Abstract
Simple Summary
In this study, we identified germline mutations that contribute to prostate cancer development in men who have multiple relatives with prostate cancer (and other cancers). We correlated the genetic mutations found in each patient with the resulting prostate cancer characteristics. Mutations found in ATM and CHEK2 genes were associated with aggressive prostate cancer. More data is needed to confirm the prevalence and impact of these germline mutations in prostate cancer.
Abstract
Background: Germline mutations in BRCA2 are associated with aggressive prostate cancer. Additional information regarding the clinical phenotype of germline pathogenic variants in other prostate cancer predisposition genes is required. Clinical testing has been limited by evidence, further restricting knowledge of variants that contribute to prostate cancer development. Objective: Prostate cancer patients who were first- and second-degree relatives from multi-case prostate cancer families underwent a gene panel screen to identify novel (non-BRCA) germline pathogenic variants in cancer predisposition genes and define clinical phenotypes associated with each gene. Methods: The germline genomic DNA (gDNA) of 94 index cases with verified prostate cancer from families with a minimum of two verified prostate cancer cases was screened with an 84-cancer-gene panel. Families were recruited for multi-case breast/ovarian cancer (n = 66), or multi-case prostate cancer (n = 28). Prostate cancer characteristics associated with each gene were compared with prostate cancer cases of confirmed non-mutation carriers (BRCAX), also from multi-case prostate cancer families (n = 111), and with data from the Prostate Cancer Outcomes Registry (PCOR). Results: Ninety-four prostate cancer index cases underwent gene panel testing; twenty-two index cases (22/94; 23%) were found to carry a class 4–5 (C4/5) variant. Six of twenty-two (27%) variants were not clinically notifiable, and seven of twenty-two (31.8%) variants were in BRCA1/2 genes. Nine of twenty-two (40.9%) index cases had variants identified in ATM (n = 4), CHEK2 (n = 2) and HOXB13G84 (n = 3); gDNA for all relatives of these nine cases was screened for the corresponding familial variant. The final cohort comprised 15 confirmed germline mutation carriers with prostate cancer (ATM n = 9, CHEK2 n = 2, HOXB13G84 n = 4). ATM and CHEK2-associated cancers were D’Amico intermediate or high risk, comparable to our previously published BRCA2 and BRCAX prostate cancer cohort. HOXB13G84 carriers demonstrated low- to intermediate-risk prostate cancer. In the BRCAX cohort, 53.2% of subjects demonstrated high-risk disease compared with 25% of the PCOR cohort. Conclusions: ATM and CHEK2 germline mutation carriers and the BRCAX (confirmed non-mutation carriers) cohort demonstrated high risk disease compared with the general population. Targeted genetic testing will help identify men at greater risk of prostate-cancer-specific mortality. Data correlating rare variants with clinical phenotype and familial predisposition will strengthen the clinical validity and utility of these results and establish these variants as significant in prostate cancer detection and management.
Keywords: prostate cancer, germline, hereditary, mutations, pathogenic variants
1. Introduction
Efforts to identify groups of men at risk of developing clinically significant prostate cancer indicate that those with a family history are at significantly higher risk compared with the general population [1,2,3]. For these individuals and their families, discovering the genetic aetiology of hereditary prostate cancer and clinical phenotype associated with each variant has broad clinical utility. Gene-specific risk stratification, targeted screening, early intervention and novel therapeutic targets are realisable outcomes. These advances are likely to reduce the burden on health systems and individuals compared with managing disease at advanced stages [4,5]. Recent studies have highlighted the benefits and feasibility of dedicated clinics to serve high-risk prostate cancer patients with pathogenic mutations in PC susceptibility genes, and clinics dedicated to identifying these high-risk individuals in a streamlined fashion [6,7]. Modified protocols for BRCA2 mutation carriers have resulted in improved clinical outcomes [8,9]. The potential for treatment response to poly (ADP-ribose) polymerase (PARP) inhibitors in mutation carriers with prostate cancer has been strengthening support for screening of relevant familial cohorts [10,11].
Gene panel sequencing is increasingly utilised to predict cancer risk in patients from multi-case cancer families. However, in the familial prostate cancer setting, the clinical utility of panel testing is limited by our current understanding of which germline pathogenic variants are associated with aggressive disease. Although research conducted in patients with prostate cancer unselected for family history, stage of disease or age at diagnosis has contributed to prevalence estimates of mutations [12], a lack of comprehensive clinical records impedes the clinical associations required to inform clinical management. In this study, we aim to identify novel germline variants in prostate cancer predisposition genes and describe their associated clinical phenotype in the setting of multi-case prostate cancer families and multi-case breast/ovarian cancer families. We provide evidence that novel germline mutations in ATM and CHEK2 are associated with aggressive prostate cancer. This finding will contribute evidence to support broader genetic screening and development of gene-targeted protocols, with the aim of improving outcomes in these high-risk prostate cancer patients.
2. Patients and Methods
2.1. Ethics Statement
Signed consent was obtained from all participants on recruitment to the Kathleen Cunningham Familial Cancer Consortium (kConFab) (www.kconfab.org (accessed on 31 March 2021)). This project was approved by the Peter MacCallum Cancer Centre, Melbourne, Australia, Human Research Ethics Committee, protocol #97/27.
