Abstract
Thunberg fritillary bulb (the dry bulbs of Fritillaria thunbergii Miq.), a traditional Chinese Medicine, is widely applied as an expectorant and antitussive. In this investigation, the primary metabolites of bulbs, flowers, leaves, and stems of F. thunbergii were analyzed by gas chromatography–mass spectrometry. Principal component analysis, partial least squares-discriminate analysis, orthogonal projection to latent structures-discriminate analysis, and heat map analysis showed that there were dissimilar metabolites, and a negative correlation between amino acids and saccharides in different analytes. Furthermore, carbodiimide, tryptophan, glucose-6-phosphate, xylose, 2-piperidinecarboxylic acid, monoamidomalonic acid, phenylalanine, and histidine were found to play an important role in the plant metabolism net of F. thunbergii.
Keywords: Fritillaria thunbergii Miq., gas chromatography-mass, spectrometry, metabolomics
1. Introduction
Fritillaria thunbergii Miq. is a perennial herbaceous plant, mainly distributed in Zhejiang, Jiangsu, and Anhui provinces in China [1]. In Chinese medical clinical practice, Thunberg fritillary bulb (Zhebeimu), the dry bulb of F. thunbergii, is often utilized in the treatment of cough caused by wind-heat and phlegm-heat in Traditional Chinese Medicine, and bronchitis, inflammation, hypertension, gastric ulcer, diarrhea, and bacterial infection [2]. Additionally, Zhebeimu is nowadays extensively used to treat drug resistant leukemia [3].
Comparing to bulbs of F. thunbergii, phytochemical researches have shown that the flowers, stems, and leaves also contain various kinds of chemical constituents including flavonoids, essential oils, saponins, and alkaloids [4,5], which are potential medicinal resources.
Gas chromatography–mass spectrometry (GC-MS) was utilized for biomarkers screening in primary metabolites as a metabolomic technique [6] for its reliable and versatile characters comparing with nuclear magnetic resonance and liquid chromatography–MS [7]. In this study, metabolomic multivariable analysis of GC-MS data, covering principal component analysis (PCA), partial least squares-discriminate analysis (PLS-DA), orthogonal projection to latent structures-discriminate analysis (OPLS-DA), heat map analysis, and total metabolites correlation analysis were applied to reveal the metabolic regularity of F. thunbergii. Metabolic distinctions between the different parts of the plant were identified. This work will contribute to the metabolic composition and comprehensive usage of F. thunbergii.
2. Methods
2.1. Samples and reagents
The bulbs (FT0001–0006), flowers (FT 0007–0012), stems (FT 0013–0018), and leaves (FT0019–0024) of F. thunbergii were collected in March 2015 from Yinzhou District, Ningbo City, Zhejiang Province of China, the place of origin of F. thunbergii. The harvested samples were identified by Professor Jian-Wei Chen at Nanjing University of Chinese Medicine, Nanjing, China. The standard substances (ribitol) were supplied by Sigma-Aldrich Trading Co., Ltd. (St. Louis, Missouri, USA). Solvents and reagents (methyl alcohol, chloroform analytical or chemical grade) were supplied by Aladdin Industrial Corporation (Shanghai, China). Pyridine solution was ordered from TCI, Tokyo, Japan. Calibration solutions (C8–C20, C21–C40) were ordered from Fluka Chemika (Buchs, Switzerland).
2.2. Sample preparation
Experimental procedure of extract preparation [2]: samples (bulbs, leaves, stems, and flowers of F. thunbergii) were accurately weighted (100 mg for each sample); then they were rapidly frozen and grinded in liquid nitrogen. Samples were transferred to 10 mL centrifuge tubes, and 1.4 mL of 100% methanol (precooled at −20°C) was added into each of them, with vortexing for 30 seconds. Next in the process, 60 mL of ribitol (0.2 mg/mL) as interior label was added, then vortexed for 30 seconds. They were sonicated for 15 minutes, and centrifuged for 15 minutes at 1530 g. Next, chloroform (750 μL) was added and 1400 μL of dH2O was added, with vortexing. Samples were centrifuged for 15 minutes. Supernatants were transferred to glass vials. Extracts were dried with a stream of nitrogen gas in a vacuum container. Pyridine solutions (60 μL) were added, vortexed for 30 seconds, and deposited for 16 hours. Bis(-trimethylsilyl) trifluoroacetamide reagent (TCI) 60 μL was added and deposited in normal atmospheric temperature for 60 minutes [8].
2.3. GC-MS method
Agilent 7890A/5975C GC-MS (Agilent, J&W Scientific, Folsom, CA, USA) was used for analysis. Gas chromatographic conditions were as follows: HP-5MS apillary column (5% phenyl methyl silox: 30 m × 250 μm, 0.25 μm; Agilent J&W Scientific). Split sampling was with injection volume 1 μL and split ratio 20:1. The injection, ion source, and interface temperatures were set at 280°C, 250°C, and 150°C, respectively. The oven temperature raising procedure was set to 80°C for 5 minutes, and then increased by 20°C/min, then to 300°C for 6 minutes. The total run time was set at a 22 minutes measurement period. The carrier gas was helium (1.0 mL/min). Mass spectrum conditions: electrospray ionization source, electron energy 70 eV; mass data collected in a full-scan mode (m/z 35–780).
