Abstract
Wastewater surveillance of SARS-CoV-2 RNA has become an important tool for tracking the presence of the virus and serving as an early indicator for the onset of rapid transmission. Nevertheless, wastewater data are still not commonly used to predict the number of infected individuals in a sewershed. The main objective of this study was to calibrate a susceptible-exposed-infectious-recovered (SEIR) model using RNA copy rates in sewage (i.e., gene copies per liter times flow rate) and the number of SARS-CoV-2 saliva-test-positive infected individuals in a university student population that was subject to repeated weekly testing during the Spring 2021 semester. A strong correlation was observed between the RNA copy rates and the number of infected individuals. The parameter in the SEIR model that had the largest impact on calibration was the maximum shedding rate, resulting in a mean value of 7.72 log10 genome copies per gram of feces. Regressing the saliva-test-positive infected individuals on predictions from the SEIR model based on the RNA copy rates yielded a slope of 0.87 (SE=0.11), which is statistically consistent with a 1:1 relationship between the two. These findings demonstrate that wastewater surveillance of SARS-CoV-2 can be used to estimate the number of infected individuals in a sewershed.
Keywords: Wastewater-based epidemiology, COVID-19, SARS-CoV-2, SEIR model, pandemic
Graphical Abstract
INTRODUCTION
Over the course of the Coronavirus Disease 2019 (COVID-19) pandemic, wastewater-based epidemiology (WBE) has been implemented as a tool to monitor community disease incidence at hundreds of locations. WBE measures the concentration of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) RNA fragments in wastewater.1 Compared to clinical testing, WBE surveillance can be faster and more cost-effective as a long-term monitoring strategy at the community level.2
There are three primary applications of WBE for COVID-19.3 The first is as a qualitative indicator of SARS-CoV-2 presence or absence.3 SARS-CoV-2 RNA copies detected in wastewater tend to increase prior to a surge in clinical cases, thus serving as a leading indicator of community disease burden. For a specific community, an increase in viral RNA copies can then signal the need for more widespread clinical testing. To date WBE has been employed in many venues ranging from small-scale communities, such as hospitals, nursing homes, airlines, and college campuses to larger-scale populations, such as large towns and cities.4–13
The second application is as a quantitative estimate of disease prevalence and trends,3 and requires an assumption about the appropriate lag or lead time that maximizes the correlation between wastewater SARS-CoV-2 RNA concentrations and reported COVID-19 cases. However, lead times vary based on numerous factors including fecal shedding rates, disease time course, testing access and processing time, and attitudes towards seeking healthcare. Therefore, rather than a precise leading indicator of disease prevalence, this application may be better interpreted as an independent measure of population-level disease prevalence.14
The third application is to estimate rapid changes in disease incidence.3 Identifying “bursts” of disease in places such as long term care facilities, hospitals, and college campuses is valuable for determining when to increase clinical testing. This application is more complex because it goes beyond detecting presence/absence or identifying trends over several weeks, instead aiming to quantify changes in trends over a matter of days. The challenge for validating this approach is obtaining a gold standard of the “true” incidence of infection compared to the predicted incidence from WBE.
Despite significant advances that have been made to reap the benefits associated with WBE for managing the COVID-19 pandemic, further refinements are needed to fully exploit this tool. The prevailing view of many in this field, supported by the U.S. Centers for Disease Control and Prevention, is that the large number of variables which impact the correlation between a wastewater concentration and the number of infected individuals mean that WBE data cannot be used to predict the number of infected individuals in a sewershed.15 Despite the challenges posed by uncertainty in the parameters needed to make robust estimates of disease prevalence using wastewater surveillance (e.g., the impact of dilution from stormwater runoff), models are available for making predictions that account for this uncertainty.
We have previously developed and published a susceptible-exposed-infectious-recovered (SEIR) model for estimating numbers of infected in a sewershed based on RNA copy rates in wastewater, as opposed to concentrations.16 Using copy rates (i.e., RNA copies per liter times flow rates) overcomes the problems associated with using concentrations alone and is more direct than methods such as concentration normalization using fecal indicators.17 When the SEIR model was published, however, we had a limited amount of data that we could use for calibration purposes, due to a limited amount of clinical testing data. Following implementation of a comprehensive clinical testing strategy,18 an opportunity became available to perform a more robust level of calibration using a unique data set from Clemson University. During the Spring 2021 semester (1/6/21 to 5/6/21), Clemson University’s mandatory testing strategy required all students, faculty, and staff to be tested for COVID-19 weekly as a condition for accessing university buildings. Individuals who tested positive were not allowed to access the campus for 10 days. Students who lived in dormitories were allowed to isolate for this period on campus. The University’s wastewater treatment plant services almost exclusively the campus; therefore, the only individuals contributing SARS-CoV-2 to the campus wastewater was a known population of infected students. The main objective of this study was to calibrate the SEIR model using the RNA copy rates in the sewage and the number of infected students identified through weekly personal testing.
Several WBE studies have reported known numbers of infected individuals. A number of these studies were performed on college campuses and correlated SARS-CoV-2 RNA concentrations to reported cases.19–23 Of these, Karthikeyan et al.19 provided a model to quantitatively link RNA concentrations to numbers of cases. They employed an infinite impulse response filter with wastewater signal as an input. However, their model is time dependent, limiting its broad applicability. Here, we validate the time-independent SEIR model that may be used to predict the number of infected individuals in a sewershed based on SARS-CoV-2 RNA copy rates in wastewater.
During the period when this study was performed, the Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1) variants of concern (VOC) became prevalent in the US. It was unclear at the time whether these variants changed the shedding rate of the virus. Thus, a second objective of this study was to determine the impact of the VOCs on the SEIR model, presumably via changes in the shedding rate.
METHODS
Individual Testing and Isolation Data.
Beginning on January 1, 2021, Clemson University required all students and employees, regardless of vaccination status, to be tested for COVID-19 on a weekly basis as a condition for access to university buildings. Individuals came to a centralized facility where saliva samples (~1 mL per person) were collected in 50 mL centrifuge tubes. The tubes were taken daily to the on-campus clinical lab (CLIA number 42D2193465) where a diagnostic RT-qPCR test was performed to determine whether an individual was positive for COVID-19.24 The TigerSaliva multiplex RT-qPCR test targets the N gene of SARS-CoV2 for diagnosis24; it is a version of the EUA-approved SalivaDirect protocol.25 Details of the methods are available elsewhere.24–28 Briefly, saliva is heat treated to 95 °C for 30 min. Then 2 μl of sample are loaded using open-source sample handlers (Opentrons OT-2) into prepared plates with enzyme mix (New England Biolabs M3006B, M3002B), primers, and probes. The assay measures the N1 sequence of SARS-CoV-2 (nCOV_N1 forward primer: Integrated DNA Technologies (IDT) 10006830, reverse primer IDT 10006831, probe IDT 10006832) and uses Hs_RPP30 as a human control gene (RNAse P forward primer IDT 10006836, reverse primer 10006837, probe IDT 10007062). The plates are loaded into standard thermocycler (Bio-Rad CFX 384) systems. From the standard curve, a Ct of 33 on the assay is equivalent to 1 viral copy per microliter.
