Abstract
Bacillus thuringiensis produces large amounts of various pesticidal proteins during the stationary phase. In order to achieve a high yield and form crystals, some pesticidal proteins require the presence of other proteins. Helper protein P20 is required for efficient production of both the Cyt1A and Cry11A crystal proteins in B. thuringiensis subsp. israelensis. Although full-length Cry1 protoxins are usually independent in terms of expression and crystallization in B. thuringiensis, in this study P20 significantly enhanced production of Cry1Ac protoxin (133 kDa) in an acrystalliferous and plasmid-negative strain. In the presence of P20, the yield of Cry1Ac protoxin increased 2.5-fold, and on average the resulting crystals were 1.85 μm long and 0.85 μm wide, three times the size of the crystals formed in the control lacking P20. Correspondingly, the recombinant strain that coexpressed P20 and Cry1Ac exhibited higher toxicity against Heliothis armigera larvae than the control. Furthermore, serious degradation of Cry1Ac in vivo was observed, which has seldom been reported previously. Actually, most protein was completely degraded during synthesis, and after synthesis about one-third of the expressed protoxins were degraded further before crystallization. In this process, P20 protected only nascent Cry1Ac from degradation, indicating that it acted as a molecular chaperon. In addition, spores were smaller and rounder and had a thinner exosporium layer when they were produced in the presence of P20. In summary, Cry1Ac was severely degraded during synthesis; this degradation was effectively relieved by P20, which resulted in enhanced production. Our results indicated that P20 is an effective tool for optimizing protein production in vivo.
Bacillus thuringiensis, an extensively exploited pesticidal bacterium, produces various pesticidal proteins during the stationary phase. These proteins usually accumulate in the form of parasporal crystals and make up the main components of B. thuringiensis agents that are active against insects and nematodes (8, 16, 30). More than 70 B. thuringiensis subspecies have been identified, and all of them are characterized by crystal formation, which distinguishes this species from its close genetic relative Bacillus cereus. A high level of production is one prerequisite for forming a crystal. Several factors may be involved in the high levels of expression of cry genes in B. thuringiensis; these factors include strong promoters, stable mRNA, high copy number, and protein crystallization (2, 5). However, the mechanism that B. thuringiensis uses to overexpress and crystallize Cry proteins is still not fully understood.
Crystallization occurs with most, but not all, pesticidal proteins of B. thuringiensis (8). This process not only packages more proteins in the limited intracellular space but also protects the proteins from proteolysis by proteases in vivo, and it also results in a high level of production. The formation of Cry protein crystals in B. thuringiensis presumably depends on the special protein structure, which facilitates autoassembly, or it depends on the assistance of some accessory proteins (2, 5). For example, large protoxins, such as Cry1 and Cry4, autoassemble on their conserved C-terminal halves via interchain disulfide bonds (2, 9). The Cry3 protein probably autoassembles on its four intramolecular salt bridges (2), while some smaller protoxins, such as Cry2 and Cyt1A, rely on other accessory proteins (1, 7, 13, 34, 36, 37). Thus, in B. thuringiensis crystallization of different toxins occurs in different ways.
Recently, some researchers have successfully increased the yields of different B. thuringiensis toxins at the transcriptional or posttranscriptional level. The combination of dual sporulation-dependent promoters and the STAB-SD stabilizing sequence of cry3A significantly increases the Cry3A yield (26). With Cry2A, ORF2 encoded by the cry2A operon acts as a scaffold during Cry2A crystallization and is necessary for full expression of the process (13). On the other hand, in order to avoid proteolytic degradation of protoxins after cell lysis, the nprA gene encoding neutral protease A has been deleted from the B. thuringiensis chromosome, and this also increases production of the Cry1Bb and Cry3Bb full-length proteins (11).
Helper protein P20 is an accessory protein in B. thuringiensis subsp. israelensis (10). This protein was first detected during a study of Cyt1A expression (23). Later, other investigations concentrated on the role of P20 in Cyt1A expression and proved that it is necessary for Cyt1A crystallization and thus for host cell viability (1, 34, 36, 37). Moreover, P20 also increased the production of other B. thuringiensis subsp. israelensis toxins (those encoded by the cry4A and cry11A genes) and even truncated Cry1C (28, 34, 37, 38). All of these proteins are poorly expressed without P20 because of structural defects, and most previous reports have focused mainly on improving the production of toxins that are poorly expressed in nature (13, 34, 36, 37). In contrast, full-length Cry1 proteins are usually expressed well and crystallize in the C-terminal halves of their molecules. They do not need the help of proteins such as P20. However, can P20 also work with them? These proteins occur frequently in many B. thuringiensis isolates and play an important role in biocontrol of lepidopterans. A probable increase in their production in the presence P20 prompted us to start this investigation.
In this study, we examined the effects of P20 on the production of full-length Cry1Ac (133 kDa), which occurs in most strains of B. thuringiensis subsp. kurstaki. Cry1Ac is highly toxic to some agriculturally important pests, such as Heliothis armigera, Heliothis virescens, and Manduca sexta (8, 25). The primary goal of this study was to construct an improved strain that potentially could be used against these pests. During this study, we found that there is serious degradation of Cry1Ac in vivo, which has seldom been reported previously, and we obtained more information about the role of P20 in protein expression. In this study, P20 doubled the production of full-length Cry1Ac in an engineered B. thuringiensis strain and also increased the toxicity Cry1Ac for H. armigera. Some changes in the spore size and structure of the engineered strain were also observed.
