Abstract
Phaleria macrocarpa, known as “Mahkota Dewa”, is a widely used medicinal plant in Malaysia. This study focused on the characterization of α-glucosidase inhibitory activity of P. macrocarpa extracts using Fourier transform infrared spectroscopy (FTIR)-based metabolomics. P. macrocarpa and its extracts contain thousands of compounds having synergistic effect. Generally, their variability exists, and there are many active components in meager amounts. Thus, the conventional measurement methods of a single component for the quality control are time consuming, laborious, expensive, and unreliable. It is of great interest to develop a rapid prediction method for herbal quality control to investigate the α-glucosidase inhibitory activity of P. macrocarpa by multicomponent analyses. In this study, a rapid and simple analytical method was developed using FTIR spectroscopy-based fingerprinting. A total of 36 extracts of different ethanol concentrations were prepared and tested on inhibitory potential and fingerprinted using FTIR spectroscopy, coupled with chemometrics of orthogonal partial least square (OPLS) at the 4000–400 cm−1 frequency region and resolution of 4 cm−1. The OPLS model generated the highest regression coefficient with R2Y = 0.98 and Q2Y = 0.70, lowest root mean square error estimation = 17.17, and root mean square error of cross validation = 57.29. A five-component (1+4+0) predictive model was build up to correlate FTIR spectra with activity, and the responsible functional groups, such as −CH, −NH, −COOH, and −OH, were identified for the bioactivity. A successful multivariate model was constructed using FTIR-attenuated total reflection as a simple and rapid technique to predict the inhibitory activity.
Keywords: α-glucosidase inhibitory activity, Fourier transform infrared spectroscopy, metabolomics, orthogonal partial least squares, Phaleria macrocarpa
1. Introduction
Phaleria macrocarpa (Scheff.) Boerl, also known as Mahkota dewa, belongs to Thymelaeaceae family and is one of the most familiar and widely used traditional medicines in South Asian countries. This plant is used for the prevention of liver disease, diabetes mellitus, gout, kidney disorders, cancer, heart disease, impotence, allergies, insect bites, hemorrhoids, strokes, blood-related disease, migraines, and acne and skin ailments [1]. In addition, P. macrocarpa also possess various types of metabolites, including carbohydrates, saponins, tannins, alkaloids, flavonoids, polyphenolic glycosides, palmitic acid, phenols, terpenoids, dodecanoic acid, lignans, ethyl stearate, and polyphenolic compounds. Especially in Malaysia and Indonesia, a large number of people use this plant in processed or unprocessed forms, such as tea and juice, as antidiabetic remedies. However, only a few compounds, such as phalerin [1], carbohydrates (structure is suspected analogous with myglitol or acarbose) [2], and magniferin [3], have been reported for the α-glucosidase inhibitory activity of P. macrocarpa based on bioassay-guided traditional method. There are several antidiabetic activities of P. macrocarpa that have been reported recently, such as the highest α-glucosidase inhibitory activity was observed in the methanol extract of the flesh and n-hexane extract of the stem of P. macrocarpa [4] and reduction of plasma glucose level in the Sprague Dawley rats after the treatment with methanolic extract of P. macrocarpa [5].
Metabolomics is a novel, holistic, and rapidly emerging field that provides a comprehensive chemical analysis of all existing metabolites, their variation, and metabolic pathway in a biological system ensuring the production of robust and high-quality data in an unbiased way [6].
Assuring the identity, safety, and quality of herbal products is a growing concern for the extravagantly complicated and unknown mechanism of disease treatment. In a herbal material, there are many active components present in meager amounts, and they cannot be isolated easily because of their variability. Consequently, the classical isolation and measurement methods are laborious, expensive, time consuming, unreliable, and involve toxic chemicals for the quality control of these herbs [7]. Therefore, there is a need to use more crude extracts (polytherapy). Moreover, only a few isolated pharmacologically active principles cannot represent the bioactivity of complex herbal extracts, and the activity of herbal preparation arises from combined interactions of thousands of constituents and their synergist/antagonist consequences [8,9]. Therefore, it is important to develop a rapid and accurate analytical method for ensuring bioactivity and safety of herbal materials. To overcome this limitation, metabolomics combined with several common analytical techniques have become an important tool for multicomponent analysis of herbal extracts where multiple components can be assessed in a single analysis [10].
There are a variety of spectroscopic techniques for herbal fingerprint analysis, such as gas chromatography mass spectrometry, LC–MS, and nuclear magnetic resonance (NMR) spectroscopy [11]. Although these are the principal analytical methods applied in plant metabolomics, they have some advantages, such as comprehensiveness, sample throughput, and precision. Additionally, these systems are not constantly available in developing countries where natural products are generally well known. The application of most available, simple, and rapid methods like Fourier transform infrared spectroscopy (FTIR) could be an efficient alternative for herb quality control [12]. FTIR spectroscopy, used to determine functional groups, is reagentless, cost effective, rapid, nondestructive, and suitable for investigations that ensure high reproducibility, specificity, easy, and minimal sample preparation [13]. In addition, the functional groups absorb the incoming FTIR radiation at specific wavelength and chemical bonds vibrate in characteristic ways, such as bending or stretching, leading to a spectrum or “fingerprint” [14,15]. Recently, researches have inserted this idea to manage big data sets to characterize samples. For instance, metabolomics together with projection-based multivariate data analysis was successfully applied for obtaining reliable results [16,17].
The objective of this study was to develop a rapid multivariate predictive model based on orthogonal partial least square (OPLS) method combined with FTIR technique by analyzing a metabolic fingerprint and chemical changes in the different ratios of ethanol–water extracts of P. macrocarpa. The model has been established from a correlation between the α-glucosidase inhibitory activity and FTIR spectra of P. macrocarpa. To the best of our knowledge, this is the first attempt to develop a rapid predictive model by fingerprinting the compositions of this plant under different solvent treatments using FTIR and metabolomics approach.
