Abstract
In an attempt to profile the metabolites of three different varieties of germinated rice, specifically black (GBR), red, and white rice, a 1H-nuclear-magnetic-resonance-based metabolomics approach was conducted. Multivariate data analysis was applied to discriminate between the three different varieties using a partial least squares discriminant analysis (PLS-DA) model. The PLS model was used to evaluate the relationship between chemicals and biological activities of germinated rice. The PLS-DA score plot exhibited a noticeable separation between the three rice varieties into three clusters by PC1 and PC2. The PLS model indicated that α-linolenic acid, γ-oryzanol, α-tocopherol, γ-aminobutyric acid, 3-hydroxybutyric acid, fumaric acid, fatty acids, threonine, tryptophan, and vanillic acid were significantly correlated with the higher bioactivities demonstrated by GBR that was extracted in 100% ethanol. Subsequently, the proposed biosynthetic pathway analysis revealed that the increased quantities of secondary metabolites found in GBR may contribute to its nutritional value and health benefits.
Keywords: germinated rice extract, metabolic pathway, metabolite profiling, multivariate data analysis, rice variety
1. Introduction
Rice (Oryza sativa L.) is the most popular cereal crop, and more than half of the global population consumes it as a staple food. While white rice or common rice is consumed most often, there are also special rice varieties where the rice grain contains pigmented substances with colors such as red, purple, brown, or black. Recently, the germination process for cereal grains, in particular rice, has become a new research interest. Germinated rice (GR) is produced by soaking rice grains in water and allowing them to bud. During the germination process, endogenous hydrolytic enzymes are activated to break down starch, fibers, and proteins, which results in the modification of nutritional and physiological properties as well as textural characteristics [1–3]. Furthermore, GR has many beneficial pharmacological properties including use as antihyperlipidemic and antihypertensive agents and the capability of reducing the risk of some chronic diseases, such as diabetes, cancer, cardiovascular diseases, and Alzheimer’s disease [4]. Moreover, pigmented rice has been reported to possess a potent antioxidant activity [5,6].
The benefits and values of consuming functional or nutraceutical foods on human health are prominent areas of research in the food science field. Several studies have reported that pigmented rice is a potent source of bioactive compounds, such as phenolic compounds, vitamins, and minerals [7–9]. Germinated rice is considered an important source of γ-aminobutyric acid (GABA), a nonprotein amino acid with significant biological activity. In addition, a previous study reported that γ-oryzanol, tocopherols, and tocotrienols are major bioactive compounds in GR [2]. These metabolites can differ from one variety to another. To date, some studies have profiled metabolites in GR using analytical methods [2–4]. Recent studies have demonstrated the successful isolation and elucidation of metabolites from rice using different analytical techniques [10–12]. Therefore, these findings necessitate the need for a comparison of the metabolite constituents in the other varieties of GR extract (GRE).
Nuclear magnetic resonance (NMR) is a powerful technique used to elucidate structural information and isolate the metabolites within complex systems. NMR spectrometry offers several advantages such as rapidity, reproducibility, and simplicity of sample preparation [13]. Recently, NMR techniques and multivariate data analysis (MVA) have been widely used for metabolite profiling and for the determination of differences between samples [14–17]. In addition, NMR spectrometry was also used to identify and characterize the composition of various types of plants, foods, and tissues [18–21].
In the present study, three varieties of GR were extracted using different solvents. The variability in the chemical composition and discrimination among the different GREs was elucidated using a combination of NMR-based metabolomics and compared with nongerminated rice (NGR). The correlation between metabolites and biological activities, including 2,2-diphenyl-1-picrylhydrazyl (DPPH) scavenging and α-glucosidase inhibitory activities were also examined. The information gathered may help more efficiently in the use of GREs in medicinal products and foods.
2. Materials and methods
2.1. Chemicals
Absolute ethanol, sodium carbonate, Folin–Ciocalteu reagent, gallic acid, phosphate buffer, deuterated methanol-d4 (CH3OH-d4), nondeuterated potassium dihydrogen phosphate (KH2PO4), deuterium oxide (D2O), and trimethylsilyl propionic acid-d4 sodium salt (TSP) were purchased from Merck (Darmstadt, Germany). The other chemicals, including p-nitrophenyl-α-d-glucopyranose (PNPG), glycine, α-glucosidase enzyme, and DPPH were supplied by Sigma–Aldrich (Germany).
2.2. Plant materials
Three varieties of paddy rice (O. sativa L.) were obtained from the Organic Agriculture Project, Sukhothai Airport, Thailand. The paddy rice samples included O. sativa L. cv. Hom Mali 105 (WR), O. sativa L. cv. Hom Deang Sukhothai 1 (RR), and O. sativa L. cv. Hom Dam Sukhothai 2 (BR). The samples were packed in an aluminum foil bag and stored at 4°C in the refrigerator until use.
2.3. Germination process and sample preparation
Paddy rice samples (500 g) were soaked in water at room temperature at a ratio of 1:5 (w/v). The water was changed every 8 hours and decanted after 24 hours. Paddy rice samples were distributed on double layers of cotton cloth and placed in five plastic baskets to give the five biological replicates and allowed to germinate at room temperature for 7 days. GR samples were collected and dried for 24 hours in an oven at 45°C. Samples from each basket were randomly collected for the extraction with both solvent ratios. The dried samples were ground using a grinder to obtain a fine powder. The ground samples were packed in polyethylene bags and stored in a chiller at 4°C until use. The NGR was used as a control.
2.4. Extraction
Five grams of ground GR and NGR (from three varieties) were immersed in 50 mL 70% or 100% ethanol. The mixtures were sonicated in an ultrasonic bath at room temperature for 1 hour and then filtered through filter paper (Whatman No.1). The residues were re-extracted with 50 mL of the same solvent under the same conditions for sonication, and the solutions of each extract were combined. The extraction solvents were removed under vacuum using rotary evaporation at 40°C. The extracts were stored in a freezer at −20°C for further analysis.
2.5. DPPH radical scavenging
DPPH radical scavenging activity was investigated using a 96-well microplate as previously described [22]. A 50-μL aliquot of each of the tested samples was put into a well, followed by the addition of 100 μL DPPH (80 mg/L). The mixtures were then left in the dark to incubate for 30 minutes. The absorbance at 517 nm was measured using a microplate reader (SPECTRAmax PLUS; Molecular devices, LLC, Sunnyvale, CA, USA). The DPPH scavenging ability was calculated as % inhibition = [(Ao − As)/Ao] × 100, where Ao is the absorbance of the reagent blank and As is the absorbance of the test samples. The results are expressed in percentage (%) DPPH scavenging ability.
