Abstract
Background
Platelet-derived growth factor receptor alpha (PDGFRA) is the second most frequently mutated tyrosine kinase receptor in glioblastoma (GBM). However, the prognostic impact of PDGFRA amplification on GBM patients remains unclear. Herein, we evaluated this impact by retrospectively analyzing outcomes of patients with IDH wild-type GBM.
Methods
Using a custom-made oncopanel, we evaluated PDGFRA gain/amplification in 107 GBM samples harboring wild-type IDH, along with MGMT promoter (MGMTp) methylation status.
Results
We detected PDGFRA gain/amplification in 31 samples (29.0%). PDGFRA gain/amplification predicted poor prognosis (P = .003). Compared to unamplified PDGFRA, PDGFRA gain/amplification in GBM was associated with higher patient age (P = .031), higher Ki-67 score (P = .019), and lower extent of surgical resection (P = .033). Unmethylated MGMTp also predicted poor prognosis (P = .005). As PDGFRA gain/amplification and unmethylated MGMTp were independent factors for poor prognosis in multivariate analyses, we grouped GBM cases based on PDGFRA and MGMTp status: poor (PDGFRA gain/amplification and unmethylated MGMTp), intermediate (PDGFRA gain/amplification or unmethylated MGMTp), and good (PDGFRA intact and methylated MGMTp) prognosis. The Kaplan-Meier survival analysis indicated that these groups significantly correlated with the OS of GBM patients (P < .001).
Conclusions
Here we report that PDGFRA gain/amplification is a predictor of poor prognosis in IDH wild-type GBM. Combining PDGFRA gain/amplification with MGMTp methylation status improves individual prognosis prediction in patients with IDH wild-type GBM.
Keywords: glioblastoma, IDH wild-type, MGMTp, PDGFRA gain/ amplification, prognostic markers
Key Points.
The median OS varies between GBM patients with and without PDGFRA gain/amplification.
PDGFRA and MGMTp statuses determine patient prognoses in GBM.
Importance of the Study.
Recently, it has been reported that IDH wild-type glioblastoma has different molecular subgroups that have distinct clinical features and prognoses. Although PDGFRA is the second most frequently mutated tyrosine kinase receptor in glioblastoma, the prognostic impact of its gain/amplification in glioblastoma patients remains unclear. Here, we demonstrated that PDGFRA gain/amplification is associated with poor prognosis in IDH wild-type glioblastoma. Moreover, using multivariate analysis, we determined that PDGFRA gain/amplification and MGMTp methylation status were independent prognostic markers. We hypothesized that these markers could improve the risk stratification of IDH wild-type glioblastoma. Additionally, we determined that glioblastomas could be subdivided into 3 groups based on the status of PDGFRA and MGMTp: poor (both PDGFRA gain/amplification and unmethylated MGMTp), intermediate (either PDGFRA gain/amplification or unmethylated MGMTp), and good (both PDGFRA intact and methylated MGMTp) prognosis groups. Such stratification will likely provide precise information to patients and can help influence their bedside decisions.
The gene encoding platelet-derived growth factor receptor alpha (PDGFRA) is present on chromosome 4q12. PDGFRA is the second most frequently mutated tyrosine kinase receptor-encoding gene, following EGFR, in glioblastoma (GBM)1,2 and plays an important role in oligodendrocyte differentiation.3 However, amplification of PDGFRA is associated with oligodendroglial morphology and malignancy.4 PDGFRA is a transmembrane receptor comprising 5 immunoglobulin-like extracellular domains and an intracellular tyrosine kinase domain. The PDGFR signaling pathway activates intracellular signaling pathways, such as the RAS/MAPK and PI3K/AKT pathways, that are involved in cell proliferation, migration, survival, and oncogenesis.5,6 Furthermore, the concept of PDGFRA as a possible drug target for GBM has been gaining attention. In fact, several PDGFRA-targeting antitumor agents, such as imatinib, sorafenib, nilotinib, and sunitinib, have already been developed.7 In the clinical scenario, pediatric GBM cases have a higher incidence of PDGFRA amplification than adult cases,8 and PDGFRA amplification is associated with the involvement of carpus callosum in GBM.9,10 In addition, it has been associated with poor patient survival in diffuse H3K27M-mutant midline gliomas.11 However, the prognostic value of PDGFRA amplification in GBM remains controversial, despite its relatively high frequency of occurrence in patients with GBM.
In this study, we aimed to identify the potential clinically distinct subgroups of IDH wild-type GBMs by examining the correlation between PDGFRA gain/amplification and patient survival.