2.2. Study Population
Families containing at least two male first- or second-degree relatives with verified prostate cancer were identified from kConFab [13]. kConFab recruitment of these families was from surgical and genetics services on the basis of multi-case prostate cancer (n = 28) or multi-case breast/ovarian cancer (n = 66). Males were eligible for inclusion if (a) their prostate cancer could be verified with a pathology report and/or doctor’s notes, (b) treatment reports were available and (c) a gDNA sample was available for mutation testing. An index case (n = 94) was selected for mutation testing from each family based on high-grade pathological features and/or youngest age at diagnosis. All cancers were prostatic adenocarcinoma with index cases diagnosed between 1994 and 2019. Affected females in the breast/ovarian cancer families had previously undergone a seven-cancer-gene panel screening (BRCA1, BRCA2, ATM 7271, PALB2, TP53, PTEN, CHEK2 1100delC) prior to this study, with no C4/5 variant identified.
2.3. Clinical and Pathological Data
Age, stage at diagnosis and prostate-specific antigen (PSA) immediately prior to biopsy was recorded. Gleason score (GS) at diagnosis (first biopsy) and radical prostatectomy were converted to ISUP Grade Group (GG) [14] for analysis. Treatment modality, cause and date of death and presence of other primary verified malignancies were recorded. D’Amico risk stratification [15] was derived from PSA, GG and stage at diagnosis.
Formalin-fixed paraffin-embedded archival 5μm prostate cancer (biopsy or prostatectomy) tissue specimens were haematoxylin and eosin stained and re-reviewed by a uropathologist to standardise and provide a contemporary histopathological assessment including cribriform and intraductal carcinoma of the prostate (IDCP). Results were reported according to the ISUP guidelines [14] and structured reporting guidelines of the Royal College of Pathologists of Australasia (RCPA).
2.4. Mutation Detection
For each prostate cancer index case, 2.5 ug of gDNA underwent an 84-hereditary-cancer-gene panel screening (https://www.invitae.com/en/providers/test-catalog/test-01101 (accessed on 31 March 2021)). Variant classification was undertaken according to the five-tier ENIGMA system (http://www.enigmaconsortiumorg/ (accessed on 31 March 2021)). C1/2 variants were not reported. Relatives (male and female, cancer-affected and unaffected) of index cases in whom a C4/5 clinically notifiable, non-BRCA pathogenic variant was identified underwent segregation analysis to determine family-specific mutation status and identify obligate carriers.
Prostate cancer index cases with no C4/5 variants detected and their male relatives (majority genetically unscreened) with verified prostate cancer formed the BRCAX control cohort (n = 111). Clinical characteristics of the PCOR (n = 39,953) and a BRCA2 cohort (n = 40) [16] were obtained for comparison. PCOR is a whole-population (Australia and New Zealand) prostate cancer registry [17].
2.5. Statistical Analysis
Descriptive statistical methods defined the clinical characteristics of each gene. Prostate-cancer-specific survival for ATM mutation carriers and the BRCAX cohort were analysed using the Kaplan–Meier method and compared using log-rank (LR) test. Odds ratios (OR) were calculated using Fisher’s exact test to compare clinical characteristics of ATM and HOXB13 mutations, and the BRCAX cohort. This analysis was not performed for CHEK2 carriers due to low carrier numbers. Student’s t-test was used to evaluate differences between C3 mutation carriers and non-carriers within the BRCAX group. These analyses were performed using Stata 16.0.
A modified segregation analysis was used to obtain an estimate of prostate cancer risk (hazard ratio, HR) for each gene separately (ATM, CHEK2), based on genotyping of family members using MENDEL version 3.2. Estimates were adjusted for clinic-based ascertainment using the retrospective likelihood method [18]. For each gene, the cumulative risk (penetrance) to a given age was calculated as one minus the exponential of minus the cumulative incidence for carriers, which itself was the sum from age zero to the given age of the estimated HR multiplied by the non-carriers’ prostate cancer age-specific incidence [19]. Non-carrier incidences were set equal to the age-specific population incidence rates for Australia in the period 1998–2002 [20]. The HR was assumed to be constant across all ages, and all ages were truncated at 80 years. p-values were two-sided and based on the likelihood ratio test, and a threshold of 0.05 was used to define statistical significance.
3. Results
Ninety-four prostate cancer index cases were screened, yielding twenty-two (23%) C4/5 mutations across ten different genes. Fourteen of twenty-two (64%) mutations were from multi-case breast/ovarian cancer families and eight of twenty-two (36%) from multi-case prostate cancer families. Six of twenty-two variants were not clinically notifiable according to kConFab clinical notification protocol (n = 1 BARD1, NBN, RECQL4, WRN; n = 2 PTCH1). Seven of twenty-two variants were newly identified in BRCA1 (n = 3) and BRCA2 (n = 4) genes. Nine of twenty-two variants were identified in ATM (n = 4), CHEK2 (n = 2) and HOXB13G84E (n = 3). gDNA segregation analysis confirmed a further six prostate-cancer-affected relatives as carriers; a final cohort for analysis contained fifteen patients (ATM n = 9, CHEK2 n = 2 and HOXB13G84E n = 4). Table 1 shows a summary of gene nomenclature, prostate-cancer-affected carriers and non-carriers of the family-specific mutation for each gene and recruitment stream (multi-case breast/ovarian and prostate cancer families). Variants were unique to individual families except the HOXB13G84E variant which was found in three families.
Table 1.