Students and employees who tested positive and resided off-campus were required to isolate for 10 days prior to returning to campus. Students who tested positive and resided on campus were given the option of isolating on campus or moving to an off-campus location. Those who opted to stay on campus were presumed to be the only contributors to the virus in the campus wastewater. The implications of this assumption are discussed in the section on Limitations. Inconclusive results comprised ~0.2% of all the saliva samples analyzed. Individuals who received an inconclusive result were advised to be retested; most did within two days because their campus access status was turned off if they did not retest.
To track students who entered into isolation and quarantine, a management system consisting of several software components, including Atlassian Jira, was utilized to facilitate housing workflows. Atlassian Jira has been used in similar applications by others.29 Internally developed applications were used for COVID-19 test result processing, case reporting, isolation and quarantine management, and contact tracing (Supporting Information, Fig. SI-1). When a student tested positive, the following processes were implemented: 1) Medical personnel were notified of the positive result, the duration of isolation was established, and temporary on-campus room space was offered to the student; 2) When a student required space on campus to isolate, medical personnel notified housing administrators; 3) Housing personnel coordinated room space for the students, either in-place, on-campus, or at off-campus locations. Students were allowed to check out early to isolate at their homes, or in complete isolation in the provided campus space; 4) When isolation was completed, medical personnel notified the student and housing administrators; and 5) Once cleared from isolation, students who had isolated in campus space checked out of their temporary arrangements.
Isolation records were maintained to manage this process, including the following data: isolation start date, isolation end date, the date housing administrators were notified, the date a student’s stay was completed, and the location of the stay. The data for on-campus isolation duration were obtained by querying the date housing administrators were notified and the date a student’s stay was completed at locations on campus. The complete data set is provided in the Supporting Information (Table SI-1). Ethical review for this study was obtained by the Institutional Review Board of Clemson University (IRB # 2021-043-02).
Wastewater Testing.
The Clemson University campus is serviced by its own wastewater treatment plant, with a rated capacity of 1.8 million gallons per day (6.8 × 106 L/d). The campus has dormitory space for ~7,000 students. The treatment plant handles that flow along with flow generated by students who live off-campus (~18,000), faculty (~1,400), and staff (~3,500) who visit the campus on a daily basis. Wastewater flow data for the period of this study are in the Supporting Information (Table SI-2). The average time for sewage to reach the treatment plant was assumed to be 1.1 h, based on the residence times cited in Kapo et al.30 for systems with flow rates of about 3.8 × 106 L per day.
Composite samples (500 mL collected in new, factory-clean Nalgene bottles) were collected two times per week (Monday→Tuesday and Wednesday→Thursday) and shipped overnight on ice to SiREM Laboratory (Knoxville, TN) for quantification of SARS-CoV-2 using RT-qPCR of the N and S protein genes and a target in the ORF 1ab region of the viral genome (the exact sequence of the targets is held as proprietary by Thermo Fisher). Sample quality and integrity was maintained through the use of location dedicated sampling equipment, single use factory clean items including gloves, sealable plastic bags, and new bottles opened only to receive the wastewater samples. Precautions against cross contamination between samples shipped to the lab included using watertight, virus-free bottles and packing each bottle in a separate sealable polyethylene bag so that a leaking or broken bottle would not contaminate other bottles in the same shipment. Upon receipt at the lab, each sample container (cooler), each plastic bag, and each bottle was decontaminated from outside to minimize cross contamination of the sample as well as incidental exposure to the virus by lab staff. SARS-CoV-2 contamination arising from laboratory procedures was evaluated with each RT-qPCR run by including control tests of PCR reagents and blank samples processed along with the actual samples. SARS-CoV-2 RNA was not detected in the blank control samples.
Positive controls included N-gene amplifications to confirm performance of the RT-qPCR reagents and to check the instrument calibration. Briefly, all RT-qPCR wastewater samples were spiked with bacteriophage MS2 RNA to confirm the RT-qPCR reaction ran without interference from wastewater components that may have carried through the virus recovery and RNA extraction process to affect the RT-qPCR reaction. Interferences were occasionally observed, meaning MS2 did not amplify as expected. Corrective actions such as rerunning the sample at a two-fold dilution often eliminated the interference such that MS2 amplified as expected. This corrective action doubled the detection limit but allowed the reporting of results in most cases where interferences were noted. The typical detection limit was 370 copies L−1. Additional details on the protocol are presented in McMahan et al.,16 including quality assurance/quality control.
The data reported in this study cover the Spring 2021 semester (January 1 to May 6, 2021). Individual testing was also required throughout the Summer and Fall 2021 semesters. However, the concentration of the virus in the campus wastewater was close to or below detection during the Summer of 2021 and only went above 2 × 104 copies per liter once during the Fall 2021 semester, so that data were not useful for the purposes of this study.
Starting in January, samples from the Clemson diagnostic lab were sequenced to measure the prevalence of different variants in the clinical tests.31 Beginning with the March 2, 2021 samples, SiREM estimated the relative quantity of VOCs using RT-qPCR targeting specific S gene mutation sites. The assay targeted the N501Y and A580D mutation sites. Alpha (B.1.1.7) has both mutations while Beta (B1.351) and Gamma (P.1) have the N501Y but not the A580D mutation. Thus, this assay cannot distinguish between Beta and Gamma strains. Results were reported as a percentage of the total virus (e.g., percent Alpha and percent combined Beta and Gamma). Details on the methodology for quantifying the variants are provided in the Supporting Information.
SEIR Modeling and Correlation to the Infected Population.
