Abstract
Background
The ovipositors of some insects are external female genitalia, which have their primary function to deliver eggs. Drosophila suzukii and its sibling species D. subpulchrella are known to have acquired highly sclerotized and enlarged ovipositors upon their shifts in oviposition sites from rotting to ripening fruits. Inside the ovipositor plates, there are scale-like polarized protrusions termed “oviprovector scales” that are likely to aid the mechanical movement of the eggs. The size and spatial distribution of the scales need to be rearranged following the divergence of the ovipositors. In this study, we examined the features of the oviprovector scales in D. suzukii and its closely related species. We also investigated whether the scales are single-cell protrusions comprised of F-actin under the same conserved gene regulatory network as the well-characterized trichomes on the larval cuticular surface.
Results
The oviprovector scales of D. suzukii and D. subpulchrella were distinct in size and spatial arrangement compared to those of D. biarmipes and other closely related species. The scale numbers also varied greatly among these species. The comparisons of the size of the scales suggested a possibility that the apical cell area of the oviprovector has expanded upon the elongation of the ovipositor plates in these species. Our transcriptome analysis revealed that 43 out of the 46 genes known to be involved in the trichome gene regulatory network are expressed in the developing female genitalia of D. suzukii and D. subpulchrella. The presence of Shavenbaby (Svb) or svb was detected in the inner cavity of the developing ovipositors of D. melanogaster, D. suzukii, and D. subpulchrella. Also, shavenoid (sha) was expressed in the corresponding patterns in the developing ovipositors and showed differential expression levels between D. suzukii and D. subpulchrella at 48 h APF.
Conclusions
The oviprovector scales have divergent size and spatial arrangements among species. Therefore, these scales may represent a rapidly diversifying morphological trait of the female reproductive tract reflecting ecological contexts. Furthermore, our results showed that the gene regulatory network underlying trichome formation is also utilized to develop the rapidly evolving trichomes on the oviprovectors of these flies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12862-022-02046-1.
Keywords: Trichome, Gene regulatory network, Ovipositor, Oviprovector scale, shavenbaby, shavenoid
Background
The morphology of male external genitalia has been known to diverse rapidly among species and for this reason, it has been used for practical purposes as one of the taxonomic characters for species identification [1–3]. However, recent studies have revealed some cases of rapidly diversifying female genital traits [4–6]. In particular, the ovipositors of some insects, including Drosophilidae, are involved in genital coupling during copulation as external genitalia with the primary function to deliver eggs onto the food source for the emerging larvae [7, 8]. Thus, the genital structures involved in egg deposition are constantly under selective pressure to fulfill this function and can experience rapid morphological evolution upon occasional shifts in oviposition sites.
Such cases are well-documented in the species of Hawaiian Drosophila and Scaptomyza, which utilize a wide range of host plants in which their larvae grow [9, 10]. As predicted, a large diversity of the size and shape of the ovipositors have evolved in these species [11, 12]. In addition, another type of diverse trait in the internal structures of these ovipositors is noticeable. On the surface of the oviprovector (the membrane surrounding the vulva), there are scale-like polarized protrusions [12], termed as the “oviprovector scale” [13]. Although the functional role of these scales is not clear, it is likely to aid the transport and delivery of eggs onto the oviposition substrate [14]. Thus, the spatial arrangement of the scales should change following the morphological divergence of the ovipositors to support the mechanical movement of the eggs effectively. Therefore, these scales are likely to have gone through frequent rearrangement and may represent a rapidly diversifying morphological trait of the female genital organ.
The shape of the observed oviprovector scales resembles the well-studied trichomes on the larval cuticular surface [15–17]. Similar trichomes are found on various parts of the body cuticle and wings of Drosophilid flies; however, those on the oviprovector have been overlooked. The gene regulatory network underlying trichome formation is well-characterized by intensive studies on the larval denticles (reviewed in [18]). If the oviprovector scales are single-cell protrusions comprised of F-actin like other epidermal trichomes, it is intriguing to examine whether the same conserved gene regulatory network has been deployed.
Among species of non-Hawaiian Drosophila, D. suzukii and its sibling species D. subpulchrella are known to have acquired highly sclerotized and enlarged ovipositors with stiff bristles lined up on the edge upon their shifts in oviposition sites from rotting to ripening fruits [8, 19, 20]. Therefore, it would be insightful to depict how the characteristics of the oviprovector scales have changed in these drastically elongated ovipositors. Moreover, if the trichome-producing gene regulatory network is involved, there should be changes in the expression patterns of some genes in the network associated with the differential scale arrangements.
A recent study on the developing pupal ovipositors of D. suzukii and D. melanogaster showed that the timing of cell proliferation and the final cell number are similar between the two species [21]. By tracking the cell shape, the authors conclude that the expansion of the apical area of the cells is responsible for the development of the elongated ovipositor plates (hypogynial valves) in D. suzukii. Given these observations on the surface cell layer of the ovipositor plates, we can ask whether the cell size expansion has also taken place in the simultaneously elongated oviprovector in this species.
In this study, we first show the divergence of the size and distribution of the oviprovector scales in D. suzukii and D. subpulchrella and their closely related species, D. biarmipes and D. melanogaster. Next, we indicate that the oviprovector scales are single-cell protrusions derived from the cells that express the trichome-associated transcription factor Shavenbaby (Svb) and provide inferences about the cell size changes of the egg duct membrane. Finally, we report that an effector gene, shavenoid (sha), involved in the actin-bundling during the trichome development, is differentially expressed between the developing ovipositors of the two species, resembling the locations where the trichomes align in the later stages. From these results, we aim to emphasize the importance of the diversifying inner ovipositor structures in response to the rapid ovipositor shape adaptation and report for the first time that a trichome network gene is involved in the development of the divergent oviprovector scales.
Results
Divergent oviprovector scales
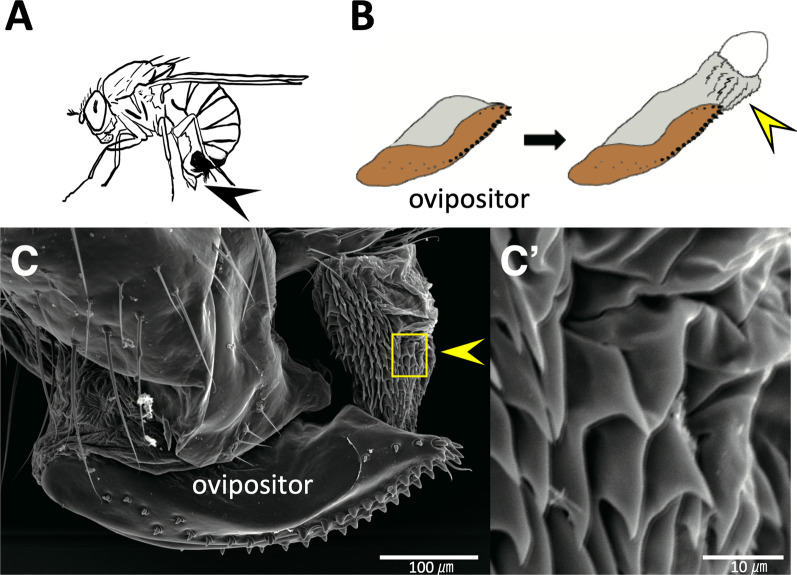
The oviprovector scales reside on the inner surface of the oviprovector (egg duct) in Drosophila species [13] and presumably aid egg deposition (Fig. 1A, B). The Scanning electron micrograph (SEM) image of the partially inverted oviprovector (inside exposed outward) of D. suzukii depicted the scales covering the inner surface of the duct (Fig. 1 C, C’) as in similar SEM images shown previously by Hamby et al. [22]. Since the scales are aligned parallel to the movement of the eggs through the duct and are likely to guide the eggs upon oviposition, they may have been rearranged during the rapid elongation of the ovipositors in D. suzukii and D. subpulchrella.
