Abstract
The JAK2-V617F mutation is the most common cause of myeloproliferative neoplasms. While experiments have shown that this gain-of-function mutation is associated with myeloid blood cell expansion and increased production of white cells, red cells and platelets, the transcriptional consequences of the JAK2-V617F mutation in different cellular compartments of the bone marrow have not yet been fully elucidated. To study the direct effects of JAK2-V617F on bone marrow cells in myeloproliferative neoplasm patients, we performed joint single-cell RNA sequencing and JAK2 genotyping on CD34+ enriched cells from 8 patients with newly diagnosed essential thrombocythemia or polycythemia vera. We found that the JAK2-V617F mutation increases the expression of interferon-response genes (e.g., HLAs) and the leptin receptor in hematopoietic progenitor cells. Furthermore, we sequenced a population of CD34-bone marrow monocytes and found the JAK2 mutation increased expression of intermediate monocyte genes and the fibrocyte-associated surface protein SLAMF7 in these cells.
Keywords: myeloproliferative neoplasm, single-cell sequencing, JAK2-V617F
INTRODUCTION
The JAK2-V617F mutation is a somatic mutation found in the majority of patients with myeloproliferative neoplasms (MPNs)1. The mutation causes constitutive JAK-STAT pathway activation in hematopoietic stem and progenitor cells (HSPCs), leading to overproduction of red blood cells, platelets, white blood cells, and/or bone marrow fibrosis. Previous work characterizing JAK2-mutant mouse models2,3 and MPN patient samples4 have shown that JAK2-V617F increases the fitness of hematopoietic stem cells (HSCs) and promotes megakaryocyte-erythroid differentiation. However, it is unclear what molecular mechanisms connect JAK-STAT pathway activation with the observed changes in differentiation and cell division. Furthermore, it is unclear which cell types within the bone marrow are directly affected by the mutation and which cells are affected by cell-non-autonomous factors. Previous studies have shown that JAK2-V617F HSPCs have increased JAK-STAT signaling compared to their wild-type (WT) counterparts in the same MPN patient5,6. However, these studies generally focused on the mutation’s effects on HSCs and megakaryocyte erythrocyte progenitors (MEPs).
Here, we used joint scRNA-seq and JAK2 genotyping of bone marrow from polycythemia vera (PV) and essential thrombocythemia (ET) patients to determine the impact of the JAK2-V617F mutation in an unbiased way on multiple bone marrow cell types. In addition to affecting MEPs, we show that the JAK2-V617F mutation directly affects bone marrow monocytes, changing their surface phenotype and increasing expression of SLAMF7, a marker associated with fibrocyte differentiation.
MATERIALS AND METHODS
Experimental procedures
Cell isolation, scRNA-seq with specific amplification of JAK2, and preprocessing and cell type identification using marker genes were performed using the same methods previously described4. All patients were newly diagnosed and treatment-naive at the time of sampling and harbored the JAK2-V617F mutation in their peripheral blood. Bone marrow samples from four healthy donors were also collected and scRNA-seq was performed without JAK2 amplification using the same protocol as used on the MPN patients.
scRNA-seq data analysis
Preprocessing of the scRNA-seq data and identification of cells with mutant or wild-type JAK2 amplicon transcripts was performed as previously published4. Differential expression analysis comparing cells with and without the JAK2-V617F mutation was performed in scanpy using the Wilcoxon rank sum test. All raw p-values were combined between patients using Fisher’s method and adjusted for multiple comparisons using the Benjamini-Hochberg method. Gene set enrichment analysis was performed using GSEApy to find enriched KEGG biological processes and ChEA/ENCODE transcription factor target groups.
After integrating and clustering data from all patients as previously published, classical, intermediate, and nonclassical subsets were identified in the monocyte population by Louvain clustering the monocyte population only. We used the expression levels of marker genes (classical: CD14, nonclassical: CD16, intermediate: CD74, CD64, HLA-DRA)7 to manually assign each cluster to a subtype.