The wastewater RNA copy rates (i.e., 24-hour flow rate times the SARS-CoV-2 copies per liter in the composite samples) were aligned with the output of the SEIR model described by McMahan et al.16 Both the SEIR model and the copy rates are summarized in the Supporting Information. Of the various parameters used in the model, calibration for this study focused on the mean of the distribution (μ) that governs the log10 maximum RNA copies per gram of feces being shed per person (denoted φij in McMahan et al.16). To calibrate, the SEIR model was run at a grid of candidate log10 values ranging from 7.3 to 7.9 and each of these values was used to predict the number of infected individuals. These predictions were then aligned with the smoothed wastewater RNA levels (see below) and the sum of the squared errors between them was computed for every considered value of μ. The calibrated value of μ was chosen to be the value that minimized the sum of the squared errors.
Assuming each gene copy corresponds to a single virus with an average mass of 1 fg32 (i.e., 10−15 g), the copy rate can be converted to a mass rate. Mass rate is the terminology used in our prior description of the SEIR model, 16 while in this study we use the more direct descriptor of copy rate.
Measuring the RNA copy rate in wastewater is an inherently error-prone process that is highly variable. Thus, to reduce the variability in the raw wastewater measurements, we view the wastewater measurements collected during the Spring 2021 semester as a time-series ordered by date of collection and we fit a B-spline model to these data, treating time as the independent variable. In fitting this model, the degree of the splines was set to three and we explored the use of different interior knot sets. In particular, we considered a sequence of knot sets of increasing cardinalities with knot placement being governed by the default settings of the bs(.) function in the spline package in R. The final model, which was identified according to the Bayesian Information Criterion (BIC), made use of one interior knot.
The data received from positive COVID-19 tests and students entering isolation were adjusted to account for the fact that individuals become and test positive on different days. In particular, the vast majority of the population under study was being tested weekly. Thus, if an individual tested positive at a given time, the point of contracting COVID-19 for that individual would have to be on one of the days between the current and the previous testing period. To account for this, we used a multiple imputation approach, i.e., we assigned, at random, a day between the two testing periods for each individual testing positive as the day that they contracted SARS-CoV-2. We then compiled a daily time series of the number of infected individuals residing on campus. To reduce variability associated with random assignments, this process was repeated 500 times and the resulting 500 time series of case counts were subsequently averaged at a daily level yielding a daily adjusted estimate of the number of infected individuals (Supporting Information, Fig. SI-2).
RESULTS AND DISCUSSION
Regression models.
To examine the relationship between the RNA copy rates and the adjusted number of saliva-test-positive infected individuals in the University’s sewershed, we fit a linear regression model to these data (Table SI-2). The first three time points (January 6, 11, and 13) were removed because individual testing had not yet ramped up to cover all students (resulting in underestimates of the number of infected individuals in comparison to predictions from the wastewater RNA copy rates). Removing the first three time points resulted in a regression model with an R2 value of 0.63 (p = 5.54 × 10−7) for the observed RNA copy rates in the wastewater (Fig. 1A).
Figure 1.
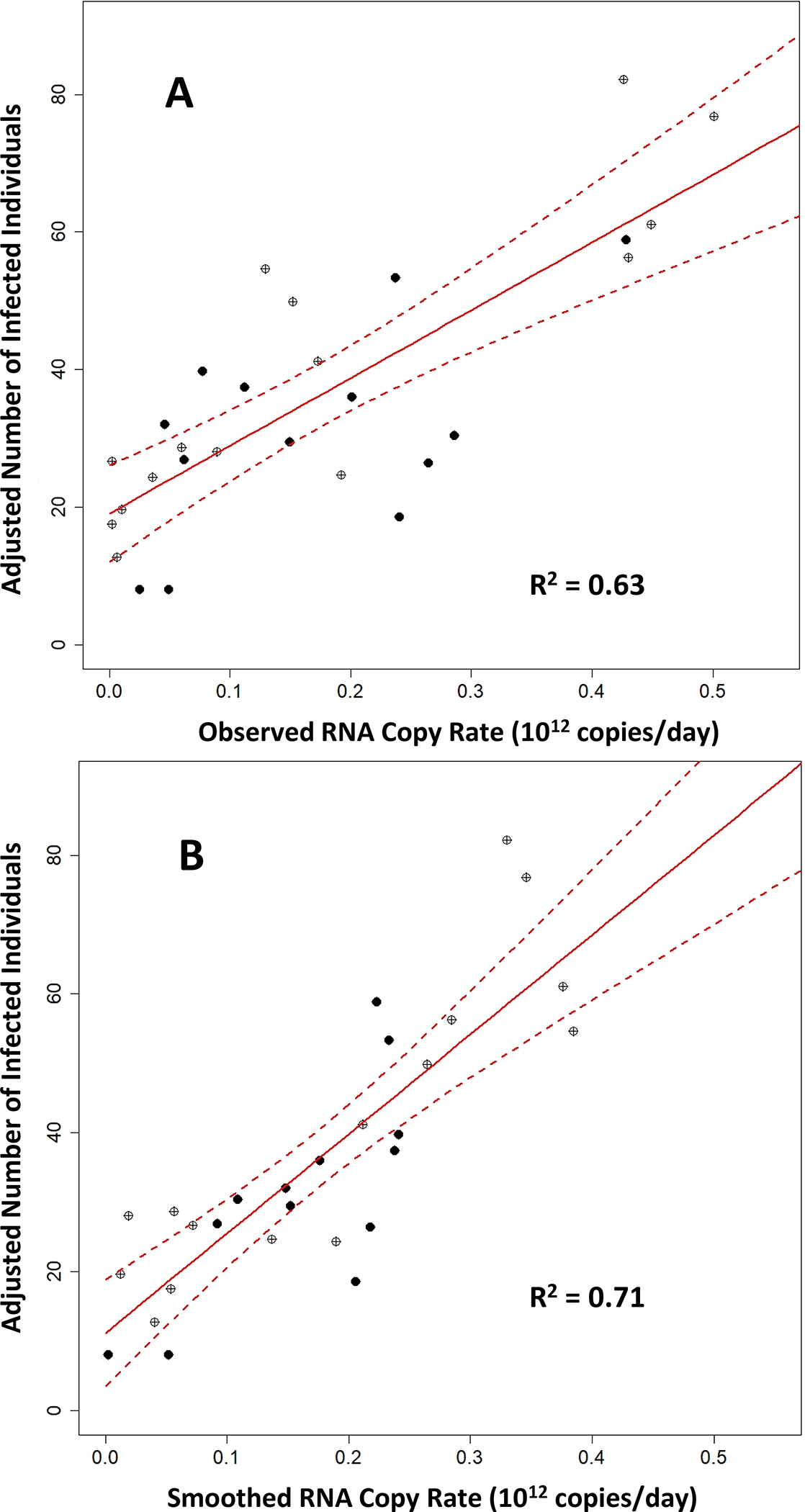
Regression model (A) for the adjusted number of saliva-test-positive infected individuals versus the observed RNA copy rates in Clemson University wastewater and (B) the smoothed wastewater data from Figure 2. Open circles are for sampling events when the Alpha and Beta/Gamma variants were not detected; filled circles represent samples when one of the variants comprised at least 25% of the total virus concentration. The red line is the best fit and the dashed lines are the 95% confidence intervals.