2.4. Data preprocessing
Raw data of GC-MS from Agilent MSD ChemStation was transformed into Channel definition format (CDF) format files (Net CDF) by Xcalibur (Thermo Fisher Scientific Inc., Waltham, MA, USA). Peaks were automatically detected and aligned by XCMS software (www.bioconductor.org/) [9]. XCMS is freely available under an open-source license at http://metlin.scripps.edu/download/.
Besides the default parameters, the parameters of XCMS were adjusted as follows: xcmsSet [fwhm = 3, snthresh = 3, mzdiff = 0.5, step = 0.1, steps = 2, max = 300), retcor method = “obiwarp”, plottype = c (“deviation”)], bandwidth (bw) = 2, minfrac = 0.3.
A series of results including data tables, RT-m/z pair, observations, and variables were listed in a *.t file, and then they were normalized in Microsoft Office Excel 2007 (Microsoft Corporation, Redmond, WA, USA) ahead of multivariate analyses.
2.5. Statistical analysis
Data was statistically analyzed by SIMCA-P 11.0 (Umetrics AB, Umea, Sweden) in Suzhou BioNovoGene Company (Suzhou, China), with variates mean-centered of PCA, PLS-DA, and OPLS-DA. Firstly, outliers were dislodged according to the cluster analysis results by means of PCA which is an unsupervised method. Secondly, PLS-DA, as a supervised method, was utilized to analyze the sample data, preventing from over-fitting with permutation test. Then, OPLS-DA was utilized to filter out the noise and increase the discrimination. Lastly, variant metabolites were found in Student t test (p < 0.05) and variable importance plot (VIP) values (it is significant if VIP > 1) of the first principal component. R 3.0.3 (www.r-project.org) was utilized for the t test.
Further quantitative analysis of metabolome was conducted on the basis of Metabolome Alkane Retention Index on the Golm Metabolome Database (http://gmd.mpimp-golm.mpg.de/). Derivatization of metabolites can produce a series of trimethylsilyl substances. One typical trimethylsilyl substance was chosen for the analysis. After normalization, raw data was processed with an internal standard method (ribitol) [10].
If a separation among bulb, stem, leaf, and flower groups was detected in the PCA scores plot, OPLS-DA would underline the differences. Possible biomarkers for group division were consequently recognized by investigating the S-plot, which plots covariances (p) against correlations [p(corr)]. As biomarkers, the influence on the model expressed in p and reliability expressed in p(corr) were supposed to be significant; therefore, probable biomarkers were identified on the outer ends of the S-shaped point swarm. Cut-off values of p j0.03j and p(corr) j0.5j were utilized.
3. Results
3.1. Identification of metabolins
A total of 293 variables from matrixes containing retention time, mass to charge ratio (M/Z), and peak intensity, were detected in total ion current in a 22-minute measurement period. However, 85 different metabolins can be identified as listed in Table 1, including 29 organic acids, 15 amino acids, 11 polyols, eight amines, seven sugars, five phosphoric acids, four fatty acids, and six others. The other signals were very weak or even not included in the database.
Table 1.
Metabolins of Fritillaria thunbergii based gas chromatography-mass spectrometry (GC-MS).
| No. | Name | Type | RT |
|---|---|---|---|
| 1 | 1,1-Dimethyl-2-propyl-hydrazine | Others | 6.10 |
| 2 | 1,4-Butanediol | Polyol | 8.09 |
| 3 | 1-Monohexadecanoylglycerol | Polyol | 16.32 |
| 4 | 2,2-Dimethyl-butanedioic acid | Organic acid | 11.03 |
| 5 | 2,3-Dihydroxybutanedioic acid | Organic acid | 11.71 |
| 6 | 2-Aminobutyric acid | Organic acid | 4.19 |
| 7 | 2-Aminoethanol | Polyol | 9.21 |
| 8 | 2-Ketosuccinic acid | Organic acid | 11.35 |
| 9 | 2-Oxobutyric acid | Organic acid | 8.61 |