2. Methods
2.1. Plant materials
The fruits of P. macrocarpa were purchased from the farm at Kota Bharu, Kelantan, Malaysia. The fruits were identified by voucher specimens (PIIUM 230-1) prior to being deposited in the herbarium of the Pharmacy Department, International Islamic University, Pahang, Malaysia.
2.2. Chemicals
The organic solvent ethanol (analytical grade), which was supplied by R & M chemicals (Essex, UK), was used for the extraction of the plant material. For enzymatic assay, enzyme (α-glucosidase) was purchased from Megazyme (Wicklow, Ireland). The substrate 4-nitrophenyl-α-D-glucopyranoside and quercetin were supplied by Sigma-Aldrich (St. Louis, MO, USA).
2.3. Sample preparation
First, the collected fruits were washed to avoid any debris. The seeds were separated, and the fruits were cut into small pieces and dried at room temperature for 1 week. The dried sample was ground to fine powder using grinding machine (Fritsch Universal Cutting Mill-115 PULVERISETTE 19; Fritsch, Germany) and stored in refrigerator at −10°C for FTIR-ATR and enzyme analyses. A total of 36 samples, resulted from six different concentrations of ethanol with six replications, were prepared for analysis. About 5.0 g of each grinded sample was immersed in 150 mL of ethanol and water at varying concentrations (100%, 80%, 60%, 40%, 20%, and 0%) and sonicated for 30 minutes. The solids were separated from the extracts by centrifugation (D-78532 tuttlingen, Universal 32R; Hettich Zentrifugen, Germany) at 20°C temperature, 4000 rpm, and 15 minutes, and the extraction solvent was evaporated using a rotary vacuum evaporator (Buchi, B-490, IL, USA) at 40°C. For further analyses, the sample was placed in a freeze dryer (Crist, Alpha 1-4 LD) to remove the remaining solvent and stored at −80°C.
2.4. α-glucosidase inhibition assay
The inhibitory activity of P. macrocarpa extract was measured using Collins et al [18] method with some modifications. To prepare the stock solution, 1 mg of extract was immersed in 1 mL of dimethyl sulfoxide. In addition, 1 mg quercetin was dissolved in 1 mL of dimethyl sulfoxide to make a positive control [19]. To dilute the prepared stock solution, 30 mM phosphate buffer (pH 6.5) was added to get a final concentration of 40 μg/mL [20]. α-glucosidase enzyme (α-glucosidase type 1 from Saccharomyces cerevisiae; Megazyme 130301) was dissolved in 50 mM potassium phosphate buffer (pH 6.5) and used as an enzyme solution. The highest concentration (0.02 U/well) was used and incubated for 5 minutes at room temperature (20°C). Next, 20 mL of 50 mM phosphate buffer (pH 6.5) was added in 6.0 mg of substrate (p-nitrophenyl-α-D-glucopyranose; Sigma-Aldrich, N1377-5G) and allowed to incubate at room temperature for 15 minutes. After incubation, the reaction was ceased by adding 50 μL of 2M glycine (pH 10). The yellow color was produced for p-nitrophenol (released from the substrate) after addition of glycine. According to the aforesaid procedure, the 30 mM phosphate buffer, fruit extract, quercetin (positive control), α-glucosidase (enzyme), substrate, and stopping reagent (glycine) were put in a 96-well microplate, with a final volume of 250 μL. The absorbance (Abs) was analyzed at 405 nm using a spectrophotometer (Nanoquant/Tecan, Infinite M 200) [20]. The α-glucosidase inhibition was measured by the following equation (Eq. 1) of Wang et al [21].
| (1) |
All experiments were conducted in triplicates. Metabolites with the highest inhibitory activity were further characterized by analyzing the concentration needed to inhibit 50% of the α-glucosidase activity (IC50) under the assay conditions, which were ascertained graphically. Different parts of the plant were compared on the basis of their IC50 values calculated from the dose response curves.
2.5. Instrumentation and FTIR-ATR measurements
The FTIR spectra of the samples were read at room temperature (20°C) using attenuated total reflectance (ATR) and an internal reflection element made of diamond using Spectrum Two instrument (Perkin Elmer Inc., USA). The spectral region was 4000–400 cm−1 at a resolution of 4 cm−1. The spectra were measured by Spectrum 10.03.09.0139 software (Perkin Elmer Inc., USA). ATR scan technique was used directly for all the samples. The freeze-dried extracts were analyzed, without any preparation, directly on the diamond ATR crystal. The ATR crystal was carefully cleansed between the measurement using solvent and acetone.
2.6. Data preprocessing and calibration model development
Each FTIR spectrum was baseline corrected using Spectrum software (Perkin Elmer, Inc.) to minimize the differences between spectra because of baseline shifts. The spectra were then exported to ASCII file that reduced the FTIR spectra automatically, and excel files were prepared for chemometric analysis. The multivariate calibration (OPLS model) for the data of FTIR spectra was produced using the SIMCA-P software version 13 (Umetrics, Umeå, Sweden). Investigation was based on a 36 × 3600 data matrix gathered so that each column corresponded to the spectra data and each row represented the sample at a given wavelength. The calibration of functioning of OPLS was evaluated using the coefficient of determination values (R2Y) and root mean square error of estimation (RMSEE). In addition, Q2Y and root mean square error of cross-validation (RMSECV) were applied for the assessment of validation capability of OPLS.
The full frequency region (4000–400 cm−1) of the FTIR spectra was applied at a resolution of 4 cm−1; consequently, a total of 7189 variables were acquired. The model was constructed and validated by the 36 independent observations. The model validity was evaluated by internal cross validation according to the value of R2Y and Q2Y cumulative.
2.7. Statistical analysis
The results were assessed using Minitab 16 (Minitab Inc., State College, PA, USA) by one-way analysis of variance with Tukey's comparison test. A 95% confidence interval was used, and the differences were considered significant at p < 0.05.