Results for smoothing the wastewater RNA copy rates using a B-spline model are shown in Figure 2. The variable nature of the observed RNA copy rates is apparent. For example, the sample collected on January 18 had the lowest RNA copy rate observed in January, yet the sampling period directly before (January 13) and after (January 20) saw two of the highest RNA copy rates collected throughout the Spring semester. This feature highlights the need to denoise the observed RNA copy rates to reveal a general trend via the proposed smoothing procedure. The maximum absolute difference between the observed and smoothed values was 0.25 × 1012 copies per day (January 18) followed by 0.20 × 1012 copies per day (April 22). The median absolute difference between the observed and smoothed values was 0.06 × 1012 copies per day. The correlation between the smoothed RNA copy rates and the adjusted number of saliva-test-positive infected individuals improved to 0.84 (R2 value of 0.71; p = 2.14 × 10−8) (Fig. 1B).
Figure 2.
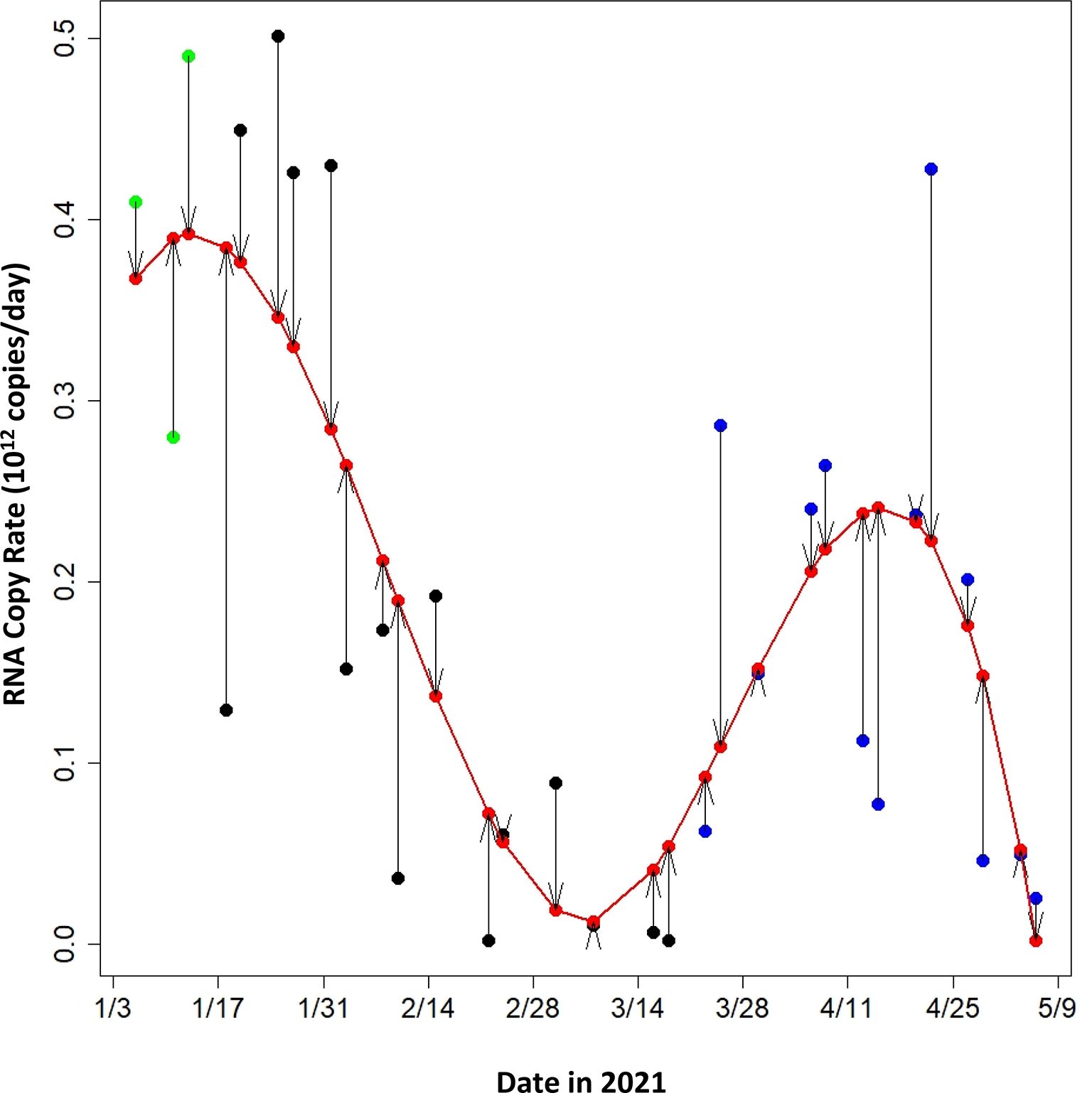
Observed RNA copy rates in wastewater measured at the Clemson University wastewater treatment plant during the 2021 Spring semester (black circles) and the smooth data based on a time series model (red line and red circles). The first three data point (green circles) were not used in the models (Figures 1 and 3) because of underreporting of infected individuals at the start of the semester. Arrows indicate the magnitude of change from the observed to the smoothed values used in the models.
The adjusted number of saliva-test-positive infected individuals contributing virus to the wastewater and the smoothed RNA copy rates for data from January 18, 2021, to May 6, 2021, were used to calibrate the SEIR model. The parameter in the SEIR model that had the largest impact on the calibration was the maximum shedding rate. Presumably most of the shedding that ends up in wastewater is attributable to feces, although other sources (e.g., respiratory fluids and urine) have been identified.33 The model presented in McMahan et al.16 used a log10 value of 7.6 genome copies per gram of feces (gc/g-feces), based on data from Wölfel et al.34 Schmidt et al.20 more recently reported a mean fecal shedding rate based on the N1 gene of 7.30 ± 0.67 log10 gc/g-feces. Using a grid of candidate values ranging from 7.3 to 7.9, the sum of the squared errors between the predicted and adjusted number of saliva-test-positive infected individuals was computed for every considered value of μ, which was minimized at a log10 value of 7.72. To further validate that the proposed calibration scheme rendered a reasonable value for μ, we predicted the number of infected individuals based on the smoothed wastewater RNA copy rates and then regressed these on the adjusted number of infected individuals. The R2 from this model fit is 0.71, with a slope of 0.87 (SE=0.11; p = 2.14 × 10−8) (Fig. 3). The 95% confidence interval for this slope (0.65 to 1.09) indicates that a 1:1 relationship between the adjusted number of saliva-test-positive infected individuals is statistically supported; a perfect relationship would have a slope of 1. After calibrating μ, similar calibration experiments were run on the other parameters in the SEIR model, including the viral load 20 days after the maximum, the time to reach the maximum viral load, the travel time of the sewage in the collection system, and the decay rate of the virus in the collection system. These results provided no evidence to suggest that the original settings for these parameters were incorrectly specified (Supporting Information, Table SI-3).
