Abstract
Cadmium (Cd) is a heavy metal that is highly toxic for plants, animals, and human beings. A better understanding of the mechanisms involved in Cd accumulation in plants is beneficial for developing strategies for either the remediation of Cd-polluted soils using hyperaccumulator plants or preventing excess Cd accumulation in the edible parts of crops and vegetables. As a ubiquitous heavy metal, the transport of Cd in plant cells is suggested to be mediated by transporters for essential elements such as Ca, Zn, K, and Mn. Identification of the genes encoding Cd transporters is important for understanding the mechanisms underlying Cd uptake, translocation, and accumulation in either crop or hyperaccumulator plants. Recent studies have shown that the transporters that mediate the uptake, transport, and accumulation of Cd in plants mainly include members of the natural resistance-associated macrophage protein (Nramp), heavy metal-transporting ATPase (HMA), zinc and iron regulated transporter protein (ZIP), ATP-binding cassette (ABC), and yellow stripe-like (YSL) families. Here, we review the latest advances in the research of these Cd transporters and lay the foundation for a systematic understanding underlying the molecular mechanisms of Cd uptake, transport, and accumulation in plants.
Keywords: Cadmium, transporters, Nramp, HMA, ABC, ZIP, YSL
1. Introduction
Cadmium (Cd) is a heavy metal that is highly toxic to animals and plants, ranking first among inorganic pollutants. Cd enters the soil–plant environment through natural processes and anthropogenic activities [1]. Natural processes include volcanic eruptions and soil erosion, and anthropogenic activities include power stations, heating systems, and urban transportation [2,3]. Soil pollution by heavy metals, including Cd, is essentially an irreversible process that may take hundreds of years to recover from. Cd accumulation in plants inhibits Fe(III) reductase activity, leading to Fe(II) deficiency that in turn affects photosynthesis [4]. Plants affected by Cd toxicity in polluted soils usually present retarded growth, chlorotic leaves, and brown root tips. Compared with other heavy metals, such as Pb, Cd is more soluble and easily absorbed by plants, and is subsequently accumulated in their edible parts, thus entering the food chain and posing a threat to humans [1]. An excessive intake of Cd in humans can damage the kidneys, leading to rhinitis, emphysema, and osteomalacia [5]. In recent years, Cd has become one of the major soil pollutants worldwide due to uncontrolled industrialization, unsustainable urbanization, and intensive agricultural practices. The itai—itai disease is the most serious chronic Cd poisoning caused by long-term oral consumption of Cd in Japan [6]. In China, Cd is the most severe pollutant in agricultural soils, with a site-level rate as high as 7.0% [7,8,9], and Cd soil pollution further shows an increasing trend from North to South China [10]. Field surveys showed that Cd concentrations in a considerable proportion of rice grains, especially in those grown in South China, exceeded the recommended food safety standard in the country [11,12,13]. One strategy to prevent Cd food contamination is to find and create more Cd low-accumulating cultivars of crops and vegetables using genetic breeding, and alleviation of Cd soil pollution can be achieved through phytoremediation utilizing high-accumulating plants. Therefore, understanding the physiological and molecular mechanisms of Cd uptake, transport, and accumulation by plants is of great significance for formulating strategies for phytoremediation of Cd-contaminated soils or prevention of Cd accumulation in crops.
An increasing number of studies have been conducted on the Cd migration pathway in plants, providing detailed information on the mechanism of Cd transport. There are four major processes that mediate Cd transport from roots to shoots: (1) root uptake; (2) loading into the root xylem; (3) long-distance translocation via the xylem and phloem pathways; (4) phloem re-translocation [14,15]. Plants absorb heavy metals by either active or passive absorption, with the root tips being the main Cd-absorbing area [16]. As a non-essential element, Cd2+ can enter the root through ion channels permeable to essential elements such as Ca2+ and K2+ [17,18]. It can also enter plant cells actively via uptake systems for essential elements such as Zn and Fe [19]. After root absorption, loading into the root xylem is one of the most critical steps for Cd transport [14]. Cd2+ or various Cd chelates can complete xylem loading through the symplast or the apoplast pathways [16]. The symplast pathway uses plasmodesmata to transport heavy metals between the cells, finally transporting them to the central column. The apoplast pathway transports water and heavy metals through the intercellular spaces or the cell wall continuum [14,20]. After Cd is loaded into the root xylem, it needs to be transported through the xylem and phloem for long-distance transport to the shoots. Phloem can serve as a major transport route for long-distance source-to-sink Cd transport via Cd–phytochelatin (PC) and Cd–glutathione complexes [21]. In addition, the phloem is primarily responsible for nutrient re-translocation, and in the Sedum alfredii Hance hyperaccumulating ecotype (HE), efficient phloem transport retransfers Cd from old to young organs [22].
Many transporter protein families are involved in the process of plant Cd uptake from the soil to be re-transported through the phloem. Clarifying the functions of these transporters regulating Cd and its chelates is important to understand the molecular mechanisms of plant responses to Cd. Thus far, the identified Cd transporters mainly include members of the natural resistance-associated macrophage protein (NRAMP), heavy metal-transporting ATPases (HMA), zinc and iron regulated transporter protein (ZIP), ATP-binding cassette (ABC), and yellow stripe-like (YSL) families.
2. Natural Resistance-Associated Macrophage Proteins
Nramps represent a class of metal transporters widely present in plants that are mainly involved in the absorption and transport of Fe2+, Mn2+, Cd2+, and other metal ions [23,24]. The involvement of Nramp genes in Cd transport was first reported in the model plant Arabidopsis thaliana. In recent years, research has been focused on food crops such as Oryza sativa, Triticum polonicum and Fagopyrum esculentum, and hyperaccumulator plants have also been explored. These proteins have also been identified in other plants.
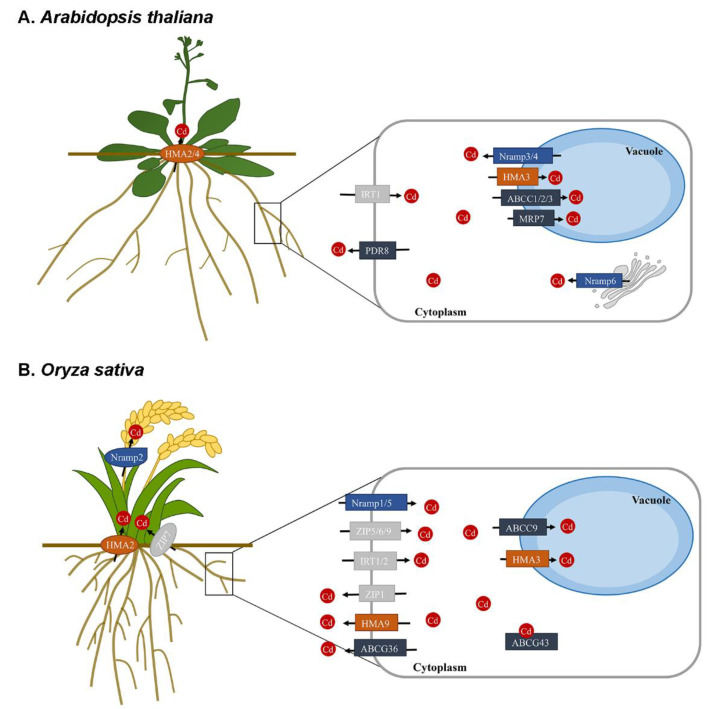
In A. thaliana, four Nramp genes have been found to be related to Cd transport. Overexpression of AtNramp1 increased Cd sensitivity and accumulation in yeast (Table 1) [25]. AtNramp3 and AtNramp4 encode tonoplast-localized proteins, and yeast expressing the two genes showed an increased sensitivity to Cd (Figure 1, Table 1). Overexpression of AtNramp3 in Arabidopsis conferred hypersensitivity to Cd [25,26,27,28], but overexpression of AtNramp4 in A. thaliana only conferred a slight hypersensitivity to Cd [25,29]; AtNramp3 and AtNramp4 can also mediate the transport of Cd out of the vacuoles in Arabidopsis [25,28]. AtNramp6 is a Cd transporter that can either transport Cd out of its storage compartment or into the toxic cellular compartment (Figure 1, Table 1) [30].
Table 1.