Gene nomenclature, affected families, carriers and non-carriers with prostate cancer (PCa) within each family and kConFab recruitment stream.
| Gene | Nomenclature | Affected Families (n) | Carriers with PCa (n) | Non-Carriers with PCa (n) | Recruitment Stream |
|---|---|---|---|---|---|
| ATM | c.6115G > A; p.Glu2039Lys | 1 | 2 | 0 | Breast/ovarian |
| c.8266A > T; p.Lys2756Ter | 1 | 2 | 0 | Breast/ovarian | |
| c.8395_8404del | 1 | 2 | 1 | Prostate | |
| c.3712_3716del; p.Leu1238Lysfs 6 | 1 | 3 | 0 | Prostate | |
| CHEK2 | c.349A > G; p.Arg117Gly | 1 | 1 | 0 | Breast/ovarian |
| c.499G > A; p.Gly167Arg | 1 | 1 | 0 | Breast/ovarian | |
| HOXB13 | G84E | 3 | 4 | 0 | Breast/ovarian |
| PTCH1 | c.3850C > T; p.Gln1284 | 1 | 1 | 0 | Breast/ovarian |
| c.290dup; p.Asn97Lysfs 43 | 1 | 1 | 0 | Breast/ovarian | |
| BARD1 | c.1921C > T; p.Arg641 | 1 | 2 | 0 | Breast/ovarian |
| NBN | c.217A>T; p.Lys73 | 1 | 1 | 0 | Breast/ovarian |
| RECQL4 | c.2269C > T; p.Gln757 | 1 | 1 | 1 | Breast/ovarian |
| WRN | c.3030_3033del; p.Thr1011Argfs 11 | 1 | 1 | Prostate | |
| BRCA1 | c.2612insT; p.PHe872Valsfs 31 | 1 | 2 | 0 | Breast/ovarian |
| c.2864C > A; p.Ser955 | 1 | 0 | 0 | Prostate | |
| c.547+1G > T (Splice donor) | 1 | 2 | 0 | Prostate | |
| BRCA2 | c.67+1G > T; (Splice donor) | 1 | 1 | 0 | Breast/ovarian |
| c.7266T > A; p.Cys2422 | 1 | 2 | 0 | Prostate | |
| c.5909C > A; p.Ser1970 | 1 | 1 | 0 | Prostate | |
| c.8756G > A; p.Gly2919Asp | 1 | 1 | 0 | Prostate |
3.1. Clinical Phenotype of Germline Mutation Carriers
3.1.1. ATM Mutation Carriers
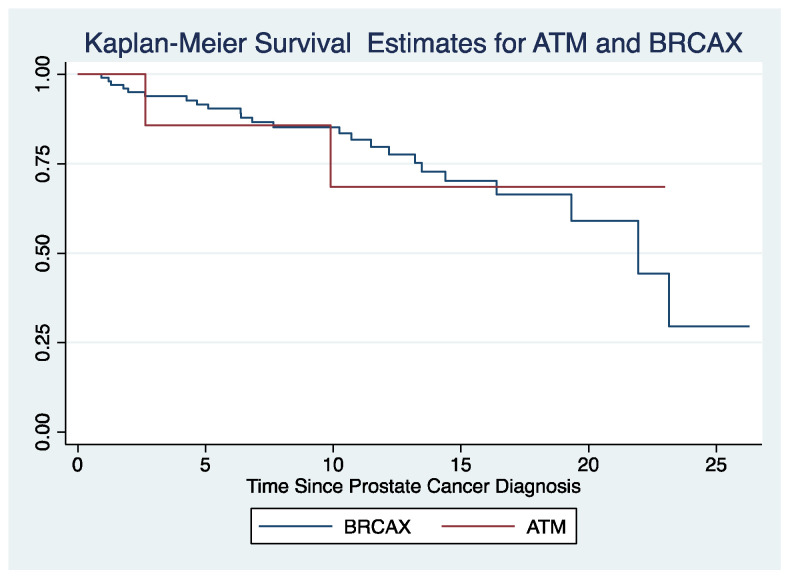
There were nine ATM mutation carriers identified with a median age at diagnosis of 65 years (mean 65, range 42–90) (Table 2). More than half (56%) of ATM mutation carriers presented with a PSA in the intermediate to high-risk range of >10 nm/mL. Eight ATM mutation carriers underwent treatment with curative intent (radiation n = 3, surgery n = 5). There were three deaths; two prostate-cancer-specific and one from myelodysplastic syndrome (MDS). The probability of prostate cancer-specific survival at five and at ten years post diagnosis was 86% (CI 33–98%) and 69% (CI 21–91%), respectively. The LR test for equality of survivor functions comparing ATM carriers and the BRCAX group was not significant p = 0.95 (Figure 1). Most ATM carriers had GG 5 disease (n = 3, 33.3%), advanced stage at diagnosis (n = 3, 33.3%) and high-grade architectural features (IDCP, cribriform structure) on pathology review (n = 4, 44.4%) (Table 2). One ATM carrier diagnosed at 42 years was stratified to the low-risk D’Amico group. The estimated HR for prostate cancer associated with ATM mutations was 3.5 (p = 0.11, 95% CI 0.69–18.2). Cumulative risks of prostate cancer approached 6% at age 50 and 20% by age 70 (Table 3). Similar to the BRCAX cohort, ATM carriers were associated with high-risk D’Amico classification and high-grade pathology (GG 4–5) with no significant statistical differences in clinical phenotype identified when comparing the two groups.
Table 2.