Calling somatic mutations in the scRNA-seq data
In the whole-genome sequencing (WGS) data, we identified somatic mutations that only occurred in the JAK2-V617F cells (220 mutations in ET 1, 398 in ET 2). Then, we detected these mutations in the scRNA-seq data by PCR amplification and sequencing (e.g., for a point mutation in UPF1 in patient ET 1) or by calling mutations in the raw 10X transcriptome reads. To call the mutations in scRNAseq data, we extracted all reads mapping to each position mutated in the WGS data using Pysam8. Only reads with unambiguous cellular and molecular barcode sequences were considered. Subsequently, we classified reads as mutant or wild type depending on whether the reads contained the mutant or wild-type allele, respectively, requiring a minimum base quality of 30. Mutations that generated false positive mutant calls in any of the 36 10X Chromium single-cell RNAseq data sets from bone marrow and peripheral blood samples from healthy individuals were considered unreliable and discarded.
Bone marrow monocyte flow cytometry
Flow cytometry of bone marrow monocytes from three patients (ET 4, PV 1, and PV 3) and three healthy donors was performed to validate some of the results of the monocyte differential expression analysis. SLAMF7 cell surface staining was done on CD14+ cells as previously published9.
RESULTS AND DISCUSSION
We analyzed joint single-cell JAK2 genotyping and scRNA-seq data from CD34-enriched bone marrow samples from 4 ET and 4 PV patients (Supplementary Fig. 1). Six of these patients were sequenced in a previous study4. Two patients (PV 2 and PV 3) also had TET2 mutations, and one patient (PV 1) had a low-frequency EZH2 mutation detected in peripheral blood by a clinical NGS assay10.
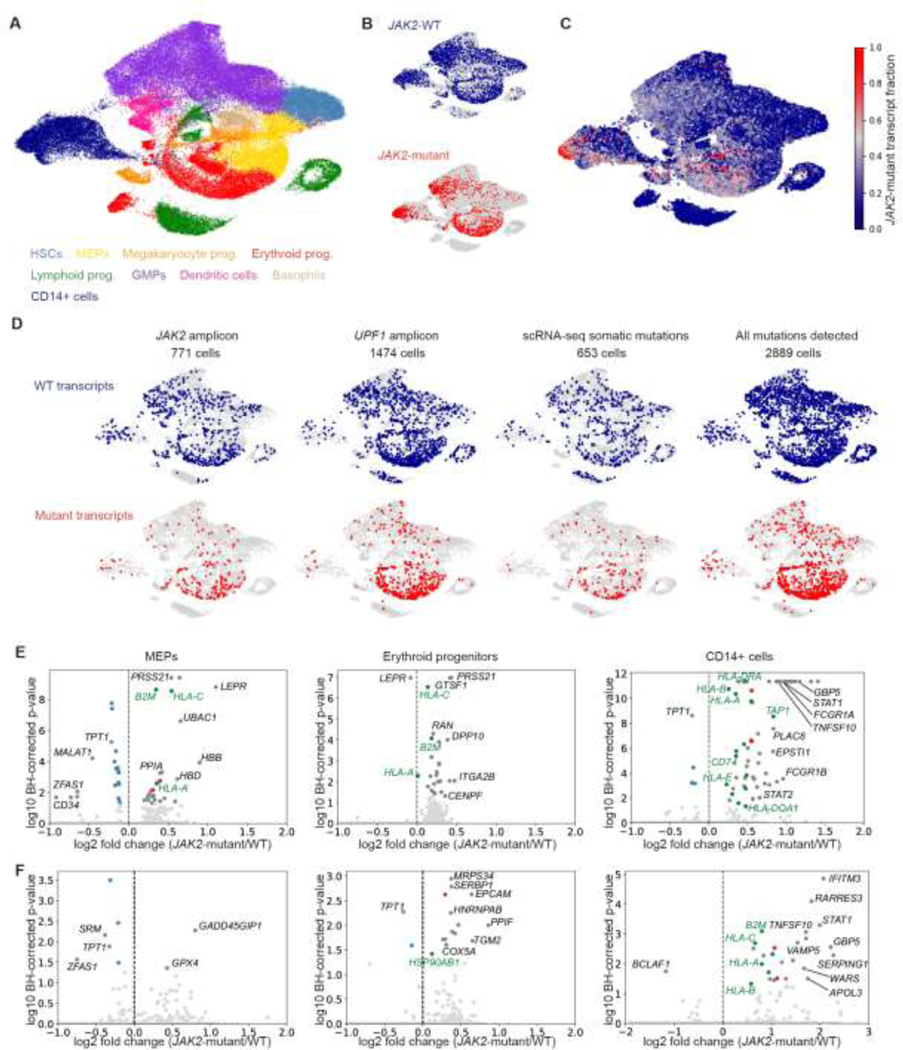
Bone marrow cells with the JAK2-V617F mutation were intermixed with the cells with WT transcripts when plotted together (Fig. 1A–C), suggesting that the mutation does not disrupt the overall structure of the differentiation hierarchy. However, as we reported previously4, we found that cells with the JAK2-V617F transcript detected were more likely to be megakaryocyte or erythroid progenitors than those with WT JAK2 transcripts (Fig. 1A–C), suggesting that JAK2-V617F induces a megakaryocyte-erythroid fate bias. We also found a substantial population of CD14+ bone marrow cells with the JAK2 mutation which do not express CD34 and therefore likely represent monocytes (Fig. 1A–C, Supplementary Fig. 1).