| 10 | 2-Piperidinecarboxylic acid | Organic acid | 10.05 |
| 11 | 3-Hydroxypyridine | Others | 6.20 |
| 12 | 3-Oxovaleric acid | Organic acid | 10.61 |
| 13 | 4,6-Dioxoheptanoic acid | Organic acid | 11.98 |
| 14 | 4-Aminobutyric acid | Amino acid | 11.22 |
| 15 | Alanine | Amino acid | 7.33 |
| 16 | Aspartic acid | Amino acid | 11.15 |
| 17 | Cadaverine | Amine | 10.55 |
| 18 | Carbodiimide | Amine | 4.57 |
| 19 | Cellobiose | Sugar | 17.10 |
| 20 | Citramalic acid | Organic acid | 10.86 |
| 21 | Citric acid | Organic acid | 12.93 |
| 22 | Dehydroascorbic acid dimer | Organic acid | 13.13 |
| 23 | Diethylene glycol | Polyol | 8.97 |
| 24 | Digalactosylglycerol | Polyol | 19.98 |
| 25 | Diisodecyl ether | Others | 6.98 |
| 26 | Ethylamine | Amine | 4.33 |
| 27 | Ethylene glycol | Polyol | 5.14 |
| 28 | Ferulic acid | Organic acid | 14.26 |
| 29 | Fructose | Sugar | 13.29 |
| 30 | Galactinol | Polyol | 18.95 |
| 31 | Galactosylglycerol | Polyol | 15.32 |
| 32 | Gentibiose | Others | 18.02 |
| 33 | Gentiobiose | Sugar | 17.70 |
| 34 | Glucose | Sugar | 13.43 |
| 35 | Glucose-6-phosphate | Phosphoric acid | 15.42 |
| 36 | Glutamine | Amino acid | 11.76 |
| 37 | Glyceric acid | Organic acid | 9.78 |
| 38 | Glycerol | Polyol | 9.29 |
| 39 | Glycerol-3-phosphate | Phosphoric acid | 12.62 |
| 40 | Glycolic acid | Organic acid | 6.92 |
| 41 | Glyoxime | Others | 12.17 |
| 42 | Gulose | Sugar | 15.70 |
| 43 | Heptanoic acid | Fatty acid | 8.21 |
| 44 | Hexadecanoic acid | Fatty acid | 13.98 |
| 45 | Histidine | Amino acid | 13.49 |
| 46 | Hydroxylamine | Amine | 8.40 |
| 47 | Isocitric acid | Organic acid | 15.36 |
| 48 | Isoleucine | Amino acid | 9.46 |
| 49 | Itaconic acid | Organic acid | 9.85 |
| 50 | Lactic acid | Organic acid | 7.84 |
| 51 | Leucine | Amino acid | 9.26 |
| 52 | Maleic acid | Organic acid | 9.53 |
| 53 | Malic acid | Organic acid | 10.94 |
| 54 | Malonic acid | Organic acid | 8.57 |
| 55 | Malonic acid methyl ester | Organic acid | 6.73 |
| 56 | Methylcitric acid | Organic acid | 13.05 |
| 57 | Monoamidomalonic acid | Organic acid | 12.37 |
| 58 | Mono-hexadecanoic acid glyceride | Fatty acid | 16.31 |
| 59 | Myo-inositol | Polyol | 14.37 |
| 60 | myo-inositol-1-phosphate | Phosphoric acid | 15.83 |
| 61 | N-Ethyl-N-vinylacetamide | Amine | 4.37 |
| 62 | Nicotinic acid | Organic acid | 9.41 |
| 63 | N-methoxy-amine | Amine | 4.10 |
| 64 | N-methyl-propanamide | Amine | 4.75 |
| 65 | Norvaline | Amino acid | 4.48 |
| 66 | Octadecanoic acid | Fatty acid | 14.88 |
| 67 | O-Methylphosphate | Phosphoric acid | 8.31 |
| 68 | Phenylalanine | Amino acid | 11.84 |
| 69 | Phosphoric acid | Phosphoric acid | 9.30 |
| 70 | Proline | Amino acid | 9.48 |
| 71 | Putrescine | Amine | 12.49 |
| 72 | Pyruvic acid | Organic acid | 6.49 |
| 73 | Ribitol | Polyol | 12.45 |
| 74 | Salicylic acid | Organic acid | 13.82 |
| 75 | Serine | Amino acid | 10.02 |
| 76 | Succinic acid | Organic acid | 9.59 |
| 77 | Sucrose | Sugar | 16.77 |
| 78 | Threonic acid | Organic acid | 11.44 |
| 79 | Threonine | Amino acid | 10.23 |
| 80 | Tryptophan | Amino acid | 14.91 |
| 81 | Tyrosine | Amino acid | 13.56 |
| 82 | Urea | Others | 8.91 |
| 83 | Valine | Amino acid | 8.71 |
| 84 | Xylose | Sugar | 12.05 |
| 85 | α-Ketoglutaric acid | Organic acid | 11.49 |
RT = retention time.
The unit of RT is minutes, and the range is 4.1–22.0 min.
The chromatogram in 0~4 min was deleted because the noise caused by program warming would have a strong impact on the data processing.
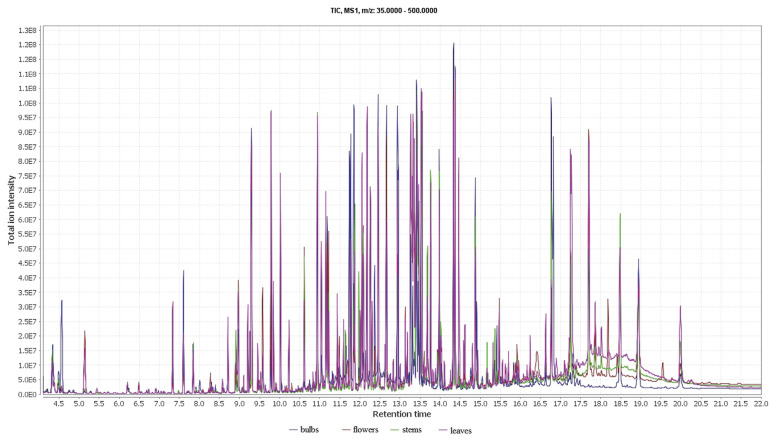
The extract of F. thunbergii was analyzed by GC-MS metabolomics. The characteristic GC-MS total ion current chromatograms of F. thunbergii are shown in Figure 1.
Figure 1.
GC–MS TIC chromatograms of bulbs, flowers, stems, and leaves.