Figure 3.
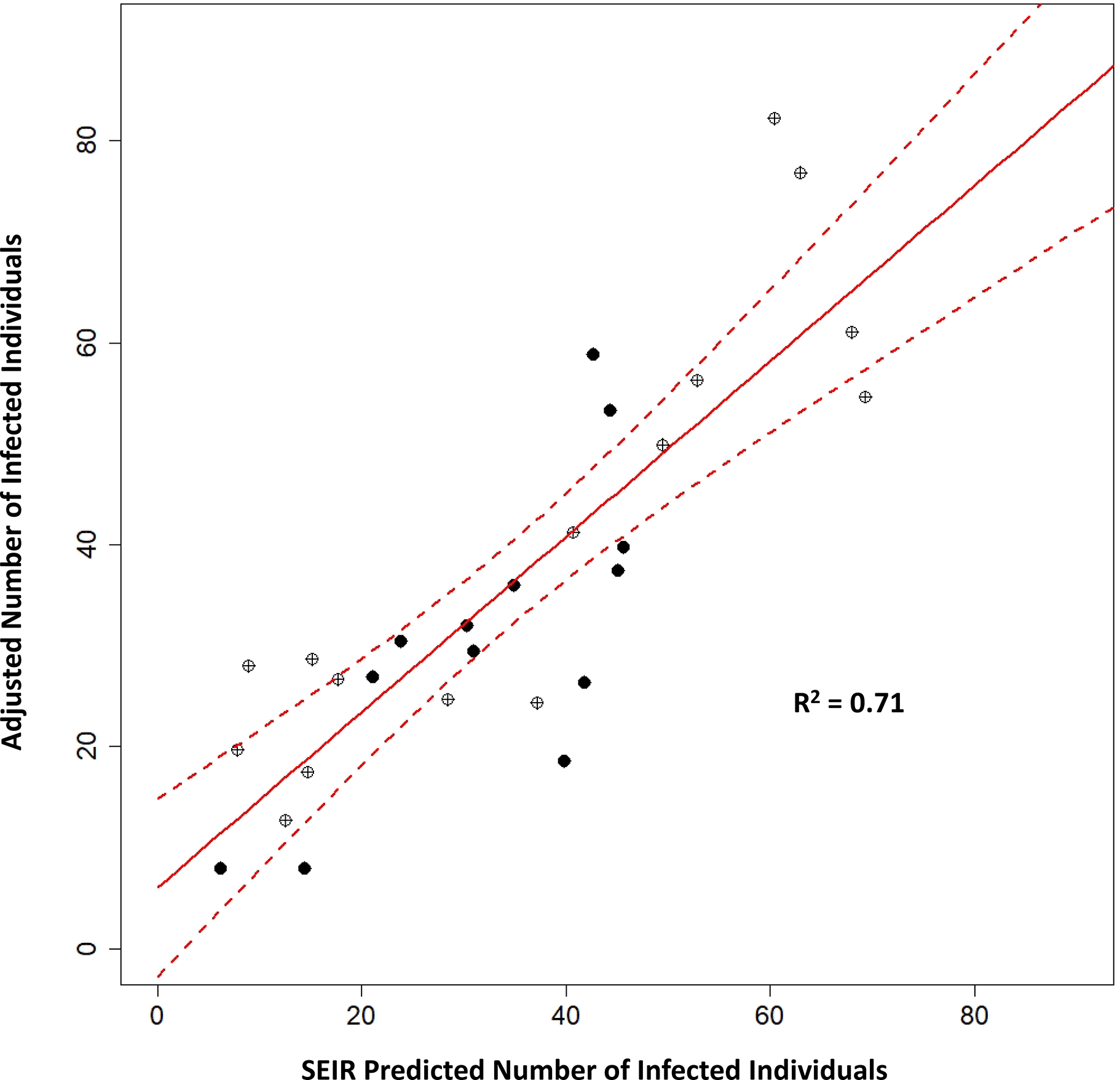
Regression model for the adjusted saliva-test-positive number of infected individuals versus the predicted number of infected individuals based on the SEIR model, using the smoothed wastewater RNA mass rate data. Open circles are for sampling events when the Alpha and Beta/Gamma variants were not detected; filled circles represent samples when one of the variants comprised at least 25% of the total virus concentration. The red line is the best fit and the dashed lines are the 95% confidence intervals.
Because the timing of when a student became infected and when they tested positive could be different by as many as six days, we estimated the number of infected individuals on campus stochastically by randomly assigning a day between the two testing periods for each individual testing positive as the day that they became infected with SARS-CoV-2. Other assignment schemes (i.e., including individuals becoming infected one to six days in advance of entering isolation) were investigated and led to no appreciable differences in the results (Supporting Information, Table SI-4).
The calibrated SEIR model provides further evidence that copy rates of SARS-CoV-2 RNA in wastewater can be used to make robust estimates of the number of infected individuals in a sewershed. By so doing, the value of WBE is elevated beyond providing information on presence or absence, or early warnings for an impending increase in reported cases. Having an estimate of the number of infected individuals will assist policymakers in deciding where limited supplies of clinical testing can be most effectively deployed. It also provides information on the magnitude of underreporting, which is often related to inadequate testing, testing hesitancy, etc.35 Because the SEIR model is publicly available, users have an opportunity to calibrate it for conditions unique to their sewershed. The goal of this work is to move wastewater surveillance out of the realm of qualitative use15 towards a more quantitative tool that can better inform public health policy.