Clinical characteristics for ATM, CHEK2, and HOXB13 germline mutations.
| ATM Mutation Carriers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Age at Diagnosis | PSA 1 | Grade Group | Stage | Architecture 2 | D’Amico | Primary Treatment | Age at Death | Cause of Death | Age at Diagnosis of Other Primary Malignancy |
| 78 | 13.6 | - | - | - | Int | Radiation | 87 | PCa | - |
| 90 | 210 | 5 | T1c | No | High | Non-curative | 93 | PCa | - |
| 57 | 160 | - | - | - | High | Radiation | 75 | MDS 3 | 70, MDS |
| 65 | 20 | 5 | T1c | Cribriform | High | Radiation | - | - | 66, Renal |
| 75 | 5 | 3 | T3aN1 | Cribriform | High | Prostatectomy | - | - | |
| 68 | 12 | 5 | T3b | IDCP | High | Prostatectomy | - | - | - |
| 49 | 2.6 | 3 | T2a | IDCP, Cribriform | Int | Prostatectomy | - | - | - |
| 42 | 6.9 | 1 | - | Low | Prostatectomy | - | - | - | |
| 61 | 12 | 2 | T2c | - | High | Prostatectomy | - | - | 83, Lung, Melanoma |
| CHEK2 Mutation Carriers | |||||||||
| 80 | - | 5 | T4 | - | High | Prostatectomy | 81 | Cardiac | 78, Bladder, Colon |
| 56 | 8.8 | 2 | T2c | - | High | Prostatectomy | - | - | 47, Breast |
| HOXB13G84E Mutation Carriers | |||||||||
| 68 | 6.7 | 1 | T1c | Low | Radiation | 92 | - | 80, Breast | |
| 69 | 5.3 | 2 | T2N0 | Int | Prostatectomy | - | - | - | |
| 47 | 3.8 | 2 | T3a | High | Prostatectomy | - | - | - | |
| 64 | - | 2 | T2a | Int | - | 89 | PCa | - | |
1 PSA immediately prior to undergoing diagnostic biopsy; 2 High-risk pathology features identified within prostatic adenocarcinoma; Abbreviations: Myelodysplastic syndrome (MDS), D’Amico Intermediate-risk (Int), intraductal carcinoma of the prostate (IDCP).
Figure 1.
Survival time in years following prostate cancer diagnosis; red line represents survival of ATM mutation carriers following diagnosis, blue line represents survival of BRCAX cohort patients.
Table 3.
Cumulative Risk of Prostate Cancer Diagnosis for ATM and CHEK2 mutation carriers.
| Percentage Cumulative Risk (95% CI) of Prostate Cancer Diagnosis | ||
|---|---|---|
| Age (Years) | ATM Mutation Carriers | CHEK2 Mutation Carriers |
| 30 | 0 (0–0) | 0 (0–0.1) |
| 40 | 0 (0–0) | 0 (0–0.4) |
| 50 | 0.5 (0.1–2.5) | 1.2 (0.1–22.4) |
| 60 | 5.7 (1.1–25.9) | 13.2 (0.7–95) |
| 70 | 20.6 (4.4–69.4) | 42.8 (2.6–100) |
| 80 | 40.9 (9.7–93.3) | 72.1 (5.8–100) |
3.1.2. CHEK2 Mutation Carriers
Two CHEK2 mutation carriers were identified who were diagnosed with prostate cancer at 80 and 56 years (Table 2). The former was diagnosed with Stage T4, GG 5 prostate cancer with IDCP after a cystoprostatectomy for concurrent primary bladder cancer. His PSA fluctuated between 9–24 for five years prior; a biopsy had not been undertaken due to comorbidities and preferences with survival <1 year following surgery. The latter was diagnosed with Stage pT2c, GG 2 disease following radical prostatectomy (D’Amico high-risk) with survival > 15 years. His cancer history included primary bilateral breast cancer (Table 2). Both patients demonstrated high-grade pathology and advanced stage at diagnosis. The estimated HR for prostate cancer associated with CHEK2 mutations was 8.6 (p = 0.29, 95% CI 0.40–182). The cumulative risk of prostate cancer for CHEK2 mutation carriers approached 1.2% at age 50 and 48% by age 70 (Table 3).
3.1.3. HOXB13G84E Mutation Carriers
There were four patients identified as HOXB13G84E mutation carriers who were identified with prostate cancer at ~66 years (mean 62, range 47–69) (Table 2). One prostate-cancer-specific death and one non-prostate-cancer-related death occurred in this group, both 25 years post diagnosis. Two carriers remain alive 13 and 12 years post diagnosis. All patients in this group underwent treatment with curative intent (surgery n = 3, radiation n = 1) (Table 2). One primary breast cancer was identified. HOXB13G84E carriers had GG 1–2 disease and were stratified to D’Amico low–intermediate risk, except one patient diagnosed with stage pT3a disease. PSA at diagnosis was <10 ng/mL for all participants. Clinical phenotype was not significantly different for HOXB13 carriers compared with the BRCAX group when comparing low-risk D’Amico (p = 0.3), patients with Grade Group 1 and 2 disease (p = 0.3), PSA (p = 0.5), age at death (p = 0.1), age at diagnosis (p = 0.8) and prostate-cancer-specific death (p = 0.4).
3.2. BRCAX Cohort Analysis
Within the cohort of 111 prostate cancer patients categorised as BRCAX (i.e., confirmed non-mutation carriers), clinical characteristics of C3 variant carriers (n = 39, Supplementary Table S1) and non-carriers (n = 72) were not significantly different. Therefore, the BRCAX cohort analysis combined these sub-groups. Mean PSA at diagnosis was 13.7 ng/mL. Twenty-four prostate-cancer-specific deaths were recorded; the probability of prostate-cancer-related mortality was 21.6% (Supplementary Figure S1). Median follow-up time was 8.6 years (mean 9.5, range 1 month to 26.3 years). A total of 64.4% (n = 67) of patients underwent curative treatment with radical prostatectomy. Other verified primary cancers were identified in 16.7% of patients (Supplementary Table S2). The BRCAX group had a lower median age at diagnosis (62 vs. 68) and a higher percentage of GG 4–5 disease (30.6% vs. 25%) compared with the PCOR cohort [17]. In the BRCAX group, 53.2% of subjects were stratified as D’Amico high risk, compared with 25% of the PCOR cohort (Table 4, Supplementary Figure S2).