MATERIALS AND METHODS
Gene, plasmid, and bacterium.
The cry1Ac10 gene has been cloned from wild-type B. thuringiensis subsp. kurstaki strain YBT-1520 and is harbored in plasmid pBMB1231 (31). The p20 gene has been cloned from B. thuringiensis subsp. israelensis; the dual promoters of cry1C, which share typical BtI and BtII promoters with other cry1 genes, were used to start p20 transcription (22, 35). Plasmids pHT304 and pHT3101 were used as Escherichia coli-B. thuringiensis shuttle vectors; these plasmids contain the same B. thuringiensis replicon, which has a copy number of about four, and have the same antibiotic resistance gene and E. coli replicon (3). E. coli mutant strain DH-5α was used for plasmid amplification. An acrystalliferous strain of B. thuringiensis subsp. kurstaki which was plasmid cured, BMB171, was used as the recipient to examine expression of cry1Ac10 in B. thuringiensis (21).
Recombinant plasmid construction.
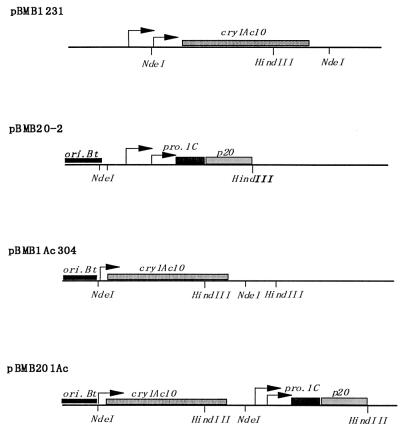
Two shuttle recombinant plasmids were obtained; one, carrying the p20 gene and cry1Ac10 in tandem in pHT3101, was designated pBMB201Ac, and the other, a control lacking p20 and carrying only cry1Ac10 in pHT304, was designated pBMB1Ac304 (Fig. 1). The 3,771-bp fragment containing the whole cry1Ac10 coding region and its flanking sequences was obtained by NdeI digestion of pBMB1231. Gene p20 (565-bp NocI/HindIII fragment) was put under control of cry1C promoters (252-bp KpnI/NocI fragment, designated pro.1C in Fig. 1) in the shuttle vector pHT3101, and this resulted in plasmid pBMB20-2. Then, the cry1Ac10 fragment was inserted into pBMB20-2 at the NdeI site, and this resulted in the recombination plasmid pBMB201Ac containing cry1Ac10 and p20 in tandem. Plasmid pBMB1Ac304 was constructed by inserting the NdeI fragment of cry1Ac10 into shuttle vector pHT304 in the same direction as pBMB201Ac. Both recombinant plasmids were transformed into the acrystalliferous strain BMB171 to examine expression of cry1Ac10. Notably, promoter BtII and the −35 region of promoter BtI were removed by NdeI digestion. BtI is a strong promoter and is active 2 to 6 h after the end of the exponential phase, while BtII is a weak promoter and is active beginning 5 h after the end of the exponential phase. As shown below, the deletion did reduce promoter activity, but this had no influence on our test to determine the role of P20. Strain BMB802 (BMB171 transformed with plasmid pBMB1231 [cry1Ac10 in pHT304] [see above]) was used as a control to observe normal expression of cry1Ac10 under control of its dual promoters.
FIG. 1.
Construction of recombinant plasmids containing the p20 gene and cry1Ac10. ori.Bt, replicon. The arrows indicate promoters. In pBMB1Ac304 and pBMB201Ac, the vectors are pHT304 and pHT3101, respectively. The two vectors share the same origin of replication and have the same copy number.
Transformation.
Recombinant plasmids were transformed into E. coli by the standard procedure (29). B. thuringiensis transformants were obtained by electroporation, and competent cells were prepared in a sucrose-glycerol buffer. After cells were premixed with a plasmid, they were electroshocked at 1.25 kV and 25 μF in 1-mm cuvettes; they were recovered in 0.8 ml of Luria-Bertani (LB) medium for 2 h and then plated on solid LB medium containing 25 μg of erythromycin per ml and incubated at 28°C overnight. Transformants were identified by their plasmid patterns after restriction enzyme digestion.
Microscopy.
A light microscope was used for cell observation, and a scanning electron microscope (SEM) was used to determine the dimensions of free crystals and spores. For light microscope observation, bacteria were cultivated on NSM medium plates (0.5% peptone, 0.3% beef paste, 0.7 mmol of CaCl2 per liter, 0.05 mmol of MnCl2 per liter, 1 mmol of MgCl2 per liter; pH 7.0) containing 25 μg of erythromycin per ml for 2 days at 28°C. The cells were stained with carbolic acid-azaleine and observed with an oil immersion lens. For SEM observation, bacteria were cultivated for 36 h at 28°C and 200 rpm in a liquid medium (ICPM medium) containing 0.6% peptone, 0.5% glucose, 0.1% CaCO3, 0.05% MgSO4, and 0.05% KH2PO4 (pH 7.0) to which 25 μg of erythromycin was added before use. Spores and crystals were collected by centrifugation and washed three times with a solution containing 1 mol of NaCl per liter and then three times with water. The mixture of spores and crystals was then resuspended in water. After pretreatment, the mixture was spread on a sample platform, coated with metal by using an ion sputter coater (IB · 5; Eiko), and observed at 40 kV with an ASID10, JEM-1200EX microscope (JEOL). To observe the spore structure, the mixture was double fixed, serially dehydrated, and embedded in resin as usual. Ultrathin sections were double stained and observed with a transmission electron microscope.