The conditions for this study approached ideal, in the sense that the population of individuals shedding viral RNA into the sewershed and the wastewater characteristics (i.e., flow rate and RNA concentrations) were well defined. Students, faculty, and staff who lived off campus were prevented from accessing campus buildings unless they had a negative PCR test. For the most part, visitors were not allowed in campus buildings and there were no large sporting events during the times when wastewater samples were collected. In communities with more transient populations (e.g., toursits36), it will be more challenging to relate the wastewater data to the number of infected individuals residing in the sewershed. Nevertheless, it should be possible to capture additional complexities in the SEIR model. This validation and calibration study is inherently stochastic, due to the imputation steps used to estimate the number of infected individuals and the stochastic nature of the SEIR model used to predict the infected population via RNA copy rates. For purposes of reproducibility, the results presented herein were obtained after setting the seed (123456) of R’s random number generator.
COVID variants.
Wastewater samples were evaluated for VOCs from March 2, 2021, through May 6, 2021. The Alpha variant was first detected on March 23, 2021, and 11 more times through May 6, 2021. The percentage of the Alpha variant ranged from 13% to 100% on two occasions (4/6/21 and 4/13/21) and remained above 13% until the total virus concentration went below detection on May 11, 2021. The Beta/Gamma variants were detected on four occasions starting on April 22, 2021, ranging from 9% to 41%. As with the Alpha variant, the Beta/Gamma variants continued to be detected until the total virus concentration went below detection on May 11, 2021.
In the diagnostic lab, the first Alpha cases were detected on February 22 and March 1, 2021.31 Over the next three weeks, the number of Alpha cases increased rapidly. On the date the Alpha variant was detected in the wastewater, Alpha cases made up 45% of the diagnosed cases in the lab. The Alpha variant represented about half of all the diagnosed cases in March (49.5%) and April (50.2%). Only four cases of the Beta variant were detected: One case of the Beta variant was found on January 20, 2021, and three cases were found between March and April. The first cases of the Gamma variant were detected in the lab on March 20, 2021 and represented about 10.5% of the diagnosed cases in March and 31.5% of the cases in April. On the day that the variant was detected in wastewater, the Gamma variant made up 44% of the samples diagnosed that day in the lab.
In Figures 1 and 3, the presence of variants detected in this study (i.e., at least 25% of the total virus concentration) is indicated by the closed circles. The approximately equal distribution of these data above and below the best fit lines suggests that individuals infected with the Alpha and Beta/Gamma variants shed the virus at similar rates to the original virus. To explore this relationship more formally, we fit a regression model to the observed RNA copy rates entering time, a period indicator, and an interaction between time and the period indicator. To capture the potential differences across the two sampling periods defined by the presence/absence of the Alpha and Beta/Gamma variants, the period indicator was defined to take a value of 1 if the sample was collected on or after March 23, 2021, and 0 otherwise. Specifying the time indicator (and interaction term) in this fashion allowed the regression model to have different slopes and intercepts over the two sampling periods, while providing a way to assess whether this is necessary through examining the significance of the period indicator and the interaction term. If either of these were significant then there would be evidence that the shedding rate differed across the two sampling periods. In fitting this model, we found that both the indicator and the interaction term were insignificant with p-values of 0.73 and 0.31, respectively. Thus, there was no evidence that the shedding rates of these two variants differ from that of the original. Additional studies are needed to determine if this assessment holds for other communities. It is yet to be determined if a similar pattern emerges for other variants, including Omicron.
Limitations.
A primary limitation of this work is the uncertainty that exists in the available data. Namely, here we assume that through Clemson University’s mandatory testing program we have been able to identify all individuals who contracted COVID-19 and that infected individuals obeyed isolation protocols; i.e., faculty, staff, and off campus students remained off campus for 10 days following testing positive. Moreover, we assume that visitors to the university did not contribute to the viral load. Both of these assumptions are unverifiable. However, based on available records we estimate testing compliance for individuals residing on campus was greater than 90% for the majority of the study period. Thus, it is unlikely that the university’s testing program failed to detect a significant number of individuals who contracted COVID-19 and subsequently contributed to the sewershed. Moreover, given the various protocols and measures put in place (e.g., denying entry, via removing identification card access, into campus facilities due to failure to comply with testing protocols) we find it highly unlikely that non-residential individuals, who were infected with COVID-19, contributed significantly to the university’s sewershed. Violations of these assumptions would lead to the under estimation of the number of active COVID-19 cases by the proposed model.
Another potential limitation in future studies aimed at estimating COVID-19 prevalence using wastewater surveillance exists in the potential confounding that could be associated with a partially vaccinated population. That is, it is possible that vaccinated individuals who become infected would shed the virus at different rates than unvaccinated individuals. However, based on available vaccination records, we found that only 8 individuals residing on campus tested positive after being vaccinated during the study period. Moreover, there were only 35 instances of such an event in the entire population participating in Clemson University’s testing program. Thus, we do not believe that uncertainties due to vaccination pose a limitation to the current study.
We have not yet evaluated the generalizability of the proposed model to other sewersheds. The population under study here consists primarily of 18–21-year-old college students who contributed to a relatively closed sewershed. Moreover, the retention time in the sewershed is approximately 1.1 hours; considerably longer times are associated with larger collections systems.30 Due to potential differences in demographics and infrastructure, our calibrated model might not generalize to other settings. However, given the necessary data, the proposed model could be recalibrated to account for shedding differences across demographics and different infrastructure according to the strategy outlined above. This could be done with existing data as long as a measure of disease burden is available that could be used to unbiasedly estimate daily case counts which could then be aligned to wastewater assessments.
Finally, the sampling protocol used here requires the sampling device to take a sample of wastewater every 15 minutes during a 24 hour period. Given the dynamics of a sewershed, it is easy to see that measuring RNA copy rates in this manner can result in variability/measurement error, as indicated in Figure 2. These sources of variability could possibly be mitigated through alternative sampling strategies. For example, taking replicate samples during a 24 hour period, and taking 24 hour samples more frequently during the study period.
CONCLUSIONS
An SEIR model was calibrated using the copy rate of SARS-CoV-2 RNA in wastewater to predict the number of infected individuals in a sewershed. A strong correlation was observed between the copy rates and a defined population of infected individuals who were the only known contributors of the virus to the Clemson University sewershed during the Spring 2021 semester. Using data from weekly testing of all students, the number of infected individuals shedding the virus into the campus sewershed was adjusted to account for the fact that individuals become and test positive on different days. The correlation was further improved by smoothing the wastewater RNA copy rates using a B-spline model. The parameter in the SEIR model that had the largest impact on calibration was the maximum shedding rate, resulting in a value of 7.72 log10 gc/g-feces. Regressing the adjusted number of saliva-test-positive infected individuals on the SEIR model-predicted number of infected individuals based on the smoothed RNA copy rates yielded a slope of 0.87 (SE=0.11), indicating that a 1:1 relationship between the two was statistically supported. This outcome suggests that wastewater surveillance of SARS-CoV-2 can be applied more quantitatively than current policy indicates is appropriate. During the Spring semester, wastewater testing indicated that the Alpha and Beta/Gamma variants became the dominant form of the virus, which was consistent with the SARS-CoV-2 sequencing performed on the clinical diagnostic samples from the university lab. An analysis relating the adjusted number of saliva-test-positive infected individuals to RNA copy rates over the two time periods suggests that the shedding rates for these variants were not different from the shedding rate of the native virus.