Materials and Methods
Glioblastoma Samples
We obtained 107 tumor tissue samples from the Central Nervous System Tumor Tissue Bank of the Kagoshima University Hospital. These samples corresponded to 107 patients with GBM with a mean age of 67 years: 61 men (57.0%) and 46 women (43.0%). This study was approved by the Institutional Review Board of Kagoshima University (approval number: 180104) and complied with the principles of the Declaration of Helsinki. Informed consent was obtained from all patients. All 107 tumor samples were grade 4 IDH wild-type GBMs and were classified according to the 2021 World Health Organization classification of tumors of the central nervous system. We prepared formalin-fixed paraffin-embedded (FFPE) tumor samples by fixing the resected tumors with phosphate-buffered 10% formalin within 24 hours of sampling. Consecutively, we routinely processed them for paraffin embedding and sectioned them for hematoxylin and eosin staining. All the tissues were histologically evaluated by board-certified pathologists who verified that the estimated tumor cell content was above 30%.
Treatments
We performed gross or subtotal tumor removal on 56 patients (52.3%) and partial tumor removal (PTR) or biopsy on 51 patients (47.7%). Additionally, we treated 105 GBM patients with temozolomide during radiotherapy as per the Stupp protocol and also performed subsequent temozolomide maintenance treatments.12 However, 3 patients were not treated because of severe clinical conditions, such as advanced age or low Karnofsky Performance Status (KPS) scores.
DNA Extraction and Quantification
We extracted DNA from the FFPE samples using the Maxwell 16 FFPE Tissue LEV DNA Purification kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions and measured the concentration using the dsDNA BR Assay kit (Life Technologies, Carlsbad, CA, USA) in the Qubit 3.0 Fluorometer. We evaluated DNA quality by diluting the extracted DNA to 5-10 ng/µL and using it as a template for polymerase chain reaction (PCR). We conducted PCR using the QIAseq DNA QuantiMIZE kit (Qiagen, Hilden, Germany).
Next-generation Sequencing
We analyzed the extracted DNA by next-generation sequencing (NGS) using an amplicon-based glioma-tailored gene panel as described previously.13 Thereafter, we identified the copy number variations (CNVs) and single nucleotide polymorphisms, including those of genes PDGFRA, TERTp, CDKN2A/B, NF1, PTEN, RB1, TP53, and EGFR. In this regard, amplicon sequences were aligned to the human reference genome GRCh37 (hg19) at the target region. Data were analyzed using the Qiagen web portal service (https://www.qiagen.com/). Based on previous reports,14,15 we defined gain as 3-5 copies and amplification as >5 copies.
Multiplex Ligation-dependent Probe Amplification
CNVs were validated using a MLPA kit (P105-2; MRC-Holland, Amsterdam, the Netherlands) containing PDGFRA-specific probes, with 6 other probes used as control probes (http://www.mlpa.com), in accordance with the manufacturer’s protocol.16 Denatured fragments were separated and quantified by electrophoresis using an ABI 3730 capillary sequencer (Applied Biosystems, Nieuwerkerk aan den IJssel, the Netherlands) and analyzed using GeneMapper (Applied Biosystems) and Coffalyser software (MRC-Holland). Based on previous publications, the CNV category was classified using the following thresholds: gain (1.2 ≤ × < 2.0), amplification (× ≥ 2.0).17
Methylation-specific Polymerase Chain Reaction
We performed bisulfite modification of the extracted genomic DNA using the EpiTect Bisulfite Kit (Qiagen). After the conversion, genomic DNA was amplified for the target O6-methylguanine-DNA methyltransferase promoter (MGMTp) region with primers specific to the methylated or unmethylated template using KOD One PCR Master Mix (TOYOBO). For methylation-specific PCR (MSP), 2 pairs of primers specific for either the methylated or the unmethylated MGMTp region were used as previously described.16 The amplification was performed with an initial denaturation at 98°C for 1 minute and 40 cycles of 98°C for 10 seconds, 64°C for 5 seconds. Analysis was performed using the Shimadzu MCE-202 MultiNA (Shimadzu) on the DNA-1000 kit. The cutoff value for methylation was >4% as previously described.18
Analyses of the Memorial Sloan Kettering Cancer Center data
We retrieved the molecular characteristics of the GBM cohort from previous studies. Subsequently, we analyzed 456 cases from the Memorial Sloan Kettering Cancer Center (MSKCC) cohort, excluding the H3F3A, IDH1/2, and BRAF V600E-mutant cases.19 All cases were conclusively diagnosed as having IDH wild-type GBM using cBioPortal for Cancer Genomics (https://cbioportal.org).
Data Analysis
We visualized and analyzed our data using the cBioPortal for Cancer Genomics tools OncoPrinter (cbioportal.org/oncoprinter).20,21 Additionally, we analyzed the data using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), a graphical user interface of the R software (The R Foundation for Statistical Computing, Vienna, Austria). We compared the risk groups and the patient characteristics using the chi-square (χ 2) test and the Kaplan-Meier log-rank test, respectively. We also performed univariate and multivariate Cox regression analyses. A difference was considered statistically significant at P < .05.