Table 4.
Clinical features of the BRCAX cohort, PCOR cohort and BRCA2 cohort.
| BRCAX (n = 111) | PCOR1 (n = 39,953) | BRCA2 (n = 40) 1 | |
|---|---|---|---|
| Age at Diagnosis | 62.7 (mean) 62 (median) 42–84 (range) 9.7 (SE) |
68 (median) | 65.9 (mean) 64.95 (median) |
| PSA pre-diagnosis, n (%) | |||
| <4 ng/mL | 17 (15.3%) | 955 (13%) | 1 (2.5%) |
| 4–10 ng/mL | 43 (38.7%) | 4111 (56%) | 10 (25%) |
| 10+ ng/mL | 25 (22.5%) | 2225 (31%) | 14 (35%) |
| Unknown | 26 (23.4%) | 1067 (14%) | 11 (27.5%) |
| ISUP Grade Group at Diagnosis, n (%) | |||
| 1 | 19 (17.1%) | 2115 (26%) | 2 (5.3%) |
| 2 | 32 (28.8%) | 2606 (32%) | 11 (28.9%) |
| 3 | 23 (20.7%) | 11,380 (7%) | |
| 4 & 5 | 34 (30.6%) | 2002 (25%) | 25 (65.8%) |
| Unknown | 3 (2.7%) | 255 (4%) | - |
| D’Amico Risk Group at Diagnosis | |||
| Low | 21 (18.9%) | 1492 (20%) | 3 (7.7%) |
| Intermediate | 31 (27.9%) | 3454 (46%) | 5 (12.8%) |
| High or very high | 59 (53.2%) | 1832 (25%) | 31 (79.5%) |
| Survival Status | |||
| Deceased n (%) | 30 (27%) | - | 23 (57.5) |
| PCa-specific death (n) | 24 (21.6%) | - | 21 (52.5%) |
| Mean, median years of follow up from diagnosis (range) | 9.5, 8.6 (0.2–26.3) | – | 4.6 years (high risk)9 years (int. risk) |
| Mean, median duration to death (years) | 9.9, 8.8 (0.9–26.3) | – | 4.5 (mean) |
| PCa-specific survival estimate | 91% 5-yr (84–96) | 95% 5-yr (2012–2016) 2 | 33% 15-yr (int. risk) |
| PCa-specific survival estimate | 85% 10-yr (76–91) | 91.3% 10-yr (2011–2015) 3 | 0% 15-yr (high risk) |
1BRCA2 cohort previously described in Bolton et al. BJU Int. 2015, 116, 207–212 [17]. 2 https://www.canceraustralia.gov.au/cancer-types/prostate-cancer/statistics (accessed on 10 September 2021); 3 https://ncci.canceraustralia.gov.au/outcomes/relative-survival-rate/10-year-relative-survival (accessed on 10 September 2021).
4. Discussion
This study demonstrates the clinical and pathological features of 15 prostate cancer patients who carry rare, pathogenic non-BRCA germline variants. Thirteen of fifteen cancers were classified as D’Amico high risk (n = 9, 60%) or intermediate risk (n = 4, 26.7%). Phenotypic differences comparing variant carriers and non-carriers in this cohort were likely diminished by the high-risk clinical features of the BRCAX group [1]. We have previously demonstrated that men with a strong family history of prostate cancer are at increased risk of developing clinically significant and aggressive disease [16]. Although germline mutations represent one mechanism through which this can occur, other mechanisms such as epigenetic gene silencing and hapolinsufficiency are also being investigated in the BRCAX cohort to explore why they demonstrate high-risk features of disease similar to the mutation-carrier groups.
ATM and CHEK2 both appear to be less penetrant than BRCA2 genes [10,21,22]. Broader panel testing than has previously been applied to families resulted in improved identification of mutations. This is demonstrated by nine patients with clinically notifiable, pathogenic variants, seven of which were not previously detected using a breast/ovarian-cancer-gene panel when testing affected female relatives.
Germline mutations in BRCA2, ATM, MLH1, PMS2, CHEK2 and HOXB13 have been identified in association with prostate cancer risk [11,21,22], and carriers of germline MSH2 and MSH6 mutations were found to have higher prostate cancer incidence than age-matched controls (4.3% vs. 3%, respectively) [21,23]. These genes combined account for only 15% of familial prostate cancer predisposition [21,24]. Genetic testing recommendations differ between guidelines due to varying levels of evidence for rarer variants [25]. Guidelines must balance the need for cost-effective recommendations within the scope of current evidence, acknowledging that carriers of rarer variants may remain unidentified. Guidelines also currently emphasise family history of breast/ovarian cancers, rather than multi-case prostate cancer history [26]. One study found that 37% of prostate-cancer-affected pathogenic mutation carriers would not have qualified for genetic testing under the NCCN genetic/familial breast and ovarian guidelines for patients with prostate cancer, based on self-reported family histories [12]. Although some testing occurs at the discretion of practitioners, this cannot be guaranteed where funding and insurance will not cover costs [7,27]. Guidelines for broader gene panel testing based on prostate cancer family history alone are recommended [26].