Fig. 1.
Oviprovector scales of Drosophila suzukii. A Adult female in an egg-laying posture. The ovipositor is indicated by an arrowhead. B Oviprovector scales on the reverted membrane (indicated by an arrowhead). C SEM image of oviprovector scales membrane (indicated by an arrowhead). C’ Enlarged image of the rectangular area in (C). Scale bar indicates 100 μm in (C) and 10 μm in (C’)
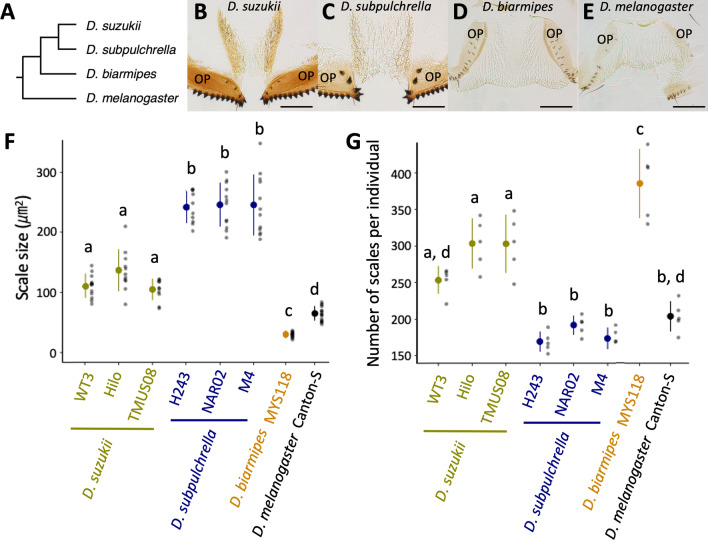
To elucidate the changes in the oviprovector scale arrangement, the ovipositors of the two species and their closely related species, D. biarmipes, and an outgroup, D. melanogaster, were dissected and mounted to visualize the scales (Fig. 2A–E, Additional file 1: Fig. S1). The degree of sclerotization and size of the scales were variable among these species. The oviprovector scales in D. suzukii and D. subpulchrella were more sclerotized than those in D. melanogaster and D. biarmipes (Fig. 2B–E). The size of the scales measured by lateral area of the protrusion was ~ 1.6–2.1 times larger in D. suzukii compared to D. melanogaster (Fig. 2F). The size was ~ 0.47 times smaller in D. biarmipes than in D. melanogaster. Between D. suzukii and D. subpulchrella, the scale size was ~ 1.8–2.3 times larger in D. subpulchrella than in D. suzukii. These measurements and the scale images of three other phylogenetically related species, D. mimetica, D. lutescens, and D. simulans (Additional file 1: Fig. S2), indicate that the drastic enlargement of the scale is likely to have taken place in the common ancestor of D. suzukii and D. subpulchrella after the split from D. biarmipes along with the elongation of the ovipositor plates. Further divergence in size occurred between the lineages leading to D. suzukii and D. subpulchrella.
Fig. 2.
Diversification of oviprovector scales in Drosophila suzukii and its closely related species. A Phylogenetic relationship of species used in this study. Tree topology is based on Suvorov et al. [57] and Finet et al. [58]. B–E Images of oviprovector scales from dissected ovipositors in different species. Ovipositor plates are indicated as OP. Scale bar indicates 100 μm. F Scale size measured from ten individuals from each strain. G Number of scales per individual counted in five individuals from each strain. Different letters indicate significant differences between strains after Tukey’s multiple comparisons (P < 0.05). Mean and standard deviation are shown as dot and error bar, respectively
The distribution of the scales on the oviprovector inner surface was also variable. The locations of the scales were confined to the narrow area at the distal opening of the oviprovector in D. melanogaster (Fig. 2E). In contrast, the scales were distributed in a broader area toward the proximal end in the other three species (Fig. 2B–D). Comparing D. suzukii and D. subpulchrella, the former showed a polarized distribution of the scales on the lateral surface of the duct (Fig. 2B), which contrasts with the uniform distribution in the latter species (Fig. 2C). The observation of the oviprovector scales in three additional strains from D. suzukii and D. subpulchrella indicated that these morphological characteristics are fixed within these species (Additional file 1: Fig. S1). The number of scales was also different among the species, largely reflecting their distribution on the oviprovector membrane (Fig. 2G, Additional file 1: Fig. S2). These observations suggest that the oviprovector scale arrangement is a rapidly diversifying trait of the female genitalia in Drosophila.
Oviprovector scales are trichomes protruded from single cells expressingsvb
The shape of the oviprovector scales resembles that of the trichomes on the larval cuticle, which are actin-rich apical extensions from the epidermal cells. To confirm that the oviprovector scales are single-cell projections like other trichomes, the immunostaining of E-cadherin combined with the rhodamine phalloidin (actin) staining was conducted on the developing ovipositor of D. melanogaster at 44 and 48 h APF. The confocal images depicted that actin projections are indeed derived from single cells (Fig. 3A, B, B’). The staining using the ovipositors of D. suzukii and D. subpulchrella at 45 h APF also showed the same structure (Fig. 3E, F). Therefore, the numbers and sizes of the oviprovector scales can be used to infer those features of trichome-forming epidermal cells of the oviprovector membrane. Furthermore, the intensity of phalloidin in the ovipositor at 44 h APF was lower than that at 48 h APF in D. melanogaster (Fig. 3A, B). This result indicated that the actin accumulates between these time points and forms scales on the oviprovector membrane.
Fig. 3.
