hurst B. L.; Kadar E. P.; Kalgutkar A. S.; Lee J. C.; Lee J.; Liu W.; Mason S. W.; Noell S.; Novak J. J.; Obach R. S.; Ogilvie K.; Patel N. C.; Pettersson M.; Rai D. K.; Reese M. R.; Sammons M. F.; Sathish J. G.; Singh R. S. P.; Steppan C. M.; Stewart A. E.; Tuttle J. B.; Updyke L.; Verhoest P. R.; Wei L.; Yang Q.; Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021, 374, 1586–1593. 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Chia C. S. B. Novel nitrile peptidomimetics for treating COVID-19. ACS Med. Chem. Lett. 2022, 13, 330–331. 10.1021/acsmedchemlett.2c00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully M. A tale of two antiviral targets – and the COVID-19 drugs that bind them. Nat. Rev. Drug Discovery 2022, 21, 3–5. 10.1038/d41573-021-00202-8. [DOI] [PubMed] [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng X.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao A.; Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Banerjee R.; Perera L.; Tillekeratne L. M. V. Potential SARS-CoV-2 main protease inhibitors. Drug Discovery Today. 2021, 26, 804–816. 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Sacco M. D.; Hurst B.; Townsend J. A.; Hu Y.; Szeto T.; Zhang X.; Tarbet B.; Marty M. T.; Chen Y.; Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.; Chen C.; Ke Z.; Wan D. C.; Chow H.; Wong K. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur. J. Med. Chem. 2013, 59, 1–6. 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y.; Zhao X.; Cui Z.; Wang M.; Wang Y.; Li L.; Sun Q.; Yang X.; Zeng D.; Liu Y.; Sun Y.; Lou Z.; Shang L.; Yin Z. Cyanohydrin as an anchoring group for potent and selective inhibitors of enterovirus 71 3C protease. J. Med. Chem. 2015, 58, 9414–9420. 10.1021/acs.jmedchem.5b01013. [DOI] [PubMed] [Google Scholar]
- Vankadara S.; Wong Y. X.; Liu B.; See Y. Y.; Tan L. H.; Tan Q. W.; Wang G.; Karuna R.; Guo X.; Tan S. H.; Fong J. Y.; Joy J.; Chia C. S. B. A head-to-head comparison of the inhibitory activities of 15 peptidomimetic SARS-CoV-2 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2021, 48, 128263. 10.1016/j.bmcl.2021.128263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D.; Procknow J. J. A new virus isolated from the human respiratory tract. Exp. Biol. Med. 1966, 121, 190–193. 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Liu D. X.; Liang J. Q.; Fung T. S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encyclopedia of Virology. 2021, 2, 428–440. 10.1016/B978-0-12-809633-8.21501-X. [DOI] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Lim H. A.; Ang M. J. Y.; Joy J.; Poulsen A.; Wu W.; Ching S. C.; Hill J.; Chia C. S. B. Novel agmatine dipeptide inhibitors against the West Nile virus NS2B/NS3 protease: A P3 and N-cap optimization study. Eur. J. Med. Chem. 2013, 62, 199–205. 10.1016/j.ejmech.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Ang M. J. Y.; Yong G. H. J.; Poulsen A.; Then S. W.; Li Z.; Joy J.; Hill J.; Chia C. S. B. Substrate-based peptidomimetic inhibitors of the Murray Valley encephalitis virus NS2B/NS3 serine protease: A P1-P4 SAR study. Eur. J. Med. Chem. 2013, 68, 72–80. 10.1016/j.ejmech.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Zhu L.; George S.; Schmidt M. F.; Al-Gharabli S. I.; Rademann J.; Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011, 92, 204–212. 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T.; Manickam M.; Namasivayam V.; Hayashi Y.; Jung S. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X. M.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; Li C.; Li Y.; Bai F.; Wang H.; Cheng X.; Cen X.; Hu S.; Yang X.; Wang J.; Liu X.; Xiao G.; Jiang H.; Rao Z.; Zhang L.; Xu Y.; Yang H.; Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020, 368, 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B.; Arutyunova E.; Khan M. B.; Lu J.; Joyce M. A.; Saffran H. A.; Shields J. A.; Kandadai A. S.; Belovodskiy A.; Hena M.; Vuong W.; Lamer T.; Young H. S.; Vederas J. C.; Tyrrell D. L.; Lemieux M. J.; Nieman J. A. Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors. RSC Med. Chem. 2021, 12, 1722–1730. 10.1039/D1MD00247C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevski N.; King L.; Pitt W. R.; Lecomte F.; Toselli F. Metabolism by aldehyde oxidase: drug design and complementary approaches to challenges in drug discovery. J. Med. Chem. 2019, 62, 10955–10994. 10.1021/acs.jmedchem.9b00875. [DOI] [PubMed] [Google Scholar]
- Gampe C.; Verma V. A. Curse or cure? A perspective on the developability of aldehydes as active pharmaceutical ingredients. J. Med. Chem. 2020, 63, 14357–14381. 10.1021/acs.jmedchem.0c01177. [DOI] [PubMed] [Google Scholar]
- Kania R. S.; Mitchell L. J.; Nieman J. A.. Anticoronaviral compounds and compositions, their pharmaceutical uses and materials for their synthesis. International Patent WO2006/061714 A2, 2006.