ATM mutations and high-risk prostate cancer have been associated in some studies through screening of patients with advanced disease [28,29], although BRCA2 remains the only gene in which pathogenic variants have been consistently linked to aggressive prostate cancer. In contrast to our findings, the PRACTICAL consortium investigated a pooled cohort of germline ATM mutation carriers, concluding that although they were associated with younger age of prostate cancer onset, variants did not conclusively predispose carriers to more aggressive prostate cancer phenotypes [10]. Conversely, a pooled analysis of loss of function (LoF) mutations, including two ATM mutations, concluded that even after adjusting for the inclusion of BRCA2 mutations in the cohort, the association between poor prognosis, aggressive disease and LoF mutations in the ATM gene remained [30]. Testing criteria for therapeutically significant somatic and/or germline mutations such as ATM has been recommended following evidence that ATM pathogenic mutations are associated with efficacy of gene-targeted therapies and early age of prostate cancer onset [10,31]. Therefore, although small, our cohort contributes significantly to the current literature correlating ATM mutation carriers and phenotype. Compared with our previously published BRCA2 cohort [16], ATM mutation carriers were diagnosed at a similar mean age (65 vs. 65.9) with similar PSA levels (ATM: 55.6% > 10 ng/mL, BRCA2: 35% > 10 ng/mL). Both carrier groups had a high proportion of GG 4–5 disease (ATM 33.3%, BRCA2 65.8%). Stage and PSA at diagnosis resulted in 66.7% of ATM carriers and 79.5% of BRCA2 carriers being classified as D’Amico high risk. Only one ATM carrier was considered low risk. However, his age at diagnosis (42 years) and family history (other related carriers diagnosed at age 49 and 61) suggest a highly penetrant variant. Importantly, ATM mutations were detected in prostate cancer index cases from two families in which close relatives with breast/ovarian cancer had previously undergone limited ATM screening for the 7271 variant. In addition, two ATM mutations were identified in screening-naïve families with no cancer-affected female relatives. These cases support prostate-specific-gene panel testing for patients from multi-case cancer families independent of female relatives.
CHEK2 carriers in this cohort also demonstrated high-risk features of prostate cancer. Brandao et al. recently evaluated the role of the CHEK2 c.349A > G variant in prostate cancer development, with results supporting its candidacy as a founder mutation associated with early onset familial prostate cancer [22]. Due to its founder variant status, Brandao et al. assert that CHEK2 may be a cost-effective addition to screening panels in select multi-case prostate cancer families [22]. CHEK2 was also associated with aggressive prostate cancer in the pooled analysis of (LoF) mutations conducted by Leongamornlert et al., which included two carriers of CHEK2 frameshift mutations [30]. Our cohort therefore contributes to evidence of prostate cancer predisposition for CHEK2 carriers and a relationship with aggressive disease, although larger numbers are required to solidify this association [22].
HOXB13G84E-associated prostate cancer demonstrated lower-risk clinical features compared with other variants. This is in keeping with the clinical profile of HOXB13 carriers previously described [5], although notably HOXB13 carriers in our cohort were not associated with younger age of onset compared with non-carriers and the cohort described by Ewing et al. [32]. This is an important contribution to the prostate cancer profile of the HOXB13G84E variant which is significantly associated with hereditary prostate cancer development.
A limitation of our study was the assumption that if the prostate cancer index case did not carry a C4-5 mutation, then the other prostate cancer cases in that family did not either. This could have affected the clinical characteristics seen in the BRCAX group and would require testing of all family members to provide confirmation. C3 variants were also common (35%) in the BRCAX group. Their association with prostate cancer heritability and risk of aggressive disease is unknown, although Mersch et al. reported that of those with sufficient evidence to be reclassified (7.7%), 91% were reclassified as benign [33]. Despite our small carrier cohort of novel mutations, the accompanying clinical information is highly informative in elucidating the clinical phenotype associated with these pathogenic variants [34].
5. Conclusions
This study contributes to the correlation of rare genetic variants with clinically significant prostate cancer. Incorporation of gene panel testing into clinical practice will allow clinicians to identify patients with a family history of prostate cancer who are at risk of aggressive disease. Confirmation of mutation status can facilitate intensive treatment rather than active surveillance for carriers. Other benefits include early disease detection and access to novel therapies. Recruitment of additional multi-case prostate cancer families to solidify the clinical validity and utility of multi-gene panel testing for these variants in patients with familial prostate cancer is required. Through broader panel testing, changes in clinical practice and clinical outcomes are realisable for this high-risk cohort.
Acknowledgments
We wish to thank Eveline Niedermayr, kConFab and Family Cancer Clinic staff and all the families who contribute to kConFab.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14153623/s1, Figure S1: Survival time in years for BRCAX cohort patients, following a diagnosis of prostate cancer; Figure S2: Kaplan Meier curve showing prostate cancer-specific survival of patients from the PCOR cohort following a diagnosis of prostate cancer; Table S1: C3 variant nomenclature identified in the BRCAX cohort; Table S2: Non-Prostate Primary Cancers verified among the BRCAX cohort.
Author Contributions
All authors contributed sufficiently to this manuscript to justify inclusion in the authorship list. Conceptualisation, D.B., D.M., R.A.T. and H.T.; Data curation, R.M., D.C. and L.K.; Formal analysis, R.M. and J.D.; Funding acquisition, R.A.T. and H.T.; Investigation, D.C. and P.S.; Methodology, J.D., L.K. and H.T.; Project administration, D.M.; Resources, D.C. and H.T.; Software, R.M. and J.D.; Supervision, D.B. and H.T.; Validation, D.M. and P.S.; Visualisation, D.B. and R.A.T.; Writing—original draft, R.M.; Writing—review & editing, D.B., D.C., P.S., R.A.T. and H.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Peter MacCallum Cancer Centre, Melbourne, Australia, Human Research Ethics Committee, protocol #97/27.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Signed consent was obtained from all participants on recruitment to the Kathleen Cunningham Familial Cancer Consortium (kConFab) (www.kconfab.org (accessed on 31 March 2021)).