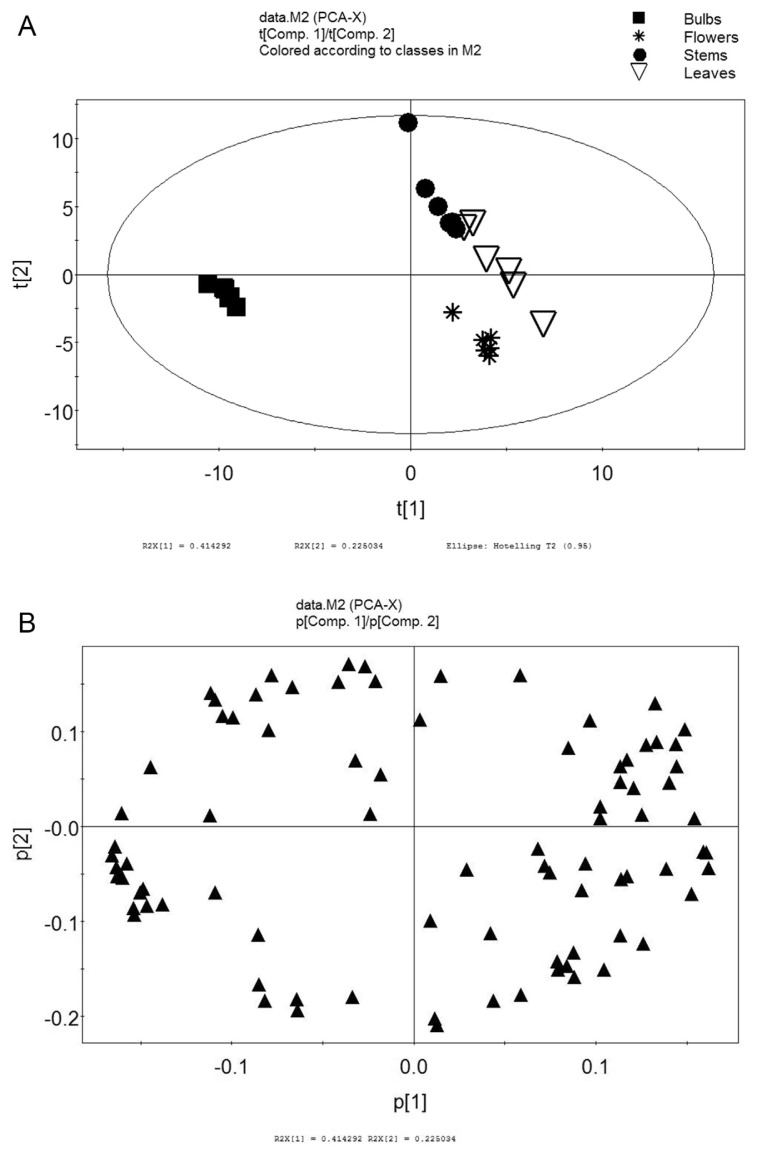
3.2. PCA
Primary metabolites of F. thunbergii were classified by PCA, an unsupervised multidimensional statistical analysis method (Figure 2) [11].
Figure 2.
(A) score plot of PCA and (B) PCA loading.
From automation simulation of PCA of all samples, our principal components were acquired (R2X = 0.896, Q2 = 0.802, which manifested the explanatory variables and predictability of the model). As shown in Figure 2A, bulbs were significantly different from others, but there were no significant differences among stems, leaves, and flowers. The model of all samples explained 63.9% of the principal components, with the principal component 1 (PC1) interpreting 41.4% and principal component 2 (PC2) interpreting 22.5%.
The PCA scatter plot indicated that the underground bulbs were remarkably separated from aerial parts (stems, leaves, and flowers) according to PC1. Bulbs were clustered by negative scores on PC1, nevertheless stems, leaves, and flowers presented positive scores on PC1, suggesting that bulbs and aerial parts were completely dissimilar in metabolic patterns. Stems and flowers were further segregated on PC2, which indicates that the metabolic pattern was also different.