Genes encoding Natural Resistance-Associated Macrophage Proteins (Nramp) for Cd transport in plants.
| Plant Species | Genes | Expression Sites | Subcellular Location | Function | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AtNramp1 | Roots | Plasma membrane | - | [25] |
| AtNramp3 | Roots and aerial parts | Tonoplast | Cd transport | [25,26,27,28] | |
| AtNramp4 | Roots and aerial parts | Tonoplast | Cd transport | [25,28,29] | |
| AtNramp6 | Seed embryo, lateral roots and young leaves | Golgi/trans-Golgi network | Cd transport | [30] | |
| Oryza sativa L. | OsNramp1 | Roots and leaves | Plasma membrane | Cd uptake and translocation | [31,32,33] |
| OsNramp2 | Embryo of germinating seeds, roots, leaf sheaths and leaf blades | Tonoplast | Cd retranslocation | [34,35] | |
| OsNramp5 | Roots epidermis, exodermis, outer layers of cortex and tissues around xylem | Plasma membrane | Cd uptake | [36,37,38,39] | |
| Triticum polonicum L. | TpNramp3 | leaf blades and roots at the jointing and booting stages, first nodes at the grain filling stage | Plasma membrane | Cd accumulation | [40] |
| TpNramp5 | Roots and basal stems of DPW | Plasma membrane | Cd accumulation | [41] | |
| Triticum turgidum L. | TtNramp6 | Roots | Plasma membrane | Cd accumulation | [42] |
| Hordeum vulgare | HvNramp5 | Roots | Plasma membrane | Cd uptake | [43] |
| Fagopyrum esculentum Moench | FeNramp5 | Roots | Plasma membrane | Cd uptake | [44] |
| Brassica napus | BnNramp1b | Vegetative tissue, flowers and siliques | - | - | [45] |
| Brassica rapa L. | BcNramp1 | Roots | Plasma membrane | Cd uptake | [46] |
| Noccaea caerulescens (Thlaspi caeulescens) | NcNramp1 | Roots and shoots | Plasma membrane | - | [47] |
| TcNramp3 | - | Tonoplast | - | [48,49] | |
| TcNramp4 | - | Tonoplast | - | [48] | |
| Sedum alfredii Hance | SaNramp1 | Young tissues of the shoots | Plasma membrane | Cd translocation | [50] |
| SaNramp3 | - | - | Cd translocation | [51] | |
| SaNramp6 | Roots | Plasma membrane | Cd uptake or translocation | [52,53] | |
| Malus xiaojinensis | MxNramp1 | Roots | Plasma membrane | Cd uptake and translocation | [54] |
| MxNramp3 | Roots and leaves | Tonoplast | Cd uptake and translocation | [54] | |
| Malus hupehensis | MhNramp1 | Roots | Cell membrane | Cd uptake | [55] |
| Spirodela polyrhiza | SpNramp1 | - | Plasma membrane | Cd accumulation | [56] |
| SpNramp2 | - | Plasma membrane | Cd accumulation | [56] | |
| SpNramp3 | - | Plasma membrane | - | [56] | |
| Crotalaria juncea | CjNramp1 | Leaves, stems, and roots | Plasma membrane | Cd uptake and translocation | [57] |
| Nicotiana tabacum | NtNRAMP1 | Roots | - | Cd uptake | [58] |
| NtNRAMP3 | Conductive tissue of leaves |
Tonoplast | Cd efflux | [59] |
Figure 1.
Uptake and transport of Cd. (A) In Arabidopsis thaliana, AtIRT1 is involved in Cd uptake by the roots. After Cd enters the root cells, it can be sequestered into the vacuole via AtHMA3, AtABCC1, AtABCC2, AtABCC3, and AtMRP7. AtNramp3 and AtNramp4 mediate the transport of Cd from the vacuole into the cytoplasm, while AtNramp6 transport Cd out of its storage compartment. AtHMA2 and AtHMA4 are involved in xylem loading to transport Cd to the shoots. Moreover, AtPDR8 mediates Cd efflux. (B) In O. sativa, OsNramp1, OsNramp5, OsZIP5, OsZIP6, OsZIP9, OsIRT1, and OsIRT2 are involved in Cd uptake by the rice roots. After Cd enters the root cells, it can be transported to the vacuoles, where it is sequestered, by OsHMA3 and OsABCC9. OsABCG43 also aids the sequestration of Cd in the roots. OsHMA2 and OsZIP7 are involved in xylem loading to transport Cd to the shoots. OsNramp2 mediates Cd re-translocation to the grains. Moreover, OsZIP1, OsHMA9, and OsABCG36 mediate Cd efflux in roots.
Nramp genes involved in the transport of Cd are mainly studied in rice among food crops. Three Nramp genes have been identified to be functionally associated with Cd. OsNramp1, a transporter localized in the plasma membrane responsible for Cd uptake and transport within plants, is mainly expressed in the roots and the leaves and is localized in all root cells except the central vasculature and in leaf mesophyll cells (Figure 1, Table 1) [32,33]. Tiwari et al. [31] observed that OsNramp1 is involved in xylem-mediated loading and that it increased the accumulation of As and Cd in plants by heterologous expression of OsNramp1 in Arabidopsis. However, Chang et al. [33] showed that OsNramp1 transported Cd and Mn when expressed in yeast but did not transport Fe or As. Overexpression of OsNramp1 in rice reduced Cd accumulation in the roots, but increased it in the leaves. Knockout of OsNramp1 resulted in decreased Cd and Mn uptake by the roots and their accumulation in the shoots and the grains [32,33].
OsNramp2 is localized in the tonoplast and mainly expressed in the embryo of germinating seeds, roots, leaf sheaths, and leaf blades (Figure 1, Table 1) [35]. The knockout of OsNramp2 significantly decreased Cd concentration in the grains, but increased it in the leaves and the straws, suggesting that it mediates Cd efflux from the vacuoles in the vegetative tissues for translocation to the grains [34,35].
OsNramp5 encodes a plasma membrane protein polarly localized at the distal side of both exodermis and endodermis cells, and responsible for the influx of Mn and Cd into root cells from external solutions (Figure 1, Table 1) [37,38]. Knockout of OsNramp5 significantly reduced Cd concentration in the roots and shoots [38,39]. In a Cd-contaminated paddy field experiment, it was found that Cd concentration in the grains of the knockout line was much lower than that of the wild-type (WT) [39]. Surprisingly, the overexpression of OsNramp5 enhanced Cd root uptake, but significantly reduced its accumulation in the shoots and grains. Xylem loading was also disturbed in OsNramp5-overexpressing plants, with a reduced translocation from the roots to the shoots [36].
In Triticum polonicum L and Triticum turgidum L, TpNramp3, TpNramp5, and TtNramp6 encode plasma membrane proteins (Table 1). Overexpression of TtNramp6 increased Cd concentration and its accumulation in the whole plant of Arabidopsis [42]. Overexpression of TpNramp3 or TpNramp5 also increased the concentrations of Cd, Co, and Mn in the whole plant [40,41]. In Hordeum vulgare, HvNramp5 encodes a plasma membrane-localized transporter required for the uptake of Cd and Mn, but not Fe (Table 1), that presents 84% identity with OsNramp5. HvNramp5 was mainly expressed in the roots, with higher expression levels in the root tips than in the basal region [43]. Knockout of HvNramp5 in barley resulted in reduced concentrations of Mn and Cd in the roots and shoots but did not change the concentrations of other metals [43]. In Fagopyrum esculentum Moench, the plasma membrane-localized transporter FeNramp5 is responsible for the uptake of Mn and Cd (Table 1). FeNramp5 can also complement the phenotype of an AtNramp1 Arabidopsis mutant in terms of growth and accumulation of Mn and Cd [44]. BnNramp1b is localized in the plasma membrane and can transport Cd (Table 1) [45]. Yue et al. demonstrated that BcNramp1 plays a role in Cd influx of Arabidopsis root cells using noninvasive microelectrode ion flux measurements (Table 1) [46].
Studies on Nramp Cd-transporting genes in hyperaccumulator plants are mainly focused on Noccaea caerulescens (Thlaspi caerulescens) and Sedum alfredii Hance. In N. caerulescens, NcNramp1 participates in the influx of Cd across the endodermal plasma membrane and thus may play an important role in the Cd flux into the stele and its root-to-shoot translocation (Table 1) [47]. TcNramp3 and TcNramp4 are localized in the tonoplast (Table 1). TcNramp3 or TcNramp4 expression rescued Cd and Zn hypersensitivity induced by the inactivation of AtNramp3 and AtNramp4 in Arabidopsis [48]. Additionally, in overexpression tobacco lines, the roots were found to be more sensitive to Cd [49]. In the S. alfredii Hance, the plasma membrane-localized SaNramp1 transporter is highly expressed in the young tissues of the shoots (Table 1), and its overexpression in tobacco significantly increased Cd concentration at this location [50]. Ectopic expression of SaNramp3 in Brassica juncea enhanced Cd root-to-shoot translocation (Table 1), thus increasing Cd accumulation in the shoots [51]. Overexpression of SaNramp6, localized in the plasma membrane, increased Cd uptake and accumulation in A. thaliana (Table 1) [52]. Employing site-directed mutagenesis and functional analysis of mutants in yeast and Arabidopsis, the conserved L157 site in SaNramp6h was found to be critical for metal transport [53].