Supplementary Material
SYNOPSIS.
The gene copy rate for SARS-CoV-2 virus measured in a campus sewershed effectively predicted the number of infected college students as measured by weekly SARS-CoV-2 saliva tests.
ACKNOWLEDGMENTS
Susan K. Peirce, Ph.D., provided editorial assistance.
FUNDING SOURCE
Clemson University provided funding for quantification of SARS-CoV-2 in the wastewater samples and the National Institutes for Health grant P20GM121342 provided funding for sequencing the clinical lab samples.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Internally developed applications for tracking COVID-19 cases; Complete data set used to determine the number of infected individuals on the Clemson University campus who were isolating in on-campus housing during the Spring 2021 semester; Compiled isolation and wastewater data used in the SEIR model; Protocol for measuring the Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1) variants; Adjusting the number of infected individuals; and Evaluation of assignment schemes; Results for calibrating the shedding parameters in the SEIR model; Evaluation of assignment schemes.
REFERENCES
- (1).Daughton CG Wastewater Surveillance for Population-Wide Covid-19: The Present and Future. Sci. Total Environ 2020, 736, 139631. 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hart OE; Halden RU Computational Analysis of SARS-CoV-2/COVID-19 Surveillance by Wastewater-Based Epidemiology Locally and Globally: Feasibility, Economy, Opportunities and Challenges. Sci. Total Environ 2020, 730, 138875. 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Olesen SW; Imakaev M; Duvallet C Making Waves: Defining the Lead Time of Wastewater-Based Epidemiology for COVID-19. Water Res 2021, 202, 117433. 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).D’Aoust PM; Graber TE; Mercier E; Montpetit D; Alexandrov I; Neault N; Baig AT; Mayne J; Zhang X; Alain T; et al. Catching a Resurgence: Increase in SARS-CoV-2 Viral RNA Identified in Wastewater 48 h before COVID-19 Clinical Tests and 96 h before Hospitalizations. Sci. Total Environ 2021, 770. 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Davó L; Seguí R; Botija P; Beltrán MJ; Albert E; Torres I; López-Fernández PÁ; Ortí R; Maestre JF; Sánchez G; et al. Early Detection of SARS-CoV-2 Infection Cases or Outbreaks at Nursing Homes by Targeted Wastewater Tracking. Clin. Microbiol. Infect 2021, 27 (7), 1061–1063. 10.1016/j.cmi.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Betancourt WQ; Schmitz BW; Innes GK; Prasek SM; Pogreba Brown KM; Stark ER; Foster AR; Sprissler RS; Harris DT; Sherchan SP; et al. COVID-19 Containment on a College Campus via Wastewater-Based Epidemiology, Targeted Clinical Testing and an Intervention. Sci. Total Environ 2021, 779, 146408. 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Acosta N; Bautista MA; Hollman J; McCalder J; Beaudet AB; Man L; Waddell BJ; Chen J; Li C; Kuzma D; et al. A Multicenter Study Investigating SARS-CoV-2 in Tertiary-Care Hospital Wastewater. Viral Burden Correlates with Increasing Hospitalized Cases as Well as Hospital-Associated Transmissions and Outbreaks. Water Res 2021, 201 (June), 117369. 10.1016/j.watres.2021.117369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Spurbeck RR; Minard-Smith A; Catlin L Feasibility of Neighborhood and Building Scale Wastewater-Based Genomic Epidemiology for Pathogen Surveillance. Sci. Total Environ 2021, 789, 147829. 10.1016/j.scitotenv.2021.147829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wu F; Xiao A; Zhang J; Moniz K; Endo N; Armas F; Bonneau R; Brown MA; Bushman M; Chai PR; et al. SARS-CoV-2 RNA Concentrations in Wastewater Foreshadow Dynamics and Clinical Presentation of New COVID-19 Cases. Sci. Total Environ 2022, 805 (August 2021), 150121. 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wright J; Driver EM; Bowes DA; Johnston B; Halden RU Comparison of High-Frequency in-Pipe SARS-CoV-2 Wastewater-Based Surveillance to Concurrent COVID-19 Random Clinical Testing on a Public U.S. University Campus. Sci. Total Environ 2022, 152877. 10.1016/j.scitotenv.2021.152877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gibas C; Lambirth K; Mittal N; Juel MAI; Barua VB; Roppolo Brazell L; Hinton K; Lontai J; Stark N; Young I; et al. Implementing Building-Level SARS-CoV-2 Wastewater Surveillance on a University Campus. Sci. Total Environ 2021, 782, 146749. 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ahmed W; Bivins A; Simpson SL; Bertsch PM; Ehret J; Hosegood I; Metcalfe SS; Smith WJM; Thomas K V; Tynan, J.; et al. Wastewater Surveillance Demonstrates High Predictive Value for COVID-19 Infection on Board Repatriation Flights to Australia. Environ. Int 2022, 158, 106938. 10.1016/j.envint.2021.106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lee BE; Sikora C; Faulder D; Risling E; Little LA; Qiu Y; Gao T; Bulat R; Craik S; Hrudey SE; et al. Early Warning and Rapid Public Health Response to Prevent COVID-19 Outbreaks in Long-Term Care Facilities (LTCF) by Monitoring SARS-CoV-2 RNA in LTCF Site-Specific Sewage Samples and Assessment of Antibodies Response in This Population: Prospective Study. BMJ Open 2021, 11 (8), 1–9. 10.1136/bmjopen-2021-052282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Reeves K; Liebig J; Feula A; Saldi T; Lasda E; Johnson W; Lilienfeld J; Maggi J; Pulley K; Wilkerson PJ; et al. High-Resolution within-Sewer SARS-CoV-2 Surveillance Facilitates Informed Intervention. Water Res 2021, 204 (January), 117613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Centers for Disease Control and Prevention. Developing a Wastewater Surveillance Sampling Strategy https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/developing-a-wastewater-surveillance-sampling-strategy.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fwastewater-surveillance%2Fdeveloping-a-wa (accessed Feb 7, 2022).