Results
Frequency of PDGFRA Gain/Amplification and MGMTp Methylation Status in IDH Wild-type GBM
A total of 107 cases of IDH wild-type GBM were examined in this study. The median overall survival (OS) was 19.7 months. Using NGS, PDGFRA copy number was assessed in all cases; it was between 1 and 3 in 76 cases (71.0%), between 3 and 5 in 11 cases (10.3%), and >5 in 20 cases (18.7%). We determined 3-5 copies as PDGFRA gain, and >5 copies as PDGFRA amplification. To validate PDGFRA gene gain/amplification performed by our NGS panel, we conducted MLPA on 15 selective GBM cases, comprising 3 defined by NGS as showing PDGFRA gain (copy number range: 3-5), 5 defined by NGS as showing PDGFRA amplification (copy number range >5), and 7 defined by NGS as lacking PDGFRA gain/amplification (copy number range: 1-3). The CNVs of our NGS analysis were consistent with those of the MLPA analysis except for 1 case (Supplementary Table 1). Moreover, to determine the cutoff value of PDGFRA CNVs corresponding to the minimal P values of log-rank test for OS, we investigated and plotted the P values depending on the various PDGFRA copy numbers (range 1-10 copies) as threshold point (Supplementary Figure 1). We determined >3 PDGFRA copies as the optimal cutoff value because P values get minimal when threshold copy number is 3. Consequently, we detected PDGFRA gain in 11 tumor samples (10.3%) and PDGFRA amplification in 20 tumor samples (18.7%); the remaining 76 samples (71.0%) harbored no PDGFRA gain/amplification.
MGMTp methylation analysis using MSP was assessed in all cases, including 12 cases as demonstrated in Supplementary Figure 2. Consequently, we detected MGMTp methylation in 58 tumor samples (54.2%); in the remaining 49 samples (45.8%), MGMTp methylation was not observed.
Genetic and Clinical Factors Influencing Prognosis
First, we analyzed whether the identified genetic markers were prognostic markers. Notably, PDGFRA gain/amplification and unmethylated MGMTp were significant predictors of poor prognosis, as determined by our univariate [HR: 2.22 (1.30-3.78), P = .003; and HR: 2.10 (1.24-3.57), P = .006, respectively] and multivariate analyses [HR: 2.52 (1.34-4.76), P = .004; and HR: 2.28 (1.28-4.07), P = .005, respectively; Table 1].
Table 1.
Genetic Prognostic Factors
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Genetic Marker | HR (95% CI) | P value | HR (95% CI) | P value |
| CDKN2A/B homdel | 1.24 (0.74-2.10) | .417 | 1.26 (0.69-2.37) | .442 |
| EGFR amp | 0.82 (0.48-1.40) | .462 | 0.89 (0.46-1.70) | .721 |
| PTEN loss and/or mut | 0.72 (0.39-1.33) | .294 | 0.58 (0.27-1.24) | .158 |
| TP53 loss and/or mut | 1.08 (0.64-1.82) | .782 | 1.04 (0.59-1.82) | .902 |
| TERTp mut | 1.16 (0.67-2.00) | .599 | 1.86 (0.87-3.96) | .110 |
| Unmethylated MGMTp | 2.10 (1.24-3.57) | .006* | 2.28 (1.28-4.07) | .005* |
| PDGFRA gain/amp | 2.22 (1.30-3.78) | .003* | 2.52 (1.34-4.76) | .004* |
amp, amplification; homdel, homozygous deletion; mut, mutation.
The symbol * indicates statistical significance.
Second, we identified the clinical prognostic factors, which included analysis of the genetic markers for PDGFRA gain/amplification and unmethylated MGMTp. Our univariate analysis revealed that age [HR: 2.53 (1.47-4.33), P < .001], PTR/biopsy [HR: 2.05 (1.20-3.48), P = .008], unmethylated MGMTp [HR: 2.10 (1.24-3.57), P = .006], and PDGFRA gain/amplification [HR: 2.22 (1.30-3.78), P = .003] were significantly associated with poor prognosis (Table 2). Thereafter, we adjusted the covariates, including sex and age and KPS score and the extent of tumor resection, in the multivariate Cox proportional hazards model. This analysis corroborated the finding that PDGFRA gain/amplification and unmethylated MGMTp were independent prognostic markers of OS in patients with IDH wild-type GBM [HR: 1.82 (1.00-3.31), P = .049; and HR: 3.00 (1.67-5.39), P < .001, respectively; Table 2].
Table 2.
Clinical and Genetic Prognostic Factors
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Prognostic Factor | HR (95% CI) | P value | HR (95% CI) | P value |
| Sex (male) | 1.60 (0.93-2.75) | .093 | 1.60 (0.91-2.83) | .104 |
| Age (>70 years) | 2.53 (1.47-4.33) | <.001* | 2.72 (1.51-4.92) | <.001* |
| KPS score (≤80 points) | 1.46 (0.81-2.64) | .210 | 1.76 (0.92-3.37) | .088 |
| PTR/biopsy | 2.05 (1.20-3.48) | .008* | 1.85 (1.03-3.32) | .041* |
| Unmethylated MGMTp | 2.10 (1.24-3.57) | .006* | 3.00 (1.67-5.39) | <.001* |
| PDGFRA gain/amp | 2.22 (1.30-3.78) | .003* | 1.82 (1.00-3.31) | .049* |
KPS, Karnofsky Performance Status; PTR, partial tumor removal.