Nramp genes have also been identified in other plants. MxNramp1 (localized in the plasma membrane) and MxNramp3 (localized in the tonoplast) can transport Cd in yeast (Table 1) [54]. In Malus hupehensis, overexpression of MhNramp1 increases Cd uptake and accumulation, thereby exacerbating cell death (Table 1) [55]. SpNramp1, SpNramp2, and SpNramp3 are plasma membrane-localized transporters in Spirodela polyrhiza (Table 1), and overexpression of SpNramp1 or SpNramp2 increased Cd accumulation [56]. Similarly, overexpression of CjNramp1 in Arabidopsis resulted in high tolerance to Cd (Table 1) [57]. Furthermore, overexpression of NtNramp1 in tobacco could promote Cd uptake and Fe transportation (Table 1) [58], and the tonoplast-localized NtNramp3 transporter was found to be involved in the regulation of Cd transport from the vacuole to the cytoplasm using CRISPR/Cas9 technology (Table 1) [59].
3. Heavy Metal Transporting ATPases
HMAs play an important role in absorbing and transporting essential metal ions, such as Cu2+, Co2+ and Zn2+, by ATP hydrolysis; they can also transport Cd2+ and Pb2+. HMAs can be divided into two classes: those transporting monovalent cations (Cu, Ag) and those transporting divalent cations (Zn, Co, Cd, Pb) [60]. First described in A. thaliana, they have been studied more in food crops and hyperaccumulator plants in recent years due to their strong capacity to transport Cd; they have also been slightly less researched in other plants.
AtHMA2, AtHMA3, and AtHMA4 are reportedly associated with Cd transport in A. thaliana. AtHMA3 encodes a tonoplast-localized transporter that plays a role in Cd, Zn, Co, and Pb detoxification (Figure 1, Table 2) [64]. Overexpression of AtHMA3 enhanced Cd tolerance and increased its accumulation [63,64]. AtHMA2 and AtHMA4, localized in the plasma membrane, are responsible for the xylem loading of Zn/Cd and play a key role in their accumulation in the shoots (Figure 1, Table 2) [61,62,65]. Ceasar et al. [66] found that the di-cysteine residues at the C-terminus of HMA4 in A. thaliana were only partially required for Cd transport. Furthermore, ectopic expression of 35S::AtHMA4 reduced Cd accumulation due to the induction of the apoplastic barrier in tobacco [67].
Table 2.
Genes encoding Heavy Metal transporting ATPases (HMAs) for Cd transport in plants.
| Plant Species | Genes | Expression Sites | Subcellular Location | Function | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AtHMA2 | - | Plasma membrane | Cd translocation | [61,62] |
| AtHMA3 | Root apex | Tonoplast | Cd sequestration | [63,64] | |
| AtHMA4 | tissues surrounding the root vascular vessels | Plasma membrane | Cd translocation | [61,65,66,67] | |
| Oryza sativa L | OsHMA2 | in the mature zone of the roots at the vegetative stage | Plasma membrane | Cd translocation | [68,69,70] |
| OsHMA3 | Roots | Tonoplast | Cd sequestration | [71,72,73,74,75,76] | |
| OsHMA9 | vascular bundles and anthers |
Plasma membrane | Cd efflux |
[77] | |
| Triticum aestivum L. | TaHMA2 | Nodes | Plasma membrane | Cd translocation | [78] |
| Glycine max | GmHAM3w | Roots | Endoplasmic reticulum (ER) | Cd sequestration | [79] |
| Sedum plumbizincicola | SpHMA1 | Leaves | Chloroplast envelope | Cd efflux |
[80] |
| SpHMA3 | Leaves | Tonoplast | Cd sequestration | [81] | |
| Sedum alfredii Hance | SaHMA3 | Shoots | Tonoplast | Cd sequestration | [82] |
| Thlaspi caerulescens | TcHMA3 | Roots and shoots |
Tonoplast | Cd sequestration | [83] |
| TcHMA4 | Roots | - | [84] | ||
| Brassica juncea | BjHMA4 | Roots, stems and leaves | Cytosol | Cd translocation | [85] |
| Iris lactea | IlHMA2 | Roots | Plasma membrane | Cd translocation | [86] |
| Populus tomentosa Carr. | PtoHMA5 | - | - | Cd translocation | [87] |
The study of the HMA family is predominantly focused on food crops. Three Cd-transport associated HMA genes were identified in the genome of rice, one of the major food crops. The plasma membrane-localized transporter OsHMA2 is involved in the root-to-shoot translocation of Zn and Cd (Figure 1, Table 2). OsHMA2 is mainly expressed in the mature zone of the roots at the vegetative stage, with the C-terminal region being essential for Zn/Cd translocation into the shoots [68,69]. Moreover, at the reproductive stage, OsHMA2 also showed a high expression in the nodes. Knockout of OsHMA2 resulted in reduced Zn and Cd concentrations in the upper nodes and reproductive organs compared with the WT, suggesting that OsHMA2 participates in the transport of Zn and Cd through the phloem to developing tissues [70]. OsHMA3 is localized in the tonoplast and sequestrates Cd into the root vacuoles to reduce its translocation, thereby mitigating Cd poisoning (Figure 1, Table 2) [71,72,73,74]. Silencing of OsHMA3 resulted in increased root-to-shoot Cd translocation, whereas OsHMA3 overexpression markedly decreased root-to-shoot Cd translocation and increased Cd tolerance, while greatly reducing its concentration in the grains [72,75]. The C-terminal region, and particularly the region containing the first 105 amino-acids, has an important role in the activity of OsHMA3 [76]. OsHMA9 encodes a heavy metal (Cd, Cu, Zn, and Pb) efflux protein present in the plasma membrane (Figure 1, Table 2). Knockout of OsHMA9 results in higher Cd accumulation in the shoots compared with that of the WT, thus making the mutant sensitive to Cd [77]. Moreover, in Triticum aestivum L., overexpression of TaHMA2 improved the root-shoot Zn/Cd translocation (Table 2) [78]. In Glycine max (soybean), GmHAM3w restricts Cd to the endoplasmic reticulum, where it is localized, and in the roots to limit translocation to the shoots (Table 2). Overexpression of GmHMA3w increased Cd concentration in the roots and decreased it in the shoots [79].
As a popular tool for the remediation of Cd-contaminated soils, there have been many studies on HMA genes with Cd transport and sequestration functions in hyperaccumulator plants in recent years. SpHMA1 is an important efflux transporter localized in the chloroplast envelope and is responsible for exporting Cd from the chloroplast (Table 2), thus preventing Cd accumulation in Sedum plumbizincicola. Significantly increased Cd concentration in chloroplasts in SpHMA1 RNAi transgenic plants and CRISPR/Cas9-induced mutants compared to WT [80]. SpHMA3, localized in the tonoplast and expressed mainly in the shoots (Table 2), plays an important role in Cd detoxification in young leaves by sequestering Cd into the vacuole [81]. In S. alfredii, the tonoplast-localized transporter SaHMA3 is mainly expressed in shoots (Table 2). Its overexpression in tobacco significantly enhanced Cd tolerance and accumulation and greatly increased Cd sequestration in the roots [82]. Increased amounts of Cd were sequestered in the roots, but not in the leaf vacuoles, probably due to the heterologous expression. TcHMA3 is a tonoplast-localized transporter responsible for Cd sequestration into the leaf vacuoles in Thlaspi caeulescens (Table 2) [83]. TcHMA4 is involved in the active efflux of a large number of different heavy metals (Cd, Zn, Pb, and Cu) out of the cell (Table 2), with the C-terminus of the TcHMA4 protein being essential for heavy metal binding [84]. Moreover, BjHMA4R can significantly improve Cd tolerance and accumulation at low heavy metal concentrations by specifically binding to Cd2+ in the cytosol (Table 2) [85]. In other plants, IlHMA2 is a plasma membrane transporter involved in Cd root-to-shoot translocation (Table 2). The genes regulating Zn homeostasis were significantly down regulated in IlHMA2-silenced lines, compared with that in WT [86]. PtoHMA5 also participates in Cd root-to-shoot translocation (Table 2) [87].
4. ATP-Binding Cassette
This protein superfamily is one of the largest known superfamilies, with over 120 members in both A. thaliana and O. sativa. ABC transporters comprise four core domains (two nucleotide-binding and two transmembrane domains) [88] and are located in the plasma, vacuolar, and mitochondrial membranes, where they facilitate the transmembrane transport of substances via active transport [89,90,91,92]. The ABC family is further divided into 13 subfamilies, according to the size and domains of their members; the subfamilies involved in the transport of Cd and its chelates include the multidrug resistance-associated protein (MRP), pleiotropic drug resistance (PDR), and ABC transporter of the mitochondrion (ATM) subfamilies [93]. The current research on these three subfamilies is mainly focused on A. thaliana and O. sativa.