Data Availability Statement
The dataset can be made available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
kConFab is currently supported by a National Breast Cancer Foundation grant, and the Prostate Cancer Foundation Australia (PIRA-1519).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thorne H., Willems A.J., Niedermayr E., Hoh I.M.Y., Li J., Clouston D., Mitchell G., Fox S., Hopper J.L., Bolton D., et al. Decreased Prostate Cancer-Specific Survival of Men with BRCA2 Mutations from Multiple Breast Cancer Families. Cancer Prev. Res. 2011;4:1002–1010. doi: 10.1158/1940-6207.CAPR-10-0397. [DOI] [PubMed] [Google Scholar]
- 2.Hunter S.M., Rowley S.M., Clouston D., Li J., Lupat R., Krishnananthan N., Risbridger G., Taylor R., Bolton D., Campbell I.G., et al. Searching for candidate genes in familial BRCAX mutation carriers with prostate cancer. Urol. Oncol. 2016;34:120.e9–120.e16. doi: 10.1016/j.urolonc.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Schaid D.J., McDonnell S.K., FitzGerald L.M., DeRycke L., Fogarty Z., Giles G.G., MacInnis R.J., Southey M.C., Nguyen-Dumont T., Cancel-Tassin G., et al. Two-stage Study of Familial Prostate Cancer by Whole-exome Sequencing and Custom Capture Identifies 10 Novel Genes Associated with the Risk of Prostate Cancer. Eur. Urol. 2021;79:353–361. doi: 10.1016/j.eururo.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark R., McAlpine K., Fleshner N. A Clinical Trial of Prophylactic Prostatectomy for BRCA2 Mutation Carriers: Is Now the Time? Eur. Urol. Focus. 2021;7:506–507. doi: 10.1016/j.euf.2021.04.018. [DOI] [PubMed] [Google Scholar]
- 5.Cheng H.H., Sokolova A.O., Schaeffer E.M., Small E.J., Higano C.S. Germline and Somatic Mutations in Prostate Cancer for the Clinician. J. Natl. Compr. Cancer Netw. 2019;17:515–521. doi: 10.6004/jnccn.2019.7307. [DOI] [PubMed] [Google Scholar]
- 6.Sessine M.S., Das S., Park B., Salami S.S., Kaffenberger S.D., Kasputis A., Solorzano M., Luke M., Vince R.A., Kaye D.R., et al. Initial Findings from a High Genetic Risk Prostate Cancer Clinic. Urology. 2021;156:96–103. doi: 10.1016/j.urology.2021.05.078. [DOI] [PubMed] [Google Scholar]
- 7.Scheinberg T., Goodwin A., Ip E., Linton A., Mak B., Smith D.P., Stockler M.R., Strach M.C., Tran B., Young A.L., et al. Evaluation of a Mainstream Model of Genetic Testing for Men with Prostate Cancer. JCO Oncol. Pr. 2021;17:e204–e216. doi: 10.1200/OP.20.00399. [DOI] [PubMed] [Google Scholar]
- 8.Castro E., Goh C., Leongamornlert D., Saunders E., Tymrakiewicz M., Dadaev T., Govindasami K., Guy M., Ellis S., Frost D., et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur. Urol. 2015;68:186–193. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Taylor R., Fraser M., Livingstone J., Espiritu S.M.G., Thorne H., Huang V., Lo W., Shiah Y.-J., Yamaguchi T.N., Sliwinski A., et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat. Commun. 2017;8:13671. doi: 10.1038/ncomms13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson Q., Brook M.N., Dadaev T., Wakerell S., Saunders E.J., Muir K., Neal D.E., Giles G.G., MacInnis R.J., Thibodeau S.N., et al. Rare Germline Variants in ATM Predispose to Prostate Cancer: A PRACTICAL Consortium Study. Eur. Urol. Oncol. 2021;4:570–579. doi: 10.1016/j.euo.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandhu S., Moore C.M., Chiong E., Beltran H., Bristow R.G., Williams S.G. Prostate cancer. Lancet. 2021;398:1075–1090. doi: 10.1016/S0140-6736(21)00950-8. [DOI] [PubMed] [Google Scholar]
- 12.Nicolosi P., Ledet E., Yang S., Michalski S., Freschi B., O’Leary E., Esplin E.D., Nussbaum R.L., Sartor O. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorne H., Mitchell G., Fox S. kConFab: A Familial Breast Cancer Consortium Facilitating Research and Translational Oncology. J. Natl. Cancer Inst. Monogr. 2011;2011:79–81. doi: 10.1093/jncimonographs/lgr042. [DOI] [PubMed] [Google Scholar]
- 14.van Leenders G., van der Kwast T.H., Grignon D.J., Evans A.J., Kristiansen G., Kweldam C.F., Iczkowski K.A. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2020;44:e87–e99. doi: 10.1097/PAS.0000000000001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Amico A.V., Whittington R., Malkowicz S.B., Schultz D., Blank K., Broderick G.A., Wein A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 16.Bolton D., Cheng Y., Willems-Jones A.J., Li J., Niedermeyr E., Mitchell G., Clouston D., Lawrentschuk N., Sliwinski A., Fox S., et al. Altered significance of D’Amico risk classification in patients with prostate cancer linked to a familial breast cancer (kConFab) cohort. Br. J. Urol. 2014;116:207–212. doi: 10.1111/bju.12792. [DOI] [PubMed] [Google Scholar]
- 17.Papa N., O’Callaghan M., James E., Millar J. Prostate Cancer in Australian and New Zealand Men: Patterns of Care within PCOR-ANZ 2015–2018. Monash University & Movember; Melbourne, VIC, Australia: 2021. [Google Scholar]