2.6. In-vitro α-glucosidase inhibition assay
The assay to detect α-glucosidase inhibition activity was carried out following the previously described protocol [23]. The enzyme reaction was performed using PNPG as a substrate dissolved in 50mM phosphate buffer (pH 6.5). The extracts were added to the 96-well microplate and incubated at room temperature for 5 minutes. Then, 75 μL of substrate PNPG was added to each well of the sample, blank substrate, negative control, and positive control, while the rest were loaded with 75 μL KH2PO4 buffer (30mM). The mixture was incubated at room temperature for 15 min. The reactions for the sample, blank, substrate, and negative control were stopped by the addition of 50 μL 2M glycine (pH 10). Meanwhile, the rest of the samples were loaded with 50 μL deionized water. Absorbance was measured at 405 nm using a spectrophotometer (SPEC-TRAmax PLUS). The α-glucosidase inhibition activity of the test sample was recorded as percentage inhibition. Percentage inhibition of the sample = [(an − as)/an] ×100, where an is the absorbance difference between the negative control and the blank substrate and as is the difference between the absorbance of the sample and the blank sample.
2.7. 1H-NMR analysis
The 1H-NMR spectrum was recorded on a 500-MHz Varian INOVA NMR spectrometer (Varian Inc., CA, USA) operating at a proton NMR frequency of 499.887 MHz and temperature of 26°C. Each 1H-NMR spectrum acquired a width of 20 ppm, which consisted of 64 scans with a 3.53-minute acquisition time. For each sample, a relaxation delay of 1.0 second was recorded. The preparation procedure was performed following the reported method [13] with some modifications. GR samples (10 mg each) were extracted by mixing with 0.375 mL CH3OH-d4 and the same volume of KH2PO4 buffer in D2O (pH 6.0) containing 0.1% TSP in a centrifuge tube (2 mL). The mixtures were vortexed for 1 minute at room temperature, sonicated for 15 minutes, and then centrifuged for 10 minutes at 1889.42 g. The clear supernatants (0.6 mL) were transferred to NMR tubes and subjected to 1H-NMR analysis. The TSP was used as a reference at δ 0.00. The metabolites were identified based on 1H-NMR spectra and comparisons with the NMR spectra of reference compounds, published literature, and Chenomx database NMR suite software. Furthermore, two dimensional J-resolved NMR was applied to assist in the identification of some metabolites with overlapping signals.
2.8. The bucketing of 1H-NMR spectra and multivariate statistical analysis
Phasing and baseline corrections were performed manually for all spectra using Chenomx software version 6.2 (Edmonton, AB, Canada). The 1H-NMR spectra were reduced to ASCII files using Chenomx profiler according to the method described by Mediani et al [22]. All spectral intensities were binned using a spectral width (δ 0.04), forming a region of δ 0.50–10.0. The regions of δ 4.70–4.90 and δ 3.23–3.36, which correspond to water and residual methanol, respectively, were removed from the analysis. Multivariate data analysis of NMR spectra data was performed with five replications after the binning process to investigate the systematic variations in the data sets. The principal component analysis (PCA) and partial least squares (PLS) models were performed by SIMCA-P software version 13.0 (Umetrics, Umeå, Sweden) using the Pareto scaling method [24].
2.9. Quantitative data analysis
Relative quantification of the identified compounds was performed according to the mean peak height of the 1H-NMR signals of interest after binning. The relative quantification of the GR varieties was performed by plotting their peak area means relatively to TSP. The 1H-NMR chemical shifts of metabolites for quantification were selected from the binned results and the assignment was based on their unique signal characteristics. The data were analyzed by analysis of variance (SPSS Inc., Chicago, IL, USA), and Tukey’s significant difference multiple-comparison test was applied to evaluate the significant differences between the red GR (GRR), black GR (GBR), and white GR (GWR) samples.
3. Results and discussion
3.1. Visual inspection of 1H-NMR spectra and metabolite identification in GR and NGR
Representative 1H-NMR spectra of the extracts obtained from GR and NGR extracted by ethanol at different solvent ratios are depicted in Figure S1. The most intense signals within each 1H-NMR spectrum were in the carbohydrate region, which ranged from δ 3.0 to δ 5.50 and corresponded to sugars such as fructose, sucrose, α-glucose, and β-glucose. There were spectra peaks found in other regions, such as the aliphatic (δ 0.50–3.0) and aromatic (δ 5.50–9.0). However, the relative intensities of the peaks in the aromatic region were lower than those of peaks in other regions. There were more peaks in the aromatic region for GBR and GRR compared to GWR. In addition, intensities of the peaks in the aromatic region were not clearly distinguished between the different solvent ratios of each rice variety. The problem of the overlapping signals in the 1H-NMR spectra was solved by utilizing two-dimensional J-resolved analysis. The signal splitting and coupling constants provided by J-resolved analysis were used for assigning and confirming the problematic metabolite signals from one-dimensional NMR (Figure S2). In general, there were more signals in the aromatic region (δ 5.50–9.0) of GR when compared to NGR.
The different varieties of GREs in either 70% or 100% ethanol demonstrated variations with regards to signal intensity, especially those in the aromatic region. The identified metabolites are listed in Table 1. It is interesting to note that among the tested rice varieties, only GWR extracted in either 70% or 100% ethanol displayed variation as depicted by the absence of organic acids (2,3-butanediol and fumaric acid), amino acids (tryptophan and phenylalanine), and phenolic compounds (sinapic acid, p-hydroxybenzoic acid, and vanillic acid). This observation was consistent with a previous finding, which demonstrated differences in the chemical compositions of different types of GREs [25].
Table 1.