Figure 1. JAK2-V617F megakaryocyte and erythroid progenitors have higher expression of pro-inflammatory and antigen presentation genes.
A. UMAP of scRNA-seq data from bone marrow from 8 MPN patients, colored by cell type classifications. B. JAK2-WT (blue) and JAK2-mutant (red) transcripts detected in single cells in MPN patient bone marrow. C. Smoothed JAK2-V617F transcript fraction for all patients combined. D. Detection of mutations associated with JAK2-V617F in patient ET 1. Additional mutations were called using targeted amplification of loci identified from WGS (e.g., UPF1) and by directly identifying somatic mutations in the scRNA-seq data. E-F. Volcano plots showing differential expression analysis results from comparing cells with mutant transcripts to cells with WT transcripts within the MEP, erythroid progenitor, and CD14+ compartments for ET patients (E) and PV patients (F). Ribosomal genes, antigen presentation genes, and proteasomal genes are colored in blue, green, and red, respectively.
We compared the transcriptomic profiles of JAK2-V617F bone marrow cells to those from WT cells to determine how the mutation changes gene expression in individual cells in MPN patients. To increase the power of our differential expression analysis, we used the published single cell WGS data4 for ET 1 and ET 2 to identify and detect somatic mutations other than JAK2-V617F that uniquely marked the JAK2-mutant clonal population in each of these patients (Fig. 1D, Supplementary Table 1). By leveraging the WGS data, we were able to increase the number of cells that could be assigned as either JAK2-mutant or JAK2-WT by more than two-fold for ET 1 and ET 2.
Using the expanded set of genotyped cells, we identified genes that were differentially expressed between JAK2-mutant and JAK2-WT bone marrow cells in ET and PV patients (Fig. 1E–F). While we found few or no significantly differentially expressed genes in HSCs or GMPs (Supplementary File 1), MHC class I antigen presentation genes (e.g., HLAs, B2M) were consistently upregulated in JAK2-V617F MEPs, erythroid progenitors, and monocytes in both ET and PV patients. Interferon signaling through STAT1 has been previously associated with increased MHC I expression11, suggesting that the observed upregulation of these genes in MPN could be caused by increased JAK-STAT activity. MHC I presentation of T-cell antigens, including JAK2-V617F itself12, could induce an adaptive immune response against MPN cells, and PD-L1-mediated immune escape through reduced T-cell activation was previously reported in JAK2-mutant MPN13. Upregulation of these inflammation-associated genes in JAK2-mutant MEPs could also contribute to platelet activation and thromboinflammation14.
We also observed that JAK2-V617F bone marrow monocytes had increased expression of proinflammatory and interferon response genes in both ET and PV patients (Fig. 1E–F). These genes are enriched for IRF and STAT targets (Supplementary Fig. 2), suggesting that their upregulation may be due to direct effects of constitutive JAK2 activation. Finally, in JAK2-V617F MEPs, we also noted increased expression of the leptin receptor (Fig. 1E), which has been reported to be a marker for long-term engrafting HSCs in mice15.