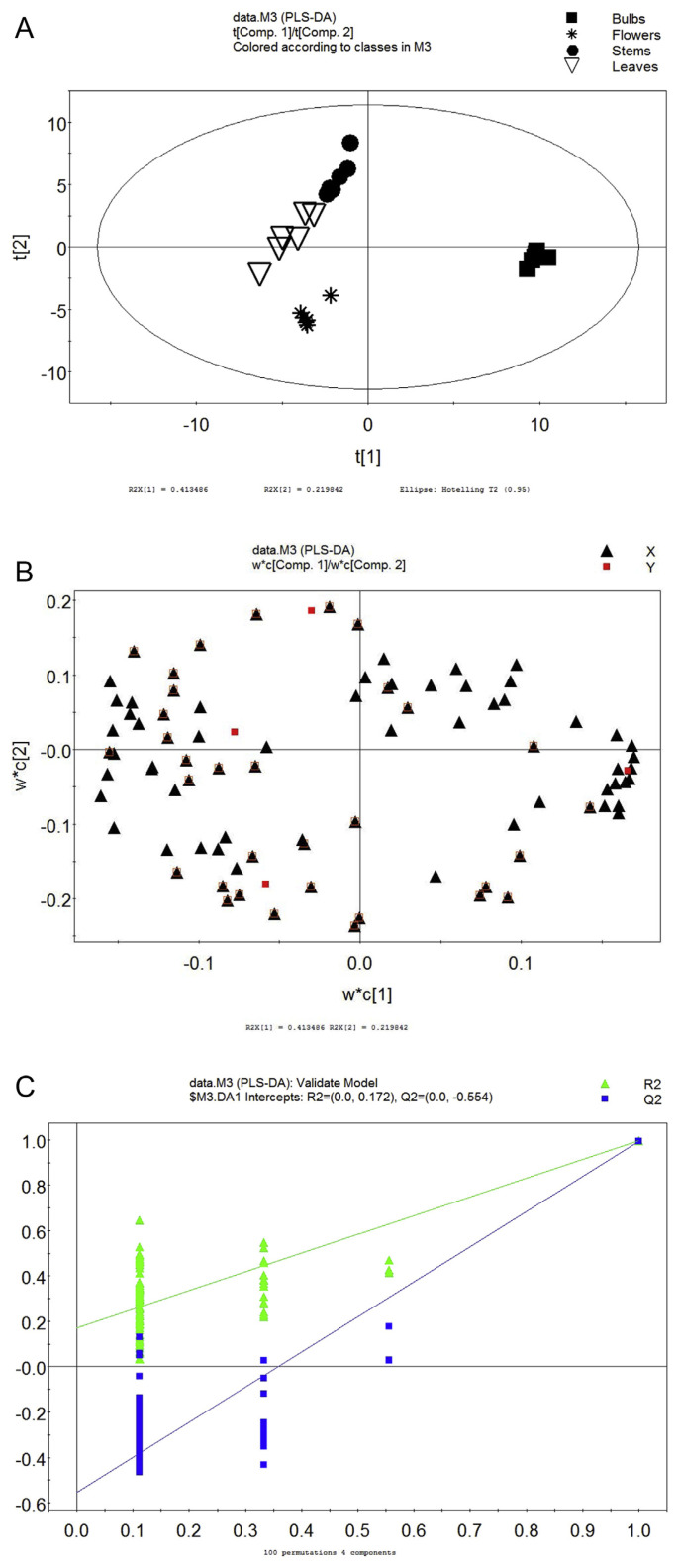
3.3. PLS-DA
Primary metabolites of F. thunbergii were further classified by PLS-DA, a supervised multidimensional statistical analysis method (as shown in Figure 3A). Compared with PLS, PLS-DA can find out the significantly differential metabolites more clearly [12]. X-axis (t [1]) indicated scores of the first principal component, while Y-axis (t [2]) indicated scores of the second principal component. R2Y = 0.986 and Q2Y = 0.967 indicated that this PLS-DA model can explain the differences between sample groups. Then, the results of permutation tests conducted 100 times showed that the Y-intercept of R2 was 0.172 and the Y-intercept of Q2 was −0.554 (Figure 3B). Accordingly, the model was valid and reliable.
Figure 3.
(A) score plot of PLS-DA; (B) PLS-DA loading; and (C) validate model of PLS-DA.
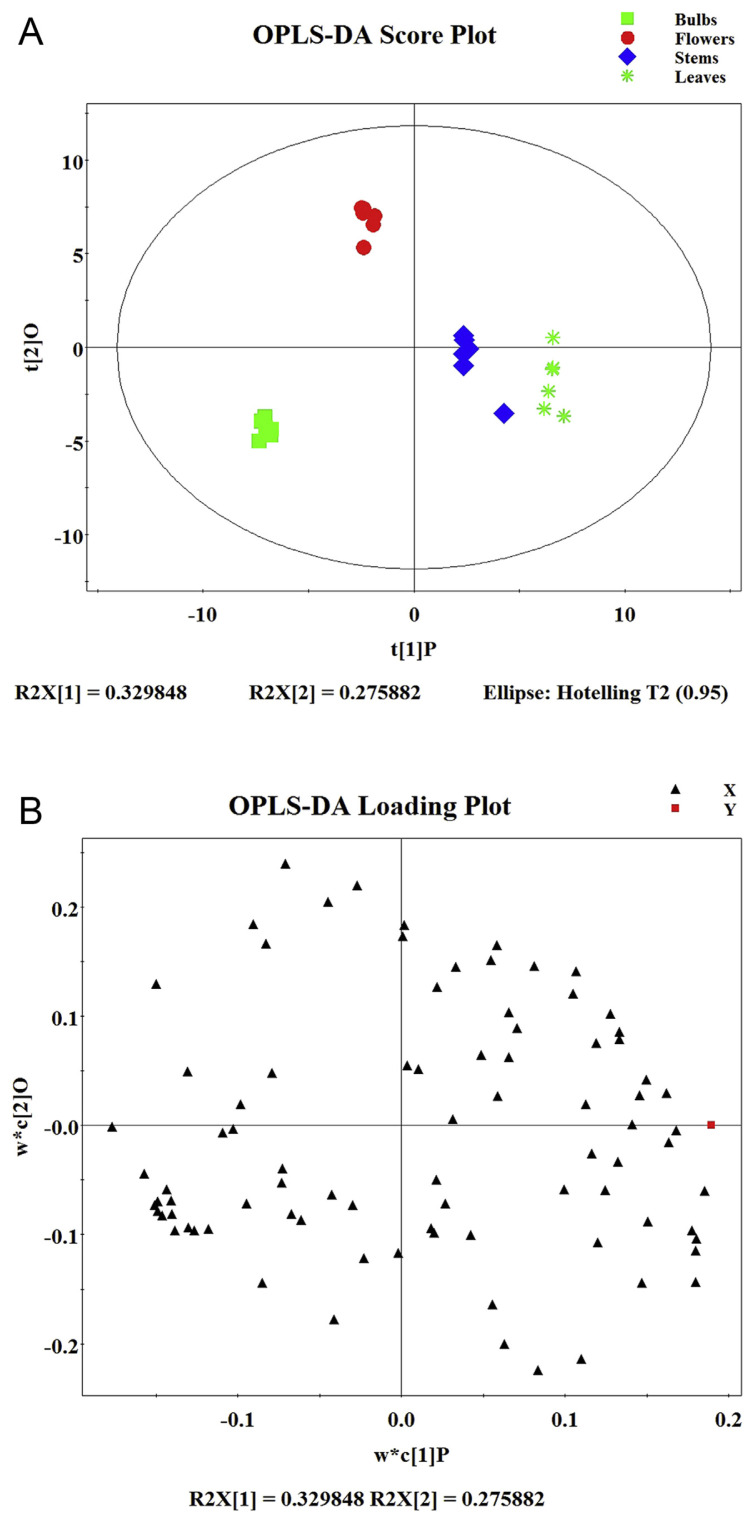
3.4. OPLS-DA
OPLS-DA, a discriminant multidimensional statistical analysis method, was consequently utilized to locate the radically differential primary metabolites in the GC-MS database among the bulbs, flowers, leaves, and stems, for OPLS-DA can filter out the noises [13].