Determination of crystal size.
The dimensions of crystals were determined by measuring 30 crystals in each sample on the screen of the SEM at a magnification of ×10,000. The length (L) was determined between the two tips of a bipyramid, so the height of a pyramid was L/2. The width (W) (i.e., the edge of the square base of a pyramid) was measured at the middle of the bipyramid. Thus, the volume of a crystal was W2 · L/3, the same formula as that deduced by Park et al. (26).
Monitoring cell growth.
Bacterial cell growth was monitored by determining the optical density at 600 nm (OD600) of the culture fluid. To compare the growth rates of strains BMB171/pBMB201Ac and BMB171/pBMB3041Ac, we inoculated equal amounts of mid-exponential-phase bacterial cells from LB medium into ICPM medium and incubated the preparations at 200 rpm and 28°C until cell lysis occurred. The OD600 was measured once every 0.5 h. The number of cells in a culture was determined by CFU counting.
SDS-PAGE and protein quantification.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (18). To quantify expression of the Cry1Ac10 protein, samples that had been subjected to different treatments were prepared in parallel. A crystal-spore mixture was obtained from an exact volume of a thoroughly suspended culture and washed as described above. The mixture was resuspended in one-half the original volume of water, and then the preparation was mixed with an equal volume of 2× sample buffer and boiled for 4 min. After a brief centrifugation, different samples were loaded by using the same volume.
The proteins in 133-kDa Cry1Ac10 bands on SDS-PAGE gels were quantified by using the densitometry program of Bioimage Systems as described by the manufacturer.
Bioassay.
Neonate larvae of H. armigera were used to examine the toxicities of different cry1Ac10 transformants that contained or did not contain p20. The insect was reared with an artificial diet. The bioassay diet included 12 g of yeast powder, 24 g of soybean powder (roasted), 1.5 g of vitamin C, 0.42 g of sodium benzoate, 3.9 ml of 36% acetic acid, and 4.5 g of agar powder in 300 ml of sterilized water and was premixed with culture fluid in a series of dilutions. Twenty larvae were used for each dilution, and there were three replicates. After 72 h of incubation at 23°C, mortalities were recorded and the 50% lethal concentration (LC50) for each treatment was calculated.
RESULTS
Bacterial growth.
Bacterial growth was monitored by determining the OD600 of the culture fluid. It was found that strains BMB171/pBMB201Ac and BMB171/pBMB3041Ac had the same growth curve. Both strains took about 10 h to reach the stationary phase, and the OD600 were 0.724 and 0.730, respectively, after the cultures were diluted with ICPM medium seven times. The two cultures contained similar numbers of CFU (about 1.08 × 107 CFU/ml). However, strain BMB171/pBMB201Ac was slower to form spores and crystals and exhibited some cell coagulation in the stationary phase and thereafter.
Enhanced Cry1Ac10 production.
Production of Cry1Ac10 by different strains was detected by SDS-PAGE. All strains were cultivated and processed in parallel. The results revealed that Cry1Ac10 production by B. thuringiensis was substantially enhanced by helper protein P20 (Fig. 2A). Quantification of the 133-kDa protein band on SDS-PAGE gels demonstrated that the amount of full-length protoxin in transformant BMB171/pBMB201Ac (Fig. 2A, lane 2) was 3.5-fold greater than the amount in control transformant BMB1Ac304 lacking P20 (lane 3) and 1.73-fold greater than the amount in another control transformant that lacked P20, BMB802, in which cry1Ac10 promoters were intact (lane 4). In the gel pattern some intermediate products larger than 60 kDa were visible; apparently, they were derived from full-length protoxins by proteolysis during the SDS-PAGE sample treatment.
FIG. 2.
Effects of helper protein P20 on expression of cry1Ac10. (A) Lane 1, standard protein markers; lane 2, expression of cry1Ac10 in BMB171/pBMB201Ac in the presence of P20; lane 3, expression of cry1Ac10 in BMB171/pBMB1Ac304 in the absence of P20; lane 4, expression of cry1Ac10 driven by its intact double promoters in BMB802. (B) Densitometry of the 130-kDa protein in panel A. The values on the ordinate are relative concentrations.
Morphological changes.
The effect of P20 on the formation of Cry1Ac10 crystals was observed with microscopes. Morphological changes in the host cells caused by P20 were also observed. As determined with a light microscope, the differences in cell morphology between the transformants with P20 and those without P20 were obvious (Fig. 3). After 2 days of cultivation on an NSM medium plate, the cells of both transformants sporulated and started to lyse. There were more free spores and crystals in the cells of control strain BMB171/pBMB1Ac304 lacking P20, and the sporulated cells were regular thin rods (Fig. 3A). In contrast, the cells of BMB171/pBMB201Ac seemed to bulge with large endogenous crystals, which occupied most of the intracellular space and pushed spores to the opposite sides of the cells (Fig. 3B). Altogether, there were more crystals that stained red in the field of view for the strain in which P20 was present than in the field of view for the control strain that lacked P20 (Fig. 3B).