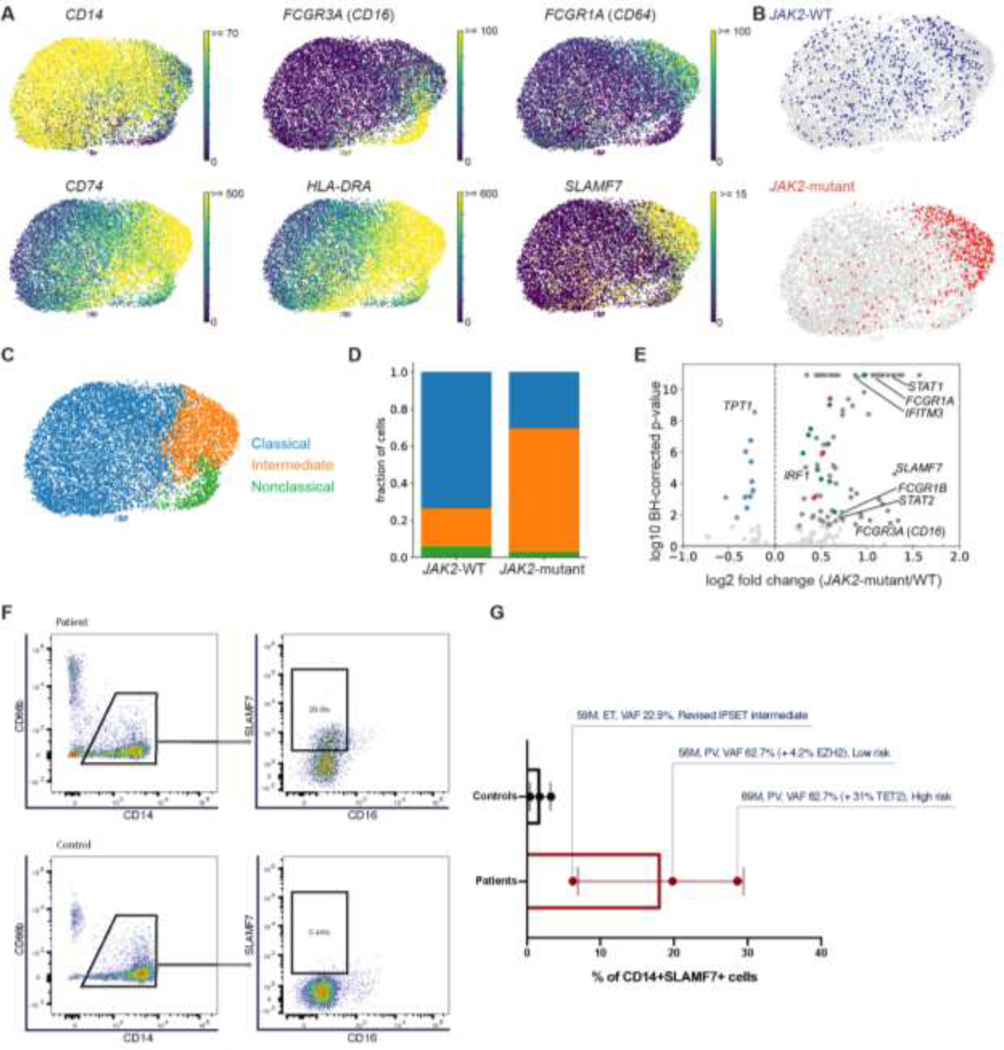
We further characterized the transcriptional phenotype of the bone marrow CD34-CD14+ cluster and found that these cells express monocyte genes (Fig. 2A). Using expression of monocyte subtype markers7, we identified classical, intermediate, and nonclassical monocyte subsets in our scRNA-seq data and found that JAK2-V617F monocytes were more likely than WT cells to have an intermediate monocyte phenotype (Fig. 2B–D). Furthermore, MPN patients had a higher fraction of intermediate monocytes overall than healthy controls (Supplementary Fig. 3). Intermediate monocytes have been shown to express high levels of antigen presentation genes, secrete both pro- and anti-inflammatory cytokines, and play a role in many infectious and autoimmune conditions7. Previous work showed that patients with JAK2-V617F myelofibrosis have more intermediate monocytes than healthy donors and exhibit dysregulation of cytokine production16. This abnormal phenotype is partially reversed by ruxolitinib treatment, suggesting that JAK-STAT signaling contributes to monocyte dysregulation16. Our single cell data suggests that JAK2-V617F acts directly on monocytes to change their phenotype, since we found that individual cells harboring the mutation are more likely to be intermediate monocytes.