3. Results
3.1. Extraction yield
Extracts of different solvent concentrations were obtained from fruit flesh of P. macrocarpa under investigation. The extraction yields of different solvent ratios of P. macrocarpa fruit flesh are illustrated in Table 1. The percentage yield of aqueous and ethanolic extracts of P. macrocarpa were significantly (p > 0.05) different from one another. The extract yield mainly depends on the solvent properties. Depending on the extraction solvent, the range of the percentage yields of the fruit extracts was 10–25%, as presented by the results. Comparing all extracts in this study, the highest extraction yield (25%) was obtained from the water extracts of P. macrocarpa. The percentage yield of the ethanol extract was found to be 10%, which was the lowest yield. This result indicates that the extraction yield depends on the solvent polarity. A more polar solvent gives a higher yield. The results demonstrated that water was the most suitable extraction solvent according to the extract's yield. The yields of different extracts exhibited the following trend: water > 20% > 40% > 60% > 80 > 100%, which was in agreement with the previous studies [22,23]. Javadi et al [22] reported that the water extract of Cosmos caudatus leaves showed maximum yield compared with the various organic solvents. Sultana et al [23] also found that the aqueous methanol extract of medicinal plant leaves was higher than the various absolute organic solvents.
Table 1.
Comparison of the extraction yield and α-glucosidase inhibitory activity of the Phaleria macrocarpa at different concentrations of ethanol.
| Extraction solvent (%) | Extraction yield (%) | α-glucosidase inhibitory activity IC50 (μg/mL) |
|---|---|---|
| Water | 25.40 ± 0.60 a | 299.24 ± 29.40 a |
| 20% Ethanol | 23.56 ± 1.15 b | 137.99 ± 31.89 b |
| 40% Ethanol | 22.83 ± 1.09 bc | 65.24 ± 16.37 c |
| 60% Ethanol | 21.47 ± 1.11 c | 32.82 ± 5.89 d |
| 80% Ethanol | 19.28 ± 1.09 d | 13.15 ± 2.96 d |
| 100% Ethanol | 10.26 ± 1.08 e | 7.42 ± 1.70 d |
| Quercetin | ND | 4.34 ± 1.08 d |
Values are presented as the mean ± standard deviation (SD), n = 6. Values in each column with different subscript letters are significantly different (p < 0.05).
ND = not determined.
3.2. Inhibition of α-glucosidase activity
The inhibition activities (values of IC50) of P. macrocarpa at different ethanolic concentrations are presented in Table 1. A lower IC50 value is desirable for higher inhibitory potential. The IC50 values of six different extracts of P. macrocarpa ranged from 7.4 μg/mL to 299.2 μg/mL. The inhibition potential of different samples was compared on the basis of their resulting IC50 values. At 100%, 80%, and 60% ethanolic extracts, there were no significant differences (p > 0.05) among the IC50 values generated for these extracts. However, the inhibition potential (IC50 value) of water and 20% and 40% ethanolic extracts showed a significant difference (p < 0.05). When the ethanol concentration of the extract was increased to 100%, it showed a very high inhibitory activity against α-glucosidase, which was significantly different (p < 0.05) when compared with aqueous extract. Absolute ethanol and 80% ethanol exhibited the highest inhibition of α-glucosidase activity with an IC50 of 7.4 μg/mL and 13.1 μg/mL, respectively, compared with an IC50 of 4.3 μg/mL estimated for quercetin (positive control). At high ethanol concentrations, there was a high recovery of the more lipophilic compounds, but a low recovery of the more hydrophilic compounds. The water extract exhibited the lowest activity (299.2 μg/mL). Ethanol is less polar than water. Therefore, ethanol recovers the more lipophilic compounds, and water recovers the more hydrophilic compounds. In this study, 100% ethanol extract having the highest activity indicates that more lipophilic compounds in P. macrocarpa may be responsible for α-glucosidase activity. This finding is in agreement with a previous study, which reported that 100% ethanol extract of air dried Phyllanthusniruri showed the highest inhibitory activity of α-glucosidase compared with water and 50% and 70% of ethanol extract of air dried Phyllanthusniruri [24]. Moreover, this is further supported by a recent study which reported that 100% of ethanol extract of Ipmoea aquatic showed the highest inhibitory activity of α-glucosidase compared with water and 20%, 50%, and 80% ethanol extract of Ipmoea aquatic [25]. This activity showed the following trend: 100% > 80% > 60% > 40% > 20% > water (Table 1). The inhibitory potential of P. macrocarpa decreased significantly with increasing the solvent polarity (water ratio), resulting in a lower potential of water solvent than organic solvents for extracting metabolites responsible for inhibitory activity [22,26]. This finding might be also for the concentrations of the extracted biologically active constituents and their variability at different solvent ratios. This result is in agreement with the other studies reporting that absolute organic solvents showed a strong biological activity compared with a mixture solvent (aqueous–organic) [22,23]. Besides that, the α-glucosidase inhibitory activity of P. macrocarpa was in accordance with a previous study as potential inhibiting carbohydrate hydrolyzing enzymes in both in vitro and in vivo effectively [3].