- Chanprapaph S.; Saparpakorn P.; Sangma C.; Niyomrattanakit P.; Hannongbua S.; Angsuthanasombat C.; Katzenmeier G. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem. Biophys. Res. Commun. 2005, 330, 1237–1246. 10.1016/j.bbrc.2005.03.107. [DOI] [PubMed] [Google Scholar]
- Phoo W. W.; Zhang Z.; Wirawan M.; Chew E. J. C.; Chew A. B. L.; Kouretova J.; Steinmetzer T.; Luo D. Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antiviral Res. 2018, 160, 17–24. 10.1016/j.antiviral.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Ullrich S.; Sasi V. M.; Mahawaththa M. C.; Ekanayake K. B.; Morewood R.; George J.; Shuttleworth L.; Zhang X.; Whitefield C.; Otting G.; Jackson C.; Nitsche C. Challenges of short substrate analogues as SARS-CoV-2 main protease inhibitors. Bioorg. Med. Chem. Lett. 2021, 50, 128333. 10.1016/j.bmcl.2021.128333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. L.; Kania R. S.; Brothers M. A.; Davies J. F.; Ferre R. A.; Gajiwala K. S.; He M.; Hogan R. J.; Kozminski K.; Li L. Y.; Lockner J. W.; Lou J.; Marra M. T.; Mitchell L. J. Jr; Murray B. W.; Nieman J. A.; Noell S.; Planken S. P.; Rowe T.; Ryan K.; Smith G. J.; Solowiej J. E.; Steppan C. M.; Taggart B. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Brindisi M.; Shahabi D.; Chapman M. E.; Mesecar A. D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020, 15, 907–932. 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Sun X.; Curth U.; Drosten C.; Sauerhering L.; Becker S.; Rox K.; Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020, 368, 409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Boland S.; Scholle M. D.; Bardiot D.; Marchand A.; Chaltin P.; Blatt L. M.; Beigelman L.; Symons J. A.; Raboisson P.; Gurard-Levin Z. A.; Vandyck K.; Deval J. Dual inhibition of SARS-CoV-2 and human rhinovirus with protease inhibitors in clinical development. Antiviral Res. 2021, 187, 105020. 10.1016/j.antiviral.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras B.; Jones R. M.; Anson B. J.; Arenson D.; Aschenbrenner L.; Bakowski M. A.; Beutler N.; Binder J.; Chen E.; Eng H.; Hammond H.; Hammond J.; Haupt R. E.; Hoffman R.; Kadar E. P.; Kania R.; Kimoto E.; Kirkpatrick M. G.; Lanyon L.; Lendy E. K.; Lillis J. R.; Logue J.; Luthra S. A.; Ma C.; Mason S. W.; McGrath M. E.; Noell S.; Obach R. S.; O’Brien M. N.; O’Connor R.; Ogilvie K.; Owen D.; Pettersson M.; Reese M. R.; Rogers T. F.; Rosales R.; Rossulek M.; Sathish J. G.; Shirai N.; Steppan C.; Ticehurst M.; Updyke L. W.; Weston S.; Zhu Y.; White K. M.; García-Sastre A.; Wang J.; Chatterjee A. K.; Mesecar A. D.; Frieman M. B.; Anderson A. S.; Allerton C. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021, 12, 6055. 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinàs-Brunet M.; Bailey M.; Déziel R.; Fazal G.; Gorys V.; Halmos T.; Maurice R.; Poirier M.; Poupart M. A.; Rancourt J.; Thibeault D.; Wernic D.; Lamarre D.; Goulet S. Studies on the C-terminal of hexapeptide inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett. 1998, 8, 2719–2724. 10.1016/S0960-894X(98)00480-6. [DOI] [PubMed] [Google Scholar]
- Dragovich P. S.; Webber S. E.; Babine R. E.; Fuhrman S. A.; Patick A. K.; Matthews D. A.; Lee C. A.; Reich S. H.; Prins T. J.; Marakovits J. T.; Littlefield E. S.; Zhou R.; Tikhe J.; Ford C. E.; Wallace M. B.; Meador J. W.; Ferre R. A.; Brown E. L.; Binford S. L.; Harr J. E.; DeLisle D. M.; Worland S. T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies. J. Med. Chem. 1998, 41, 2806–2818. 10.1021/jm980068d. [DOI] [PubMed] [Google Scholar]