The symbol * indicates statistical significance.
Genetic and Clinical Factors Associated With and Without PDGFRA Gain/Amplification
Table 3 compares the genetic and clinical factors of the patients based on their PDGFRA status. We discovered that the TP53 mutation was more common in GBMs with PDGFRA gain/amplification than in those without (P = .011). Conversely, alterations in EGFR (P < .001), and TERTp (P < .001) were more common in GBMs without PDGFRA gain/amplification than in those with the gain/amplification (Supplementary Figure 3). In our study, no case had both PDGFRA amplification and EGFR amplification (Supplementary Figure 3). Furthermore, patients with GBM with PDGFRA gain/amplification were associated with higher age (P = .031), higher Ki-67 score (P = .019), and lower extent of surgical resection (P = .033) than those without PDGFRA gain/amplification.
Table 3.
Background of Patients With and Without PDGFRA Gain/Amplification
| Prognostic Factor | All (n = 107) | PDGFRA Gain/Amp (n = 31) | PDGFRA Intact (n = 76) | P value | |
|---|---|---|---|---|---|
| Sex | Male | 61 (57.0%) | 19 (61.3%) | 42 (55.3%) | .668 |
| Female | 46 (43.0%) | 12 (38.7%) | 34 (44.7%) | ||
| Age | <70 years | 60 (56.1%) | 12 (38.7%) | 48 (63.2%) | .031* |
| >70 years | 47 (43.9%) | 19 (61.3%) | 28 (36.8%) | ||
| KPS score | >80 points | 31 (29.0%) | 10 (32.3%) | 21 (27.6%) | .645 |
| ≤80 points | 76 (71.0%) | 21 (67.7%) | 55 (72.4%) | ||
| Resection | GTR/STR | 56 (52.3%) | 11 (35.5%) | 45 (59.2%) | .033* |
| PTR/biopsy | 51 (47.7%) | 20 (64.5%) | 31 (40.8%) | ||
| Ki-67 | >35% | 56 (52.3%) | 22 (71.0%) | 34 (44.7%) | .019* |
| <35% | 51 (47.7%) | 9 (29.0%) | 42 (55.3%) | ||
| CDKN2A/B homdel | 51 (47.7%) | 16 (51.6%) | 35 (46.1%) | .672 | |
| NF1 loss and/or mut | 23 (21.5%) | 4 (12.9%) | 19 (25.0%) | .202 | |
| PTEN loss and/or mut | 73 (68.2%) | 17 (54.8%) | 56 (73.7%) | .069 | |
| RB1 loss and/or mut | 39 (36.4%) | 10 (32.3%) | 29 (38.2%) | .660 | |
| TERTp mut | 70 (65.4%) | 12 (38.7%) | 58 (76.3%) | <.001* | |
| TP53 loss and/or mut | 52 (48.6%) | 21 (67.7%) | 31 (40.8%) | .011* | |
| EGFR amp | 43 (40.2%) | 2 (6.5%) | 41 (53.9%) | <.001* | |
| Unmethylated MGMTp | 49 (45.8%) | 13 (41.9%) | 36 (47.4%) | .672 |
amp, amplification; GTR, gross tumor removal; homdel, homozygous deletion; KPS, Karnofsky Performance Status; mut, mutation; PTR, partial tumor removal; STR, subtotal tumor removal.
The symbol * indicates statistical significance.