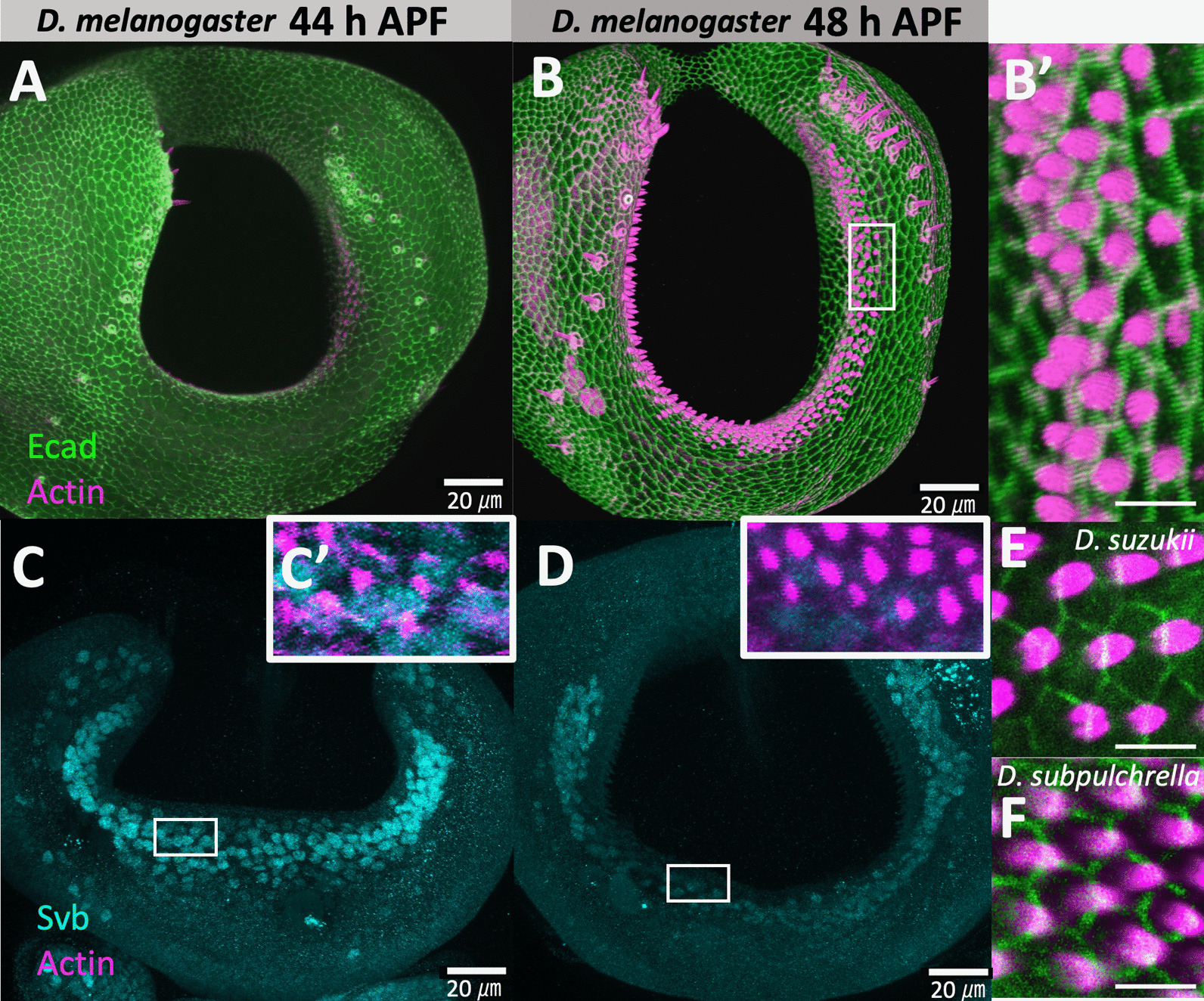
Confocal images of developing ovipositors in D. melanogaster (y, w), D. suzukii (TMUS08), and D. subpulchrella (H243). A Configuration of E-cadherin (green) and F-actin/phalloidin (magenta) at 44 h APF in D. melanogaster. B Configuration of E-cadherin (green) and F-actin/phalloidin (magenta) at 48 h APF in D. melanogaster. B’ Enlarged image of the rectangular area in (B). C Immunostaining of Svb (light blue) at 44 h APF in D. melanogaster. C’ Enlarged image of the rectangular area in (C) with F-actin/phalloidin (magenta). D Immunostaining of Svb (light blue) at 48 h APF in D. melanogaster. D’ Enlarged image of the rectangular area in D with F-actin/phalloidin (magenta). E Configuration of E-cadherin (green) and F-actin/phalloidin (magenta) in D. suzukii at 45 h APF. F Configuration of E-cadherin (green) and F-actin/phalloidin (magenta) in D. subpulchrella at 45 h APF. Scale bars indicate 20 μm in (A–D) and 5 μm in (B’), (E), and (F)
The gene regulatory network that controls trichome formation is well-characterized in D. melanogaster, and svb is known to be the master control gene regulating the network [17, 18, 23]. The antibody staining of Svb at 44 and 48 h APF in D. melanogaster revealed that this gene product is present in the cells on the inner surface of the developing ovipositor where actin bundles are forming (Fig. 3 C, C’, D, D’). This result implied that the trichome gene network is activated in those scale-producing cells.
Transcriptomic analysis of 48 h APF female genitalia fromD. suzukiiandD. subpulchrella
Motivated by the presence of Svb in the cells on the inner surface of the developing ovipositor in D. melanogaster, we next asked (1) if the known genes in the network are expressed in the developing female genitalia of D. suzukii and D. subpulchrella, and (2) if any of the trichome-related genes contribute to the morphological differences of the oviprovector scales between these species. The ovipositor morphogenesis takes place at early pupal stages in Drosophila at around 30–54 h APF [21]. A time point was taken at 48 h APF when the projection of the ovipositor plates has proceeded to the mid-step. The extracted RNA samples from the posterior tip of the abdomen (including the entire developing ovipositor) cleanly sliced off from the flash-frozen pupae at 48 h APF were subjected to RNA-seq analysis.
The transcripts of 8666 genes with more than 1 transcript per million (TPM) in were detected by mapping the reads to the reference coding sequences (CDS) generated by reciprocal re-annotation (Additional file 2: Tables S1, S2). Among them, 1573 genes were differentially expressed between D. suzukii and D. subpulchrella (false discovery rate (FDR) < 0.01) (Additional file 2: Table S2). Among the homologs of 50 genes that are known to be involved in trichome pattern formation in D. melanogaster [18], 46 genes were included in the reference CDS, and 43 genes showed detectable gene expression levels (> 1 TPM in all three replicates) in either of the two species, suggesting the involvement of the genes in the trichome regulatory network. Eleven genes showed differential expression at FDR < 0.01 (Table 1). Five genes, Cuticular protein 11 A (Cpr11A), krotzkopf verkehrt (kkv), CG10175, CG15005 and Osiris 24 (Osi24), showed higher expression in D. suzukii, and six genes, shavenoid (sha), morpheyus (mey), nyobe (nyo), neyo (neo), CG31559 and Reduction in Cnn dots 6 (Rcd6), showed higher expression in D. subpulchrella (at FDR < 0.01). The master control gene of the trichome regulatory network, shavenbaby (svb), did not show differential expression between these species at this stage.
Table 1.