- Binford S. L.; Maldonado F.; Brothers M. A.; Weady P. T.; Zalman L. S.; Meador J. W.; Matthews D. A.; Patick A. K. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 2005, 49, 619–626. 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang M. J. Y.; Lau Q. Y.; Ng F. M.; Then S. W.; Poulsen A.; Cheong Y. K.; Ngoh Z. X.; Tan Y. W.; Peng J.; Keller T. H.; Hill J.; Chu J. J. H.; Chia C. S. B. Peptidomimetic ethyl propenoate covalent inhibitors of the enterovirus 71 3C protease: a P2–P4 study. J. Enzyme Inhib. Med. Chem. 2016, 31, 332–339. 10.3109/14756366.2015.1018245. [DOI] [PubMed] [Google Scholar]
- Tan Y. W.; Ang M. J. Y.; Lau Q. Y.; Poulsen A.; Ng F. M.; Then S. W.; Peng J.; Hill J.; Hong W. J.; Chia C. S. B.; Chu J. J. H. Antiviral activities of peptide-based covalent inhibitors of the enterovirus 71 3C protease. Sci. Rep. 2016, 6, 33663. 10.1038/srep33663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Chen S.; Hsu M.; Wu J.; Tseng C. K.; Liu Y.; Chen H.; Kuo C.; Wu C.; Chang L.; Chen W.; Liao S.; Chang T.; Hung H.; Shr H.; Liu C.; Huang Y.; Chang L.; Hsu J.; Peters C. J.; Wang A. H.; Hsu M. Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006, 49, 4971–4980. 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- Patick A. K. Rhinovirus chemotherapy. Antiviral Res. 2006, 71, 391–396. 10.1016/j.antiviral.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns M. P.; von Hahn T. Novel therapies for hepatitis C – one pill fits all?. Nat. Rev. Drug Discovery 2013, 12, 595–610. 10.1038/nrd4050. [DOI] [PubMed] [Google Scholar]
- Marquis R. W.; Ru Y.; Yamashita D. S.; Oh H. J.; Yen J.; Thompson S. K.; Carr T. J.; Levy M. A.; Tomaszek T. A.; Ijames C. F.; Smith W. W.; Zhao B.; Janson C. A.; Abdel-Meguid S. S.; D’Alessio K. J.; McQueney M. S.; Veber D. F. Potent dipeptidylketone inhibitors of the cysteine protease cathepsin K. Bioorg. Med. Chem. 1999, 7, 581–588. 10.1016/S0968-0896(99)00011-5. [DOI] [PubMed] [Google Scholar]
- Dragovich P. S.; Zhou R.; Webber S. E.; Prins T. J.; Kwok A. K.; Okano K.; Fuhrman S. A.; Zalman L. S.; Maldonado F. C.; Brown E. L.; Meador J. W.; Patick A. K.; Ford C. E.; Brothers M. A.; Binford S. L.; Matthews D. A.; Ferre R. A.; Worland S. T. Structure-based design of ketone containing, tripeptidyl human rhinovirus 3C protease inhibitors. Bioorg. Med. Chem. Lett. 2000, 10, 45–48. 10.1016/S0960-894X(99)00587-9. [DOI] [PubMed] [Google Scholar]
- Konno S.; Kobayashi K.; Senda M.; Funai Y.; Seki Y.; Tamai I.; Schäkel L.; Sakata K.; Pillaiyar T.; Taguchi A.; Taniguchi A.; Gütschow M.; Müller C. E.; Takeuchi K.; Hirohama M.; Kawaguchi A.; Kojima M.; Senda T.; Shirasaka Y.; Kamitani W.; Hayashi Y. 3CL protease inhibitors with an electrophilic arylketone moiety as anti-SARS-CoV-2 agents. J. Med. Chem. 2022, 65, 2926–2939. 10.1021/acs.jmedchem.1c00665. [DOI] [PubMed] [Google Scholar]
- Thanigaimalai P.; Konno S.; Yamamoto T.; Koiwai Y.; Taguchi A.; Takayama K.; Yakushiji F.; Akaji K.; Chen S.; Naser-Tavakolian A.; Schön A.; Freire Hayashi Y.; et al. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: Design, synthesis, biological evaluation, and docking studies. Eur. J. Med. Chem. 2013, 68, 372–384. 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.