PDGFRA Gain/Amplification and Unmethylated MGMTp Are Associated With Poor Patient Prognoses
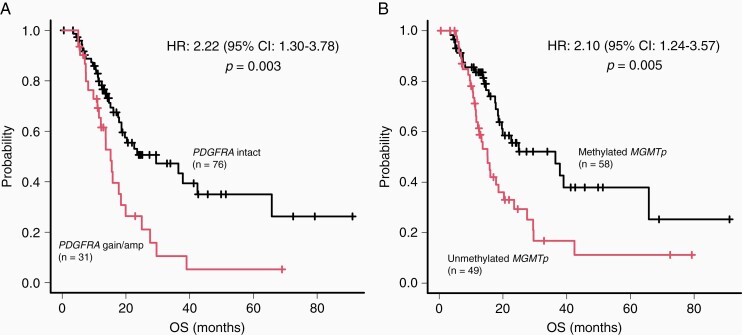
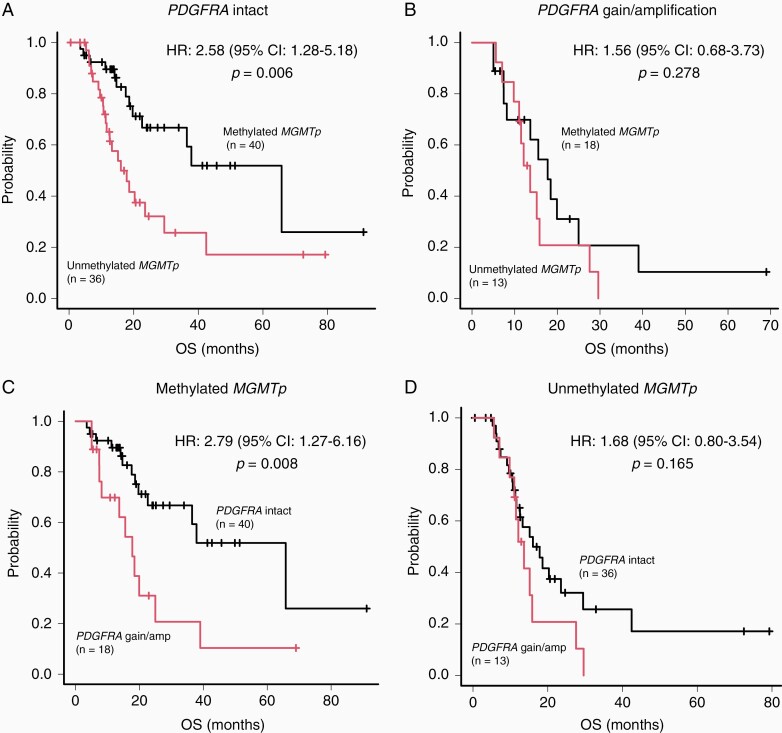
We observed a significant difference in the median OS of patients with and without PDGFRA gain/amplification (15.2 and 29.5 months, respectively; P = .003; Figure 1A). Subsequently, we validated this observation by analyzing the data of 456 patients with IDH wild-type GBM obtained from the MSKCC cohort. We observed a significant difference in the median OS of the patients with and without PDGFRA amplification (16.6 and 23.5 months, respectively; P = .017; Supplementary Figure 4). Moreover, unmethylated MGMTp was a significant predictor of poor prognosis (P = .005; Figure 1B). In patients with PDGFRA intact, unmethylated MGMTp was a significant predictor of poor prognosis (P = .006, Figure 2A), whereas in patients with PDGFRA gain/amplification, there was no significant difference between the median OS of patients with methylated MGMTp and those with unmethylated MGMTp (P = .278, Figure 2B). In addition, in patients with methylated MGMTp, PDGFRA gain/amplification was a significant predictor of poor prognosis (P = .008, Figure 2C), whereas in patients with unmethylated MGMTp, there was no significant difference between the median OS of patients with and without PDGFRA gain/amplification (P = .165, Figure 2D).
Figure 1.
Prognostic impact of PDGFRA and MGMTp status. (A) IDH wild-type glioblastoma (GBM) cases with PDGFRA gain/amplification exhibiting significantly shorter overall survival (OS) than those without PDGFRA gain/amplification. (B) IDH wild-type GBM cases with unmethylated MGMTp exhibiting significantly shorter OS than those with methylated MGMTp.
Figure 2.
Survival analysis of IDH wild-type glioblastoma (GBM) cases according to PDGFRA and MGMTp status. (A) Survival analysis of patients with IDH wild-type GBM harboring either unmethylated or methylated MGMTp without PDGFRA gain/amplification. (B) Survival analysis of patients with IDH wild-type GBM harboring either unmethylated or methylated MGMTp with PDGFRA gain/amplification. (C) Kaplan-Meier analysis for OS of patients with and without PDGFRA gain/amplification in IDH wild-type GBMs with methylated MGMTp. (D) Kaplan-Meier survival analysis of patients with and without PDGFRA gain/amplification in IDH wild-type GBMs with unmethylated MGMTp.
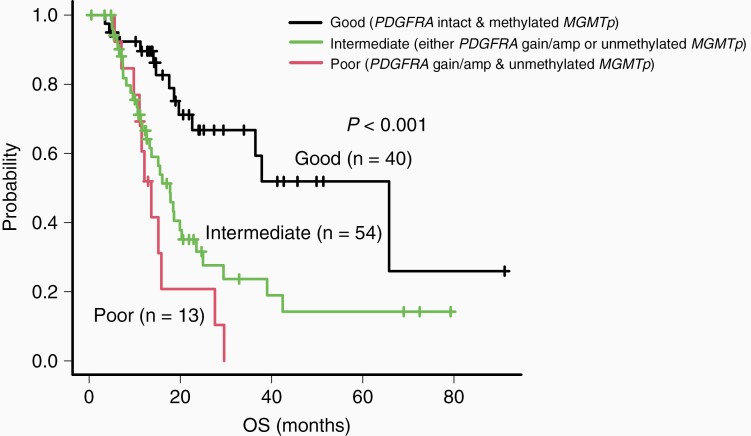
Prognostic Impact of the Combination of PDGFRA Gain/Amplification and MGMTp Methylation Status in IDH Wild-type GBM
We aimed to perform a risk stratification of IDH wild-type GBM. PDGFRA gain/amplification and unmethylated MGMTp were associated with clinical outcomes of the patients and were independent prognostic factors in the multivariate analysis. Therefore, we included these 2 molecular markers for the subsequent risk stratification. We subdivided IDH wild-type GBM into 3 groups according to the status of PDGFRA and MGMTp (Figure 3). IDH wild-type patients with GBM with PDGFRA gain/amplification and unmethylated MGMTp were assigned to the poor-prognosis group, whereas those harboring either PDGFRA gain/amplification or unmethylated MGMTp were assigned to the intermediate-prognosis group. Lastly, patients with PDGFRA intact and methylated MGMTp were assigned to the good-prognosis group. Remarkably, our Kaplan-Meier survival analysis indicated that these groups were correlated with the OS of the patients (P < .001; Figure 3).