Assignment of 1H-NMR spectral peaks of ethanol-extracted germinated rice analyzed by NMR
| Metabolite | 1H-NMR characteristic signals (δ) | Rice varieties/ethanol ratio (%) | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| White | Black | Red | |||||
|
|
|
|
|||||
| 70 | 100 | 70 | 100 | 70 | 100 | ||
| Amino acids | |||||||
| Valine (1) | δ 1.02 (d, J = 7.0 Hz) | + | + | + | + | + | + |
| δ 1.06 (d, J = 7.0 Hz) | |||||||
| Threonine (2) | δ 1.34 (d, J = 7.0 Hz) | + | + | + | + | + | + |
| δ 4.22 (m) | |||||||
| Tyrosine (3) | δ 7.18 (d, J = 8.5 Hz) | + | + | + | + | + | + |
| δ 6.84 (d, J = 8.5 Hz) | |||||||
| Alanine (4) | δ 1.46 (d, J = 7.0 Hz) | + | + | + | + | + | + |
| Glutamine (5) | δ 2.46–2.42 (m) | + | + | + | + | + | + |
| Glutamic acid (6) | δ 2.10–2.16 (m) | + | + | + | + | + | + |
| δ 1.98–2.06 (m) | |||||||
| Aspartate (7) | δ 2.82 (dd, J = 17.0 Hz, 8.5 Hz) | + | + | + | + | + | + |
| δ 2.86 (dd, J = 17.0 Hz, 4.0 Hz) | |||||||
| Leucine (8) | 1.64–1.68 (m) | + | + | + | + | + | + |
| 0.98–1.00 (m) | |||||||
| Phenylalanine (9) | δ 7.42 (d, J = 7.0 Hz) | - | - | + | + | + | + |
| δ 7.40 (t, J = 7.0 Hz) | |||||||
| δ 7.34 (t, J = 7.0 Hz) | |||||||
| Hydroxy-l-proline (10) | 4.33–4.38 (m) | + | + | + | + | + | + |
| Tryptophan (11) | δ 7.22 (t, J = 8.0 Hz) | — | — | + | + | + | + |
| δ 7.28 (t, J = 8.5 Hz) | |||||||
| δ 7.74 (d, J = 8.5 Hz) | |||||||
| Organic acids | |||||||
| 3-Hydroxybutyric acid (12) | δ 1.20 (d, J = 5.5 Hz) | + | + | + | + | + | + |
| 2,3-Butanediol (13) | δ 1.10 (d, J = 7.0 Hz) | + | + | + | + | + | + |
| Fumaric acid (14) | δ 6.62 (s) | — | — | + | + | + | + |
| γ-Aminobutyric acid (15) | δ 1.85–1.90 (m) | + | + | + | + | + | + |
| δ 2.30 (t, J = 7.5 Hz) | |||||||
| δ 3.02 (t, J = 7.5 Hz) | |||||||
| α-Aminobutyric acid (16) | 0.95–0.98 (t) | + | + | + | + | + | + |
| 1.85–1.90 (m) | |||||||
| Citric acid (17) | 2.50–2.54 (d, J = 17.0 Hz) | — | — | + | + | + | + |
| 2.68–2.71 (d, J = 17.0 Hz) | |||||||
| Oxoglutaric acid (18) | 2.42–2.44 (t, J = 7.0 Hz) | + | + | + | + | + | + |
| 2.98–3.02 (t, J = 7.0 Hz) | |||||||
| Malate (19) | 2.68 (dd, J = 13.5, 2.5 Hz) | — | — | + | + | + | + |
| 4.30 (br d) | |||||||
| Lipids | |||||||
| Fatty acids (20) | δ 0.86 (t, J = 7.0 Hz) | + | + | + | + | + | + |
| δ 1.30 (m) | |||||||
| α-Linolenic acid (21) | δ 0.98 (t, J = 7.5 Hz) | + | + | + | + | + | + |
| δ 1.26 (s) | |||||||
| Mevalonate | |||||||
| γ–Oryzanol (22) | δ 7.75 (d, J = 16.0 Hz) | + | + | + | + | + | + |
| δ 5.38 (d, J = 6.0 Hz) | |||||||
| δ 4.93 (m) | |||||||
| δ 3.94 (s) | |||||||
| δ 1.14 (q, J = 9.0 Hz) | |||||||
| α-Tocopherol (23) | δ 2.67 (br t, J = 5.0 Hz) | + | + | + | + | + | + |
| δ 1.36 (s) | |||||||
| Sugars | |||||||
| Fructose (24) | δ 4.12 (dd, J = 5.5 Hz) | + | + | + | + | + | + |
| δ 3.78–3.82 (m) | |||||||
| δ 3.86 (dd, J = 12.0 Hz, 2.5 Hz)) | |||||||
| Sucrose (25) | δ 3.54 (dd, J = 9.5 Hz, 3.5 Hz) | + | + | + | + | + | + |
| δ 3.76–3.90 (m) | |||||||
| δ 3.66 (s) | |||||||
| δ 4.04 (t, J = 8.0 Hz) | |||||||
| δ 5.42 (d, J = 3.5 Hz) | |||||||
| Glucose (26) | δ 4.62 (d, J = 8.0 Hz) | + | + | + | + | + | + |
| Xylose (27) | δ 5.18 (d, J = 4.0 Hz) | + | + | + | + | + | + |
| δ 3.22 (t, J = 9.5 Hz) | |||||||
| Mannose (28) | 5.19 (br d) | + | + | + | + | + | + |
| Galactitol (29) | 3.97 (m) | + | + | + | + | + | + |
| Phenolics | |||||||
| Sinapic acid (30) | δ 7.04 (s) | — | — | + | + | + | + |
| δ 7.55 (d, J = 17.0 Hz) | |||||||
| Ferulic acid (31) | δ 7.14 (d, J = 8.0 Hz) | + | + | + | + | + | + |
| δ 7.34 (d, 14.5 Hz) | |||||||
| δ 7.27 (s) | |||||||
| δ 3.90 (s) | |||||||
| p-Hydroxybenzoic acid (32) | δ 6.91 (t, J = 9.0 Hz) | — | — | + | + | + | + |
| δ 7.94 (d, J = 7.5 Hz) | |||||||
| Gallic acid (33) | δ 7.09 (s) | + | + | + | + | + | + |
| Vanillic acid (34) | δ 7.48 (dd, J = 8.0 Hz, 2.0 Hz) | — | — | + | + | + | + |
d = doublet; dd = doublet of doublet; m = multiplet; NMR = nuclear magnetic resonance; s = singlet; t = triplet.
3.2. Changes of bioactive components in GR and NGR
GR has been established as a functional food and its health benefits are relied on powerful method to obtain high concentrations of bioactive compounds [3]. The PCA results comparing GR extracts with NGR by NMR analysis showed that they were clearly separated by PC1 (Figure S3A). The loading column plot showed that GR extracts were separated due the highest content of some phenolics, α-tocopherol, γ-oryzanol, GABA, xylose, glucose, and mannose (Figure S3B). Germination process of rice can be involved in both physical and biochemical activities [4]. After water absorption, hydrolytic enzymes are activated and large molecules are degraded to small biomolecules in the endosperm of germinated seeds, such as biopolymers, carbohydrates, and polypeptides [2,25]. Phenolic contents also showed increase apart from nutrition level changes [6]. The finding of this study demonstrated that phenolic, α-tocopherol, γ-oryzanol, α-linolenic acid, GABA, and glucose of all the studied rice varieties significantly (p < 0.05) improved after germination as compared to that before germination. The possible explanation is that the water absorption activates the bioactive compounds and bio-activities. The reducing sugar enhancement in GR might be due to the starch hydrolysis by the activated enzymes during the germination process [25]. The α-tocopherol and γ-oryzanol are beneficial and important antioxidant compounds presented in the rice bran, which acts in the protection of rice oil from oxidation. These results were in line with previous studies [6]. In addition, the germination process was reported as the cause of the increment of antioxidant compounds [6].