Expression differences of trichome related genes between D. suzukii and D. subpulchrella
| Gene name | log2CPM | Log2Fold-change (D. suzukii/D. subpulchrella) |
FDR | Function in trichome formation |
|---|---|---|---|---|
| wg | 5.8 | − 0.51 | 5.2 × 10− 2 | Upstream genes of svb |
| abd-A | 7.2 | − 0.20 | 3.2 × 10− 1 | Upstream genes of svb |
| Ubx | 2.3 | − 0.61 | 3.8 × 10− 1 | Upstream genes of svb |
| hh | 4.9 | − 0.28 | 3.6 × 10− 1 | Upstream genes of svb |
| lin | 5.1 | − 0.14 | 6.2 × 10− 1 | Upstream genes of svb |
| Ser | 7.8 | − 0.43 | 2.0 × 10− 2 | Upstream genes of svb |
| rho | 4.0 | 0.24 | 5.8 × 10− 1 | Upstream genes of svb |
| aos | 4.0 | − 0.49 | 1.2 × 10− 1 | Upstream genes of svb |
| SoxN | Low expressiona | – | – | Upstream genes of svb |
| D | 3.2 | − 0.37 | 4.3 × 10− 1 | Upstream genes of svb |
| tal | n.a.b | – | – | Upstream genes of svb |
| ovo/svb | 5.9 | − 0.43 | 3.0 × 10− 1 | Master control gene |
| sha | 8.5 | − 0.78 | 7.6 × 10− 5 | Actin bundling |
| sn | 8.8 | − 0.02 | 9.2 × 10− 1 | Actin bundling |
| f | 9.6 | 0.02 | 9.4 × 10− 1 | Actin bundling |
| wasp | 5.8 | 0.23 | 3.2 × 10− 1 | Actin bundling |
| mwh | 8.8 | − 0.51 | 6.7 × 10− 2 | Actin bundling |
| Actn | 10.0 | 0.17 | 4.0 × 10− 1 | Actin bundling |
| y | 4.4 | − 0.76 | 2.9 × 10− 2 | Cuticle |
| ChLD3 | 5.5 | 0.43 | 2.0 × 10− 1 | Cuticle |
| Cda5 | 6.0 | 0.40 | 2.1 × 10− 1 | Cuticle |
| Cpr11A | 3.0 | 1.34 | 3.1 × 10− 4 | Cuticle |
| kkv | 9.7 | 0.92 | 2.1 × 10− 4 | Cuticle |
| Peritrophin-A | Low expressiona | – | – | Cuticle |
| m | 10.6 | − 0.14 | 6.5 × 10− 1 | Membrane matrix |
| dyl | 9.8 | 0.74 | 9.9 × 10− 2 | Membrane matrix |
| mey | 11.7 | − 0.54 | 6.3 × 10− 3 | Membrane matrix |
| nyo | 10.3 | − 0.63 | 2.0 × 10− 3 | Membrane matrix |
| tyn | 12.7 | − 0.13 | 6.4 × 10− 1 | Membrane matrix |
| neo | 12.2 | − 0.53 | 7.7 × 10− 3 | Membrane matrix |
| zye | 5.4 | 0.20 | 5.9 × 10− 1 | Membrane matrix |
| cyr | 8.6 | − 0.07 | 8.7 × 10− 1 | Membrane matrix |
| CG16798 | 7.9 | − 0.18 | 5.5 × 10− 1 | Membrane matrix |
| CG12814 | 7.2 | − 0.40 | 5.7 × 10− 2 | Membrane matrix |
| CG4702 | 11.0 | 0.14 | 6.6 × 10− 1 | Unknown |
| CG8303 | 5.7 | − 0.04 | 9.2 × 10− 1 | Unknown |
| CG10175 | 3.2 | 1.95 | 6.2 × 10− 6 | Unknown |
| CG14395 | 9.2 | − 0.65 | 4.8 × 10− 2 | Unknown |
| CG15005 | 4.9 | 1.61 | 4.7 × 10− 6 | Unknown |
| CG15022 | Low expressiona | – | – | Unknown |
| CG31559 | 7.2 | − 0.99 | 8.0 × 10− 10 | Unknown |
| CG32694 | 3.6 | − 0.10 | 8.8 × 10− 1 | Unknown |
| CG45064 | n.a.b | – | – | Unknown |
| Sox21b | n.a.b | – | – | Unknown |
| Osi24 | 6.5 | 0.98 | 3.7 × 10− 3 | Unknown |
| Spn88Ea | 7.4 | − 0.14 | 5.4 × 10− 1 | Unknown |
| Tg | n.a.b | – | – | Unknown |
| Rcd6 | 4.0 | − 0.78 | 3.3 × 10− 3 | Unknown |
| PH4αEFB | 6.0 | − 0.01 | 9.7 × 10− 1 | Unknown |
| ImpE1 | 12.5 | 0.26 | 2.7 × 10− 1 | Unknown |
Genes related to trichome formation are from Arif et al. [18]
Underline indicates FDR < 0.01
aTPM < 1 in both species
bCDS was not available in our analysis
shaandsvbare expressed in the inner cavity of the developing ovipositor
The trichome network genes were detected by the RNA-seq analysis. To confirm that the network pathway is activated in the developing oviprovector of D. suzukii and D. subpulchrella, in situ hybridizations of an effector gene, sha, and the master control gene, svb, were conducted to examine its expression at the inner cells of the developing ovipositors. sha, has been shown to be a direct target of Svb and functions in bundling actin fibers on the apical surface of the cells [16, 24]. The gene also showed a differential expression level between the two species (Table 1).
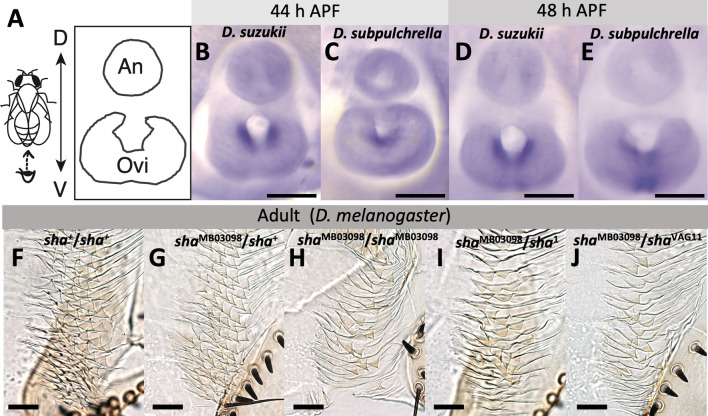
At 44 and 48 h APF, when the duct inside the ovipositor plates is still not closed at the distal end, a clear signal was detected at the inner cavity of the ovipositors in both D. suzukii and D. subpulchrella (Fig. 4B–E). While the expression domain was split into two lateral areas on the inner surface of the cavity in D. suzukii (Fig. 4B, D), only one domain connected at the ventral side of the cavity was observed in D. subpulchrella (Fig. 4C, E). The pattern resembled that of the adult oviprovector scale distributions of the two species (Fig. 2B, C), indicating that this gene in the trichome regulatory network, is expressed in the cells where trichomes develop at a later stage in these species.
Fig. 4.
sha expression in the developing ovipositor and the oviprovector scale defects in sha mutants. A Schematic image of the developing female terminalia. Abbreviations indicate analia (An) and ovipositor (Ovi). B in situ hybridization of sha at 44 h APF in D. suzukii. C in situ hybridization of sha at 44 h APF in D. subpulchrella. D in situ hybridization of sha at 48 h APF in D. suzukii. E In situ hybridization of sha at 48 h APF in D. subpulchrella. F Oviprovector scales of Canton-S (sha+/sha+). G Oviprovector scales of shaMB03098/sha+. H Oviprovector scales of shaMB03098/shaMB03098. I Oviprovector scales of shaMB03098/sha1. I Oviprovector scales of shaMB03098/shaVAG11. Scale bars indicate 100 μm in (B–E) and 20 μm in (F–J)
In contrast, in situ hybridization on the developing ovipositors of D. suzukii and D. subpulchrella at 44 and 48 h APF svb was expressed throughout the inner cavity of the ovipositors and did not show notable differences in spatial pattern between the two species (Additional file 1: Fig. S3B–E). The discordance between svb and sha expression domains in the developing ovipositors indicates that the functional changes between these species are likely to be found outside of svb transcriptional regulation.
Oviprovector scales ofD. melanogaster shamutants
To further investigate whether sha is involved in the development of trichomes on the oviprovector, mutant phenotypes were observed in D. melanogaster. In the wild type D. melanogaster, the oviprovector scales (trichomes) are triangular with hook-like tips and highly arrayed configuration (Figs. 2E and 4F, Additional file 1: Fig. S4A). The scales of a homozygous sha mutant, shaMB03098, showed a malformation lacking most of the spikes (Fig. 4H, Additional file 1: Fig. S4B). While this malformation phenotype was not observed in the shaMB03098/ sha+ heterozygote (Fig. 4G), similar defects were observed in two other sha mutant alleles that were tested in heterozygotes, sha1/shaMB03098 and shaVAG11/shaMB03098 (Fig. 4I, J). Mutants of the genes involved in the apical extracellular matrix formation, trinity (tyn) and miniature (m) [17, 25], showed a similarly arrayed but slightly smaller trichomes compared to the wild type (Additional file 1: Fig. S4C, D). These results corroborate that the gene regulatory network underlying the development of larval denticles is likely to be also deployed in the developmental system of oviprovector scales.