In A. thaliana, AtABCC1 and AtABCC2—two important tonoplast transporters—play an essential role in sequestering the PC–Cd(II) complexes to the vacuoles (Figure 1, Table 3), thereby reducing the metal concentration in the root cells and its translocation to the shoots [92]. AtABCC3 is involved in the vacuolar transport of the PC–Cd complexes (Figure 1, Table 3), with its activity being regulated by Cd and coordinated with the function of AtABCC1/AtABCC2 [94]. The expression levels of AtMRP6/AtABCC6 are significantly upregulated under Cd stress (Table 3) [95]. Overexpression of AtMRP7, which is localized in both the tonoplast and the plasma membrane (Figure 1, Table 3), increased Cd concentration in the leaf vacuoles and its retention in the roots in tobacco [96]. AtPDR8, located in the plasma membrane and the root epidermal cells, is an important efflux transporter that increases Cd tolerance by effluxing Cd2+ out of the root epidermal cells (Figure 1, Table 3). Overexpression of AtPDR8 improved Cd tolerance but did not affect its accumulation or that of Pb [91]. AtATM3 is a transporter localized in the mitochondrial membrane (Table 3), and its overexpression improved Cd tolerance and accumulation by increasing the biogenesis of Fe-S clusters and exporting them from the mitochondria into the cytosol in Arabidopsis [90]. Overexpression of AtATM3 in B. juncea conferred enhanced Cd and Pb tolerance by inducing the expression of its glutathione synthetase II (BjGSHII) and phytochelatin synthase 1 (BjPCS1) enzymes [97].
Table 3.
Genes encoding ATP-Binding Cassette (ABC) for Cd transport in plants.
| Plant Species | Genes | Expression Sites | Subcellular Location | Function | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AtABCC1 | - | Tonoplast | Cd sequestration | [92] |
| AtABCC2 | - | Tonoplast | Cd sequestration | [92] | |
| AtABCC3 | - | - | Cd sequestration | [94] | |
| AtMRP6/AtABCC6 | Xylem-opposite pericycle cells where lateral roots initiate | - | [95] | ||
| AtMRP7 | - | Plasma membrane and tonoplast | Cd sequestration | [96] | |
| AtPDR8 | Root epidermal cells | Plasma membrane | Cd efflux | [91] | |
| AtATM3 | Roots | Mitochondrial membrane | - | [90,97] | |
| Oryza sativa L. | OsABCC9 | Root stele | Tonoplast | Cd sequestration | [98] |
| OsABCG36 | Roots | Plasma membrane | Cd efflux | [99] | |
| OsABCG43 | Roots | - | Cd sequestration | [100] | |
| OsABCG48 | - | - | - | [101] | |
| Triticum aestivum | TaABCC13 | - | - | Cd uptake and transport | [102] |
| Rehmannia glutinosa | RgABCC1 | Roots | - | - | [103] |
| Populus tomentosa | PtoABCG36 | Roots | Plasma membrane | Cd efflux | [104] |
In O. sativa, OsABCC9 was predominantly expressed in the root stele after Cd treatment (Figure 1, Table 3). It mainly mediates Cd accumulation by sequestering of Cd into the root vacuoles, thereby reducing its translocation to the shoots and grains [98]. The plasma membrane-localized OsABCG36 transporter functions as a Cd extrusion pump (Figure 1, Table 3), thus increasing Cd tolerance by exporting it or its conjugates from the root cells in rice. Compared with the WT, OsABCG36 knockout had a significantly higher Cd accumulation in the root cell sap and significantly increased sensitivity to Cd [99]. Yeast heterologous expression indicated that OsABCG43 and OsABCG48 conferred Cd tolerance (Figure 1, Table 3); overexpression of OsABCG48 in rice reduced Cd concentration in the roots [100,101]. Similarly, in Triticum aestivum, TaABCC13 was reportedly involved in Cd uptake and transport (Table 3), as Cd concentration in the roots and shoots of TaABCC13:RNAi line decreased, compared with that of the WT [102].
In other plants, some ABC genes have also been found to have a Cd-transporting role. Yeast-expressed RgABCC1, found in Rehmannia glutinosa, increased Cd tolerance (Table 3) [103]. Similarly, PtoABCG36 reduced Cd concentration in plants by mediating its efflux (Table 3), thereby improving Cd tolerance [104].
5. Zinc- and Iron-Regulated Transporter Proteins
There are many members in the ZIP family, with all of them generally presenting eight transmembrane regions and metal ion-binding conserved domains that play a role in their transport. Not only can they transport essential metal ions such as Fe2+ and Zn2+, but also Cd2+ [105]. The first member of the ZIP family to be described was NcZNT1, found in N. caerulescens (Table 4) [106]. Overexpression of NcZNT1 enhanced the tolerance and accumulation of Zn and Cd in Arabidopsis, suggesting its involvement in the long-distance translocation of xylem loading from the roots to the shoots [107].
Table 4.
Genes encoding Zinc and Iron regulated transporter Protein (ZIP) and Yellow Stripe-Like proteins (YSL) for Cd transport in plants.
| Plant Species | Genes | Expression Sites | Subcellular Location | Function | References |
|---|---|---|---|---|---|
| Genes encoding Zinc and Iron regulated transporter Protein (ZIP) | |||||
| Noccaea caerulescens L. | NcZNT1 | roots and shoots | - | - | [106,107] |
| Arabidopsis thaliana | AtIRT1 | Roots | Plasma membrane | Cd uptake | [108,109] |
| Oryza sativa L. | OsIRT1 | Roots | Plasma membrane | Cd uptake | [108,109,110] |
| OsIRT2 | Roots | Plasma membrane | Cd uptake | [110] | |
| OsZIP1 | Roots | Endoplasmic reticulum (ER) and plasma membrane | Cd efflux | [111] | |
| OsZIP5 | Roots | Plasma membrane | Cd uptake | [112] | |
| OsZIP6 | Shoots and roots | - | Cd uptake | [113] | |
| OsZIP7 | parenchyma cells of vascular bundles in roots and nodes | Plasma membrane | Cd translocation | [114] | |
| OsZIP9 | Roots | Plasma membrane | Cd uptake | [112,115] | |
| Nicotiana tabacum var Xanthi | NtZIP4A/B | Leaves and roots | Plasma membrane | Cd translocation | [116,117] |
| Morus alba | MaZIP4 | - | Plasma membrane | - | [118] |
| Genes encoding Yellow Stripe-Like proteins (YSL) | |||||
| Miscanthus sacchariflorus | MsYSL1 | Stems | Plasma membrane | Cd translocation | [119] |
| Solanum nigrum | SnYSL3 | Vascular tissues and epidermal cells of the roots and stems | Plasma membrane | Cd translocation | [120] |
| Vaccinium ssp. | VcYSL6 | - | Chloroplast | - | [121] |
| Brassica juncea | BjYSL7 | Stems | Plasma membrane | Cd translocation | [122] |
In recent years, studies on the role of the ZIP family in Cd transport have mainly focused on O. sativa. OsIRT1 and OsIRT2 are the major transporters participating in Fe and Cd uptake as observed in an heterologous expression experiment in yeast (Figure 1, Table 4) [110]. The IRT1 protein, first described in A. thaliana, mediates the absorption of a variety of metals including Fe, Zn, and Cd (Figure 1, Table 4) [108,109]. Similarly, IRT1 has also been explored in pea seedlings, mulberry (Morus L.), Triticum polonicum L., and Hordeum vulgare. Overexpression of IRTI in Arabidopsis and rice increased their sensitivity to Zn and Cd [110,118,123,124,125,126]. OsZIP1, a metal efflux transporter, is localized in the endoplasmic reticulum and the plasma membrane and is mainly expressed in the roots (Figure 1, Table 4). Overexpression of OsZIP1 protects rice plants from an excess of Zn, Cu, and Cd by limiting metal accumulation in their tissues [111]. Plasma membrane-localized proteins OsZIP5 and OsZIP9 have influx transporter activity that functions synergistically in the Zn/Cd uptake in rice (Figure 1, Table 4). Overexpression of OsZIP9 markedly increased the Zn/Cd levels in the aboveground tissues in brown rice. OsZIP9 is also responsible for the uptake of Zn and Co into the root cells [112,115]. Employing electrophysiological techniques, Kavitha et al. [113] demonstrated the uptake of Cd by OsZIP6 (Figure 1, Table 4). OsZIP7 encodes a plasma membrane-localized protein responsible for Cd/Zn influx and is expressed in the parenchyma cells of vascular bundles in the roots and nodes (Figure 1, Table 4). Compared with the WT, an OsZIP7 knockout results in Zn and Cd retention in the roots and the basal ganglia, hindering their upward transmission and thus playing a role in xylem loading in the roots and inter-vascular transfer in the nodes to deliver Zn/Cd to the grains in rice [114].
ZIP genes related to Cd transport have also been reported in other plants. In Nicotiana tabacum, NtZIP4A and NtZIP4B are two copies of ZIP4, with 97.57% homology at the amino acid level. NtZIP4A/B is a plasma membrane-localized transporter that regulates Zn and Cd translocation from the roots to the shoots (Table 4) [116,117]. Similarly, MaZIP4 is also localized in the plasma membrane and has Cd transport activity (Table 4) [118].