Figure 3.
Risk stratification of IDH wild-type glioblastoma cases based on PDGFRA and MGMTp mutational status. The Kaplan-Meier overall survival curves as per risk stratification. The poor-, intermediate-, and good-prognosis groups include patients with PDGFRA gain/amplification and unmethylated MGMTp, with either PDGFRA gain/amplification or unmethylated MGMTp, and with PDGFRA intact and methylated MGMTp, respectively.
However, these groups were not associated with any clinical factors, including sex, age, the extent of surgical resection, KPS score, Ki-67 score, and other genetic factors, except for EGFR amplification (Supplementary Table 2).
Discussion
In this study, we demonstrated the impact of PDGFRA gain/amplification as a prognostic marker of IDH wild-type GBM, along with the MGMTp methylation status. Studies have detected PDGFRA amplification in approximately 8.5%-29% of GBM cases9,22–27; our results corroborated these observations. We demonstrated that PDGFRA gain/amplification is a significant predictor of poor prognoses in patients with IDH wild-type GBM; this was validated using the MSKCC dataset. Consistent with previous reports,28–30 we also identified the age and extent of surgical resection and unmethylated MGMTp as independent GBM prognostic factors, highlighting the accuracy of our study. PDGFRA gain/amplification and unmethylated MGMTp were identified as independent prognostic markers in multivariate analysis; however, the hazard ratio and P value for PDGFRA gain/amplification became less significant than those for unmethylated MGMTp when clinical factors were included. This could be attributed to the fact that PDGFRA gain/amplification is associated with higher patient age and lower extent of resection. Therefore, PDGFRA gain/amplification as a prognostic factor is confounded by age and extent of resection. To date, the prognostic value of PDGFRA gain/amplification in GBM remains controversial. While some previous studies have reported no prognostic impact of PDGFRA amplification in GBM,23,26,27,31 one study has reported poor prognostic impact of this amplification in GBM.32 These differences might be attributed to the different methods used to detect gene amplification, or intratumor heterogeneity, a notable feature of GBM.33 Studies have demonstrated that NGS panel-based identification of CNVs is more sensitive than conventional methods, including multiplex ligation-dependent probe amplification (MLPA) and PCR.34 The MLPA method renders false-negative results because of contamination by unamplified non-neoplastic and neoplastic DNA caused by intratumoral heterogeneity.35 To solve these problems, we histologically evaluated all the tissue samples by board-certified pathologists and measured the estimated tumor cell content. Following this, we extracted the tumor cell DNA, highlighting highly reliable data. In contrast, studies have linked PDGFRA amplification with the significantly worse OS of patients with IDH-mutant GBM8 and WHO grade II and III tumors,36,37 which is different from the results of the patient group analyzed in this study. Moreover, Alnahhas et al reported that PDGFRA amplification was associated with poor survival only in EGFR/ERBB-altered GBM.38 However, in our study, no case had both PDGFRA amplification and EGFR amplification, suggesting that our patient background was different from that of the patient group analyzed in their study.
Cui et al reported that PDGFRA alterations are associated with the involvement of the corpus callosum, resulting in the low extent of surgical resection values.9 Therefore, we assume that PDGFRA amplification may be associated with poor prognosis due to the low extent of surgical resection. In our study, IDH wild-type GBM with PDGFRA gain/amplification was significantly associated with older age, consistent with previous reports. Moreover, it was significantly associated with a higher Ki-67 score. Previous studies have shown that the Ki-67 score is an important prognostic factor in GBM and a marker of cell proliferation.39–41 Thus, our results demonstrated that PDGFRA gain/amplification causes poor prognoses in patients with IDH wild-type GBM by increasing the proliferative ability of tumors and increasing the rate of incomplete resection of tumors.