FIG. 3.
Morphological changes in crystals and spores caused by helper protein P20. The effect of P20 on the formation of Cry1Ac10 crystals was observed with microscopes. For light microscope observation, bacteria were cultivated on NSM medium plates for 2 days at 28°C and stained with carbolic acid-azaleine. For SEM observation, bacteria were cultivated in liquid ICPM medium; free crystals and spores were washed and ion coated. (A) BMB171/pBMB1Ac304 Cry1Ac10 in the absence of P20, observed with an oil immersion lens after staining. Magnification, ×950. (B) BMB171/pBMB201Ac Cry1Ac10 in the presence of P20, observed with an oil immersion lens after staining. Magnification, ×950. (C) Crystals and spores of BMB171/pBMB1Ac304, observed by SEM after ion coating. Magnification, ×4,800. Bar, 1 μm. (D) Crystals and spores of BMB201Ac/171, observed by SEM after ion coating. Magnification, ×4,800. Bar, 1 μm.
For SEM observation, bacteria were cultivated in liquid ICPM medium until cell lysis. Free spores and crystals were washed and ion coated. As determined by SEM, the crystals of Cry1Ac10 in both transformants were bipyramidal. However, the crystals generated in transformant BMB171/pBMB1Ac304 were smaller (Fig. 3C), only 1.4 μm long and 0.55 μm wide on average with an average volume of 0.14 μm3, as calculated with the formula described above (Table 1). In contrast, the crystals in BMB171/pBMB201Ac containing P20 were 1.85 μm long and 0.85 μm wide, and the calculated volume was 0.44 μm3, so these crystals were about three times larger than those in the control. When P20 was present, the spores were smaller and spherical, in contrast to the rod-shaped spores of control strain BMB171/pBMB1Ac304 (Fig. 3D). The former spores were 1.05 μm long, and the latter spores were 1.85 μm long, while both types of spores were 0.8 μm wide (Table 1).
TABLE 1.
Sizes of crystals and spores of transformants BMB171/pBMB201Ac and BMB171/pBMB1Ac304a
| Crystals or spores | Shape | Length (μm) | Width (μm) | Vol (μm3) |
|---|---|---|---|---|
| Cry1Ac10b | Bipyramidal | 1.40 ± 0.26 | 0.55 ± 0.09 | 0.14 |
| Cry1Ac10 + P20c | Bipyramidal | 1.85 ± 0.28 | 0.85 ± 0.11 | 0.44 |
| Spores | Long ellipsoidal | 1.80 ± 0.13 | 0.80 ± 0.04 | |
| Spores (P20) | Spherical | 1.05 ± 0.27 | 0.80 ± 0.02 |
The values are means based on two independent experiments; 30 crystals were measured by SEM in each treatment.
Cry1Ac10 crystals in BMB171/pBMB1Ac304 without P20.
Cry1Ac10 crystals in BMB171/pBMB201Ac in the presence of P20.
An ultrastructure study was performed to examine the changes in the internal structure. This study showed that both types of crystals had normal regular lattices (results not shown), while the spore structures were much different, especially in the exosporium. The spores of BMB171/pBMB1Ac304 were enclosed by a thick exosporium layer. This layer seemed to be floppy and slightly electron stained and varied from 100 to 700 nm thick; it did not have any fine structure except possible layers with different densities (Fig. 4a). This region in strain BMB171/pBMB201Ac spores was thinner (thickness, about 50 to 200 nm) and seemed tight (Fig. 4b). No obvious differences between the two strains in other regions were observed when preparations were observed at a high magnification (Fig. 4c and d). In both strains the spore coat was a frequently wrinkled multilayer structure which was about 30 nm thick and had high electron density. The inner and outer coats could not be distinguished easily. The cortexes were also similar, about 70 nm thick, and less electron stained. In addition, the results showed that whole spores of BMB171/pBMB201Ac were spherical and that all layers were concentric, whereas all parts of BMB171/pBMB1Ac304 spores were elliptical.
FIG. 4.
Changes in spore structure in the presence of helper protein P20. Samples for transmission electron microscopy were processed in a traditional way; a spore-crystal mixture was double fixed, serial dehydrated, and embedded in resin. Ultrathin sections were double stained and observed at 80 kV. The arrows indicate the spore exosporium. (a) Spores of BMB171/pBMB1Ac304 covered with a thick layer of exosporium. Magnification, ×15,000. (b) Spores of BMB201Ac/171, showing the thin exosporium. Magnification, ×15,000. (c and d) High magnifications of spores of BMB171/pBMB1Ac304 and BMB201Ac/171, respectively. There were no obvious differences in spore coat (the wrinkled black layer under the exosporium) and cortex (the white layer under the coat). Magnification, ×100,000.
Bioassay results.
Bioassay results showed that the transformant containing P20 exhibited greater toxicity against neonate larvae of H. armigera than controls lacking P20 exhibited (Table 2). Bacteria were cultivated in ICPM liquid medium, and the bioassay was conducted with a serially diluted crude culture. BMB171/pBMB201Ac exhibited the greatest toxicity and its LC50 was 2.63 μl/ml, while the LC50s of BMB171/pBMB1Ac304 and BMB802 were 6.58 and 4.83 μl/ml, respectively (Table 2).