This model separated bulbs, flowers, stems, and leaves from each other, along the discriminating t [1] (Figure 4A). The loading S-plots of OPLS-DA identified the underlying biomarkers of each part, with the loading values of the variables related to the PC1 exhibited in Figure 4B. The loading S-plot indicated that tryptophan, gentiobiose, xylose, phenylalanine, and 3-oxovaleric acid were the most contributive principles distinguishing bulbs from flowers; 2,2-dimethyl-butanedioic acid, 2,3-dihydroxybutanedioic acid, 2-ketosuccinic acid, 2-oxobutyric acid, and 2-piperidinecarboxylic acid were the most contributive principles discriminative bulbs from stems; 1-monohexadecanoylglycerol, 2,2-dimethyl-butanedioic acid, 2,3-dihydroxybutanedioic acid, 2-oxobutyric acid, and 2-piperidinecarboxylic acid were the most contributive primary metabolites discriminative bulbs from leaves.
Figure 4.
(A) OPLS-DA score plot and (B) OPLS-DA loading plot.
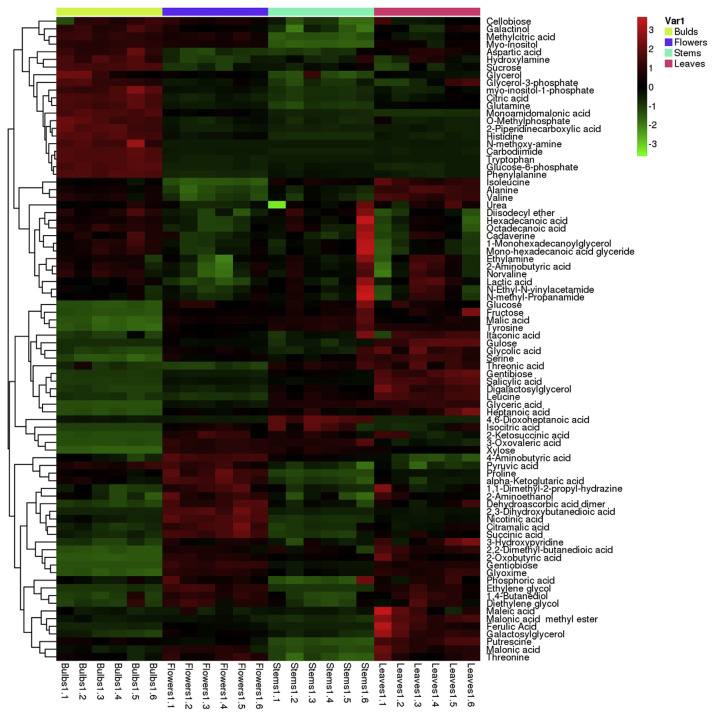
3.5. Heat map analysis
Metabolites of F. thunbergii were analyzed by cluster analysis and thermal map analysis (Figure 5 colors indicated the contents, with red meaning a high content, and green meaning a low content). Samples of bulbs were clustered in a category, while samples of flowers, stems, and leaves were clustered in another category. This indicated that the primary metabolites of flowers, stems, and leaves were similar, while those of the bulbs were different. Bulbs had a high content of amino acids (phenylalanine, tryptophan, histidine, glutamine, aspartic acid, etc.), phosphoric acids (glucose-6-phosphate, O-methylphosphate, myo-inositol-1-phosphate), fatty acids (heptanoic acid, octadecanoic acid, mono-hexadecanoic acid glyceride), and amines (N-methoxy-amine, cadaverine, ethylamine, N-ethyl-N-vinyl-acetamide, N-methyl-propanamide).
Figure 5.
Heat map analysis.
Furthermore, flowers and leaves were clustered in the same bracket with a high content of organic acids (pyruvic acid, nicotinic acid, succinic acid, maleic acid, ferulic acid, etc.) and polyols (ethylene glycol, 1,4-butanediol, diethylene glycol, galactosylglycerol), while stems were clustered in another bracket with a high content of threonic acid, gentibiose, hexadecanoic acid, especially 4,6-dioxoheptanoic acid.