3.3. Analysis of FTIR spectra
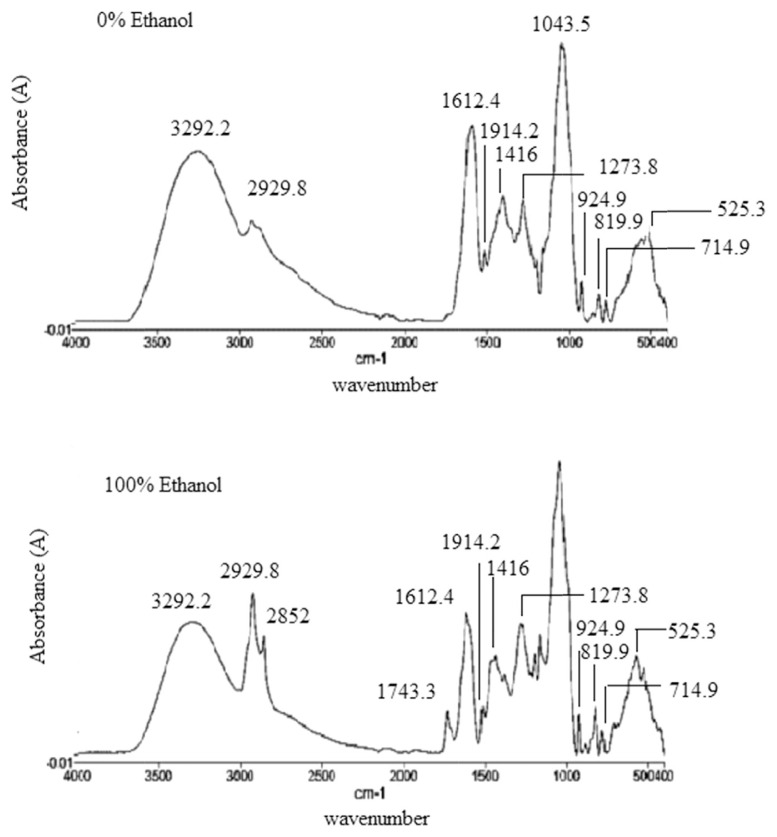
FTIR spectroscopy has been widely used and is one of the advanced techniques that could be used for analysis of bioactive compounds of natural products. The importance of IR spectroscopy for the qualitative analysis arises from its properties, especially as fingerprint technique, meaning that there are no two bioactive compounds having the same FTIR spectra. Figure. 1 shows typical spectra of six different extracts of P. macrocarpa, and the functional group assignment is summarized in Table 2. The whole range of FTIR raw spectra looks very similar by observation because of the similarity in chemical composition. That is, significant relationship within the FTIR spectra and α-glucosidase inhibition was not detected by raw evaluation. The obvious differences between spectra of the two samples, spectra 1 and 6, were due to the intensity at wave numbers of 2852 cm−1, 1734.3 cm−1, 1368.6 cm−1, 1162 cm−1, and 572.72 cm−1. The polarity of pure water and ethanol are 1.000 and 0.654, respectively. The dissimilarities observed between spectra 1 and 6 at 2852 cm−1, 1734.3 cm−1, 1368.6 cm−1, 1162 cm−1, and 572.72 cm−1 are due to the slightly different solvating properties of ethanol and water. The peak at 3292.2 cm−1 is for O–H bonds of alcohols and phenols (H–bonded hydroxyl group) and carboxylic acids. The peaks at 2929.8 cm−1 and 2852 cm−1 are assigned to the sp3 and sp2 stretching of C–H bonds of methyl and aldehyde groups [27]. The peak at 1734.3 cm−1 is for C‗O stretching due to the presence of aldehyde group. The peak at 1612.4 cm−1 indicates the presence of C‗C stretching of alkenes and N–H bending of amines and amides. The peak at 1514.2 cm−1 is assigned to the vibration of N‗O in nitro (R–NO2) and stretching of C‗C in aromatics. The presence of N‗O vibration in nitro group and C–H bending in alkanes (–CH3) are indicated by the peak at 1416 cm−1 [28]. The bands at 1368.6 cm−1, 1273.8 cm−1, 1199.2 cm−1, 1162 cm−1, and 1043.5 cm−1 represent S‗O vibration of sulfoxide, C–O stretching of alcohols and carboxylic acids, and C–N vibration of amines [29]. The band at 924.93 cm−1 is associated with C–H out of plane bending vibration of mono-substituted aliphatic alkenes, where 819.94 cm−1, 779.3 cm−1, and 714.96 cm−1 are for C–H out of plane bending vibrations of meta and para-substituted of aromatics. The absorbance at about 714.96 cm−1 is assigned to the C–H bond of long chain alkanes [28].
Figure 1.
Typical FTIR spectra of extracts of 100% ethanol extract (active) and 0% ethanol extract (nonactive).
Table 2.
Functional groups and modes of vibration in ethanolic and aqueous extracts.
| Wavenumber (cm−1) | Functional group | Vibration mode |
|---|---|---|
| 3292 | O–H | Stretching (sym) |
| 2929 and 2852 | −C–H (CH2) | Asymmetric & symmetric stretching vibration of methylene (−CH2) group |
| 2852 | −C–H (CHO) | Stretching (sym) |
| 1734 | C‗O (CHO, COOCH3) | Carbonyl (C‗O) functional group from the ester linkage & aldehyde |
| 1612 | N–H (NH2), C‗C (alkene) | Bending, stretching |
| 1514 | N‗O (NO2), C‗C (aromatic) | Stretching |
| 1416 | −C–H (CH3) | Bending |
| 1368 | S‗O (SO2), C–X, N‗O (NO2) | Stretching |
| 1273–1043 | C–X, S‗O (SO2), C–N (NH2), C–O (OH, COOH) | Stretching |
| 924 | −C–H (alkene) | Bending & out of plane vibration |
| 819 | −C–H (aromatic) | bending & out of plane vibration |
| 779–540 | C–X (halogen) | Vibration of halogen & out of plane vibration of disubstituted aromatics |
3.4. OPLS and statistical validation
The OPLS model was established from 36 observations (samples) and 7189 (X = 7188, Y = 1) variables (spectra) using one predictive and four orthogonal (1+4+0) components with R2Y = 0.98 and Q2Y = 0.70.
The OPLS, a new analog of partial least square (PLS), is able to rotate the projection so that the model focuses on the effect of interest that is often masked by other unwanted variation, and the first OPLS component shows the difference between the classes. In addition, PLS considers only component 1 and component 2, but OPLS considers orthogonal component also and multiple data set are incorporated by OPLS [30]. This phenomenon was successfully used to obtain new information of a particular combination of metabolite, protein, and transcript correlation [29]. Therefore, OPLS can incorporate a large data set for better identification and interpretation of relevant information.