- 18.Kraft P., Thomas D.C. Bias and Efficiency in Family-Based Gene-Characterization Studies: Conditional, Prospective, Retrospective, and Joint Likelihoods. Am. J. Hum. Genet. 2000;66:1119–1131. doi: 10.1086/302808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen-Dumont. T., Dowty J.G., Steen J.A., Renault A.L., Hammet F., Mahmoodi M., Theys D., Rewse A., Tsimiklis H., Winship I.M., et al. Population-Based Estimates of the Age-Specific Cumulative Risk of Breast Cancer for Pathogenic Variants in CHEK2: Findings from the Australian Breast Cancer Family Registry. Cancers. 2021;13:1378. doi: 10.3390/cancers13061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curado M.P., Edwards B., Shin H.R., Storm H., Ferlay J., Heanue M., Boyle P. Cancer Incidence in Five Continents Volume IX. IARC Press; Lyon, France: 2007. [Google Scholar]
- 21.Vietri M., D’Elia G., Caliendo G., Resse M., Casamassimi A., Passariello L., Albanese L., Cioffi M., Molinari A. Hereditary Prostate Cancer: Genes Related, Target Therapy and Prevention. Int. J. Mol. Sci. 2021;22:3753. doi: 10.3390/ijms22073753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandão A., Paulo P., Maia S., Pinheiro M., Peixoto A., Cardoso M., Silva M.P., Santos C., Eeles R.A., Kote-Jarai Z., et al. The CHEK2 Variant C.349A>G Is Associated with Prostate Cancer Risk and Carriers Share a Common Ancestor. Cancers. 2020;12:3254. doi: 10.3390/cancers12113254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bancroft E.K., Page E.C., Brook M.N., Thomas S., Taylor N., Pope J., McHugh J., Jones A.-B., Karlsson Q., Merson S., et al. A prospective prostate cancer screening programme for men with pathogenic variants in mismatch repair genes (IMPACT): Initial results from an international prospective study. Lancet Oncol. 2021;22:1618–1631. doi: 10.1016/S1470-2045(21)00522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen-Dumont T., Dowty J., MacInnis R., Steen J., Riaz M., Dugué P.-A., Renault A.-L., Hammet F., Mahmoodi M., Theys D., et al. Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Cancers. 2021;13:1495. doi: 10.3390/cancers13071495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geary J., Majumder M., Guerrini C., Cook-Deegan R. Development of an Open Database of Genes Included in Hereditary Cancer Genetic Testing Panels Available From Major Sources in the US. JAMA Oncol. 2022;8:7639. doi: 10.1001/jamaoncol.2021.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giri V.N., Knudsen K.E., Kelly W.K., Cheng H.H., Cooney K.A., Cookson M.S., Dahut W., Weissman S., Soule H.R., Petrylak D.P., et al. Implementation of Germline Testing for Prostate Cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J. Clin. Oncol. 2020;38:2798–2811. doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andoni T., Wiggins J., Robinson R., Charlton R., Sandberg M., Eeles R. Half of germline pathogenic and likely pathogenic variants found on panel tests do not fulfil NHS testing criteria. Sci. Rep. 2022;12:2507. doi: 10.1038/s41598-022-06376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhen J.T., Syed J., Nguyen K.A., Leapman M.S., Agarwal N., Ms K.B., Llor X., Hofstatter E., Shuch B. Genetic testing for hereditary prostate cancer: Current status and limitations. Cancer. 2018;124:3105–3117. doi: 10.1002/cncr.31316. [DOI] [PubMed] [Google Scholar]
- 29.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leongamornlert D., Collaborators T.U., Saunders E., Dadaev T., Tymrakiewicz M., Goh C., Jugurnauth-Little S., Kozarewa I., Fenwick K., Assiotis I., et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br. J. Cancer. 2014;110:1663–1672. doi: 10.1038/bjc.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(MSAC) MSAC. 1618—Testing of Tumour Prostate Tissue to Detect BRCA1/2 Pathogenic Gene Variants in Men with Metastatic Castration-Resistant Prostate Cancer to Help Determine Eligibility for PBS Olaparib 2022. [(accessed on 16 April 2022)]; Updated 21 February 2022. Available online: http://www.msac.gov.au/internet/msac/publishing.nsf/Content/1618-public.
- 32.Ewing C.M., Ray A.M., Lange E.M., Zuhlke K.A., Robbins C.M., Tembe W.D., Wiley K.E., Isaacs S.D., Johng D., Wang Y., et al. Germline Mutations in HOXB13 and Prostate-Cancer Risk. N. Engl. J. Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mersch J., Brown N., Pirzadeh-Miller S., Mundt E., Cox H.C., Brown K., Aston M., Esterling L., Manley S., Ross T. Prevalence of Variant Reclassification Following Hereditary Cancer Genetic Testing. JAMA. 2018;320:1266–1274. doi: 10.1001/jama.2018.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall M.J., Bernhisel R., Hughes E., Larson K., Rosenthal E.T., Singh N.A., Lancaster J.M., Kurian A.W. Germline Pathogenic Variants in the Ataxia Telangiectasia Mutated (ATM) Gene are Associated with High and Moderate Risks for Multiple Cancers. Cancer Prev. Res. 2021;14:433–440. doi: 10.1158/1940-6207.CAPR-20-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset can be made available upon request.