6. Yellow Stripe-Like Proteins
The YSL family mediates the transmembrane transport of metal ions and chelates formed by metal ions and nicotinamide in plants, as well as the long-distance transport from the roots to the shoots [105]. YSL proteins were first reported to have a role in Fe transport, and then were subsequently found to participate in the transport of Cu, Zn, Cd, and Mn [127]. Members of this family involved in Cd transport include YSL1, YSL3, YSL6, and YSL7. MsYSL1 and SnYSL3 are plasma membrane-localized transporters responsible for long-distance Cd translocation from the roots to the shoots (Table 4). An excess of Cd reportedly stimulated their expression. Overexpression of MsYSL1 or SnYSL3 in Arabidopsis increased the Cd translocation ratio under Cd stress [119,120]. VcYSL6 is located in the chloroplast, and its expression is up-regulated under Cd induction (Table 4). Overexpression of VcYSL6 in tobacco increased Cd concentrations in the leaves [121]. BjYSL7 encodes a plasma membrane-localized metal–nicotinamide transporter (Table 4). The concentrations of Cd and Ni in the shoots of BjYSL7-overexpressing transgenic tobacco plants are significantly higher than that of WT plants, suggesting a role of BjYSL7 in Cd translocation from the roots to the shoots [122].
7. Conclusions and Further Perspectives
In this review, we outlined the role of transporters in the uptake and transport of Cd by plants. After long-term evolution, plants have formed a set of complex mechanisms to cope with Cd stress. The key role of transporters in it has also been confirmed by multiple studies, and excellent progress has been made in determining the localization, specific expression, and function of each protein family member. However, the regulatory network for Cd uptake and transport in plants is extremely large and involves multiple genes. For example, in O. sativa, OsZIP5 and OsZIP9 are tandem duplicates and act synergistically in Cd uptake [112]. OsNRAMP1 and OsNRAMP5 are involved in Cd uptake via roots and knocking out both these genes resulted in large decreases in the uptake of Cd, compared to the case for the knockout of either one of genes [33]. However, the functions of many genes and the relationships between them are still unknown. Therefore, the traditional way of examining a single gene can no longer meet the requirements of the post-genomic era, and the mutual synergy between functional genes should be explored further in future research. Moreover, unknown genes related to plant Cd uptake and transport and the synergistic relationship between these genes can be further explored by constructing mutants and using molecular biology techniques in future studies. This would contribute to our understanding of the vast regulatory network of genes involved in Cd uptake, translocation and accumulation. In addition, studying the functions of various genes and the mechanisms underlying these functions would help cultivate Cd-tolerant plants using transgenic technology, which would further be helpful to restore Cd-contaminated soil.
Author Contributions
Conceptualization, J.T. and L.L.; Data Curation, J.T.; Writing—Original Draft Preparation, J.T.; Writing—Review & Editing, L.L.; Visualization, L.L.; Supervision, L.L.; Project Administration, L.L.; Funding Acquisition, L.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Nature Science Foundation of China, grant number 41977130 and 31672235, and projects from the Natural Science 624 Foundation of Zhejiang Province, grant number LZ22C150004, and by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences, grant number DE-AC02-76SF00515.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sarwar N., Saifullah , Malhi S.S., Zia M.H., Naeem A., Bibi S., Farid G. Role of mineral nutrition in minimizing cadmium accumulation by plants. J. Sci. Food Agric. 2010;90:925–937. doi: 10.1002/jsfa.3916. [DOI] [PubMed] [Google Scholar]
- 2.Sanita L., Gabbrielli R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999;41:105–130. doi: 10.1016/S0098-8472(98)00058-6. [DOI] [Google Scholar]
- 3.Li Y., Kuang H., Hu C., Ge G. Source Apportionment of Heavy Metal Pollution in Agricultural Soils around the Poyang Lake Region Using UNMIX Model. Sustainability. 2021;13:5272. doi: 10.3390/su13095272. [DOI] [Google Scholar]
- 4.Ghori N.H., Ghori T., Hayat M.Q., Imadi S.R., Gul A., Altay V., Ozturk M. Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 2019;16:1807–1828. doi: 10.1007/s13762-019-02215-8. [DOI] [Google Scholar]
- 5.Edelstein M., Ben-Hur M. Heavy metals and metalloids: Sources, risks and strategies to reduce their accumulation in horticultural crops. Sci. Hortic. 2018;234:431–444. doi: 10.1016/j.scienta.2017.12.039. [DOI] [Google Scholar]
- 6.Inaba T., Kobayashi E., Suwazono Y., Uetani M., Oishi M., Nakagawa H., Nogawa K. Estimation of cumulative cadmium intake causing Itai-itai disease. Toxicol. Lett. 2005;159:192–201. doi: 10.1016/j.toxlet.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Khan M.A., Khan S., Khan A., Alam M. Soil contamination with cadmium, consequences and remediation using organic amendments. Sci. Total Environ. 2017;601–602:1591–1605. doi: 10.1016/j.scitotenv.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q., Li Z., Lu X., Duan Q., Huang L., Bi J. A review of soil heavy metal pollution from industrial and agricultural regions in China: Pollution and risk assessment. Sci. Total Environ. 2018;642:690–700. doi: 10.1016/j.scitotenv.2018.06.068. [DOI] [PubMed] [Google Scholar]
- 9.Zhao F.J., Ma Y., Zhu Y.G., Tang Z., McGrath S.P. Soil contamination in China: Current status and mitigation strategies. Environ. Sci. Technol. 2015;49:750–759. doi: 10.1021/es5047099. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Duan X., Wang L. Spatial distribution and source analysis of heavy metals in soils influenced by industrial enterprise distribution: Case study in Jiangsu Province. Sci. Total Environ. 2020;710:134953. doi: 10.1016/j.scitotenv.2019.134953. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y., Cheng H., Tao S. The Challenges and Solutions for Cadmium-contaminated Rice in China: A Critical Review. Environ. Int. 2016;92–93:515–532. doi: 10.1016/j.envint.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 12.Wang P., Chen H., Kopittke P.M., Zhao F.J. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 2019;249:1038–1048. doi: 10.1016/j.envpol.2019.03.063. [DOI] [PubMed] [Google Scholar]
- 13.Zheng S., Wang Q., Yuan Y., Sun W. Human health risk assessment of heavy metals in soil and food crops in the Pearl River Delta urban agglomeration of China. Food Chem. 2020;316:126213. doi: 10.1016/j.foodchem.2020.126213. [DOI] [PubMed] [Google Scholar]
- 14.Song Y., Jin L., Wang X. Cadmium absorption and transportation pathways in plants. Int. J. Phytoremediat. 2017;19:133–141. doi: 10.1080/15226514.2016.1207598. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Luo N., Li Y.W., Cai Q.Y., Li H.Y., Mo C.H., Wong M.H. Cadmium in rice: Transport mechanisms, influencing factors, and minimizing measures. Environ. Pollut. 2017;224:622–630. doi: 10.1016/j.envpol.2017.01.087. [DOI] [PubMed] [Google Scholar]
- 16.Lux A., Martinka M., Vaculik M., White P.J. Root responses to cadmium in the rhizosphere: A review. J. Exp. Bot. 2011;62:21–37. doi: 10.1093/jxb/erq281. [DOI] [PubMed] [Google Scholar]
- 17.Li L.Z., Tu C., Wu L.H., Peijnenburg W.J., Ebbs S., Luo Y.M. Pathways of root uptake and membrane transport of Cd(2+) in the zinc/cadmium hyperaccumulating Plant Sedum plumbizincicola. Environ. Toxicol. Chem. 2017;36:1038–1046. doi: 10.1002/etc.3625. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Ouyang Y., Fan Y., Qiu B., Zhang G., Zeng F. The pathway of transmembrane cadmium influx via calcium-permeable channels and its spatial characteristics along rice root. J. Exp. Bot. 2018;69:5279–5291. doi: 10.1093/jxb/ery293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L.L., Tian S.K., Yang X.E., Li T.Q., He Z.L. Cadmium uptake and xylem loading are active processes in the hyperaccumulator Sedum alfredii. J. Plant Physiol. 2009;166:579–587. doi: 10.1016/j.jplph.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Seregin I.V., Kozhevnikova A.D. Roles of root and shoot tissues in transport and accumulation of cadmium, lead, nickel, and strontium. Russ. J. Plant Physiol. 2011;55:1–22. doi: 10.1134/S1021443708010019. [DOI] [Google Scholar]