Discussion
The oviprovector scales in D. suzukii and D. subpulchrella were distinct in size and distribution from those of D. biarmipes and other closely related species (Fig. 2, Additional file 1: Fig. S2) as anticipated from their uniquely enlarged ovipositor plates [8, 20, 21]. The number of the scales also varied among the species. D. suzukii and D. subpulchrella have shifted their oviposition sites from rotting to ripening fruits, and the shift has promoted the evolution in various traits including the ovipositor morphology as well as the mechanosensory and chemosensory perception [8, 20, 21, 26–30]. The ovipositor is an essential device for the flies to deposit eggs to the appropriate substrate; therefore, the changes in ovipositor shape should accompany an adjustment of oviprovector morphology to support egg-laying effectively. The exact function has not been explored in detail. However, the arrayed trichomes tilting towards the distal direction might effectively reduce the friction between the surfaces of the egg and the duct when the egg is moving toward the distal opening. Alternatively, they might be functioning as small hooks to grab and push the egg toward the vulva opening by reversing the membrane [14, 22]. Since similar oviprovector scales are present in other insect species (e.g., [14, 31–33]), it is possible that they share a common functional role in oviposition.
The ovipositor plates of D. suzukii are less curved and become narrower at the distal tip than those of D. subpulchrella [8, 20]. The change in shape could have affected the mechanics of egg deposition and provoked a change in the number and distribution of scales on the oviprovector. We speculate that applying forces from the lateral direction by a pair of plates to push the eggs along the longitudinal direction of the ovipositor required the alignment of scales on the lateral surface of the oviprovector in D. suzukii. Whereas due to the curved ovipositor plates of D. subpulchrella, the egg might be slightly pushed towards the ventral side of the ovipositor. Thus, the scales on the ventral surface of the oviprovector might effectively reduce the surface friction or grab the egg by the spiny tips. These speculations require rigorous validation by a detailed biophysical analysis. Nevertheless, such fine-tuning of the egg transporting mechanics upon the changes in external morphology of the ovipositors might have enforced frequent rearrangements of the oviprovector scales representing a rapidly diversifying morphological trait of the female reproductive tract.
The gene regulatory network underlying trichome development is shared in many body regions, such as in the denticle belts of the larval cuticle and on the surface of the wings and adult cuticle (reviewed in [18, 34]). We have shown by the immunostaining of Svb, in situ hybridizations of sha and svb, and the RNA-seq analysis using the developing female genitalia that the svb-regulated pathway is likely to be involved in the formation of the trichomes on the oviprovector membrane. The expression patterns of sha at 44 and 48 h APF in D. suzukii and D. subpulchrella resembled the corresponding patterns of the trichome distribution on the adult oviprovectors of these species (Fig. 4A–E). It is yet to be pursued whether the causal mutations of the interspecific differences in the ovipositor scale characteristics reside in the cis-regulatory regions of the effector gene sha or in the upstream genes. Our in situ hybridization results so far did not indicate that a change in svb expression pattern is responsible for the interspecific differences in the expression pattern of sha in the developing ovipositors of the two species.
Some components of the trichome gene regulatory network differ between the larval and leg trichomes [35]. In contrary to various factors regulating svb in the embryonic and larval cuticle, the development of trichomes is suppressed by microRNA-92a in a cuticular patch, “naked valley”, in the leg (T2 femurs) by regulating the translation of sha [36]. In our analysis, it was intriguing to see that svb did not show differential expression between the two species despite the significant expression level differences (1.7-fold lower in D. suzukii) of sha in the developing female genitalia (Table 1). Thus, a possibility that sha is directly regulated by microRNAs independently from Svb remains to be investigated. mey, nyo, and neo also showed significantly lower expression (1.4–1.5-fold lower) in D. suzukii than in D. subpulchrella (Table 1). These genes are suggested to act together to form apical extracellular matrix during the development of the larval denticles [25], therefore, they may be differentially regulated simultaneously in the developing oviprovector scales between the two species. Nevertheless, it should be noted that per nucleus transcript abundance cannot be accurately compared in our data because the number of trichome-producing cells in the oviprovector are different between the two species (Fig. 2G). Further investigation of the detailed expression domains of these genes may reveal some differences in the gene regulatory network components between the oviprovector and other parts of the fly body.
During the ovipositor morphogenesis at the pupal stage, Green et al. [21] have analyzed the size and number of the external cell layer of the ovipositor plate. They concluded that an accelerated cell size expansion instead of an enhanced cell division causes the enlargement of the ovipositors in D. suzukii. Since the trichomes on the oviprovector were shown to be single-cell extensions as in the cuticle trichomes (Fig. 3), the differences in the size of the trichomes may serve as an indirect measurement to infer differences in apical cell area between species. The lateral areas of the scales in D. suzukii and D. subpulchrella were roughly two to five times larger than that of D. biarmipes. These size differences imply a possibility that the expansion of apical cell areas in D. suzukii and D. subpulchrella took place at the inner membrane of the oviprovector, at least in the scale producing cells.
It is also interesting to note that the arrayed alignment and the direction of the actin bundles in the developing ovipositors imply planar cell polarity. Mechanisms underlying such polarity are well-studied in other actin-based external bristles and hairs on the adult cuticle and wing (reviewed in [37, 38]) as well as in the multiciliated cells of the mammalian oviduct [39–41]. Further analyses of the changes in cell size and polarity during morphogenesis may uncover the details of the cell dynamics underlying differential degrees of the oviprovector rearrangement upon the ecological niche exploitation in these species.
Conclusions
The oviprovector scales inside the recently enlarged ovipositors of D. suzukii and D. subpulchrella were distinct in size and distribution pattern compared to those in a closely related species. Although indirectly, the size of the scales suggested the possibility that the apical cell area of the oviprovector has expanded upon the elongation of the ovipositor plates in the two species. The number of the scales also varied greatly among species. Our transcriptome analysis revealed that most of the known genes in the trichome gene regulatory network underlying larval denticle formation are expressed in the developing female genitalia of D. suzukii and D. subpulchrella. We confirmed the presence of Svb in the inner cavity of the developing ovipositors of D. melanogaster and the sha expression in the similar regions of D. suzukii and D. subpulchrella, suggesting that the network is also utilized to develop the ovipositor trichomes. Taken together, in parallel to the well-documented cases of rapidly evolving genital structures in Drosophila males (e.g., [42–45]), our study opens an opportunity to explore the evolution of a rapidly diversifying morphological trait of the female genitalia. Our study also highlights the gene regulatory network of a functionally distinct and divergent trichome system associated with ecological niche exploitation.
Methods
Fly strains
Wild-derived strains of D. suzukii (TMUS08, TMUS09, TMUS18, WT3, and Hilo), D. subpulchrella (H243, M4, NAR02, and NAR07), D. mimetica (CNX324), and D. lutescens (MYK13) were maintained at 20 °C unless stated otherwise. A wild-derived strain of D. biarmipes (MYS118) was maintained at 25 °C. TMUS08, TMUS09, and TMUS18 were collected in Hachioji, Japan, in 2015. Hilo was collected in Hilo, Island of Hawai‘i, USA, in 2017. WT3 strain was obtained from the Drosophila Species Stock Center. H243 strain was collected in Hiratsuka, Japan, in 1979. M4 was collected in Matsumoto, Japan, in 1982. NAR02 and NAR07 were collected in Narusawa-mura, Japan, in 2016. MYS118 was collected in Mysore, India, in 1981. CNX324 was collected in Chiangmai, Thailand, in 1981. MYK13 was collected in Miyake-jima, Japan, in 1978. D. simulans w501 was maintained at 20 °C. D. melanogaster Canton-S, y1 w1, and other mutant strains were maintained at 25 °C. y1w1 (#1495), sha1 (#107623), and m1 (#105713) were obtained from KYOTO Stock Center (DGRC). shaVAG11 (#32097), shaMB03098 (#24445), and tynEP1421 (#10277) were obtained from Bloomington Drosophila Stock Center. All strains were kept under a 12 h light:12 h dark light cycle.