TABLE 2.
Toxicities of different cry1Ac10 transformants in an acrystalliferous plasmid-cured strain against H. armigera larvae
| Strain | LC50 (μl/ml)a |
|---|---|
| BMB171/pBMB201Ac | 2.63 (1.97–3.25) |
| BMB171/pBMB1Ac304 | 6.58 (5.15–8.43) |
| BMB802 | 4.83 (3.55–6.16) |
The results obtained after 72 h of infection are shown. ICPM medium was used to cultivate the bacteria. The LC50 was the amount of fermented medium per milliliter of infected diet. The values in parentheses are the 95% confidence intervals.
Role of P20.
The role of P20 in Cry1Ac10 synthesis was reflected by the variation in the 133-kDa protein concentration from the start of synthesis to crystal formation compared to the variation in a control lacking P20 (Fig. 5). Apparently, in the absence of P20, Cry1Ac protein seriously degraded during the whole process, especially during synthesis (Fig. 5A, lanes 1 and 2, and B), in contrast to the preparations containing P20 (Fig. 5A, lanes 7 and 8, and B). From 13 to 16 h of cultivation, the Cry1Ac10 protoxin concentration reached its maximum value, but this value was only one-half the value obtained for strain BMB171/pBMB201Ac containing P20, which was greatest from 16 to 19 h (Fig. 5B). The increase in both cases indicated that protein synthesis was occurring. This period lasted about 4 h and was concurrent with the active time of the BtI promoter of cry1Ac10 (2 to 6 h after the end of the exponential phase). Therefore, what P20 saved was the nascent peptides of Cry1Ac10 during synthesis. Afterwards, however, degradation to the protoxins occurred in both cases; the concentration of full-length Cry1Ac protoxin declined continuously from the end of synthesis to the emergence of crystals in sporulated cells (25 h) (Fig. 5B). Consequently, about one-third of the once-expressed protoxin was degraded in both cases during this period. This indicated that P20 could not protect expressed full-length Cry1Ac10 from degradation. From these results we concluded that the 20-kDa protein exerted its effects only on the nascent peptides, not on the mature proteins.
FIG. 5.
Variation in the concentration of Cry1Ac10 during its expression and the role of helper protein P20. (A) Lanes 1 to 5, expression of Cry1Ac10 in BMB171/pBMB1Ac304 after 13, 16, 19, 22, and 25 h of cultivation, respectively; lanes 6 to 10, expression of Cry1Ac10 in BMB171/pBMB201Ac after 13, 16, 19, 22, and 25 h of cultivation, respectively. (B) Densitometry of the 130-kDa bands in panel A.
Figure 5 shows that the times that expression of Cry1Ac10 started in two strains were not the same, although the actual times should be a little earlier than the times detected by SDS-PAGE because the samples were prepared at 3-h intervals. In strain BMB171/pBMB201Ac containing P20, Cry1Ac10 protoxin was first detected after 16 h of incubation (Fig. 5A, lane 7), about 3 h later than it was detected in the control lacking P20 (lane 1). Although the two strains had the same growth rate, when P20 was present the appearance of crystals in mother cells occurred some time later, as did the lysis of sporulated cells. This was more obvious in a more nutritious medium (NSM) than in ICPM.
DISCUSSION
Generally, expression of pesticidal proteins in B. thuringiensis is directly related to their toxicity. Several ways have been proposed to achieve a high yield of B. thuringiensis toxins in vivo; these include putting the cry gene under control of an effective promoter, increasing the gene copy number, promoting gene coexpression, and avoiding intracellular protease degradation (6, 9, 11, 26). In this study, we enhanced production of the 133-kDa protoxin of Cry1Ac by using helper protein P20 from B. thuringiensis subsp. israelensis. Usually, the bipyramidal crystals of Cry1 protoxins are approximately 1 to 1.5 μm long (4). In the presence of P20, the yield of Cry1Ac protein increased 2.5-fold (Fig. 2), and the protein formed larger crystals (average length, 1.85 μm); in contrast, the average length of the crystals in the control lacking P20 was 1.45 μm (Table 1). As expected, recombinant strain BMB171/pBMB201Ac harboring cry1Ac10 and p20 exhibited greater toxicity against H. armigera larvae than strain BMB171/pBMB1Ac304 lacking p20 exhibited (Table 2).
P20 has been shown to promote expression of several other proteins of B. thuringiensis subsp. israelensis and is supposed to act as a molecular chaperon during expression of other proteins (see below). P20 was first discovered during a study of cyt1A expression and is essential for efficient production of Cyt1A (1, 34, 36). Other genes that coexist with p20, such as cry4A and cry11A, are also expressed more abundantly with the help of P20 in E. coli (34, 38). Furthermore, when the p20 gene is driven by the stronger promoters of cry1Ac (other than those naturally harbored in the cry11A operon control region as the third open reading frame), larger crystals of Cry11A.are obtained (10, 37). Although Chang et al. hypothesized that the high level of expression of Cyt1A- and Cry11A-encoding genes in B. thuringiensis does not require P20 but requires other coexpressed proteins (6), later reports confirmed that P20 does play a unique role in the production of the two B. thuringiensis subsp. israelensis proteins (36, 37). The discrepancy in the findings may have been due to the effects of another helper protein, P19, which had not been discovered yet (2, 10).