- 21.Mendoza-Cozatl D.G., Butko E., Springer F., Torpey J.W., Komives E.A., Kehr J., Schroeder J.I. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008;54:249–259. doi: 10.1111/j.1365-313X.2008.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y., Tian S., Foyer C.H., Hou D., Wang H., Zhou W., Liu T., Ge J., Lu L., Lin X. Efficient phloem transport significantly remobilizes cadmium from old to young organs in a hyperaccumulator Sedum alfredii. J. Hazard. Mater. 2019;365:421–429. doi: 10.1016/j.jhazmat.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 23.Hall J.L., Williams L.E. Transition metal transporters in plants. J. Exp. Bot. 2003;54:2601–2613. doi: 10.1093/jxb/erg303. [DOI] [PubMed] [Google Scholar]
- 24.Socha A.L., Guerinot M.L. Mn-euvering manganese: The role of transporter gene family members in manganese uptake and mobilization in plants. Front. Plant Sci. 2014;5:106. doi: 10.3389/fpls.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomine S., Wang R., Ward J.M., Crawford N.M., Crawford J.I. Cadmium and iron transport by members of a Plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Wang L., Zheng L., Wang Y., Chen X., Zhang W. A Functional Study Identifying Critical Residues Involving Metal Transport Activity and Selectivity in Natural Resistance-Associated Macrophage Protein 3 in Arabidopsis thaliana. Int. J. Mol. Sci. 2018;19:1430. doi: 10.3390/ijms19051430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomine S., Lelievre F., Debarbieux E., Schroeder J.I., Barbier-Brygoo H. AtNRAMP3, a multispecic vacuolar metal transporter involved in Plant responses to iron deciency. Plant J. 2003;34:685–695. doi: 10.1046/j.1365-313X.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 28.Pottier M., Oomen R., Picco C., Giraudat J., Scholz-Starke J., Richaud P., Carpaneto A., Thomine S. Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium. Plant J. 2015;83:625–637. doi: 10.1111/tpj.12914. [DOI] [PubMed] [Google Scholar]
- 29.Lanquar V., Lelièvre F., Barbier-Brygoo H., Thomine S. Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci. Plant Nutr. 2004;50:1141–1150. doi: 10.1080/00380768.2004.10408587. [DOI] [Google Scholar]
- 30.Cailliatte R., Lapeyre B., Briat J.F., Mari S., Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem. J. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- 31.Tiwari M., Sharma D., Dwivedi S., Singh M., Tripathi R.D., Trivedi P.K. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant Cell Environ. 2014;37:140–152. doi: 10.1111/pce.12138. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi R., Ishimaru Y., Senoura T., Shimo H., Ishikawa S., Arao T., Nakanishi H., Nishizawa N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang J.D., Huang S., Yamaji N., Zhang W., Ma J.F., Zhao F.J. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020;43:2476–2491. doi: 10.1111/pce.13843. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J., Yang W., Zhang S., Yang T., Liu Q., Dong J., Fu H., Mao X., Liu B. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice. 2018;11:61. doi: 10.1186/s12284-018-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang J.-D., Xie Y., Zhang H., Zhang S., Zhao F.-J. The vacuolar transporter OsNRAMP2 mediates Fe remobilization during germination and affects Cd distribution to rice grain. Plant Soil. 2022:1–17. doi: 10.1007/s11104-022-05323-6. [DOI] [Google Scholar]
- 36.Chang J.D., Huang S., Konishi N., Wang P., Chen J., Huang X.Y., Ma J.F., Zhao F.J. Overexpression of the manganese/cadmium transporter OsNRAMP5 reduces cadmium accumulation in rice grain. J. Exp. Bot. 2020;71:5705–5715. doi: 10.1093/jxb/eraa287. [DOI] [PubMed] [Google Scholar]
- 37.Ishimaru Y., Takahashi R., Bashir K., Shimo H., Senoura T., Sugimoto K., Ono K., Yano M., Ishikawa S., Arao T., et al. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012;2:286. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki A., Yamaji N., Yokosho K., Ma J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24:2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang L., Mao B., Li Y., Lv Q., Zhang L., Chen C., He H., Wang W., Zeng X., Shao Y., et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017;7:14438. doi: 10.1038/s41598-017-14832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng F., Wang C., Cheng Y., Kang H., Fan X., Sha L., Zhang H., Zeng J., Zhou Y., Wang Y. Cloning and Characterization of TpNRAMP3, a Metal Transporter from Polish Wheat (Triticum polonicum L.) Front. Plant Sci. 2018;9:1354. doi: 10.3389/fpls.2018.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng F., Wang C., Zhu J., Zeng J., Kang H., Fan X., Sha L., Zhang H., Zhou Y., Wang Y. Expression of TpNRAMP5, a metal transporter from Polish wheat (Triticum polonicum L.), enhances the accumulation of Cd, Co and Mn in transgenic Arabidopsis plants. Planta. 2018;247:1395–1406. doi: 10.1007/s00425-018-2872-3. [DOI] [PubMed] [Google Scholar]
- 42.Wang C., Chen X., Yao Q., Long D., Fan X., Kang H., Zeng J., Sha L., Zhang H., Zhou Y., et al. Overexpression of TtNRAMP6 enhances the accumulation of Cd in Arabidopsis. Gene. 2019;696:225–232. doi: 10.1016/j.gene.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Wu D., Yamaji N., Yamane M., Kashino-Fujii M., Sato K., Feng Ma J. The HvNramp5 Transporter Mediates Uptake of Cadmium and Manganese, But Not Iron. Plant Physiol. 2016;172:1899–1910. doi: 10.1104/pp.16.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokosho K., Yamaji N., Ma J.F. Buckwheat FeNramp5 Mediates High Manganese Uptake in Roots. Plant Cell Physiol. 2021;62:600–609. doi: 10.1093/pcp/pcaa153. [DOI] [PubMed] [Google Scholar]
- 45.Meng J.G., Zhang X.D., Tan S.K., Zhao K.X., Yang Z.M. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus. Biometals. 2017;30:917–931. doi: 10.1007/s10534-017-0057-3. [DOI] [PubMed] [Google Scholar]
- 46.Yue X., Song J., Fang B., Wang L., Zou J., Su N., Cui J. BcNRAMP1 promotes the absorption of cadmium and manganese in Arabidopsis. Chemosphere. 2021;283:131113. doi: 10.1016/j.chemosphere.2021.131113. [DOI] [PubMed] [Google Scholar]
- 47.Milner M.J., Mitani-Ueno N., Yamaji N., Yokosho K., Craft E., Fei Z., Ebbs S., Clemencia Zambrano M., Ma J.F., Kochian L.V. Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J. 2014;78:398–410. doi: 10.1111/tpj.12480. [DOI] [PubMed] [Google Scholar]
- 48.Oomen R.J., Wu J., Lelievre F., Blanchet S., Richaud P., Barbier-Brygoo H., Aarts M.G., Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 49.Wei W., Chai T., Zhang Y., Han L., Xu J., Guan Z. The Thlaspi caerulescens NRAMP homologue TcNRAMP3 is capable of divalent cation transport. Mol. Biotechnol. 2009;41:15–21. doi: 10.1007/s12033-008-9088-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J., Zhang M., Song H., Zhao J., Shabala S., Tian S., Yang X. A novel plasma membrane-based NRAMP transporter contributes to Cd and Zn hyperaccumulation in Sedum alfredii Hance. Environ. Exp. Bot. 2020;176 doi: 10.1016/j.envexpbot.2020.104121. [DOI] [Google Scholar]
- 51.Feng Y., Wu Y., Zhang J., Meng Q., Wang Q., Ma L., Ma X., Yang X. Ectopic expression of SaNRAMP3 from Sedum alfredii enhanced cadmium root-to-shoot transport in Brassica juncea. Ecotoxicol. Environ. Saf. 2018;156:279–286. doi: 10.1016/j.ecoenv.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 52.Chen S., Han X., Fang J., Lu Z., Qiu W., Liu M., Sang J., Jiang J., Zhuo R. Sedum alfredii SaNramp6 Metal Transporter Contributes to Cadmium Accumulation in Transgenic Arabidopsis thaliana. Sci. Rep. 2017;7:13318. doi: 10.1038/s41598-017-13463-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Z., Chen S., Han X., Zhang J., Qiao G., Jiang Y., Zhuo R., Qiu W. A Single Amino Acid Change in Nramp6 from Sedum Alfredii Hance Affects Cadmium Accumulation. Int. J. Mol. Sci. 2020;21:3169. doi: 10.3390/ijms21093169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zha Q., Xiao Z., Zhang X., Han Z., Wang Y. Cloning and functional analysis of MxNRAMP1 and MxNRAMP3, two genes related to high metal tolerance of Malus xiaojinensis. S. Afr. J. Bot. 2016;102:75–80. doi: 10.1016/j.sajb.2015.05.031. [DOI] [Google Scholar]
- 55.Zhang W., Yue S., Song J., Xun M., Han M., Yang H. MhNRAMP1 from Malus hupehensis Exacerbates Cell Death by Accelerating Cd Uptake in Tobacco and Apple Calli. Front. Plant Sci. 2020;11:957. doi: 10.3389/fpls.2020.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y., Li G., Yang J., Zhao X., Sun Z., Hou H. Role of Nramp transporter genes of Spirodela polyrhiza in cadmium accumulation. Ecotoxicol. Environ. Saf. 2021;227:112907. doi: 10.1016/j.ecoenv.2021.112907. [DOI] [PubMed] [Google Scholar]