3.3. Comparison of DPPH scavenging ability and α-glucosidase inhibition of GRE samples
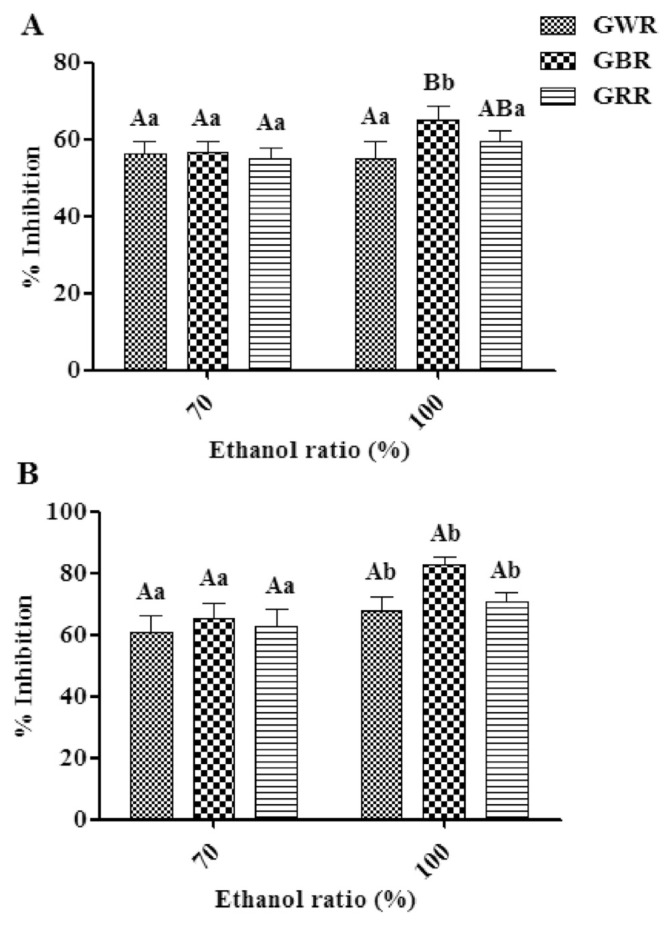
The percentage inhibition calculations for the DPPH radicals of three GRE extracts using 70% ethanol were not significantly different (p > 0.05) and varied from 55.22% to 56.78%. These were in the same range as GRE extracts in 100% ethanol (55.13–65.10%) (Figure 1A). The highest percentage inhibition for a DPPH radical was observed in the GBR100 sample. This result was consistent with a previous study demonstrating that pigmented rice has greater DPPH scavenging power than white rice [26]. In addition, the results showed that the DPPH inhibitions for GREs extracted with 100% ethanol were higher than those with 70% ethanol for all varieties, and GBR100 sample was the only exhibited significantly different (p < 0.05) (Figure 1A). This result was in agreement with a previous study that showed that pure ethanol extract exhibited the greatest bioactivities of Limnophila aromatica [27]. This may be due to several factors including plant matrix, number of hydroxyl groups, and polarity index of the used solvent.
Figure 1.
2,2-Diphenyl-1-picrylhydrazyl radical scavenging ability (A) and the α-glucosidase inhibitory activity (B) of various germinated rice samples extracted with different ratios of ethanol. The different capital letters signify the different rice varieties that were extracted using the same ethanol ratio (n =5). Different lowercase letters were used to distinguish between ethanol ratios used for the extraction of the same rice variety (n =5). DPPH =2,2-diphenyl-1-picrylhydrazyl; GBR =germinated black rice; GRR =germinated red rice; GWR =germinated white rice.
The in vitro α-glucosidase inhibitory activity of the GREs is shown in Figure 1B. The percent inhibition of α-glucosidase in GREs at a concentration of 1.0 mg/mL ranged from 60.95% to 65.44% and 68.02% to 82.87% for 70% and 100% ethanol extracts, respectively. There were no significant differences in the percentage inhibition of α-glucosidase between GREs except for GBR100 sample (p > 0.05). In addition, the highest α-glucosidase inhibition was shown for GREs for all varieties extracted with 100% ethanol. This was in agreement with a previous study showing that the most significant α-glucosidase inhibition observed for Cosmos caudatus extracts was in samples extracted with absolute ethanol [28]. This phenomenon was attributed to the fact that organic solvents are more efficient in extracting bioactive compounds that possess α-glucosidase inhibitory activity than water [29].
3.4. Correlation between biological activity and metabolite variation
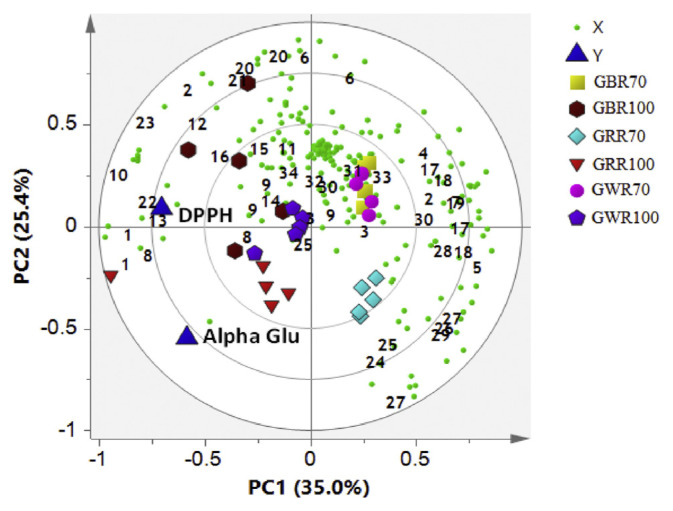
The PLS analysis was performed to assess the internal association between metabolites and biological activities. The chemical shift of the metabolites was represented by X variables, while biological activities were represented by Y variables. The biological activities of the samples were predicted from MVA by utilizing PLS data. In this study, a single graphical representation, which combined the score and loading plots, was created as shown in the PLS biplot. From the PLS biplot (Figure 2), a distinct separation was observed between the different groups of solvent ratios, but good discrimination was not achieved among the three rice varieties using the same solvent ratios. All GRE samples extracted by 100% ethanol were separated by PC1 and were strongly correlated with both biological activities. It was indicated that Y variables (DPPH scavenging and α-glucosidase inhibitory activities) were positioned near to all rice varieties extracted with 100% ethanol (Figure 2). Among the metabolites that might be contributing towards the higher DPPH scavenging and α-glucosidase inhibitory activities associated with these GREs extracted with 100% ethanol are α-linolenic acid, γ-oryzanol, α-tocopherol, GABA, α-aminobutyric acid, glutamic acid, leucine, hydroxyl-l-proline, 3-hydroxybutyric acid, 2,3-butanediol, fumaric acid, fatty acids, vanillic acid, phenylalanine, and valine. Therefore, it can be suggested that these metabolites might be important to the biological activities associated with these GREs. An earlier study showed that the antioxidant activity of tomato was strongly associated with the amount of phenolics in the tomato [30]. In a similar case, higher phenolic compounds in these GREs extracted with 100% ethanol may play a role in the high DPPH radical scavenging activity. Furthermore, α-linolenic acid, vitamin E (α-tocopherol), and other assigned phenolic compounds were identified and previously reported to be the potential α-glucosidase inhibitor in C. caudatus [28].
Figure 2.