To select the most beneficial model, some parameters such as regression coefficient (R2Y), RMSEE, and RMSECV were evaluated. A value of R2Y presents the goodness of fit for the components, whereas Q2Y indicates the predictive ability. R2Y is the percentage of variation of the response explained by the model; Q2Y is the percentage of the variation of the response predicted by the model according to cross validation. The model fitness and predictive ability are regarded as effective when the cumulative values of R2Y and Q2Y are 0.5 or above [31,32]. RMSEE is a measure of average deviation of the model from the data [33]. For this model, the RMSEE value was 17.14, which is comparable to other predictive model developed by other studies [34]. Additionally, RMSECV is a measure of the predictive quality of the model for the new samples. The lower the RMSECV value, the higher the predictive accuracy for new sample. The RMSECV value was 57.29 for this developed OPLS model.
Table 3 shows the results obtained from the OPLS model in terms of R2Y, RMSEE, and RMSECV either for normal or other spectra filters such as its derivatives, multiplicative signal correction and standard normal variant. As presented in Figure 2 and Table 3, OPLS calibration in the second derivatives spectra showed the highest R2Y (0.98) and the lowest RMSEE (17.17) and RMSECV (57.29) values compared with other spectra filter.
Table 3.
Multivariate calibration for determining of α-glucosidase inhibitory metabolites using FTIR based on OPLS technique.
| Multivariate calibration | Data filter | R2Y | RMSEE | RMSECV |
|---|---|---|---|---|
| OPLS | Normal | 0.344523 | 87.6634 | 100.576 |
| 1st derivative | 0.973757 | 18.7113 | 47.2206 | |
| 2nd derivative | 0.977128 | 17.1745 | 57.2975 | |
| 3rd derivative | 0.937399 | 27.9516 | 66.2902 | |
| MSC | 0.898656 | 37.421 | 50.6524 | |
| SNV | 0.859918 | 42.5036 | 53.5465 |
MSC = multiplicative signal correction; OPLS = orthogonal partial least square; R2Y = regression coefficient; RMSECV = root mean square error of cross-validation; RMSEE = root mean square error of estimation; SNV = standard normal variant.
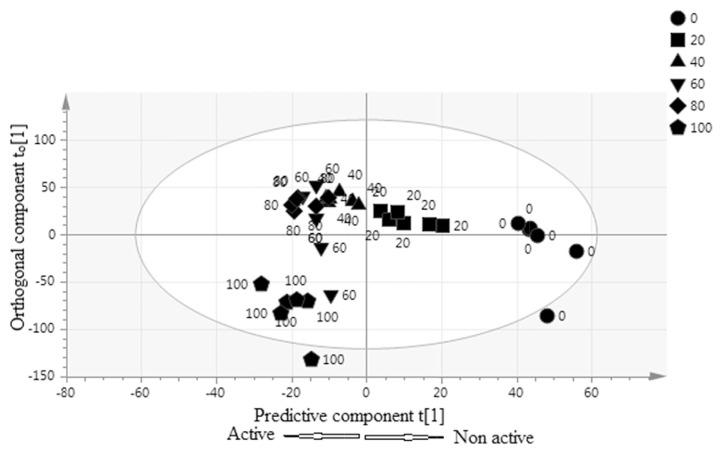
Figure 2.
Orthogonal partial least squares score scatter plot of different ratio of Phaleria macrocarpa extracts.
The first two OPLS components showed the maximum variation with a value of 35.5% and 36.9% for components 1 and 2, respectively. Figure 2 illustrates the OPLS score scatter plot produced by multivariate data analysis. The processed data obtained from this plot indicated that the P. macrocarpa extracts were well separated based on activity. A gradient shifting of the sample along the OPLS component 1 to the left side represents an increase in the activity. Based on the activity, the active extracts (100%, 80%, 60%, and 40% ethanol) were significantly separated by predictive component t [1] from the non-active extracts (20% ethanol and water). By contrast, the most active extracts (100% ethanol) were at the left of the ellipse separated significantly by the orthogonal component t0 [1] from their less active samples (water extracts) which were at the right side.
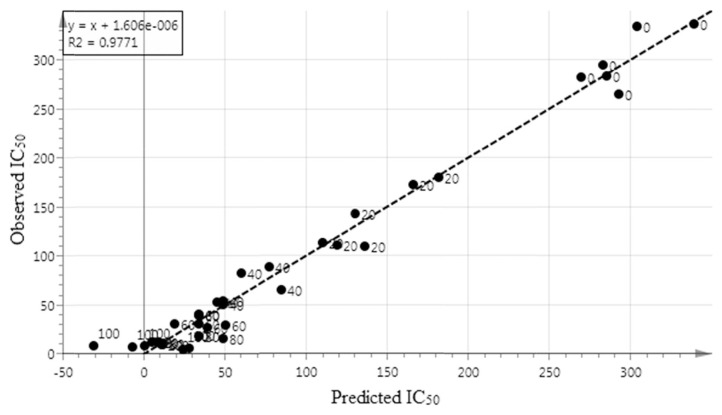
Figure 3 illustrates the coefficient of determination (R2Y) value of the regression line between the predicted and observed IC50value based on FTIR spectra. The value of R2Y in this research was 0.98, which exhibited the very good fitness and predictive ability of the regression model.
Figure 3.
Prediction versus observation IC50 values from all samples. The R2Yof the regression line indicates the goodness of fit between experimental observations and predicted model.