- 57.Nakanishi-Masuno T., Shitan N., Sugiyama A., Takanashi K., Inaba S., Kaneko S., Yazaki K. The Crotalaria juncea metal transporter CjNRAMP1 has a high Fe uptake activity, even in an environment with high Cd contamination. Int. J. Phytoremediat. 2018;20:1427–1437. doi: 10.1080/15226514.2018.1501333. [DOI] [PubMed] [Google Scholar]
- 58.Liu W., Huo C., He L., Ji X., Yu T., Yuan J., Zhou Z., Song L., Yu Q., Chen J., et al. The NtNRAMP1 transporter is involved in cadmium and iron transport in tobacco (Nicotiana tabacum) Plant Physiol. Biochem. 2022;173:59–67. doi: 10.1016/j.plaphy.2022.01.024. [DOI] [PubMed] [Google Scholar]
- 59.Jia H., Yin Z., Xuan D., Lian W., Han D., Zhu Z., Li C., Li C., Song Z. Mutation of NtNRAMP3 improves cadmium tolerance and its accumulation in tobacco leaves by regulating the subcellular distribution of cadmium. J. Hazard. Mater. 2022;432:128701. doi: 10.1016/j.jhazmat.2022.128701. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi R., Bashir K., Ishimaru Y., Nishizawa N.K., Nakanishi H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal Behav. 2012;7:1605–1607. doi: 10.4161/psb.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong C.K.E., Cobbett C.S. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009;181:71–78. doi: 10.1111/j.1469-8137.2008.02638.x. [DOI] [PubMed] [Google Scholar]
- 62.Wong C.K.E., Jarvis R.S., Sherson S.M., Cobbett C.S. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 2009;181:79–88. doi: 10.1111/j.1469-8137.2008.02637.x. [DOI] [PubMed] [Google Scholar]
- 63.Gravot A., Lieutaud A., Verret F., Auroy P., Vavasseur A., Richaud P. AtHMA3, a Plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004;561:22–28. doi: 10.1016/S0014-5793(04)00072-9. [DOI] [PubMed] [Google Scholar]
- 64.Morel M., Crouzet J., Gravot A., Auroy P., Leonhardt N., Vavasseur A., Richaud P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verret F., Gravot A., Auroy P., Leonhardt N., David P., Nussaume L., Vavasseur A., Richaud P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and Plant metal tolerance. FEBS Lett. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 66.Ceasar S.A., Lekeux G., Motte P., Xiao Z., Galleni M., Hanikenne M. di-Cysteine Residues of the Arabidopsis thaliana HMA4 C-Terminus Are Only Partially Required for Cadmium Transport. Front. Plant Sci. 2020;11:560. doi: 10.3389/fpls.2020.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siemianowski O., Barabasz A., Kendziorek M., Ruszczynska A., Bulska E., Williams L.E., Antosiewicz D.M. HMA4 expression in tobacco reduces Cd accumulation due to the induction of the apoplastic barrier. J. Exp. Bot. 2014;65:1125–1139. doi: 10.1093/jxb/ert471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satoh-Nagasawa N., Mori M., Nakazawa N., Kawamoto T., Nagato Y., Sakurai K., Takahashi H., Watanabe A., Akagi H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012;53:213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi R., Ishimaru Y., Shimo H., Ogo Y., Senoura T., Nishizawa N.K., Nakanishi H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012;35:1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- 70.Yamaji N., Xia J., Mitani-Ueno N., Yokosho K., Feng Ma J. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 2013;162:927–939. doi: 10.1104/pp.113.216564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyadate H., Adachi S., Hiraizumi A., Tezuka K., Nakazawa N., Kawamoto T., Katou K., Kodama I., Sakurai K., Takahashi H., et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189:190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 72.Sasaki A., Yamaji N., Ma J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014;65:6013–6021. doi: 10.1093/jxb/eru340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ueno D., Yamaji N., Kono I., Huang C.F., Ando T., Yano M., Ma J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA. 2010;107:16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H.Q., Xuan W., Huang X.Y., Mao C., Zhao F.J. Cadmium Inhibits Lateral Root Emergence in Rice by Disrupting OsPIN-Mediated Auxin Distribution and the Protective Effect of OsHMA3. Plant Cell Physiol. 2021;62:166–177. doi: 10.1093/pcp/pcaa150. [DOI] [PubMed] [Google Scholar]
- 75.Lu C., Zhang L., Tang Z., Huang X.Y., Ma J.F., Zhao F.J. Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 2019;126:619–626. doi: 10.1016/j.envint.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Kumagai S., Suzuki T., Tezuka K., Satoh-Nagasawa N., Takahashi H., Sakurai K., Watanabe A., Fujimura T., Akagi H. Functional analysis of the C-terminal region of the vacuolar cadmium-transporting rice OsHMA3. FEBS Lett. 2014;588:789–794. doi: 10.1016/j.febslet.2014.01.037. [DOI] [PubMed] [Google Scholar]
- 77.Lee S., Kim Y.Y., Lee Y., An G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007;145:831–842. doi: 10.1104/pp.107.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan J., Wang J., Chai T., Zhang Y., Feng S., Li Y., Zhao H., Liu H., Chai X. Functional analyses of TaHMA2, a P(1B)-type ATPase in wheat. Plant Biotechnol. J. 2013;11:420–431. doi: 10.1111/pbi.12027. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y., Wang C., Liu Y., Yu K., Zhou Y. GmHMA3 sequesters Cd to the root endoplasmic reticulum to limit translocation to the stems in soybean. Plant Sci. 2018;270:23–29. doi: 10.1016/j.plantsci.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Zhao H., Wang L., Zhao F.J., Wu L., Liu A., Xu W. SpHMA1 is a chloroplast cadmium exporter protecting photochemical reactions in the Cd hyperaccumulator Sedum plumbizincicola. Plant Cell Environ. 2019;42:1112–1124. doi: 10.1111/pce.13456. [DOI] [PubMed] [Google Scholar]
- 81.Liu H., Zhao H., Wu L., Liu A., Zhao F.J., Xu W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017;215:687–698. doi: 10.1111/nph.14622. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J., Zhang M., Shohag M.J., Tian S., Song H., Feng Y., Yang X. Enhanced expression of SaHMA3 plays critical roles in Cd hyperaccumulation and hypertolerance in Cd hyperaccumulator Sedum alfredii Hance. Planta. 2016;243:577–589. doi: 10.1007/s00425-015-2429-7. [DOI] [PubMed] [Google Scholar]
- 83.Ueno D., Milner M.J., Yamaji N., Yokosho K., Koyama E., Clemencia Zambrano M., Kaskie M., Ebbs S., Kochian L.V., Ma J.F. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011;66:852–862. doi: 10.1111/j.1365-313X.2011.04548.x. [DOI] [PubMed] [Google Scholar]
- 84.Papoyan A., Kochian L.V. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004;136:3814–3823. doi: 10.1104/pp.104.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J., Liang S., Xiang W., Dai H., Duan Y., Kang F., Chai T. A repeat region from the Brassica juncea HMA4 gene BjHMA4R is specifically involved in Cd(2+) binding in the cytosol under low heavy metal concentrations. BMC Plant Biol. 2019;19:89. doi: 10.1186/s12870-019-1674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo Q., Tian X., Mao P., Meng L. Functional characterization of IlHMA2, a P1B2-ATPase in Iris lactea response to Cd. Environ. Exp. Bot. 2019;157:131–139. doi: 10.1016/j.envexpbot.2018.10.008. [DOI] [Google Scholar]
- 87.Wang X., Zhi J., Liu X., Zhang H., Liu H., Xu J. Transgenic tobacco plants expressing a P1B-ATPase gene from Populus tomentosa Carr. (PtoHMA5) demonstrate improved cadmium transport. Int. J. Biol. Macromol. 2018;113:655–661. doi: 10.1016/j.ijbiomac.2018.02.081. [DOI] [PubMed] [Google Scholar]
- 88.Grafe K., Schmitt L. The ABC transporter G subfamily in Arabidopsis thaliana. J. Exp. Bot. 2021;72:92–106. doi: 10.1093/jxb/eraa260. [DOI] [PubMed] [Google Scholar]
- 89.Verrier P.J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E., et al. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Kim D.Y., Bovet L., Kushnir S., Noh E.W., Martinoia E., Lee Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006;140:922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim D.Y., Bovet L., Maeshima M., Martinoia E., Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- 92.Park J., Song W.Y., Ko D., Eom Y., Hansen T.H., Schiller M., Lee T.G., Martinoia E., Lee Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012;69:278–288. doi: 10.1111/j.1365-313X.2011.04789.x. [DOI] [PubMed] [Google Scholar]