The biplot obtained from partial least squares data describing the variations between all of the germinated rice samples. 1, valine; 2, threonine; 3, tyrosine; 4, alanine; 5, glutamine; 6, glutamic; 7, aspartate; 8, leucine; 9, phenylalanine; 10, hydroxy-l-proline; 11, tryptophan; 12, 3-hydroxybutyric acid; 13, 2,3-butanediol; 14, fumaric acid; 15, γ-aminobutyric acid; 16, α-aminobutyric acid; 17, citric acid; 18, oxoglutaric acid; 19, malate; 20, fatty acids; 21, α-linolenic acid; 22, γ–oryzanol; 23, α-tocopherol; 24, fructose; 25, sucrose; 26, glucose; 27, xylose; 28, mannose; 29, galactitol; 30, sinapic acid; 31, ferulic acid; 32, p-hydroxybenzoic acid; 33, gallic acid; and 34, vanillic acid. 70 =extracted with 70% ethanol; 100 =extracted with 100% ethanol; Alpha Glu = alpha glucosidase; DPPH =2,2-diphenyl-1-picrylhydrazyl; GBR =germinated black rice; GRR =germinated red rice GWR =germinated white rice; PC = principal component.
Our results also showed that all samples extracted with 100% ethanol were positioned on the left side of the PLS biplot and were more active compared to those of samples extracted with 70% ethanol (Figure 2). According to the loading plots, this separation was due to more sugars content in GRR70 compared to GRR100. This is because water has a higher dielectric constant than ethanol. Therefore, 70% ethanol may be more efficient at extracting sugars compared to absolute ethanol. This is expected since the sugars are polar compounds.
The generated PLS models were validated using the relationship between observed and predicted values (Figure S4). The permutation tests were also generated as an internal validation of the model (Figure S5) as well as the cross validation analysis (Table S1). The results showed that Y-axis intercepts of R2 and Q2 of DPPH were 0.075 and −0.216 and for α-glucosidase inhibitory activity, and the values were 0.014 and −0.259, respectively (Table 2). Thus, the generated PLS biplot was valid and no overfitting issues were detected based on the rule of thumb, where Y-axis intercepts are R2 < 0.3 and Q2 < 0.05 and the R line is not horizontal [31]. The root mean square error of crossvalidation and root mean square of estimation were low with excellent regression coefficient (R > 0.9). The same trend in permutation testing was used to validate the PLS biplot. Therefore, these data suggest that the PLS model was appropriate for assessing the biological activities (DPPH and α-glucosidase inhibitory activities) of rice samples based on their respective NMR spectra. The variable importance to projections (VIP) was also checked to validate the metabolites contributing to the bioactivities (Table S2).
Table 2.
PLS model validation
| Biological assay | No. of components | R2Y | Q2Y | R2Y Intercepts | Q2Y Intercepts | RMSEE | RMSECV |
|---|---|---|---|---|---|---|---|
| DPPH | 2 | 0.60 | 0.50 | 0.075 | −0.216 | 2.826 | 2.967 |
| α-Glucosidase | 2 | 0.60 | 0.50 | 0.014 | −0.259 | 4.886 | 5.374 |
DPPH = 1,1-diphenyl-2-picryl-hydrazyl; Q2Y = fraction of the variation of the Y variables predicted by the model; R2Y = fraction of the variation of the Y variables explained by the model; RMSECV = root mean square error of crossvalidation; RMSEE = root mean square of estimation.
3.5. Classification of GREs by PLS discriminant analysis
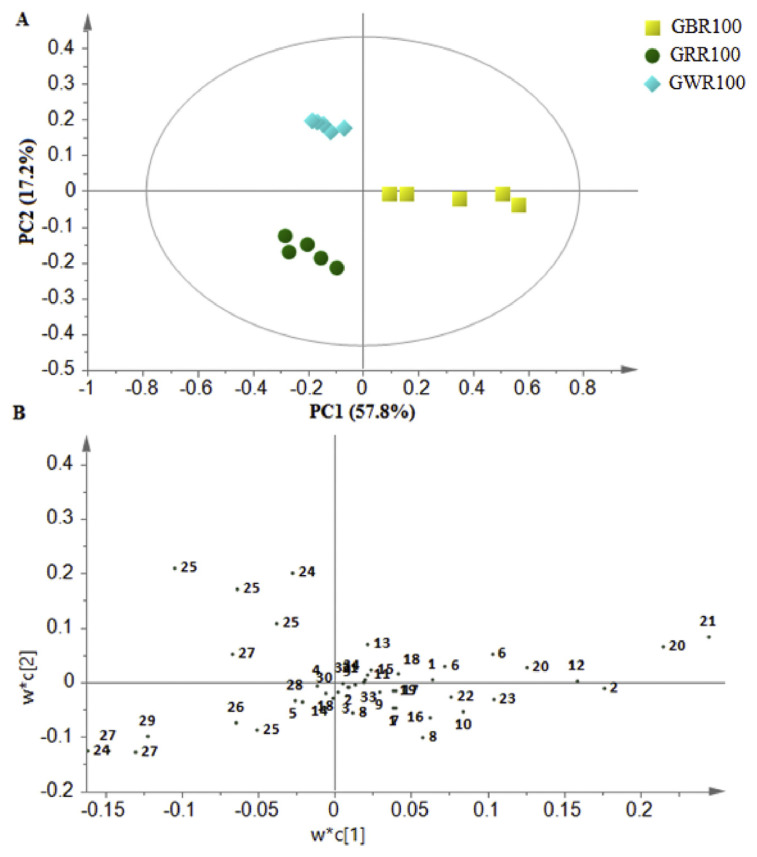
Variation among the metabolites in the GREs was further evaluated using MVA. The PLS discriminant analysis (PLS-DA) score plot was performed to evaluate the variations between the extracts of three GR varieties in 100% ethanol, while a loading plot was applied to identify the metabolites that contributed to the discrimination among GRE samples in PLS-DA score plots. The first two PCs accounted for 75.0% of the total variance (Figure 3A). PC1 showed the most sample variation and described 57.8% of the total variance, followed by PC2, accounting for 17.2% of total variance. There was a clear discrimination among the three GREs. The GBR can be distinguished from GRR and GWR by PC1, while GWR was clustered separately from the others by PC2. In addition, the loading plot showed metabolites that influenced the separation of each sample (Figure 3B). The PLS-DA score plot was matched with the loading plot, where the positions of any clusters in the PLS-DA score plot consistent with the X variable (NMR chemical shifts) can be found in the same location in the loading plot.
Figure 3.
Partial least squares discriminant analysis score plot (PC1 vs. PC2) (A) and the loading plot PC1 (B) of 1H-nuclear magnetic resonance data representing the three varieties of germinated rice extracted by 100% ethanol. 1, valine; 2, threonine; 3, tyrosine; 4, alanine; 5, glutamine; 6, glutamic; 7, aspartate; 8, leucine; 9, phenylalanine; 10, hydroxy-l-proline; 11, tryptophan; 12, 3-hydroxybutyric acid; 13, 2,3-butanediol; 14, fumaric acid; 15, γ-aminobutyric acid; 16, α-aminobutyric acid; 17, citric acid; 18, oxoglutaric acid; 19, malate; 20, fatty acids; 21, α-linolenic acid; 22, γ –oryzanol; 23, α-tocopherol; 24, fructose; 25, sucrose; 26, glucose; 27, xylose; 28, mannose; 29, galactitol; 30, sinapic acid; 31, ferulic acid; 32, p-hydroxybenzoic acid; 33, gallic acid; and 34, vanillic acid. GWR100 =germinated white rice extracted with 100% ethanol; GBR100 =germinated black rice extracted with 100% ethanol; GRR100 =germinated red rice extracted with 100% ethanol; PC = principal component.