Observation of ovipositor scale and morphological measurement
Adult female flies (7–14 days after eclosion) were collected and stored in 70% ethanol before dissection. Dissected ovipositors were mounted in 50% Hoyer’s medium and placed at 68 °C overnight. Images were captured using an Olympus IX71 inverted microscope equipped with a DP73 camera (Olympus) at × 200 and × 640 magnifications for the entire distribution and single scale measurement, respectively.
SEMs were taken by a JEOL JSM-6510 scanning electron microscope after being coated with gold using a Hitachi E101 ion sputter (Hitachi Ltd., Tokyo, Japan). For this, the oviprovector was everted by gently pushing the abdomen of virgin female samples with fine forceps in absolute ethanol. The ethanol was then substituted with t-butanol, and the specimens were sublimation dried.
The number of ovipositor scales was manually counted under Olympus IX71 inverted microscope. To measure the size, a single scale on the lateral side of the oviprovector membrane was chosen randomly from each dissected image. The size was measured using Fiji version 2.3 [46] and the statistical tests were conducted by using R version 4.1 [47].
RNA extraction and sequencing
Three independent biological replicates of RNA-seq libraries for TMUS08 and H243 were generated from abdominal tip tissue dissected from 20 female pupae at 48 ± 1 h after puparium formation (h APF). Female files were collected at the white pupal stage by sorting gonad size and placed in a humid chamber at 25 °C. Staged pupae were flash-frozen by placing them on a cooled metal plate with a cake of dry ice. The posterior tip of the abdomen, including the entire developing ovipositor, was cut at the A6–7 tergite by a blade on the plate. Total RNA was extracted using TRIzol Plus RNA Purification Kit (Life Technologies), and the samples were treated by DNase I (Invitrogen) to avoid DNA contamination. RNA quality was examined using a High Sensitivity RNA ScreenTape for TapeStation (Agilent Technologies).
Sequencing libraries were generated using the KAPA Stranded mRNA-Seq Kit (KAPA Biosystems) and an Adapter Kit (FastGene) using 200 ng of total RNA. Indexed libraries were sent to Macrogen Japan for sequencing HiSeq X Ten (Illumina), producing 150-bp paired-end reads.
Differential gene expression analysis
Raw fastq files were adaptor trimmed and quality controlled by fastp ver. 0.20.0 [48] with the following criterion: minimum length 50 bp, Q-score per base ≥ 20, the average Q-score ≥ 30 and maximum number of N bases per read = 1. A reciprocal re-annotation pipeline [49] was applied for generating a total of 13,220 reference CDS from D. suzukii WT3 [50] and D. subpulchrella RU33 [29] CDS (Additional file 3). D. melanogaster ortholog gene was assigned from reciprocal best hit to D. melanogaster gene products (Flybase: dmel-all-translation-r6.43). After mapping by bowtie2 ver. 2.4.4 [51] the number of reads were counted by samtools ver. 1.10 [52]. Differential gene expression analysis was conducted by edgeR [53]. Differentially expressed genes (DEGs) were detected at the significance level of FDR < 0.01. Genes were considered to be expressed if TPM > 1 in all three biological replicates. Those that were not expressed in both species were excluded from the analysis.
Immunohistochemistry and in situ hybridization
Antibody staining was conducted as in a previous study with minor modification [54]. In brief, newly emerged female pupae were incubated at 25 ºC and fixed at 45 h APF for D. suzukii and D. subpulchrella and at 44 and 48 h APF for D. melanogaster. Rat anti-E-cadherin 1:200 (DCAD2, DSHB) and donkey anti-rat Alexa 488 1:200 (Thermo Fisher Scientific) antibodies were used. Rhodamine phalloidin 1:200 (Thermo Fisher Scientific) staining was performed with a secondary antibody. The antibody for Shavenbaby (Svb) was a gift from Julia Zeitlinger and was used at 1:10. Images of the developing ovipositors of D. suzukii and D. subpulchrella were captured by using a C2+ confocal microscope (Nikon) at × 200 magnification. Those of D. melanogaster were captured at × 63 on a Leica TCS SP8 confocal microscope. Images were processed by using Fiji version 2.3 [46], MorphoGraphX [55] and Photoshop version 23.1 (Adobe).
For in situ hybridization, newly emerged female pupae were incubated at 25 ºC for 44 and 48 h. Staged pupae were collected, fixed, and then proceeded to in situ hybridization as in the previous study with minor modification [56]. A 974-bp sha probe were generated using the following primer set with the addition of T7 linker sequence (GATCACTAATACGACTCACTATAGGG); CAAAGTTTGGCGACCAAGGG (sha-fwd), and TCCATCTCCTCCTCGAGTGG (sha-rev). Images were captured using an Olympus SZX16 light microscope with a DP73 camera (Olympus) at × 16 magnification.
Supplementary information
Additional file 1: Figure S1. Oviprovector scales from additional strains. (A–C) Oviprovector scales from dissected ovipositors in three additional strains, Hilo, TMUS09, and TMUS18, of D. suzukii. (D–F) Oviprovector scales from dissected ovipositors in three additional strains, M4, NAR02, and NAR07 of D. subpulchrella. Scale bar indicates 50 μm. Figure S2. Diversification of oviprovector scales in D. suzukii and its related species. . (A–C) Oviprovector scales from dissected ovipositors in three additional strains, Hilo, TMUS09, and TMUS18, of D. suzukii. (D–F) Oviprovector scales from dissected ovipositors in three additional strains, M4, NAR02, and NAR07 of D. subpulchrella. Scale bar indicates 50 μm. Figure S3. svb expression in the developing ovipositor. (A) Schematic image of the developing female terminalia. Abbreviations indicate analia (An) and ovipositor (Ovi). (B) in situ hybridization of svb at 44 h APF in D. suzukii. (C) in situ hybridization of svb at 44 h APF in D. subpulchrella. (D) in situ hybridization of svb at 48 h APF in D. suzukii. (E) in situ hybridization of svb at 48 h APF in D. subpulchrella. Scale bars indicate 100 μm. Figure S4. Oviprovector scales of mutants of trichome-related genes in D. melanogaster. (A) Wildtype Canton-S. (B–C) Mutant phenotype of trichome patterning genes, sha (B), m (C) and tyn (D). Scale bar indicates 20 μm.
Additional file 2: Table S1. Mapping results of RNA-seq using D. suzukii and D. subpulchrella female terminalia at 48 h APF. Table S2. Differentially expressed genes between D. suzukii and D. subpulchrella female terminalia at 48 h APF.
Additional file 3. Reciprocally re-annotated CDS. The .zip folder containing reciprocally re-annotated reference CDS used for mapping D. suzukii and D. subpulchrella RNA-seq reads.
Acknowledgements
We are grateful to Julia Zeitlinger for providing the Svb antibody. We also thank Koichiro Tamura, KYOTO Stock Center (DGRC), and Bloomington Drosophila Stock Center for fly stocks and Sawako Shimizu for technical support.
For EcoEvoDevo collection of BMC Ecology and Evolution (https://www.biomedcentral.com/collections/eed).
Abbreviations
- h APF
Hour after puparium formation
- SEM
Scanning electron micrograph
- CDS
Coding sequence
- DEG
Differentially expressed gene
- FDR
False discovery rate
- TPM
Transcripts per million
Author contributions
KMT, KT, and AT conceived and designed the study. GR and MR analyzed D. melanogaster genitalia. YK obtained SEM images. KMT and KT performed RNA-seq. KMT, KT, and AT wrote the manuscript. KMT and KT contributed equally to this work. All authors have read and approved the manuscript.