Figure 2. JAK2-V617F induces SLAMF7 expression and an intermediate monocyte phenotype in MPN patients.
A. Marker gene expression UMAPs of the bone marrow monocyte compartment measured by scRNA-seq in 8 MPN patients. B. JAK2-WT (blue) and JAK2-V617F (red) transcripts detected in monocytes. C. UMAP of scRNA-seq data from monocytes colored by transcriptionally defined monocyte subset classifications. D. Fraction of CD14+ cells with a JAK2-WT or JAK2-mutant transcript detected by scRNA-seq that are classical, intermediate, or nonclassical monocytes. The monocyte subset definitions and color scheme are the same as in C. E. Differential expression analysis of monocytes from all 8 MPN patients, comparing cells with at least one mutant transcript detected to cells with a WT transcript detected. Ribosomal genes, antigen presentation genes, and proteasomal genes are colored in blue, green, and red, respectively. F-G. Flow cytometry and SLAMF7 staining of CD14+ cells from three MPN patients and three healthy controls. Gating scheme to identify SLAMF7 is shown in F, and the proportion of CD14+ cells expressing SLAMF7 is shown in G.
Another way monocytes may contribute to MPN pathogenesis is by differentiating into fibrocytes and contributing to bone marrow fibrosis. In JAK2-V617F myelofibrosis patients, CD14+ monocytes have been previously shown to preferentially differentiate into fibrocytes in vitro and express elevated levels of the cell surface marker SLAMF79. After combining our monocyte scRNA-seq data from both ET and PV patients, we found that SLAMF7 is significantly more highly expressed in JAK2-V617F monocytes than WT monocytes (Fig. 2E), although this difference could be due to the higher intermediate monocyte fraction in JAK2-mutant monocytes.
Flow cytometry revealed that ET and PV patients have a higher fraction of SLAMF7+ bone marrow monocytes than healthy donors (Fig. 2F–G), suggesting that SLAMF7+ monocytes may play a pathogenic role even in MPN subtypes not defined by bone marrow fibrosis. Consistent with this, none of the patients in our study had evidence of significant reticulin fibrosis in the bone marrow (four patients had MF grade 0, one patient (ET 4) had MF grade 0 –1 and three patients (ET 2, PV 1, and PV 3) had MF grade 1; Supplementary Fig. 1A). Inhibition of SLAMF7 with the monoclonal antibody drug elotuzumab has been shown to suppress fibrocyte differentiation and prevent progression in in vitro and in vivo models of myelofibrosis9. Our results also suggest that presence of JAK2-mutant monocytes could be investigated as an early biomarker of myelofibrosis risk in ET and PV patients.
In summary, we found that the JAK2-V617F mutation increases the expression of STAT signaling targets (e.g., antigen presentation and other pro-inflammatory genes) in HSPCs as well as monocytes. Our results suggest that the JAK2 mutation could lead to a pathogenic pro-inflammatory, pro-fibrotic phenotype in bone marrow monocytes, and that this population should be further investigated to determine what role they play in the clinical manifestations of ET and PV and in progression to myelofibrosis.
Supplementary Material
Highlights.
Joint scRNA-seq and JAK2 genotyping in myeloproliferative neoplasm (MPN) patients.
JAK2-V617F hematopoietic progenitors express interferon response genes more highly.
Monocytes with JAK2-V617F have a pro-inflammatory, intermediate monocyte phenotype.
JAK2-V617F monocytes express SLAMF7, which is associated with fibrosis in MPNs.
ACKNOWLEDGMENTS
We thank the individuals who participated in our study. Portions of this research were conducted on the O2 High Performance Compute Cluster, supported by the Research Computing Group, at Harvard Medical School (https://it.hms.harvard.edu/our-services/research-computing/).