Based on the loading plot (Figure 3B), the metabolites, including fatty acids, linoleic acid, amino acids (aspartate, valine, tryptophan, alanine, threonine, and GABA), fumaric acid and phenolic compounds (sinapic acid, vanillic acid, and ferulic acid), could influence the separation of GBR100 from the other tested rice samples. These metabolites were positioned in the right quadrant, which was consistent with GBR100 as shown in the PLS-DA score plot. Furthermore, sugars (sucrose, glucose, and fructose) could also be contributing to the discrimination of GRR100 and GWR100 from GBR100. These findings are consistent with a previous study [25], which displayed variation in the sum of the sugars in a different type of GR. Thus, the different varieties of GR used in this study may vary in terms of their sugar quantities.
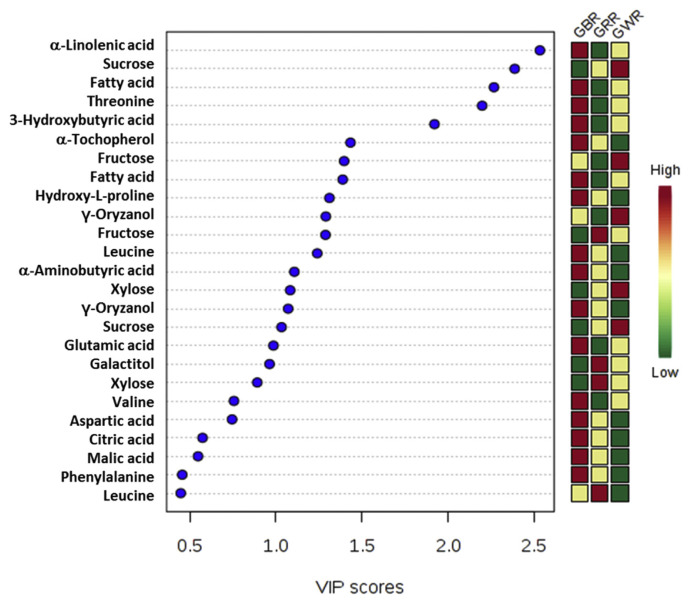
The significance of the variables was further examined by determining VIP. The VIP values yielded, which are > 0.5, are considered essential towards the observed results [22]. Based on Figure 4, α-linoleic acid, fatty acids, threonine, 3-hydroxybutyric acid, valine, aspartate, γ-oryzanol, and glutamate significantly influenced the discrimination of all GR varieties extracts. From the misclassification table, which was checked to confirm the discernment features between different rice varieties, the results displayed that all extracts have been classified with 100% accuracy (Table S3). The permutation analysis was also performed for the PLS-DA model (Figure S6). The results showed that Y-axis intercepts of R2 and Q2 of GBR were 0.497 and −0.45 and for GRR, the values were 0.472 and −0.469, respectively. The Y-axis intercepts of R2 and Q2 of GWR showed similar values of 0.494 and −0.421, respectively.
Figure 4.
VIP values of the major metabolites used for the separation of GRE derived from partial least squares discriminant analysis data plots for all germinated rice samples. GBR =germinated black rice; GRR =germinated red rice; GWR =germinated white rice; VIP =variable importance to projections.
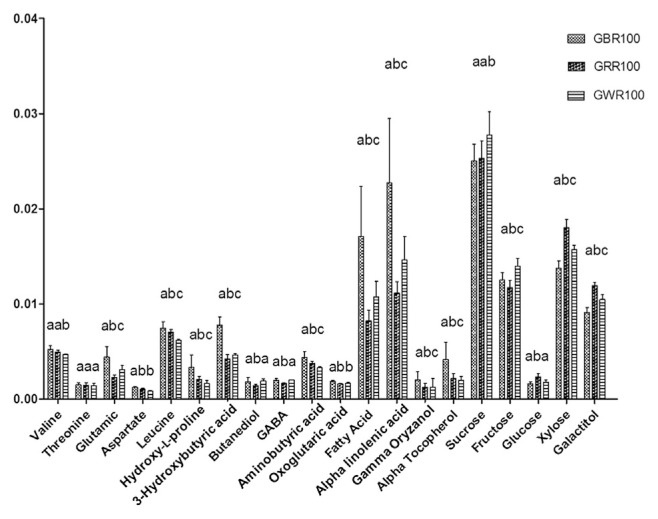
3.6. Relative quantification and metabolic pathway differences in GREs
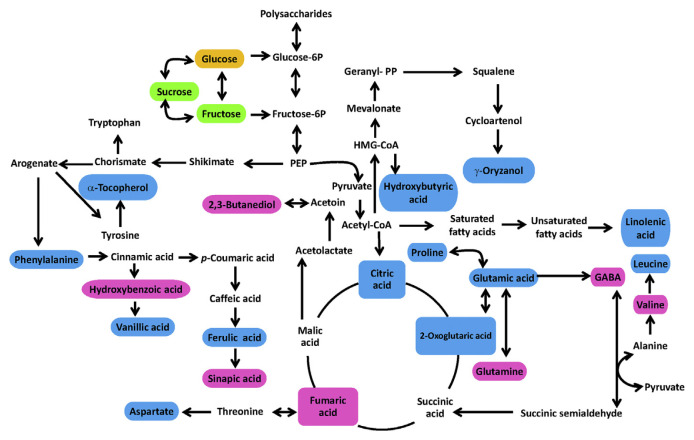
The levels of metabolites responsible for the differences between GR varieties were determined to gain insight into their variances (Figure 5). Based on the analysis of variance test, there were significant differences between GBR100, GRR100, and GWR100. The GBR100 had higher content of γ-oryzanol, α-tocopherol, linolenic acid, and other phenolics as previously shown in PLS-DA results. Figure 6 represents the simplified pathway for metabolites identified by NMR and their differences among the three GR varieties. The metabolic pathways were determined according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, which was used to better understand the metabolite changes depicted by the NMR results. For GBR, our findings showed low quantities of sugars and amino acids (p < 0.05) generated from pyruvate and high levels of organic acids (p < 0.05) stemming from the tricarboxylic acid cycle. For the secondary metabolites, phenylalanine is the main intermediate in the generation of phenolic compounds in the phenylpropanoid pathway [30]. Phenylalanine is converted to cinnamic acid, which is the synthesis precursor of hydroxybenzoic, vanillic, ferulic, and sinapic acids. All of these metabolites are reported to have many biological activities, including antioxidant and α-glucosidase inhibitory activities [32]. Alkaline hydrolysis is also associated with phenolic compounds and was increased in GBR samples; this could be explained by cell wall dismantling during germination.
Figure 5.