- 93.Rea P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- 94.Brunetti P., Zanella L., De Paolis A., Di Litta D., Cecchetti V., Falasca G., Barbieri M., Altamura M.M., Costantino P., Cardarelli M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015;66:3815–3829. doi: 10.1093/jxb/erv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gaillard S., Jacquet H., Vavasseur A., Leonhardt N., Forestier C. AtMRP6/AtABCC6, an ATP-binding cassette transporter gene expressed during early steps of seedling development and up-regulated by cadmium in Arabidopsis thaliana. BMC Plant Biol. 2008;8:22. doi: 10.1186/1471-2229-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wojas S., Hennig J., Plaza S., Geisler M., Siemianowski O., Sklodowska A., Ruszczynska A., Bulska E., Antosiewicz D.M. Ectopic expression of Arabidopsis ABC transporter MRP7 modifies cadmium root-to-shoot transport and accumulation. Environ. Pollut. 2009;157:2781–2789. doi: 10.1016/j.envpol.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 97.Bhuiyan M.S.U., Min S.R., Jeong W.J., Sultana S., Choi K.S., Lee Y., Liu J.R. Overexpression of AtATM3 in Brassica juncea confers enhanced heavy metal tolerance and accumulation. Plant Cell Tissue Organ. Cult. 2011;107:69–77. doi: 10.1007/s11240-011-9958-y. [DOI] [Google Scholar]
- 98.Yang G., Fu S., Huang J., Li L., Long Y., Wei Q., Wang Z., Chen Z., Xia J. The tonoplast-localized transporter OsABCC9 is involved in cadmium tolerance and accumulation in rice. Plant Sci. 2021;307:110894. doi: 10.1016/j.plantsci.2021.110894. [DOI] [PubMed] [Google Scholar]
- 99.Fu S., Lu Y., Zhang X., Yang G., Chao D., Wang Z., Shi M., Chen J., Chao D.Y., Li R., et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019;70:5909–5918. doi: 10.1093/jxb/erz335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oda K., Otani M., Uraguchi S., Akihiro T., Fujiwara T. Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci. Biotechnol. Biochem. 2011;75:1211–1213. doi: 10.1271/bbb.110193. [DOI] [PubMed] [Google Scholar]
- 101.Cai X., Wang M., Jiang Y., Wang C., Ow D.W. Overexpression of OsABCG48 Lowers Cadmium in Rice (Oryza sativa L.) Agronomy. 2021;11:918. doi: 10.3390/agronomy11050918. [DOI] [Google Scholar]
- 102.Bhati K.K., Alok A., Kumar A., Kaur J., Tiwari S., Pandey A.K. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 2016;67:4379–4389. doi: 10.1093/jxb/erw224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Y.H., Wang C.J., Li R.F., Yi Y.J., Zeng L., Yang H., Zhang C.F., Song K.Y., Guo S.J. Transcriptome-based identification and expression characterization of RgABCC transp.porters in Rehmannia glutinosa. PLoS ONE. 2021;16:e0253188. doi: 10.1371/journal.pone.0253188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H., Liu Y., Peng Z., Li J., Huang W., Liu Y., Wang X., Xie S., Sun L., Han E., et al. Ectopic Expression of Poplar ABC Transporter PtoABCG36 Confers Cd Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019;20:3293. doi: 10.3390/ijms20133293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Verbruggen N., Hermans C., Schat H. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2009;181:759–776. doi: 10.1111/j.1469-8137.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- 106.Pence N.S., Larsen P.B., Ebbs S.D., Letham D.L.D., Lasat M.M., Garvin D.F., Eide D., Kochian L.V. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA. 2000;97:4956–4960. doi: 10.1073/pnas.97.9.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin Y.F., Hassan Z., Talukdar S., Schat H., Aarts M.G. Expression of the ZNT1 Zinc Transporter from the Metal Hyperaccumulator Noccaea caerulescens Confers Enhanced Zinc and Cadmium Tolerance and Accumulation to Arabidopsis thaliana. PLoS ONE. 2016;11:e0149750. doi: 10.1371/journal.pone.0149750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Connolly E.L., Fett J.P., Guerinot M.L. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Korshunova Y.O., Eide D., Clark W.G., Guerinot M.L., Pakrasi H.B. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999;40:37–44. doi: 10.1023/A:1026438615520. [DOI] [PubMed] [Google Scholar]
- 110.Nakanishi H., Ogawa I., Ishimaru Y., Mori S., Nishizawa N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006;52:464–469. doi: 10.1111/j.1747-0765.2006.00055.x. [DOI] [Google Scholar]
- 111.Liu X.S., Feng S.J., Zhang B.Q., Wang M.Q., Cao H.W., Rono J.K., Chen X., Yang Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019;19:283. doi: 10.1186/s12870-019-1899-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan L., Qu M., Zhu Y., Peng C., Wang J., Gao D., Chen C. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 Function Synergistically in Zinc/Cadmium Uptake. Plant Physiol. 2020;183:1235–1249. doi: 10.1104/pp.19.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kavitha P.G., Kuruvilla S., Mathew M.K. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.) Plant Physiol. Biochem. 2015;97:165–174. doi: 10.1016/j.plaphy.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 114.Tan L., Zhu Y., Fan T., Peng C., Wang J., Sun L., Chen C. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 2019;512:112–118. doi: 10.1016/j.bbrc.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 115.Yang M., Li Y., Liu Z., Tian J., Liang L., Qiu Y., Wang G., Du Q., Cheng D., Cai H., et al. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 2020;103:1695–1709. doi: 10.1111/tpj.14855. [DOI] [PubMed] [Google Scholar]
- 116.Barabasz A., Palusinska M., Papierniak A., Kendziorek M., Kozak K., Williams L.E., Antosiewicz D.M. Functional Analysis of NtZIP4B and Zn Status-Dependent Expression Pattern of Tobacco ZIP Genes. Front. Plant Sci. 2018;9:1984. doi: 10.3389/fpls.2018.01984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maslinska-Gromadka K., Barabasz A., Palusinska M., Kozak K., Antosiewicz D.M. Suppression of NtZIP4A/B Changes Zn and Cd Root-to-Shoot Translocation in a Zn/Cd Status-Dependent Manner. Int. J. Mol. Sci. 2021;22:5355. doi: 10.3390/ijms22105355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan W., Liu C., Cao B., Qin M., Long D., Xiang Z., Zhao A. Genome-Wide Identification and Characterization of Four Gene Families Putatively Involved in Cadmium Uptake, Translocation and Sequestration in Mulberry. Front. Plant Sci. 2018;9:879. doi: 10.3389/fpls.2018.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen H., Zhang C., Guo H., Hu Y., He Y., Jiang D. Overexpression of a Miscanthus sacchariflorus yellow stripe-like transporter MsYSL1 enhances resistance of Arabidopsis to cadmium by mediating metal ion reallocation. Plant Growth Regul. 2018;85:101–111. doi: 10.1007/s10725-018-0376-6. [DOI] [Google Scholar]
- 120.Feng S., Tan J., Zhang Y., Liang S., Xiang S., Wang H., Chai T. Isolation and characterization of a novel cadmium-regulated Yellow Stripe-Like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2016;36:281–296. doi: 10.1007/s00299-016-2079-7. [DOI] [PubMed] [Google Scholar]
- 121.Chen S., Liu Y., Deng Y., Liu Y., Dong M., Tian Y., Sun H., Li Y. Cloning and functional analysis of the VcCXIP4 and VcYSL6 genes as Cd-regulating genes in blueberry. Gene. 2019;686:104–117. doi: 10.1016/j.gene.2018.10.078. [DOI] [PubMed] [Google Scholar]
- 122.Wang J.W., Li Y., Zhang Y.X., Chai T.Y. Molecular cloning and characterization of a Brassica juncea yellow stripe-like gene, BjYSL7, whose overexpression increases heavy metal tolerance of tobacco. Plant Cell Rep. 2013;32:651–662. doi: 10.1007/s00299-013-1398-1. [DOI] [PubMed] [Google Scholar]
- 123.Cohen C.K., Garvin D.F., Kochian L.V. Kinetic properties of a micronutrient transporter from Pisum sativum indicate a primary function in Fe uptake from the soil. Planta. 2004;218:784–792. doi: 10.1007/s00425-003-1156-7. [DOI] [PubMed] [Google Scholar]
- 124.Jiang Y., Chen X., Chai S., Sheng H., Sha L., Fan X., Zeng J., Kang H., Zhang H., Xiao X., et al. TpIRT1 from Polish wheat (Triticum polonicum L.) enhances the accumulation of Fe, Mn, Co, and Cd in Arabidopsis. Plant Sci. 2021;312:111058. doi: 10.1016/j.plantsci.2021.111058. [DOI] [PubMed] [Google Scholar]
- 125.Lee S., An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 126.Pedas P., Ytting C.K., Fuglsang A.T., Jahn T.P., Schjoerring J.K., Husted S. Manganese efficiency in barley: Identification and characterization of the metal ion transporter HvIRT1. Plant Physiol. 2008;148:455–466. doi: 10.1104/pp.108.118851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zang J., Huo Y., Liu J., Zhang H., Liu J., Chen H. Maize YSL2 is required for iron distribution and development in kernels. J. Exp. Bot. 2020;71:5896–5910. doi: 10.1093/jxb/eraa332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.