3.5. Bioactivity of spectral characteristic and representing functional groups
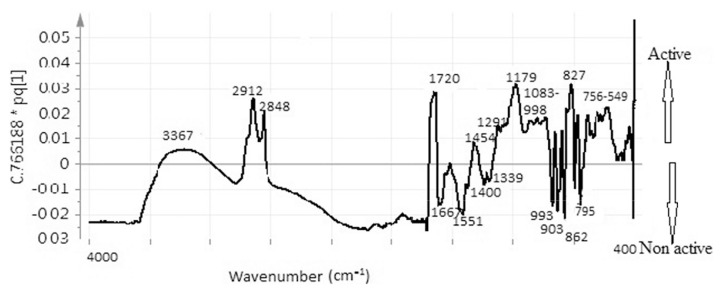
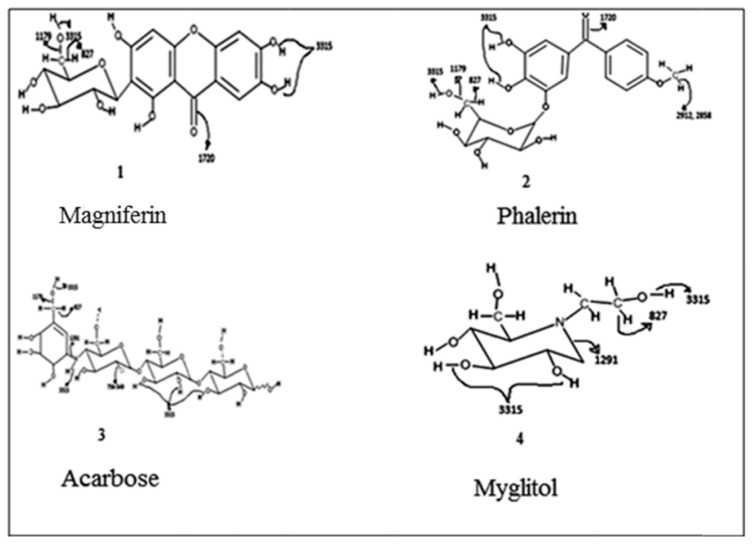
Figure 4 illustrates the line loading plot obtained from multivariate data analysis of the extracts of P. macrocarpa. This plot can make the recognition of the possible biological activity of peaks from the FTIR spectrum easier. This plot assesses the spectral characteristics that accelerate or decelerate the α-glucosidase inhibition. The inhibitory activity with a positive pq [1] value has been presented in the rightmost point of the loading plot. The wave number with increased pq [1] values accelerate the inhibitory activity. The reported α-glucosidase inhibitory activity containing compounds (Figure 5) of P. macrocarpa are phalerin [1], carbohydrate (structure is suspected analogous with myglitol or acarbose) [2], and magniferin [3]. The peaks at 2912 cm−1, 2848 cm−1, 1720 cm−1, 1179 cm−1, and 827 cm−1 have the highest pq [1] value and 3367 cm−1, 1454 cm−1, 1291 cm−1, 1083–998 cm−1, 756–549 cm−1 also have a positive pq [1] value, representing the signal from the α-glucosidase inhibitory compound. The peaks at 2912 cm−1 and 2848 cm−1 are assigned to the stretching of C–H of methyl group in phalerin and aldehyde groups, whereas 1720 cm−1 is for the vibration of C‗O in ketone (magniferin and phalerin), aldehyde, and carboxylic acid group. The peak at 1179 cm−1 may be assigned to the stretching of C–O of alcoholic group present in phalerin, magniferin, myglitol, and acarbose and carboxylic acid, whereas 827 cm−1 was for C–H out of plane bending in alkenes and aromatics. The absorption at 3315 cm−1 also leads to the inhibition potential because of the −OH (hydrogen bonded) of alcoholic and phenolic group present in all reported compounds and −NH of amines group present in acarbose, 1291 cm−1 is for the stretching of C–N of amines group observed in myglitol, whereas the absorbance at 756–549 cm−1 is for C–Cl bond which was observed in acarbose. The reported compounds such as carbohydrate, phalerin, and magniferin in P. macrocarpa have −CH, −CO, − N‗, −NH, and −OH stretching. However, aldehyde and carboxylic acid group containing compound has not yet been reported in P. macrocarpa. However, in this study, it has been seen that the aldehyde and carboxylic acid groups also induce the α-glucosidase inhibitory activity. The findings are in agreement with the previous study reported by Javadi et al [22], indicating that carboxylic acid group (α-linolenic acid) might induce inhibitory activity. The contribution of aldehyde group to inhibitory activity has not yet been reported. This group may accelerate the α-glucosidase inhibitory activity. Flavonoids can inhibit the activity of α-glucosidase, the unsaturated C ring, 3–OH, 4–CO, the linkage of the B ring at the position 3, and the hydroxyl substitution of the B ring [35]. Meanwhile, the polyphenol shows its α-glucosidase inhibitory activity by the presence of free hydroxyl group at the 3′-position of theaflavin as well as the esterification of theaflavin with a mono-galloyl group. Thus, the existence of polyphenols, flavonoids, carbohydrates, sterols, amines, and terpenes might contribute to the α-glucosidase inhibition potential of the P. macrocarpa extract. Nevertheless, other peaks obtained from this OPLS line loading plot do not have the positive contributions to the α-glucosidase inhibition, and this plot can also provide the negative contributing peaks. For instance, the spectra at 1667 cm−1, 1551 cm−1, 1400 cm−1, 1339 cm−1, 933 cm−1, 903 cm−1, 862 cm−1, 795 cm−1 reduced the α-glucosidase inhibitory activity. The band at 1667 cm−1 is for the stretching of C‗N of imines and oximes groups, whereas the peaks at 1551 cm−1 and 1400 cm−1 are assigned to the asymmetric and symmetric stretch of N‗O in −NO2, indicating that nitro, imine, and oxime groups containing compounds might contribute in the reduction of the inhibitory activity. The outcome of this study disagrees with the report by Damazio et al [36]. These results suggest that the effect of chalcones on serum glucose lowering seems to be a consequence of insulin secretion, and these chalcones represent potential compounds with strong α-glucosidase inhibitory properties. However, it has been also reported that the presence of nitro group and their position in the phenyl rings are responsible for the α-glucosidase inhibitory activity of chalcones. Therefore, the position of nitro group may be responsible for reducing the activity in P. macrocarpa. The peaks at 1339 cm−1 and 933 cm−1 are assigned to the −SO, − PH, or −PO functional groups containing compounds that may also reduce the inhibitory activity. Another phase in this research is the insertion of the entire frequency region of the FTIR spectrum in model development instead of using a part of spectrum, as reported previously [27]. The advantage of incorporating the full wave length region with that a portion of spectrum might lead to over-fitting the model. By contrast, the selection of a part of spectrum is arbitrary that might lead to ambiguity in the interpretation. Therefore, this research demonstrates the success of containing the whole FTIR spectral bands to produce a predictive multivariate calibration model (OPLS) for α-glucosidase inhibition of the extract of P. macrocarpa as a rapid alternative model for the investigation of biological activity.