Interestingly, P20 also promotes expression of Cry proteins from B. thuringiensis subspecies other than B. thuringiensis subsp. israelensis. For instances, Ge et al. reported that P20 increases production of Cry2A by 15%, although it does not have any effect on formation of Cry2A crystals, which is dependent on another accessory protein, ORF2, encoded by the cry2A operon (13). Similar to Cry2A, a number of Cry1C proteins whose toxic moieties are mutated which are unable to express and form crystals normally also have improved expression when P20 is present (27, 28). In our study, the effect of P20 on Cry1Ac was quite magnificent. P20 doubled the yield of Cry1Ac protoxin and resulted in bigger crystals in the engineered strain. However, in another case, P20 increased expression of Cry1Ab in the same recipient strain, BMB171, but no obvious crystals were observed (39).
Although P20 promoted production of most of the proteins tested, it did not do so for all proteins. At the very beginning of their study on the effect of P20, Visick and Whiteley reported that P20 improved expression of Cyt1A and Cry11A (CryIVD) at the posttranslational level, but it did not have similar effects on Cry1B and mutated Cry1A in E. coli (34). Recently, Lee and Gill reported that P20 failed to have a promoting effect on Cry20Aa, an 86-kDa mosquitocidal protein of B. thuringiensis subsp. fukuokaensis (20).
The manifest enhancement of Cry1Ac production by P20 implied that Cry1Ac underwent substantial degradation when P20 was absent, but how does this happen? Large protoxins, such as Cry1A, which are different from small protoxins, such as Cyt1A and Cry2Aa, are usually thought to have no problems in expression. It is well known that Cry1 protoxins usually have high yield and form large crystals (Cry1Ab is the exception). This is largely due to their highly conserved C-terminal halves, which take part in autoassembly (2, 5), while Cry2 protoxins without these regions need direct participation of ORF2 to form small cubic crystals. In this study, through monitoring the variations in Cry1Ac concentration during sporulation, we found that serious degradation of Cry1Ac occurred during its synthesis and even afterwards (Fig. 5). Actually, most Cry1Ac peptides are lost before crystallization, and the degradation during peptide synthesis is much more serious than that after synthesis; i.e., the nascent peptides of Cry1Ac are more vulnerable than the mature proteins. In nature, prompt crystallization is an effective way to preserve the expressed products. It is evident that the degradation in this case was the result of intracellular proteases. The highest endoprotease activity in vivo occurs concurrent with expression of toxins in the growth phase of B. thuringiensis (19). This most likely meets the nutrient requirement in secondary metabolism for accumulation of Cry proteins, but paradoxically, it results in proteolysis of Cry proteins as a side effect.
To reduce the negative effects of proteases on B. thuringiensis protoxin accumulation, an alternative strategy is to curb or modulate protease activity. Partial degradation of full-length protoxins by intracellular proteases has been reported in several subspecies, such as B. thuringiensis subsp. tenebrionis, B. thuringiensis subsp. isrealensis, and B. thuringiensis subsp. kurstaki (10, 17, 24, 33). The size of B. thuringiensis subsp. tenebrionis protoxins decreased during sporulation, and proteolysis was prevented by a protease inhibitor (24). In another case, intracellular proteases in sporulated cells of B. thuringiensis subsp. kurstaki were identified as the enzymes responsible for generating a 66-kDa fragment derived from 130-kDa protoxins (17); later, three proteases were identified as enzymes that were involved in this process, and they were significantly inhibited by 1,10-phenanthroline (32). These proteases were likely to be metalloproteases in B. thuringiensis subsp. kurstaki, as recently reviewed by Oppert (24). To avoid the negative effects of proteases, deleting the neutral protease A gene was tried, which resulted in a high concentration of full-length protoxins of Cry1Bb and Cry3Bb. The neutral protease A-deficient culture yielded approximately 1.3 times more Cry1Bb protein than the culture containing neutral protease A (11). Recently, deletion of an alkaline protease gene resulted in similar effects on Cry1Bb production (33). However, the deletion might retard bacterial growth, especially in a complex medium, and thus possibly counteract the effect of the deletion on final production. Although we are not sure which kind of protease was responsible for the serious degradation of Cry1Ac in our study, the degradation may have been the result of a combination of multiple proteases. In any case, expression of some kind of key protease inhibitor in the cellular plasma might help further enhance production of crystal proteins in a recombinant strain.
In this study, P20 not only enhanced Cry1Ac production but also affected sporulation of the engineered strain. We found that when P20 was present, the appearance of spores in cells was delayed by several hours. Probably, this was an effect of P20 on proteins related to sporulation. Increased concentrations of certain regulatory factors might keep cells at one stage longer. In addition, the presence of P20 resulted in morphological changes in spores, which were round instead of elliptical and smaller (Fig. 3 and 4). Whether this was an effect of P20 on expression of morphological proteins or just due to space limitations in sporangia containing large crystals is not known. However, in a previous report, it was found that crystal proteins can be deposited on the spore surface in B. thuringiensis and that many acrystalliferous mutants of B. thuringiensis subsp. kurstaki lack a well-defined spore coat, although they still have the potential to form a complete spore coat under certain conditions (4). In our study, no obvious changes in the spore coats were observed in strains that differed in Cry1Ac production. However, the thickness of the exosporium of the strain containing P20 was remarkably reduced (Fig. 4). More recently, it was found that the prominent component of B. thuringiensis exosporium is a glycoprotein multimer composed of a 54-kDa protein and at least three species of oligosaccharides (12). The exosporium is relatively hydrophobic, and a reduction in the thickness of this layer might result in a reduced ability to survive in an adverse environment.