Funding
This study was supported by JSPS KAKENHI (Grant No. JP19H03276) awarded to YK and AT and by the National Institutes of Health (Grant No. R21HD104956) to MR. The funders had no role in conceptualizations and study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Raw fastq files are deposited at DDBJ under the accession number DRA009998 and DRA009999 for D. suzukii TMUS08 and D. subpulchrella H243, respectively. Fly stocks can be provided upon request. No custom code was used, and all analyses are described in the main text. The authors can provide additional information upon request.
Declarations
Ethics approval and consent to participate
No ethics approval or consent to participate were needed for the species used in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kentaro M. Tanaka and Kanoko Takahashi contributed equally to this work
References
- 1.Eberhard WG. Sexual selection and animal genitalia. Cambridge, MA: Harvard University Press; 1985. [Google Scholar]
- 2.Hosken DJ, Stockley P. Sexual selection and genital evolution. Trends Ecol Evol. 2004;19:87–93. doi: 10.1016/j.tree.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Simmons LW. Sexual selection and genital evolution. Austral Entomol. 2014;53:1–17. doi: 10.1111/aen.12053. [DOI] [Google Scholar]
- 4.Ah-King M, Barron AB, Herberstein ME. Genital evolution: why are females still understudied? PLOS Biol. 2014;12:e1001851. doi: 10.1371/journal.pbio.1001851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simmons LW, Fitzpatrick JL. Female genitalia can evolve more rapidly and divergently than male genitalia. Nat Commun. 2019;10:1312. doi: 10.1038/s41467-019-09353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genevcius BC, Baker J, Bianchi FM, Marvaldi AE. Female-driven intersexual coevolution in beetle genitalia. J Evol Biol. 2020;33:957–65. doi: 10.1111/jeb.13627. [DOI] [PubMed] [Google Scholar]
- 7.Kamimura Y. Significance of constraints on genital coevolution: Why do female Drosophila appear to cooperate with males by accepting harmful matings? Evolution. 2016;70:1674–83. doi: 10.1111/evo.12955. [DOI] [PubMed] [Google Scholar]
- 8.Muto L, Kamimura Y, Tanaka KM, Takahashi A. An innovative ovipositor for niche exploitation impacts genital coevolution between sexes in a fruit-damaging Drosophila. Proc R Soc B. 2018;285:20181635. doi: 10.1098/rspb.2018.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery SL. Comparative breeding site ecology and the adaptative radiation of picture-winged Drosophila (Diptera: Drosophilidae) in Hawaii. Proc Hawaii Entomol Soc. 1975;22:65–103. [Google Scholar]
- 10.Magnacca KN, Foote D, O’Grady PM. A review of the endemic Hawaiian Drosophilidae and their host plants. Zootaxa. 2008;1728(1 SE-Article):1–58. doi: 10.11646/zootaxa.1728.1.1. [DOI] [Google Scholar]
- 11.Craddock EM, Kambysellis MP, Franchi L, Francisco P, Grey M, Hutchinson A, et al. Ultrastructural variation and adaptive evolution of the ovipositor in the endemic Hawaiian Drosophilidae. J Morphol. 2018;279:1725–52. doi: 10.1002/jmor.20884. [DOI] [PubMed] [Google Scholar]
- 12.Peláez JN, Gloss AD, Ray JF, Charboneau JLM, Verster KI, Whiteman NK. Evolution and genetic basis of the plant-penetrating ovipositor, a key adaptation in herbivorous Drosophilidae. bioRxiv. 2021 doi: 10.1101/2020.05.07.083253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McQueen E, Afkhami M, Atallah J, Gompel N, Heifetz Y, Kamimura Y, et al. A standardized nomenclature and atlas of the female terminalia of Drosophila melanogaster. Fly. 2022;16:128–51. doi: 10.1080/19336934.2022.2058309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin AD, Browning TO. A mechanism for movement of eggs along insect ovipositors. Int J Insect Morphol Embryol. 1981;10:93–108. doi: 10.1016/S0020-7322(81)80015-3. [DOI] [Google Scholar]
- 15.Sucena É, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc Natl Acad Sci USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren N, He B, Stone D, Kirakodu S, Adler PN. The shavenoid gene of Drosophila encodes a novel Actin cytoskeleton interacting protein that promotes wing hair morphogenesis. Genetics. 2006;172:1643–53. doi: 10.1534/genetics.105.051433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S. Shavenbaby couples patterning to epidermal cell shape control. PLOS Biol. 2006;4:e290. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arif S, Kittelmann S, McGregor AP. From shavenbaby to the naked valley: trichome formation as a model for evolutionary developmental biology. Evol Dev. 2015;17:120–6. doi: 10.1111/ede.12113. [DOI] [PubMed] [Google Scholar]
- 19.Walsh DB, Bolda MP, Goodhue RE, Dreves AJ, Lee J, Bruck DJ, et al. Drosophila suzukii (Diptera: Drosophilidae): Invasive pest of ripening soft fruit expanding its geographic range and damage potential. J Integr Pest Manage. 2011;2:G1–7. doi: 10.1603/IPM10010. [DOI] [Google Scholar]
- 20.Atallah J, Teixeira L, Salazar R, Zaragoza G, Kopp A. The making of a pest: the evolution of a fruit-penetrating ovipositor in Drosophila suzukii and related species. P Proc R Soc B. 2014;281:20132840. doi: 10.1098/rspb.2013.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green JE, Cavey M, Médina Caturegli E, Aigouy B, Gompel N, Prud’homme B. Evolution of ovipositor length in Drosophila suzukii is driven by enhanced cell size expansion and anisotropic tissue reorganization. Curr Biol. 2019;29:2075–2082. doi: 10.1016/j.cub.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamby KA, Bellamy E, Chiu D, Lee JC, Walton JC, Wiman VM, et al. Biotic and abiotic factors impacting development, behavior, phenology, and reproductive biology of Drosophila suzukii. J Pest Sci. 2016;89:605–19. doi: 10.1007/s10340-016-0756-5. [DOI] [Google Scholar]
- 23.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–51. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chanut-Delalande H, Ferrer P, Payre F, Plaza S. Effectors of tridimensional cell morphogenesis and their evolution. Semin Cell Dev Biol. 2012;23:341–9. doi: 10.1016/j.semcdb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes I, Chanut-Delalande H, Ferrer P, Latapie Y, Waltzer L, Affolter M, et al. Zona pellucida domain proteins remodel the apical compartment for localized cell shape changes. Dev Cell. 2010;18:64–76. doi: 10.1016/j.devcel.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Karageorgi M, Bräcker LB, Lebreton S, Minervino C, Cavey M, Siju KP, et al. Evolution of multiple sensory systems drives novel egg-laying behavior in the fruit pest Drosophila suzukii. Curr Biol. 2017;27:847–53. doi: 10.1016/j.cub.2017.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crava CM, Zanini D, Amati S, Sollai G, Crnjar R, Paoli M, et al. Structural and transcriptional evidence of mechanotransduction in the Drosophila suzukii ovipositor. J Insect Physiol. 2020;125:104088. doi: 10.1016/j.jinsphys.2020.104088. [DOI] [PubMed] [Google Scholar]
- 28.Kienzle R, Groß LB, Caughman S, Rohlfs M. Resource use by individual Drosophila suzukii reveals a flexible preference for oviposition into healthy fruits. Sci Rep. 2020;10:3132. doi: 10.1038/s41598-020-59595-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durkin SM, Chakraborty M, Abrieux A, Lewald KM, Gadau A, Svetec N, et al. Behavioral and Genomic Sensory Adaptations Underlying the Pest Activity of Drosophila suzukii. Mol Biol Evol. 2021;38:2532–46. doi: 10.1093/molbev/msab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato A, Tanaka KM, Yew JY, Takahashi A. Drosophila suzukii avoidance of microbes in oviposition choice. R Soc Open Sci. 2021;8:201601. doi: 10.1098/rsos.201601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimaldi DA. Phylogenetics and taxonomy of Zygothrica (Diptera: Drosophilidae) Bull Am Mus of Nat Hist. 1987;186:104–268. [Google Scholar]