Figure 4.
Orthogonal partial least squares line loading plots of different ratio of Phaleria macrocarpa extracts.
Figure 5.
Reported α-glucosidase inhibitory compounds from Phaleria macrocarpa.
4. Conclusion
A successful multivariate model was developed using FTIR-ATR together with OPLS algorithm as a simple and rapid method for the analysis of α-glucosidase inhibitory activity of P. macrocarpa extracts. Further, the model has been used to identify the contribution to the bioactivity of the functional groups. This study reported that the functional groups, such as −CHO, −COOH, −NO2, −NH, and −OH, induced the inhibitory activity, whereas −SH, −PH, and −PO reduced the bioactivity. The constructed model having highly significant correlation with R2Y of 0.98 could be applied for quality control process in pharmaceutical industry.
Acknowledgments
This study was financially supported by the Fundamental Research Grant Scheme: FRGS/1/2014/SKK02/UIAM/01/1 of the Ministry of Education, Malaysia. The authors extend their appreciation to the International Scientific Partnership Program (ISPP) at King Saud University, Riyadh, Saudi Arabia, for funding this research work through ISPP# 0026.
Funding Statement
This study was financially supported by the Fundamental Research Grant Scheme: FRGS/1/2014/SKK02/UIAM/01/1 of the Ministry of Education, Malaysia.
Footnotes
Conflicts of interest
There is no conflicts among the authors and institutes.
REFERENCES
- 1. Altaf R, Asmawi MZ, Dewa A, Sadikun A, Umar MI. Phytochemistry and medicinal properties of Phaleria macrocarpa (Scheff.) Boerl, extracts. Phcog Rev. 2013;7:73–80. doi: 10.4103/0973-7847.112853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sugiwati S, Setiasih S. Anti-diabetic activity of Mahkota Dewa [Phaleria macrocarpa (Sheff.) Boerl.] stem extract as an inhibitor of α-Glucosidase. J Indo Med Plant. 2010;3:2. [Google Scholar]
- 3. Rabyah BA, Atangwho IJ, Kuar N, Ahmad M, Mahmud R, Asmawi MZ. In vitro and in vivo effects of standardized extract and fractions of Phaleria macrocarpa fruits pericarp on lead carbohydrate digesting enzymes. BMC Complem Alternat Med. 2013;13:39. doi: 10.1186/1472-6882-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabina E, Zaidul IS, Ghafoor K, Jaffri JM, Sahena F, Babiker EE, Perumal V, Hamed M, Amid M, Khatib A. Screening of various parts of Phaleria macrocarpa plant for α-glucosidase inhibitory activity. J Food Biochem. 2015;40:201–10. [Google Scholar]
- 5. Yanti AR, Radji M, Mun’im A, Suyatna FD. Antioxidant effects of methanolic extract of Phaleria macrocarpa (Scheff.) boerl in fructose 10%-induced rats. Int J Pharm Tech Res. 2015;8:41–7. [Google Scholar]
- 6. Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics-A review in human disease diagnosis. Anal Chim Acta. 2009;659:23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 7. Beek TA, Tetala KKR, Koleva II, Dapkevicius A, Exarchou V, Jeurissen SMF, Claassen FW, Klift EJC. Recent developments in the rapid analysis of plants and tracking their bioactive constituents. Phytochem Rev. 2009;8:387–99. [Google Scholar]
- 8. Bankova V. Chemical diversity of propolis and the problem of standardization. J Ethnopharmacol. 2005;100:114–7. doi: 10.1016/j.jep.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 9. Liang YZ, Xie P, Chan K. Quality control of herbal medicines. J Chromatogr B. 2004;812:53–70. doi: 10.1016/j.jchromb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 10. Cozzolino D. Near infrared spectroscopy in natural products analysis. Planta Med. 2009;75:746–56. doi: 10.1055/s-0028-1112220. [DOI] [PubMed] [Google Scholar]
- 11. Oksman-Caldentey KM, Saito K. Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr Opin Biotechnol. 2005;16:174–9. doi: 10.1016/j.copbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12. Sharif KM, Rahman MM, Ajmir J, Khatib A, Hadijah S, Mohamed A, Sahena F, Zaidul ISM. Orthogonal partial least squares model for rapid prediction of antioxidant activity of P. bleo by Fourier transform infrared spectroscopy. Anal let. 2014;47:2061–71. [Google Scholar]
- 13. Roggo Y, Chalus P, Maurer L, Martinez CL, Edmond A, Jent N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J Pharm Biomed Anal. 2007;44:683–700. doi: 10.1016/j.jpba.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 14. Allwood JW, Ellis DI, Goodacre R. Metabolomic technologies and their application to the study of plants and plant–host interactions: a review. Physiologia Plantarum. 2008;132:117–35. doi: 10.1111/j.1399-3054.2007.01001.x. [DOI] [PubMed] [Google Scholar]