FUNDING
S.H. acknowledges funding from NIH NIGMS R00GM118910 and NIH NHLBI R01HL158269, the DFCI BCB Fund Award, the Jayne Koskinas Ted Giovanis Foundation, The William F. Milton Fund at Harvard University, an AACR-MPM Oncology Charitable Foundation Transformative Cancer Research grant, and Gabrielle’s Angel Foundation for Cancer Research. S.H. and A.M. acknowledge funding from the Claudia Adams Barr Program in Cancer Research. A.M. acknowledges funding from NIH NHLBI (R01HL131835) and the MPN Research Foundation. A.M. is a Scholar of The Leukemia & Lymphoma Society. C.R.R acknowledges funding from NIH NHLBI T32HL116324. D.V.E. acknowledges funding from the NSF-Simons Center for Mathematical and Statistical Analysis of Biology at Harvard, award number 1764269, and the Harvard Quantitative Biology Initiative. D.V.E and F.M. acknowledge support from the Ludwig Center at Harvard and the Dana-Farber Cancer Institute Physical Science-Oncology Center (NIH U54CA193461 to F.M.). I.C.-C. and M.K. acknowledge funding from EMBL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lundberg P, Karow A, Nienhold R, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014;123(14):2220–2228. [DOI] [PubMed] [Google Scholar]
- 2.Mullally A, Lane SW, Ball B, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 2010;17(6):584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shide K, Shimoda HK, Kumano T, et al. Development of ET, primary myelofibrosis and PV in mice expressing JAK2 V617F. Leukemia 2008;22(1):87–95. [DOI] [PubMed] [Google Scholar]
- 4.Van Egeren D, Escabi J, Nguyen M, et al. Reconstructing the Lineage Histories and Differentiation Trajectories of Individual Cancer Cells in Myeloproliferative Neoplasms. Cell Stem Cell 2021;28(3):514–523.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen E, Beer PA, Godfrey AL, et al. Distinct Clinical Phenotypes Associated with JAK2V617F Reflect Differential STAT1 Signaling. Cancer Cell 2010;18(5):524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong J, Sun T, Ma S, et al. Hematopoietic Stem Cell Heterogeneity Is Linked to the Initiation and Therapeutic Response of Myeloproliferative Neoplasms. Cell Stem Cell 2021;28(3):502–513.e6. [DOI] [PubMed] [Google Scholar]
- 7.Kapellos TS, Bonaguro L, Gemünd I, et al. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front Immunol;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maekawa T, Kato S, Kawamura T, et al. Increased SLAMF7high monocytes in myelofibrosis patients harboring JAK2V617F provide a therapeutic target of elotuzumab. Blood 2019;134(10):814–825. [DOI] [PubMed] [Google Scholar]
- 10.Kluk MJ, Lindsley RC, Aster JC, et al. Validation and Implementation of a Custom Next-Generation Sequencing Clinical Assay for Hematologic Malignancies. The Journal of Molecular Diagnostics 2016;18(4):507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christova R, Jones T, Wu P-J, et al. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNγ. Journal of Cell Science 2007;120(18):3262–3270. [DOI] [PubMed] [Google Scholar]
- 12.Holmström MO, Hjortsø MD, Ahmad SM, et al. The JAK2 V617F mutation is a target for specific T cells in the JAK2 V617F-positive myeloproliferative neoplasms. Leukemia 2017;31(2):495–498. [DOI] [PubMed] [Google Scholar]
- 13.Prestipino A, Emhardt AJ, Aumann K, et al. Oncogenic JAK2V617F causes PD-L1 expression, mediating immune escape in myeloproliferative neoplasms. Science Translational Medicine 2018;10(429):eaam7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marin Oyarzún CP, Heller PG. Platelets as Mediators of Thromboinflammation in Chronic Myeloproliferative Neoplasms. Front Immunol 2019;101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trinh T, Ropa J, Aljoufi A, et al. Leptin receptor, a surface marker for a subset of highly engrafting long-term functional hematopoietic stem cells. Leukemia 2021;35(7):2064–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barone M, Catani L, Ricci F, et al. The role of circulating monocytes and JAK inhibition in the infectious-driven inflammatory response of myelofibrosis. Oncoimmunology 2020;9(1):1782575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.