Concerning the role of P20 in protein expression, our results support the molecular chaperon hypothesis. In the early studies of P20, Mclean and Whiteley found that P20 acts either during synthesis of the Cyt1A protein or posttranslationally but not on a transcriptional level (23). Later, Visick and Whiteley reported that P20 could be coimmunoprecipitated with Cyt1A protein, and this occurred only during Cyt1A synthesis (34). This indicates that P20 acts only on the nascent Cyt1A peptide (34). During Cry1Ac synthesis in this study, P20 saved more nascent peptides to mature molecules, which resulted in double production of Cry1Ac, whereas after synthesis the protoxin concentration decreased continuously as the amount of the control lacking P20 decreased (Fig. 5). Therefore, P20 acted only on the nascent peptides, not on the full-length protoxins. This supports the hypothesis that P20 is a molecular chaperon in Cry1Ac expression. Generally, nascent peptides need this kind of molecule to help them fold correctly and promptly to avoid attacks by intracellular proteases during the process of translation (14, 15). Molecular chaperons are usually conserved and are ubiquitous in the cells of various organisms. In B. thuringiensis subspecies other than B. thuringiensis subsp. israelensis, chaperons that resemble P20 might be present and contribute to the high levels of expression of Cry proteins. Interestingly, we repeatedly identified the specific fragment of the p20 gene by PCR in different B. thuringiensis strains, but unfortunately, we failed to prove its presence in the genome by Southern blotting (data not shown).
At this point, it is not clear how P20 interacts with its targets and what kind of structural feature P20 prefers to interact with. However, based on all of the results obtained, we believe that P20 has wide adaptability to other crystal proteins. Considering its marked effect on Cry1Ac production, we believe that P20 has great potential for improving the production of heterogeneous proteins in B. thuringiensis or in other organisms.
ACKNOWLEDGMENTS
This work was supported in part by the Chinese National Project for the Development of Science & High Technology (item 101-03-01-01) and by the Open Foundation of Key Laboratory of Agro-Microbiology of the Chinese Agricultural Ministry.
We are grateful to John Smith (Foreign Language College, Shandong Agricultural University) and Patrick McGlinchey (Texas Christian University, Fort Worth, Tex.), who kindly read the manuscript. We also thank to Xu Shijin for her help with the insect bioassay and Guo Yankui (Life Science College, Shandong Agricultural University) for his assistance with SEM.
REFERENCES
- 1.Adams L F, Visick J E, Whiteley H R. A 20-kilodalton protein is required for efficient production of the Bacillus thuringiensis subsp. israelensis 27-kilodalton crystal protein in Escherichia coli. J Bacteriol. 1989;171:521–530. doi: 10.1128/jb.171.1.521-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agaisse H, Lereclus D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 4.Aronson A, Bechman W, Dunn P. Bacillus thuringiensis and related insect pathogens. Microbiol Rev. 1986;50:1–24. doi: 10.1128/mr.50.1.1-24.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum J A, Malvar T. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol. 1995;18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x. [DOI] [PubMed] [Google Scholar]
- 6.Chang C, Yu Y M, Dai S M, Law S K, Gill S S. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore N, Ellar D J. Improvement of a possible chaperon in the efficient expression of a cloned CryIIA δ-endotoxin gene in Bacillus thuringiensis. Mol Microbiol. 1992;6:1533–1537. doi: 10.1111/j.1365-2958.1992.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 8.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:808–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delecluse A, Poncet S, Klier A, Rapoport G. Expression of cryIVA and cryIVB genes independently or in combination in a crystal-negative strain of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1993;59:3922–3927. doi: 10.1128/aem.59.11.3922-3927.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dervyn E, Poncet S, Klier A, Rapoport G. Transcriptional regulation of the cryIVD gene operon from Bacillus thuringiensis subsp. israelensis. J Bacteriol. 1995;177:2283–2291. doi: 10.1128/jb.177.9.2283-2291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan W P, Tan Y, Slaney A C. Cloning of the nprA gene for neutral protease A of Bacillus thuringiensis and effect of in vivo deletion of nprA on insecticidal crystal protein. Appl Environ Microbiol. 1997;63:2311–2317. doi: 10.1128/aem.63.6.2311-2317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Patrone M, Tandecarz J S. A glycoprotein multimer from Bacillus thuringiensis sporangia: dissociation into subunits and sugar composition. Mol Cell Biochem. 1995;145:29–37. doi: 10.1007/BF00925710. [DOI] [PubMed] [Google Scholar]
- 13.Ge B X, Bideshi D, Moar W J, Federici B A. Differential effects of helper proteins encoded by the cry2A and cry11A operons on the formation of Cry2A inclusions in Bacillus thuringiensis. FEMS Microbiol Lett. 1998;165:35–41. doi: 10.1111/j.1574-6968.1998.tb13124.x. [DOI] [PubMed] [Google Scholar]