- 15. Beekes M, Lasch P, Naumann D. Analytical applications of Fourier transform-infrared (FT-IR) spectroscopy in microbiology and prion research. Veteri Microbiol. 2007;123:305–19. doi: 10.1016/j.vetmic.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 16. Yuliana ND, Khatib A, Verpoorte R, Choi YH. Comprehensive extraction method integrated with NMR metabolomics: a new bioactivity screening method for plants, adenosineA1 receptor binding compounds in Orthosiphonstamineus Benth. Anal Chem. 2001;83:6902–6. doi: 10.1021/ac201458n. [DOI] [PubMed] [Google Scholar]
- 17. Mediani A, Abas F, Khatib A, Maulidiani H, Shaari K, Choi YH, Lajis NH. 1H-NMR based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res Intl. 2012;49:763–70. [Google Scholar]
- 18. Collins RA, Ng TB, Fong WP, Wan CC, Yeung HW. Inhibition of glycohydrolase enzymes by aqueous extracts of Chinese medicinal herbs in a microplate format. IUBMB Life. 1997;42:1163–9. doi: 10.1080/15216549700203631. [DOI] [PubMed] [Google Scholar]
- 19. Subramanian R, Asmawi MZ, Sadikun A. In vitro α-glucosidase and α-amylase enzyme inhibitory effects of Andrographispaniculata extract and andrographolide. Acta Biochim Polo. 2008;55:391–8. [PubMed] [Google Scholar]
- 20. Deutschläander MS, van de Venter M, Roux S, Louw J, Lall N. Hypoglycaemic activity of four plant extracts traditionally used in South Africa for diabetes. J Ethnopharmcol. 2009;124:619–24. doi: 10.1016/j.jep.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 21. Wang H, Du YJ, Song HC. α-Glucosidase and α-amylase inhibitory activities of guavaleaves. Food Chem. 2010;123:6–13. [Google Scholar]
- 22. Javadi N, Abas F, Hamid AA, Simoh S, Shaari K, Ismail IS, Mediani A, Khatib A. GC-MS-based metabolite profiling of Cosmos caudatus leaves possessing α-glucosidase inhibitory activity. J Food Sci. 2014;79:6. doi: 10.1111/1750-3841.12491. [DOI] [PubMed] [Google Scholar]
- 23. Sultana B, Anwar F, Ashaf M. Effect of extraction solvent/ technique on the antioxidant activity of selected medicinal plant extracts. Molecules. 2009;14:2167–80. doi: 10.3390/molecules14062167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mediani A, Abas F, Khatib A, Tan CP, Ismail IS, Shaari K, Ismail A, Lajis NH. Relationship between metabolites composition and biological activities of Phyllanthusniruri extracts prepared by different drying methods and solvents extraction. Plant Foods Hum Nutr. 2015;70:184–92. doi: 10.1007/s11130-015-0478-5. [DOI] [PubMed] [Google Scholar]
- 25. Sajak AA, Abas F, Ismail A, Khatib A. Effect of different drying treatments and solvent ratios on phytochemical constituents of Ipomoea aquatica and correlation with α-glucosidase inhibitory activity. Int J Food Prop. 2016;19:2817–31. [Google Scholar]
- 26. Ryu HW, Cho JK, Curtis-Long MJ, Yuk HJ, Kim YS, Jung S, Kim YS, Lee BW, Park KH. α-glucosidase inhibition and α-glucosidase inhibitory activity of prenylated xanthones from Garcinia mangostana. Phytochem. 2011;72:2148–54. doi: 10.1016/j.phytochem.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 27. Lu X, Rasco BA. Determination of antioxidant content and antioxidant activity in foods using infrared spectroscopy and chemometrics: a review. Crit Rev Food Sci Nutr. 2012;52:853–75. doi: 10.1080/10408398.2010.511322. [DOI] [PubMed] [Google Scholar]
- 28.Günzler H, Gremlich HU. IR spectroscopy: an introduction. Weinheim Germany: Wiley-vcnVerlag GmbH; 2002. [Google Scholar]
- 29.Socrates G. Infrared and raman characteristic group frequencies: tables and charts Chichester, England: J. Wiley and Sons; 2004. [Google Scholar]
- 30. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemometr. 2002;16:119–28. [Google Scholar]
- 31. Bylesjö M, Daniel E, Miyako K, Thomas M, Johan T. Data integration in plantbiology: the O2PLS method for combined modeling of transcript and metabolite data. Plant J. 2007;52:1181–91. doi: 10.1111/j.1365-313X.2007.03293.x. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi and megavariate data analysis. Umeå, Sweden: Umetrics AB; 2006. [Google Scholar]
- 33. Yeniay Ö, Göktas A. A comparison of partial least squares regression with other prediction methods. Hacet J Math Stat. 2002;31:13. [Google Scholar]
- 34. Tistaert C, Dejaegher B, Chataigné G, Rivière C, Hoai NN, Van MC, Leclercq JQ, Heyden YV. Potential antioxidant compounds in Mallotus species fingerprints. Part II: fingerprint alignment, data analysis and peak identification. Anal Chim Acta. 2012;721:35–43. doi: 10.1016/j.aca.2012.01.058. [DOI] [PubMed] [Google Scholar]
- 35. Tadera K, Minami Y, Takamatsu K, Matsuoka T. Inhibition of α.-glucosidase and α-amylase by flavonoids. J Nutr Sci Vitaminol. 2006;52:149–53. doi: 10.3177/jnsv.52.149. [DOI] [PubMed] [Google Scholar]
- 36. Damazio RG, Zanatta AP, Cazarolli LH, Chiaradia LD, Mascarello A, Nunes AJ, Yunes RA, Silva FRMB. α-Glucosidase inhibitory activity of naphthyl chalcones. J Med Chem. 2010;45:1332–7. doi: 10.1016/j.ejmech.2009.12.017. [DOI] [PubMed] [Google Scholar]