- (16).McMahan CS; Self S; Rennert L; Kalbaugh C; Kriebel D; Graves D; Colby C; Deaver JA; Popat SC; Karanfil T; et al. COVID-19 Wastewater Epidemiology: A Model to Estimate Infected Populations. Lancet Planet. Heal 2021, 5 (12), e874–e881. 10.1016/S2542-5196(21)00230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Feng S; Roguet A; McClary-Gutierrez JS; Newton RJ; Kloczko N; Meiman JG; McLellan SL Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities. ACS ES&T Water 2021, 1 (8), 1955–1965. [Google Scholar]
- (18).Rennert L; Mcmahan C Risk of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection in a University Student Population. Clin Infect Dis 2022, 74 (4), 719–722. 10.1093/cid/ciab454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Karthikeyan S; Nguyen A; McDonald D; Zong Y; Ronquillo N; Ren J; Zou J; Farmer S; Humphrey G; Henderson D; et al. Rapid, Large-Scale Wastewater Surveillance and Automated COVID-19 Cases on a University Campus. Am. Soc. Microbiol 2021, 6 (4), e00793–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schmitz BW; Innes GK; Prasek SM; Betancourt WQ; Stark ER; Foster AR; Abraham AG; Gerba CP; Pepper IL Enumerating Asymptomatic COVID-19 Cases and Estimating SARS-CoV-2 Fecal Shedding Rates via Wastewater-Based Epidemiology. Sci. Total Environ 2021, 801, 149794. 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fahrenfeld NL; Morales Medina WR; D’Elia S; Modica M; Ruiz A; McLane M Comparison of Residential Dormitory COVID-19 Monitoring via Weekly Saliva Testing and Sewage Monitoring. Sci. Total Environ 2021, No. xxxx, 151947. 10.1016/j.scitotenv.2021.151947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Scott LC; Aubee A; Babahaji L; Vigil K; Tims S; Aw TG Targeted Wastewater Surveillance of SARS-CoV-2 on a University Campus for COVID-19 Outbreak Detection and Mitigation. Environ. Res 2021, 200 (March), 111374. 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Layton B; Kaya D; Kelly C; Williamson K; Bachhuber S; Banwarth P; Bethel J; Carter K; Dalziel B; Dasenko M; et al. Wastewater-Based Epidemiology Predicts COVID-19 Community Prevalence. Prepr. (Version 1) available Res. Sq [ 10.21203/rs.3.rs-690031/v1] 2022. [DOI] [Google Scholar]
- (24).Ham RE; Smothers AR; King KL; Napalitano JM; Swann TJ; Pekarek LG; Blenner MA; Dean D Efficient SARS-CoV-2 Quantitative Reverse Transcriptase PCR Saliva Diagnostic Strategy Utilizing Open-Source Pipetting Robots. J. Vis. Exp 2022, 2 (180), 1–20. 10.3791/63395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Vogels CBF; Watkins AE; Harden CA; Brackney DE; Shafer J; Wang J; Caraballo C; Kalinich CC; Ott IM; Fauver JR; et al. SalivaDirect: A Simplified and Flexible Platform to Enhance SARS-CoV-2 Testing Capacity. Med 2021, 2 (3), 263–280.e6. 10.1016/J.MEDJ.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Plumb EV; Ham RE; Napolitano JM; King KL; Swann TJ; Kalbaugh CA; Rennert L; Dean D Implementation of a Rural Community Diagnostic Testing Strategy for SARS-CoV-2 in Upstate South Carolina. Front. Public Heal 2022, 0, 660. 10.3389/FPUBH.2022.858421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).King KL; Wilson S; Napolitano JM; Sell KJ; Rennert L; Parkinson CL; Dean D SARS-CoV-2 Variants of Concern Alpha and Delta Show Increased Viral Load in Saliva. medRxiv 2022. 10.1101/2022.02.10.22270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ham RE; Smothers AR; Che R; Sell KJ; Peng CA; Dean D Identifying SARS-CoV-2 Variants of Concern through Saliva-Based RT-QPCR by Targeting Recurrent Mutation Sites. medRxiv Prepr. Serv. Heal. Sci 2022. 10.1101/2022.03.02.22271785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Judson TJ; Odisho AY; Young JJ; Bigazzi O; Steuer D; Gonzales R; Neinstein AB Implementation of a Digital Chatbot to Screen Health System Employees during the COVID-19 Pandemic. J. Am. Med. Informatics Assoc 2020, 27 (9), 1450–1455. 10.1093/jamia/ocaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kapo KE; Paschka M; Vamshi R; Sebasky M; Mcdonough K Estimation of U. S. Sewer Residence Time Distributions for National-Scale Risk Assessment of down-the-Drain Chemicals. Sci. Total Environ 2017, 603–604, 445–452. 10.1016/j.scitotenv.2017.06.075. [DOI] [PubMed] [Google Scholar]
- (31).King KL; Wilson S; Napolitano JM; Sell KJ; Rennert L; Parkinson CJ; Dean D SARS-CoV-2 Variants of Concern Alpha and Delta Show Increased Viral Load in Saliva. medRxiv 2022. 10.1101/2022.02.10.22270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Bar-On YM; Flamholz A; Phillips R; Milo R SARS-CoV-2 (COVID-19) by the Numbers. eLife 2020, 9, e57309. https://elifesciences.org/articles/57309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Crank K; Chen W; Bivins A; Lowry S; Bibby K Contribution of SARS-CoV-2 RNA Shedding Routes to RNA Loads in Wastewater. Sci. Total Environ 2022, 806, 150376. 10.1016/j.scitotenv.2021.150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wölfel R; Corman VM; Guggemos W; Seilmaier M; Zange S; Müller MA; Niemeyer D; Jones TC; Vollmar P; Rothe C; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581 (7809), 465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- (35).Lau H; Khosrawipour T; Kocbach P; Ichii H; Bania J; Khosrawipour V Evaluating the Massive Underreporting and Undertesting of COVID-19 Cases in Multiple Global Epicenters. Pulmonology 2021, 27 (2), 110–115. 10.1016/j.pulmoe.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Weidhaas J; Aanderud ZT; Roper DK; VanDerslice J; Gaddis EB; Ostermiller J; Hoffman K; Jamal R; Heck P; Zhang Y; et al. Correlation of SARS-CoV-2 RNA in Wastewater with COVID-19 Disease Burden in Sewersheds. Sci. Total Environ 2021, 775, 145790. 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