- 14.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 15.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;318:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 16.Hofte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar N S, Venkateswerlu G. Analysis of 66 KDA toxin from Bacillus thuringiensis subsp. kurstaki reveals differential amino terminal processing of protoxin by endogenous protease(s) Biochem Mol Biol Int. 1998;45:769–774. doi: 10.1080/15216549800203182. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli L K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lecadet M M, Lescourret M, Klier A. Characterization of an intercellular protease isolated from Bacillus thuringiensis sporulating cells and able to modify RNA polymerase. Eur J Biotechnol. 1977;79:329–338. doi: 10.1111/j.1432-1033.1977.tb11813.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee H K, Gill S S. Molecular cloning and characterization of a novel mosquitocidal protein gene from Bacillus thuringiensis subsp. fukuokaensis. Appl Environ Microbiol. 1997;63:4664–4670. doi: 10.1128/aem.63.12.4664-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Yang C, Liu Z, Li F, Yu Z. Screening of acrystalliferous mutants from Bacillus thuringiensis and their transformation properties. Acta Microbiol Sin. 2000;40:85–90. [PubMed] [Google Scholar]
- 22.Liu Z-D, Sun M, Chen Y-H, Yu Z-N. The influence of the 20kDa protein from Bacillus thuringiensis subsp. israelensis on the cytolytic activity of CytA. Acta Genet Sin. 1999;26:81–86. [PubMed] [Google Scholar]
- 23.Mclean K, Whiteley H R. Expression in Escherichia coli of a cloned crystal protein gene of Bacillus thuringiensis subsp. israelensis. J Bacteriol. 1987;169:1017–1023. doi: 10.1128/jb.169.3.1017-1023.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oppert B. Protease interaction with Bacillus thuringiensis insecticidal toxins. Arch Insect Biochem Physiol. 1999;42:1–12. doi: 10.1002/(SICI)1520-6327(199909)42:1<1::AID-ARCH2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 25.Padidam M. The insecticidal protein CryIA(c) from Bacillus thuringiensis is highly toxic for Heliothis armigera. J Invertebr Pathol. 1992;59:109–111. doi: 10.1016/0022-2011(92)90121-j. [DOI] [PubMed] [Google Scholar]
- 26.Park H W, Ge B-X, Bauer L S, Federici B A. Optimization of Cry3A yields in Bacillus thuringiensis by use of sporulation-dependent promoters in combination with the STAB-SD mRNA sequence. Appl Environ Microbiol. 1998;64:3932–3938. doi: 10.1128/aem.64.10.3932-3938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H W, Bideshi D K, Federici B A. Molecular genetic manipulation of truncated Cry1C protein synthesis in Bacillus thuringiensis to improve stability and yield. Appl Environ Microbiol. 2000;66:4449–4455. doi: 10.1128/aem.66.10.4449-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rang C, Bes M, Lullien-Pellerin V, Wu D, Federici B A, Frutos R. Influence of the 20kDa protein from Bacillus thuringiensis ssp. israelensis on the rate of production of truncated Cry1C proteins. FEMS Microbiol Lett. 1996;141:261–264. doi: 10.1111/j.1574-6968.1996.tb08395.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun M, Liu Z-D, Yu Z-N. Characterization of the insecticidal crystal protein genes of Bacillus thuringiensis YBT-1520. Acta Microbiol Sin. 2000;41:365–371. [PubMed] [Google Scholar]
- 32.Suresh N, Venkateswerlu G. Intracellular proteases in sporulated Bacillus thuringiensis subsp. kurstaki and their role in protoxin activation. FEMS Microbiol Lett. 1998;166:377–382. [Google Scholar]
- 33.Tan Y, Donovan W P. Deletion of aprA and nprA genes for alkaline protease A and neutral protease A from Bacillus thuringiensis: effect on insecticidal crystal proteins. J Biotechnol. 2001;84:67–72. doi: 10.1016/s0168-1656(00)00328-x. [DOI] [PubMed] [Google Scholar]
- 34.Visick J E, Whiteley H R. Effects of a 20-kilodalton protein from Bacillus thuringiensis subsp. israelensis on production of CytA protein by Escherichia coli. J Bacteriol. 1991;173:1748–1756. doi: 10.1128/jb.173.5.1748-1756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong H C, Schnepf H E, Whiteley H R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983;258:1960–1967. [PubMed] [Google Scholar]
- 36.Wu D, Federici B A. A 20-kilodalton protein preserves cell viability and promotes CytA crystal formation during sporulation in Bacillus thuringiensis. J Bacteriol. 1993;175:5276–5280. doi: 10.1128/jb.175.16.5276-5280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu D, Federici B A. Improved production of the insecticidal CryIVD protein in Bacillus thuringiensis using cryIA(c) promoters to express the gene for an associated 20-kDa protein. Appl Microbiol Biotechnol. 1995;42:697–702. doi: 10.1007/BF00171947. [DOI] [PubMed] [Google Scholar]
- 38.Yoshisue H, Yoshida K, Sen K, Sakai H, Komano T. Effects of Bacillus thuringiensis 20-kDa protein on production of the Bti 130-kDa crystal protein in Escherichia coli. Biosci Biotechnol Biochem. 1992;56:1429–1433. doi: 10.1271/bbb.56.1429. [DOI] [PubMed] [Google Scholar]
- 39.Zheng R. M.S. thesis. Wuhan, People's Republic of China: Huazhong Agricultural University; 1998. [Google Scholar]