- 32.Matushkina NA. Ovipositor internal microsculpture in the relic silverfish Tricholepidion gertschi (Insecta: Zygentoma) Psyche. 2011;2011:563852. [Google Scholar]
- 33.Eggs B, Birkhold AI, Röhrle O, Betz O. Structure and function of the musculoskeletal ovipositor system of an ichneumonid wasp. BMC Zool. 2018;3:12. doi: 10.1186/s40850-018-0037-2. [DOI] [Google Scholar]
- 34.Kittelmann S, Preger-Ben Noon E, McGregor AP, Frankel N. A complex gene regulatory architecture underlies the development and evolution of cuticle morphology in Drosophila. Curr Opin Genet Dev. 2021;69:21–7. doi: 10.1016/j.gde.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Kittelmann S, Buffry AD, Franke FA, Almudi I, Yoth M, Sabaris G, et al. Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLOS Genet. 2018;14:e1007375. doi: 10.1371/journal.pgen.1007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arif S, Murat S, Almudi I, Nunes MDS, Bortolamiol-Becet D, McGregor NS, et al. Evolution of mir-92a underlies natural morphological variation in Drosophila melanogaster. Curr Biol. 2013;23:523–8. doi: 10.1016/j.cub.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maung SMTW, Jenny A. Planar cell polarity in Drosophila. Organogenesis. 2011;7:165–79. doi: 10.4161/org.7.3.18143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler MT, Wallingford JB. Planar cell polarity in development and disease. Nat Rev Mol Cell Biol. 2017;18:375–88. doi: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi D, Komatsu K, Hirao M, Toyooka Y, Koyama H, Tissir F, et al. Celsr1 is required for the generation of polarity at multiple levels of the mouse oviduct. Development. 2014;141:4558–68. doi: 10.1242/dev.115659. [DOI] [PubMed] [Google Scholar]
- 40.Shi D, Usami F, Komatsu K, Oka S, Abe T, Uemura T, et al. Dynamics of planar cell polarity protein Vangl2 in the mouse oviduct epithelium. Mech Dev. 2016;141:78–89. doi: 10.1016/j.mod.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Koyama H, Shi D, Fujimori T. Biophysics in oviduct: Planar cell polarity, cilia, epithelial fold and tube morphogenesis, egg dynamics. Biophys Physicobiol. 2019;16:89–107. doi: 10.2142/biophysico.16.0_89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onuma M, Kamimura Y, Sawamura K. Genital coupling and copulatory wounding in the Drosophila auraria species complex (Diptera: Drosophilidae) Biol J Linn Soc. 2022;135:195–207. doi: 10.1093/biolinnean/blab134. [DOI] [Google Scholar]
- 43.Kamimura Y. Twin intromittent organs of Drosophila for traumatic insemination. Biol Lett. 2007;3:401–4. doi: 10.1098/rsbl.2007.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masly JP. 170 Years of “Lock-and-Key”: Genital morphology and reproductive isolation. Int J Evol Biol. 2012;2012:247352. doi: 10.1155/2012/247352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice GR, David JR, Gompel N, Yassin A, Rebeiz M. Resolving between novelty and homology in the rapidly evolving phallus of Drosophila. J Exp Zool Part B. 2021;2012:247352. doi: 10.1002/jez.b.23113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. URL https://www.R-project.org/
- 48.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres-Oliva M, Almudi I, McGregor AP, Posnien N. A robust (re-)annotation approach to generate unbiased mapping references for RNA-seq-based analyses of differential expression across closely related species. BMC Genomics. 2016;17:392. doi: 10.1186/s12864-016-2646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paris M, Boyer R, Jaenichen R, Wolf J, Karageorgi M, Green J, et al. Near-chromosome level genome assembly of the fruit pest Drosophila suzukii using long-read sequencing. Sci Rep. 2020;10:11227. doi: 10.1038/s41598-020-67373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith SJ, Davidson LA, Rebeiz M. Evolutionary expansion of apical extracellular matrix is required for the elongation of cells in a novel structure. eLife. 2020;9:e55965. doi: 10.7554/eLife.55965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Reuille PB, Routier-Kierzkowska A-L, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:e05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagen JFD, Mendes CC, Blogg A, Payne A, Tanaka KM, Gaspar P, et al. tartan underlies the evolution of Drosophila male genital morphology. Proc Natl Acad Sci. USA. 2019;116:19025. doi: 10.1073/pnas.1909829116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’Agostino ERR, et al. Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol. 2022;32:111–123.e5. doi: 10.1016/j.cub.2021.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finet C, Kassner VA, Carvalho AB, Chung H, Day JP, Day S, et al. DrosoPhyla: Resources for drosophilid phylogeny and systematics. Genome Biol Evol. 2021;13:evab179. doi: 10.1093/gbe/evab179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Oviprovector scales from additional strains. (A–C) Oviprovector scales from dissected ovipositors in three additional strains, Hilo, TMUS09, and TMUS18, of D. suzukii. (D–F) Oviprovector scales from dissected ovipositors in three additional strains, M4, NAR02, and NAR07 of D. subpulchrella. Scale bar indicates 50 μm. Figure S2. Diversification of oviprovector scales in D. suzukii and its related species. . (A–C) Oviprovector scales from dissected ovipositors in three additional strains, Hilo, TMUS09, and TMUS18, of D. suzukii. (D–F) Oviprovector scales from dissected ovipositors in three additional strains, M4, NAR02, and NAR07 of D. subpulchrella. Scale bar indicates 50 μm. Figure S3. svb expression in the developing ovipositor. (A) Schematic image of the developing female terminalia. Abbreviations indicate analia (An) and ovipositor (Ovi). (B) in situ hybridization of svb at 44 h APF in D. suzukii. (C) in situ hybridization of svb at 44 h APF in D. subpulchrella. (D) in situ hybridization of svb at 48 h APF in D. suzukii. (E) in situ hybridization of svb at 48 h APF in D. subpulchrella. Scale bars indicate 100 μm. Figure S4. Oviprovector scales of mutants of trichome-related genes in D. melanogaster. (A) Wildtype Canton-S. (B–C) Mutant phenotype of trichome patterning genes, sha (B), m (C) and tyn (D). Scale bar indicates 20 μm.
Additional file 2: Table S1. Mapping results of RNA-seq using D. suzukii and D. subpulchrella female terminalia at 48 h APF. Table S2. Differentially expressed genes between D. suzukii and D. subpulchrella female terminalia at 48 h APF.
Additional file 3. Reciprocally re-annotated CDS. The .zip folder containing reciprocally re-annotated reference CDS used for mapping D. suzukii and D. subpulchrella RNA-seq reads.
Data Availability Statement
Raw fastq files are deposited at DDBJ under the accession number DRA009998 and DRA009999 for D. suzukii TMUS08 and D. subpulchrella H243, respectively. Fly stocks can be provided upon request. No custom code was used, and all analyses are described in the main text. The authors can provide additional information upon request.