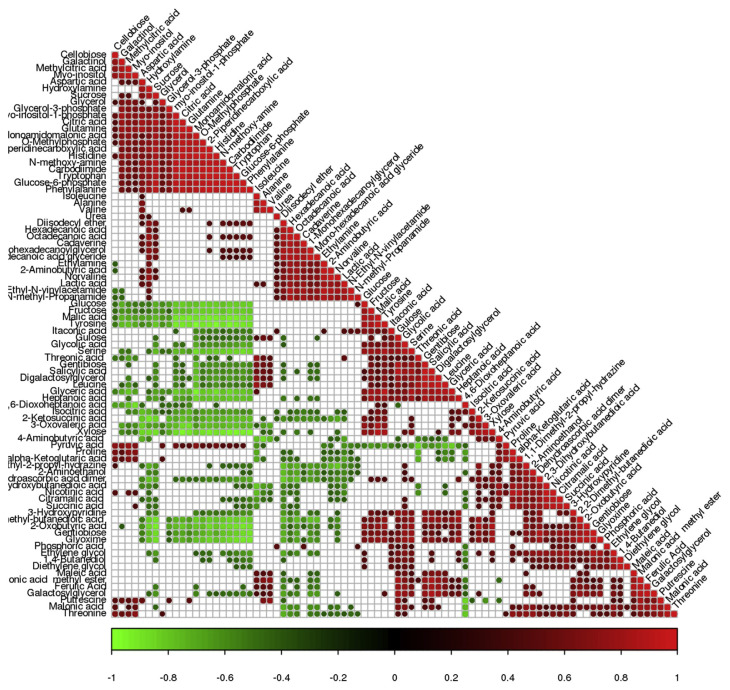
3.6. Total metabolites correlation analysis
Correlations including statistical significances of all the metabolites were analyzed by using a Pearson correlation coefficients method and incidence matrix was protracted by a p-heatmap bag in R language. Positive and negative correlations were represented by 1 or −1 (correlation coefficients). As shown in Figure 6, a negative correlation was found between amino acids and saccharides.
Figure 6.
Total metabolites correlation analysis
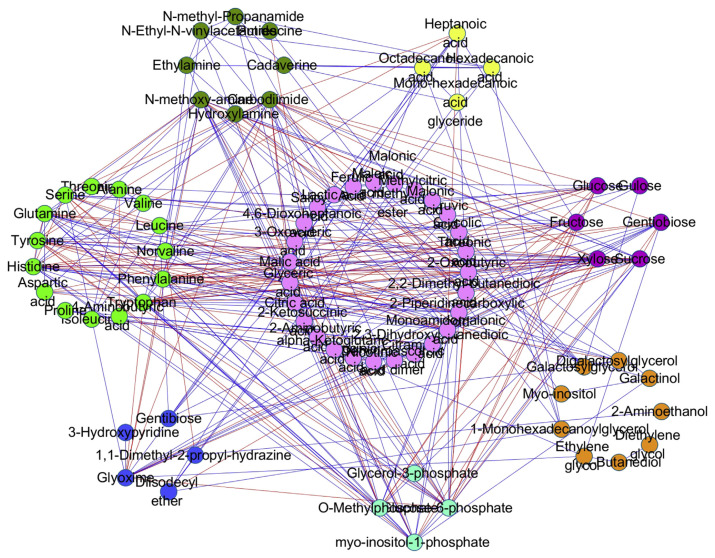
3.7. Association network analysis of metabolites
The correlation coefficients of metabolites were calculated by means of Pearson correlation coefficients. Correlation coefficients and p values were calculated by cor() or cor.test() of R language. P.adjust() was utilized to control the final false positive rates (FDR < 0.05, r2 > 0.64). Cytoscape 3.02 (http://www.cytoscape.org/) was utilized to map the network [14].
As shown in Figure 7, in the metabolic network analysis, red stroke lines represent upregulation, while blue lines represent downregulation. Red nodes represent nucleotide, purple represents sugar, light blue represents phosphate, light green represents amino, dark green represents amine, orange represents polyol, pink represents organic acid, and dark blue represents others.
Figure 7.
Association network analysis of metabolites.
Nodes degree can be defined as the number of one metabolite connecting with others, which indicated the importance of the metabolins. Carbodiimide, tryptophan, glucose-6-phosphate, xylose, 2-piperidinecarboxylic acid, monoamidomalonic acid, phenylalanine, and histidine all played an import role in the primary metabolic process of F. thunbergii.
4. Discussion
Primary metabolites, such as saccharides, lipids, and amino acids can affect plants’ own growth and reproduction by involvement in the biosynthesis of some necessary materials [15]. It is worthy to investigate primary metabolites for that they are not sensitive to environmental conditions, and exiting in plants from beginning to end [16].
Multivariate analysis of data indicated that the contents of the primary metabolites of the leaves, flowers, stems, and bulbs were disparate. The bulbs were clustered in a category, while the flowers, stems, and leaves were clustered in another category, which indicated the primary metabolites of the bulbs were different from the others. Furthermore, the flowers and leaves were clustered in the same subcategory, while stems were in another subcategory. It was also found that carbodiimide, tryptophan, glucose-6-phosphate, xylose, 2-piperidinecarboxylic acid, monoamidomalonic acid, phenylalanine, and histidine played a significant role in plant metabolism net of F. thunbergii.