Relative quantification of the identified metabolites in different rice varieties based on the mean peak area of the 1H nuclear magnetic resonance signals. Different lower case letters show significant differences between rice varieties (n =5). GABA =γ-aminobutyric acid; GWR100 =germinated white rice extracted with 100% ethanol; GBR100 =germinated black rice extracted with 100% ethanol; GRR100 =germinated red rice extracted with 100% ethanol.
Figure 6.
Metabolic networks and the metabolites change of all germinated rice samples. Blue color indicates significantly higher metabolite in black rice, green color indicates significantly higher metabolite in white rice, and orange color indicates significantly higher metabolite in red rice. The pink color shows not significantly different metabolite between rice varieties. Fructose-6P =fructose 6-phosphate; GABA =γ-aminobutyric acid; Glucose-6P =glucose 6-phosphate; HMG-CoA =3-hydroxy-3-methylglutaryl-coenzyme A; PEP =phospoenolpyruvate
Germination may also raise the availability of hydrolyzable insoluble phenolic compounds, which tend to increase insoluble phenolic compounds in black rice. GBR samples contained significantly higher levels (p < 0.05) of ferulic, sinapic, p-hydroxybenzoic, gallic, and vanillic acids compared to other samples. This increase is associated with a decrease in sucrose and other sugars because sucrose and other sugars are essential intermediate metabolites in phenolic metabolism. Furthermore, the shikimate pathway is responsible for the production of phenolic acids. Hence, changes in the levels of sugars and amino acids could be a general metabolic variable amongst the different germinated rice varieties. In addition, increased sugar concentrations could be a result of a rise in the levels of phenylpropanoids.
The γ-oryzanol is a triterpene-like compound that stems from the mevalonate pathway and is linked to ferulic acid by an ester bond. It is also a known antioxidant compound [33]. Other antioxidant compounds, such as α-tocopherol, were also found in GR. α-Tocopherol is synthesized via two pathways, shikimate and nonmevalonate. The precursor for the head group is derived from the shikimate pathway (synthesized via tyrosine) and the phytyl tail group is derived from geranylgeranyl diphosphate in the nonmevalonate pathway. Fatty acids are widespread in plants. Formation of fatty-acid-derived volatile compounds starts with linoleic and linolenic acids. These volatile compounds create odor and flavor in rice. This study showed that γ-oryzanol was higher (p < 0.05) in GWR than GRR but not GBR, whereas α-tocopherol and α-linolenic acid were significantly (p < 0.05) higher in GBR (Figure 6). Proline is the main precursor for the synthesis of GABA prior to its conversion to glutamic acid. GABA, as a stress-induced amino acid transporter, can accumulate under environmental stress [34,35]. This accumulation leads to the production of succinic acid and other organic acids. Our findings showed that the levels of GABA were slightly higher in GBR but not significantly different from GWR, whereas GRR contained the lowest levels of GABA among all tested rice varieties. GR is rich in GABA, which in mammals acts as a neurotransmitter [33].
4. Conclusion
Applying chemometric methods to the 1H-NMR spectra of the tested rice samples enabled the identification of metabolites responsible for variations among GBR, GRR, and GWR using different ethanol ratios. There were 34 metabolites identified in this study. The GBR samples varied from other samples due to their higher content of α-linolenic acid, γ-oryzanol, α-tocopherol, GABA, 3-hydroxybutyric acid, fumaric acid, fatty acids, threonine, tryptophan, and vanillic acid. The PLS analysis showed a significant correlation between GBR100 samples and both DPPH radical scavenging and α-glucosidase inhibitory activities as well as the metabolites contributing to the bioactivities. The PLS-DA score plot showed a good separation of the three tested GREs. Higher levels of amino acids, organic acids, and some phenolic compounds were found in GBR than in the other GREs. This study may help provide guidance in the development of pharmacological products or functional foods from GREs.
Acknowledgments
This research was financially supported by the joint funding between the Thailand Research Fund and Neresuan University through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0163/2554) to Mr. Phaiwan Pramai and Associate Professor Dr. Sudarat Jiamyangyuen. The additional financial assistance from Naresuan University is acknowledged.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jfda.2016.11.023.
Funding Statement
This research was financially supported by the joint funding between the Thailand Research Fund and Neresuan University through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0163/2554) to Mr. Phaiwan Pramai and Associate Professor Dr. Sudarat Jiamyangyuen. The additional financial assistance from Naresuan University is acknowledged.
Footnotes
Conflicts of interest
All authors have no conflicts of interest to declare.
REFERENCES
- 1. Hunt JR, Johnson LK, Juliano BO. Bioavailability of zinc from cooked philippine milled, undermilled, and brown rice, as assessed in rats by using growth, bone zinc, and zinc-65 retention. J Agric Food Chem. 2002;50:5229–35. doi: 10.1021/jf020222b. [DOI] [PubMed] [Google Scholar]
- 2. Kim HY, Hwang IG, Kim TM, Woo KS, Park DS, Kim JH, Kim DJ, Lee J, Lee YR, Jeong HS. Chemical and functional components in different parts of rough rice (Oryza sativa L.) before and after germination. Food Chem. 2012;134:288–93. [Google Scholar]
- 3. Komatsuzaki N, Tsukahara K, Toyoshima H, Suzuki T, Shimizu N, Kimura T. Effect of soaking and gaseous treatment on GABA content in germinated brown rice. J Food Eng. 2007;78:556–60. [Google Scholar]
- 4. Wu F, Yang N, Touré A, Jin Z, Xu X. Germinated brown rice and its role in human health. Cr Rev Food Sci Nutr. 2013;53:451–63. doi: 10.1080/10408398.2010.542259. [DOI] [PubMed] [Google Scholar]
- 5. Huang YP, Lai HM. Bioactive compounds and antioxidative activity of colored rice bran. J Food Drug Anal. 2016;24:564–74. doi: 10.1016/j.jfda.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen S, Wang Y, Li M, Xu F, Chai L, Bao J. The effect of anaerobic treatment on polyphenols, antioxidant properties, tocols and free amino acids in white, red, and black germinated rice (Oryza sativa L.) J Funct Food. 2015;19:641–8. [Google Scholar]
- 7. Yawadio R, Tanimori S, Morita N. Identification of phenolic compounds isolated from pigmented rices and their aldose reductase inhibitory activities. Food Chem. 2007;101:1616–25. [Google Scholar]