MGMTp methylation is a well-established favorable prognostic marker for survival and predicts the response to temozolomide in patients with GBM.28,42 Since PDGFRA gain/amplification and unmethylated MGMTp were independent prognostic markers, we investigated the potential interaction between PDGFRA gain/amplification and unmethylated MGMTp in patients with GBM and hypothesized that these markers improved the risk stratification of IDH wild-type GBM. Consequently, our finding that the subset of GBMs with PDGFRA gain/amplification and unmethylated MGMTp have the poorest prognosis and GBMs with PDGFRA intact and methylated MGMTp have the most favorable prognosis has important clinical implications. We demonstrated that such stratification, surprisingly, is independent of clinical factors, including age, sex, the extent of resection, KPS score, and other genetic factors except for EGFR amplification. Our most striking finding was that the prognostic impact of PDGFRA gain/amplification is one of the most powerful predictors of survival in patients with GBM, along with the MGMTp methylation status. Furthermore, PDGFRA gain/amplification in combination with the MGMTp methylation status improves individual prognosis in patients with IDH wild-type GBM.
This study has several limitations. First, it was a retrospective study with a small sample size. Second, differences in molecular biology techniques should be considered. We identified CNVs using NGS, whereas other studies have used MLPA, PCR, or fluorescent in situ hybridization for this purpose.22,23,25–27,36
Conclusions
We report that PDGFRA gain/amplification is a predictor of poor prognosis in IDH wild-type GBM. Our study illustrates the potential use of molecular markers for a refined stratification of IDH wild-type GBM. We recommend the incorporation of PDGFRA gain/amplification and MGMTp in the molecular stratification of IDH wild-type GBM. Such a stratification will likely provide precise information to patients and help influence their bedside decisions.
Supplementary Material
Contributor Information
Nayuta Higa, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Toshiaki Akahane, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan; Center for Human Genome and Gene Analysis, Kagoshima University Hospital, Kagoshima-City, Kagoshima, Japan.
Seiya Yokoyama, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Hajime Yonezawa, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Hiroyuki Uchida, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Tomoko Takajo, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Ryosuke Otsuji, Department of Neurosurgery, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Taiji Hamada, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Kei Matsuo, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Mari Kirishima, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Nobuhiro Hata, Department of Neurosurgery, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Ryosuke Hanaya, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan.
Akihide Tanimoto, Department of Pathology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan; Center for Human Genome and Gene Analysis, Kagoshima University Hospital, Kagoshima-City, Kagoshima, Japan.
Koji Yoshimoto, Department of Neurosurgery, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima-City, Kagoshima, Japan; Department of Neurosurgery, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Funding
No funding was received for this study.
Ethical approval and consent to participate
The study was approved by the Institutional Review Board of Kagoshima University (approval number: 180104) and complied with the Helsinki Declaration. Informed consent was obtained from all patients.
Conflict of interest statement. The authors declare that they have no competing interests.
Authorship statement. Conception and design: N.H., A.T., and K.Y. Development of methodology: T.A., T.H., S.Y., R.O., and T.T. Acquisition of data (eg, acquired and managed patients and provided facilities): N.H., H.Y., H.U., and M.K. Analysis and interpretation of data (eg, statistical analysis, biostatistics, and computational analysis): N.H., T.A., T.H., and S.Y. Manuscript writing, review, and/or revision: N.H., A.T., and K.Y. Administrative, technical, or material support (eg, reporting or organizing data and constructing databases): N.H., T.T., H.Y., H.U., T.A., T.H., K.M., and S.Y. Study supervision: R.H., N.H., A.T., and K.Y.
Data Availability
All data used and analyzed in the current study are available from the corresponding author upon reasonable request.
References
- 1. Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brennan CW, Verhaak RGW, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richardson WD, Pringle N, Mosley MJ, et al. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53(2):309–319. [DOI] [PubMed] [Google Scholar]
- 4. Smith JS, Wang XY, Qian J, et al. Amplification of the platelet-derived growth factor receptor-A (PDGFRA) gene occurs in oligodendrogliomas with grade IV anaplastic features. J Neuropathol Exp Neurol. 2000;59(6):495–503. [DOI] [PubMed] [Google Scholar]
- 5. Nazarenko I, Hede SM, He X, et al. PDGF and PDGF receptors in glioma. Ups J Med Sci. 2012;117(2):99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. [DOI] [PubMed] [Google Scholar]
- 7. Roskoski R Jr. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;129:65–83. [DOI] [PubMed] [Google Scholar]
- 8. Phillips JJ, Aranda D, Ellison DW, et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23(5):565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui M, Gao X, Chi Y, et al. Molecular alterations and their correlation with the survival of glioblastoma patients with corpus callosum involvement. Front Neurosci. 2021;15:701426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen S, Feng S, Liu H, et al. Associations of histological and molecular alterations with invasion of the corpus callosum in gliomas. Acta Neurochir. 2020;162(7):1691–1699. [DOI] [PubMed] [Google Scholar]