The results indicated that more kinds of nutrition are stored in the bulbs for growth of palingenetic leaves and stems, such as amino acids (phenylalanine, tryptophan, histidine, glutamine, and aspartic acid, etc.), phosphoric acids (glucose-6-phosphate, O-methylphosphate, myo-inositol-1-phosphate), fatty acids (heptanoic acid, octadecanoic acid, mono-hexadecanoic acid glyceride), and amines (N-methoxy-amine, cadaverine, ethylamine, N-ethyl-N-vinylacetamide, N-methyl-propanamide). It can be hypothesized that this is the reason why flowers, tip leaves, and tip stems are removed in March or April to boost the production of bulbs.
It is worth mentioning that tryptophan was structurally similar to indole-3-acetic acid, which is an important precursor in plant auxin biosynthesis widespread in higher plants [17].
This investigation suggested that metabolite profiling with GC-MS could be applied as a powerful technique for assessing the quality, distinguishing different medical parts, and researching metabolic regulation mechanisms of medical plants.
Acknowledgments
Programs were supported by the Zhejiang Provincial Natural Science Foundation of China (LY15H280009), Ningbo Natural Science Foundation (2015A610290), Traditional Chinese Medicine Science and Technology Plan Projects in Zhejiang Province (2010ZB132), Yinzhou District Agricultural Science and Social Development Projects [Yin (2015) No. 69].
Funding Statement
Programs were supported by the Zhejiang Provincial Natural Science Foundation of China (LY15H280009), Ningbo Natural Science Foundation (2015A610290), Traditional Chinese Medicine Science and Technology Plan Projects in Zhejiang Province (2010ZB132), Yinzhou District Agricultural Science and Social Development Projects [Yin (2015) No. 69].
Footnotes
Conflicts of interest
All authors declare no conflicts of interest.
REFERENCES
- 1. Hao DC, Gu XJ, Xiao PG. Phytochemical and biological research of Fritillaria medicine resources. Chin J Nat Med. 2013;11:330–44. doi: 10.1016/S1875-5364(13)60050-3. [DOI] [PubMed] [Google Scholar]
- 2.Weckwerth W, Kahl G. 1 Metabolic Profiling of Plants by GC-MS. Weinheim: Wiley-VCH Verlag GmbH & Co. KgaA; 2013. [Google Scholar]
- 3. Wang J, Chen XY, Lai ZL, Zhang YY, Hospital D. Clinical observation on effects of adjuvant chemotherapy with fritillary Thunbergii bulb complex granules on symptoms of refractory acute leukemia patients. Beijing Zhong Yi Yao Da Xue Xue Bao. 2013;32:833–5. [Google Scholar]
- 4. Cui M. Research progress on fritillary flower. Qi Lu Yao Shi. 2011;30:661–2. [Google Scholar]
- 5. Yan M, Jin X, Xu D. Studies on the chemical constituents of the stems and leaves of Thunberg fritillary (Fritillaria thunbergii) Zhong Cao Yao. 1994;25:344–6. [Google Scholar]
- 6.Shuman JL, Cortes DF, Armenta JM, Pokrzywa RM, Mendes P, Shulaev V. Plant metabolomics by GC-MS and differential analysis. Berlin: Springer; 2011. [DOI] [PubMed] [Google Scholar]
- 7. Bhatia A, Bharti SK, Tewari SK, Sidhu OP, Roy R. Metabolic profiling for studying chemotype variations in Withania somnifera (L.) Dunal fruits using GC-MS and NMR spectroscopy. Phytochemistry. 2013;93:105–15. doi: 10.1016/j.phytochem.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 8. Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–96. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 9. Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 10.Hummel J, Selbig J, Walther D, Kopka J. The Golm Metabolome Database: a database for GC-MS based metabolite profiling. Berlin Heidelberg: Springer; 2007. [Google Scholar]
- 11. Bro R, Smilde AK. Principal component analysis. Anal Methods. 2014;6:2812–31. [Google Scholar]
- 12. Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML. A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 13. Galindo-Prieto B, Eriksson L, Trygg J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS) Chemom. 2014;28:623–32. [Google Scholar]
- 14. Javadi N, Abas F, Mediani A, Hamid AA, Khatib A, Simoh S. Effect of storage time on metabolite profile and alpha-glucosidase inhibitory activity of Cosmos caudatus, leaves–GCMS based metabolomics approach. J Food Drug Anal. 2015;23:433–41. doi: 10.1016/j.jfda.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou S, Lou YR, Tzin V, Jander G. Alteration of plant primary metabolism in response to insect herbivory. Plant Physiol. 2015;169:1488–98. doi: 10.1104/pp.15.01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neilan BA, Pearson LA, Muenchhoff J, Moffitt MC, Dittmann E. Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ Microbio. 2013;15:1239–53. doi: 10.1111/j.1462-2920.2012.02729.x. [DOI] [PubMed] [Google Scholar]
- 17. Naveed M, Qureshi MA, Zahir ZA, Hussain MB, Sessitsch A, Mitter B. L-tryptophan-dependent biosynthesis of indole-3-acetic acid (IAA) improves plant growth and colonization of maize by Burkholderia phytofirmans. Ann Microbio. 2014;65:1–9. [Google Scholar]