- 8. Abas F, Shaari K, Israf DA, Syafri S, Zainal Z, Lajis NH. LC–DAD–ESI–MS analysis of nitric oxide inhibitory fractions of tenggek burung (Melicope ptelefolia Champ. ex Benth.) J Food Compos Anal. 2010;23:107–12. [Google Scholar]
- 9. Tananuwong K, Tangsrianugul N. Effects of storage conditions and cooking on colour and antioxidant activities of organic pigmented rice. Int J Food Sci Tech. 2013;48:67–73. [Google Scholar]
- 10. Yang Z, Nakabayashi R, Okazaki Y, Mori T, Takamatsu S, Kitanaka S, Kikuchi J, Saito K. Toward better annotation in plant metabolomics: isolation and structure elucidation of 36 specialized metabolites from Oryza sativa (rice) by using MS/MS and NMR analyses. Metabolomics. 2014;10:543–55. doi: 10.1007/s11306-013-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim JK, Park SY, Lim SH, Yeo Y, Cho HS, HSH Comparative metabolic profiling of pigmented rice (Oryza sativa L.) cultivars reveals primary metabolites are correlated with secondary metabolites. J Cereal Sci. 2013;57:14–20. [Google Scholar]
- 12. Frank T, Reichardt B, Shu Q, Engel KH. Metabolite profiling of colored rice (Oryza sativa L.) grains. J Cereal Sci. 2012;55:112–9. [Google Scholar]
- 13. Kim HK, Choi YH, Verpoorte R. NMR-based metabolomic analysis of plants. Nat Protoc. 2010;5:536–49. doi: 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- 14. Bollard ME, Holmes E, Lindon JC, Mitchell SC, Branstetter D, Zhang W, Nicholson JK. Investigations into biochemical changes due to diurnal variation and estrus cycle in female rats using high-resolution 1H NMR spectroscopy of urine and pattern recognition. Anal Biochem. 2001;295:194–202. doi: 10.1006/abio.2001.5211. [DOI] [PubMed] [Google Scholar]
- 15. Charlton AJ, Farrington WHH, Brereton P. Application of 1H NMR and multivariate statistics for screening complex mixtures: quality control and authenticity of instant coffee. J Agric Food Chem. 2002;50:3098–103. doi: 10.1021/jf011539z. [DOI] [PubMed] [Google Scholar]
- 16. Choi YH, Tapias EC, Kim HK, Lefeber AWM, Erkelens C, Verhoeven JT, Brzin J, Zel J, Verpoorte R. Metabolic discrimination of Catharanthus roseus leaves infected by phytoplasma using 1H-NMR spectroscopy and multivariate data analysis. Plant Phys. 2004;135:2398–410. doi: 10.1104/pp.104.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gallo V, Mastrorilli P, Cafagna I, Nitti GI, Latronico M, Longobardi F, Minoja AP, Napoli C, Romito VA, Schafer H, Schütz B. Effects of agronomical practices on chemical composition of table grapes evaluated by NMR spectroscopy. J Food Compos Anal. 2014;35:44–52. [Google Scholar]
- 18. Wishart DS. Metabolomics: applications to food science and nutrition research. Trends Food Sci Tech. 2008;19:482–93. [Google Scholar]
- 19. Wu H, Southam AD, Hines A, Viant MR. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal Biochem. 2008;372:204–12. doi: 10.1016/j.ab.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 20. Kim HS, Park SJ, Hyun SH, Yang SO, Lee J, Auh JH, Kim JH, Cho SM, Marriott PJ, Choi HK. Biochemical monitoring of black raspberry (Rubus coreanus Miquel) fruits according to maturation stage by 1H NMR using multiple solvent systems. Food Res Int. 2011;44:1977–87. [Google Scholar]
- 21. Sekiyama Y, Chikayama E, Kikuchi J. Profiling polar and semipolar plant metabolites throughout extraction processes using a combined solution-state and high-resolution magic angle spinning NMR approach. Anal Chem. 2010;82:1643–52. doi: 10.1021/ac9019076. [DOI] [PubMed] [Google Scholar]
- 22. Mediani A, Abas F, Khatib A, Maulidiani H, Shaari K, Choi YH, Lajis NH. 1H-NMR-based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res Int. 2012;49:763–70. [Google Scholar]
- 23. Lee SY, Mediani A, Nur Ashikin AH, Azliana ABS, Abas F. Antioxidant and α-glucosidase inhibitory activities of the leaf and stem of selected traditional medicinal plants. Int Food Res J. 2014;21:165–72. [Google Scholar]
- 24. Granato D, Branco GF, Faria Jde F, Cruz AG. Characterization of Brazilian lager and brown ale beers based on color, phenolic compounds, and antioxidant activity using chemometrics. J Sci Food Agr. 2011;91:563–71. doi: 10.1002/jsfa.4222. [DOI] [PubMed] [Google Scholar]
- 25. Moongngarm A, Saetung N. Comparison of chemical compositions and bioactive compounds of germinated rough rice and brown rice. Food Chem. 2010;122:782–8. [Google Scholar]
- 26. Yodpitak S, Sookwong P, Akkaravessapong P, Wongpornchai S. Changes in antioxidant activity and antioxidative compounds of brown rice after pre-germination. J Food Nutr Res. 2013;1:132–7. [Google Scholar]
- 27. Do QD, Angkawijaya AE, Tran-Nguyen PL, Huynh LH, Soetaredjo FE, Ismadji S, Ju YH. Effect of extraction solvent on total phenol content, total flavonoid content, and antioxidant activity of Limnophila aromatica. J Food Drug Anal. 2014;22:296–302. doi: 10.1016/j.jfda.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Javadi N, Abas F, Mediani A, Hamid AA, Khatib A, Simoh S, Shaari K. Effect of storage time on metabolite profile and alpha-glucosidase inhibitory activity of Cosmos caudatus leaves–GCMS based metabolomics approach. J Food Drug Anal. 2015;23:433–41. doi: 10.1016/j.jfda.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryu HW, Cho JK, Curtis-Long MJ, Yuk HJ, Kim YS, Jung S, Kim YS, Lee BW, Park KH. α-Glucosidase inhibition and antihyperglycemic activity of prenylated xanthones from Garcinia mangostana. Phytochemistry. 2011;72:2148–54. doi: 10.1016/j.phytochem.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 30. Martínez-Valverde I, Periago MJ, Provan G, Chesson A. Phenolic compounds, lycopene and antioxidant activity in commercial varieties of tomato (Lycopersicum esculentum) J Sci Food Agr. 2002;82:323–30. [Google Scholar]
- 31. Maulidiani, Abas F, Khatib A, Shitan M, Shaari K, Lajis NH. Comparison of partial least squares and artificial neural network for the prediction of antioxidant activity in extract of Pegaga (Centella) varieties from 1H nuclear magnetic resonance spectroscopy. Food Res Int. 2013;54:852–60. [Google Scholar]
- 32. Gogna N, Hamid N, Dorai K. Metabolomic profiling of the phytomedicinal constituents of Carica papaya L. leaves and seeds by 1H NMR spectroscopy and multivariate statistical analysis. J Pharmaceut Biomed Anal. 2015;115:74–85. doi: 10.1016/j.jpba.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 33. Zhang Q, Zhang J, Shen J, Silva A, Dennis D, Barrow C. A simple 96-well microplate method for estimation of total polyphenol content in seaweeds. J Appl Phycol. 2006;18:445–50. [Google Scholar]
- 34. Kinnersley AM, Turano FJ. Gamma aminobutyric acid (GABA) and plant responses to stress. Cr Rev Plant Sci. 2000;19:479–509. [Google Scholar]
- 35. Kishor PK, Sangam S, Amrutha RN, Laxmi PS, Naidu KR, Rao KRSS, Sreenivasulu N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci. 2005;88:424–38. [Google Scholar]