- 11. Dufour C, Perbet R, Leblond P, et al. Identification of prognostic markers in diffuse midline gliomas H3K27M-mutant. Brain Pathol. 2020;30(1):179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 13. Higa N, Akahane T, Yokoyama S, et al. A tailored next-generation sequencing panel identified distinct subtypes of wildtype IDH and tert promoter glioblastomas. Cancer Sci. 2020;111(10):3902–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McNulty SN, Cottrell CE, Vigh-Conrad KA, et al. Beyond sequence variation: assessment of copy number variation in adult glioblastoma through targeted tumor somatic profiling. Hum Pathol. 2019;86:170–181. [DOI] [PubMed] [Google Scholar]
- 16. Jeuken J, Cornelissen S, Boots-Sprenger S, et al. Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn. 2006;8(4):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeuken J, Sijben A, Alenda C, et al. Robust detection of EGFR copy number changes and EGFR variant Ⅲ: technical aspects and relevance for glioma diagnostics. Brain Pathol. 2009;19(4):661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adachi J, Mishima K, Wakiya K, et al. O 6-methylguanine-DNA methyltransferase promoter methylation in 45 primary central nervous system lymphomas: quantitative assessment of methylation and response to temozolomide treatment. J Neurooncol. 2012;107(1):147–153. [DOI] [PubMed] [Google Scholar]
- 19. Jonsson P, Lin AL, Young RJ, et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res. 2019;25(18):5537–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funakoshi Y, Hata N, Takigawa K, et al. Clinical significance of CDKN2A homozygous deletion in combination with methylated MGMT status for IDH-wildtype glioblastoma. Cancer Med. 2021;10(10):3177–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joensuu H, Puputti M, Sihto H, et al. Amplification of genes encoding KIT, PDGFRα and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol. 2005;207(2):224–231. [DOI] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinho O, Longatto-Filho A, Lambros MBK, et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. Br J Cancer. 2009;101(6):973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nobusawa S, Stawski R, Kim YH, et al. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: a population-based study. Neuropathology. 2011;31(6):583–588. [DOI] [PubMed] [Google Scholar]
- 27. González-Tablas M, Arandia D, Jara-Acevedo M, et al. Heterogeneous EGFR, CDK4, MDM4, and PDGFRA gene expression profiles in primary GBM: no association with patient survival. Cancers. 2020;12(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Binabaj MM, Bahrami A, ShahidSales S, et al. The prognostic value of MGMT promoter methylation in glioblastoma: a meta-analysis of clinical trials. J Cell Physiol. 2018;233(1):378–386. [DOI] [PubMed] [Google Scholar]
- 29. Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol. 2008;10(1):79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alentorn A, Marie Y, Carpentier C, et al. Prevalence, clinico-pathological value, and co-occurrence of PDGFRA abnormalities in diffuse gliomas. Neuro Oncol. 2012;14(11):1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burford A, Little SE, Jury A, et al. Distinct phenotypic differences associated with differential amplification of receptor tyrosine kinase genes at 4q12 in glioblastoma. PLoS One. 2013;8(8):e71777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sottoriva A, Spiteri I, Piccirillo SGM, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kerkhof J, Schenkel LC, Reilly J, et al. Clinical validation of copy number variant detection from targeted next-generation sequencing panels. J Mol Diagn. 2017;19(6):905–920. [DOI] [PubMed] [Google Scholar]
- 35. Minegishi K, Dobashi Y, Tsubochi H, et al. Screening of the copy number increase of AKT in lung carcinoma by custom-designed MLPA. Int J Clin Exp Pathol. 2019;12(9):3344–3356. [PMC free article] [PubMed] [Google Scholar]
- 36. Fujimoto K, Arita H, Satomi K, et al. Tert promoter mutation status is necessary and sufficient to diagnose IDH-wildtype diffuse astrocytic glioma with molecular features of glioblastoma. Acta Neuropathol. 2021;142(2):323–338. [DOI] [PubMed] [Google Scholar]
- 37. Yang RR, Shi ZF, Zhang ZY, et al. IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2020;30(3):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alnahhas I, Rayi A, Guillermo Prieto Eibl MDP, Ong S, Giglio P, Puduvalli V. Prognostic implications of epidermal and platelet-derived growth factor receptor alterations in 2 cohorts of IDHwt glioblastoma. Neurooncol Adv. 2021;3(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reavey-Cantwell JF, Haroun RI, Zahurak M, et al. The prognostic value of tumor markers in patients with glioblastoma multiforme: analysis of 32 patients and review of the literature. J Neurooncol. 2001;55(3):195–204. [DOI] [PubMed] [Google Scholar]
- 40. Scott RJ, Hall PA, Haldane JS, et al. A comparison of immunohistochemical markers of cell proliferation with experimentally determined growth fraction. J Pathol. 1991;165(2):173–178. [DOI] [PubMed] [Google Scholar]
- 41. Yoshida Y, Nakada M, Harada T, et al. The expression level of sphingosine-1-phosphate receptor type 1 is related to MIB-1 labeling index and predicts survival of glioblastoma patients. J Neurooncol. 2010;98(1):41–47. [DOI] [PubMed] [Google Scholar]
- 42. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used and analyzed in the current study are available from the corresponding author upon reasonable request.