Abstract
Immune checkpoint inhibitors are effective for advanced hepatocellular carcinoma (HCC), but there remains a need for peripheral blood biomarkers to predict the clinical response. Here, we analyzed the peripheral blood of 45 patients with advanced HCC who underwent nivolumab. During treatment, frequency of classical monocytes (CD14+CD16−) was increased on day 7, and the fold increase in the frequency on day 7 over day 0 (cMonocyteD7/D0) was significantly higher in patients with durable clinical benefit (DCB) than in patients with non-DCB (NDB). When we analyzed transcriptomes of classical monocytes, CD274, gene encoding PD-L1, was upregulated in NDB patients compared to DCB patients at day 7. Notably, gene signature of suppressive tumor-associated macrophages, or IL4l1+PD-L1+IDO1+ macrophages, was enriched after treatment in NDB patients, but not in DCB patients. Accordingly, the fold increase in the frequency of PD-L1+ classical monocytes at day 7 over day 0 (cMonocyte-PDL1D7/D0) was higher in NDB patients than DCB patients. The combined biomarker cMonocyteD7/D0/cMonocyte-PDL1D7/D0 was termed the “monocyte index”, which was significantly higher in DCB patients than NDB patients. Moreover, the monocyte index was an independent prognostic factor for survival. Overall, our results suggest that early changes of circulating classical monocytes, represented as a monocyte index, could predict clinical outcomes of advanced HCC patients undergoing anti-PD-1 therapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03258-6.
Keywords: Hepatocellular carcinoma, Anti-PD-1 therapy, Nivolumab, Biomarker, Classical monocyte
Introduction
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths worldwide [1], and the majority of HCC patients are diagnosed at a late stage that cannot be cured with locoregional therapies. Since immune checkpoint inhibitor (ICI) treatment has showed clinical success in various types of cancer, several clinical trials have evaluated the efficacy of anti-programmed cell death protein 1 (PD-1) blockade, with either nivolumab or pembrolizumab, in advanced HCC. However, the results consistently show that less than 20% of HCC patients exhibit a clinical response to anti-PD-1 therapy [2–6]. Thus, there is an urgent need to find biomarkers that can identify patients who will benefit from anti-PD-1 therapy.
In clinical trials and clinical practice with various cancers, the most common biomarkers used for ICI treatment are parameters assessed in the tumor specimen, such as tumor mutational burden, programmed death-ligand 1 (PD-L1) expression, and features of tumor-infiltrating lymphocytes [7]. Among patients with advanced HCC, a higher combined positive PD-L1 score, CD8+ T-cell infiltration, PD-1 expression in tumor, and T-cell exhaustion gene signature are reportedly associated with better outcomes after ICI treatment [8–10]. Additionally, a high density of CD38+ macrophages is correlated with prolonged survival of patients with advanced HCC following ICI treatment [11]. However, HCC diagnosis does not require pathologic confirmation and liver tumor biopsy is not available in some cases, prohibiting assessment of tumor specimen-derived biomarkers. Therefore, there is a need for biomarkers from peripheral blood, especially for HCC.
Few studies have investigated peripheral blood biomarkers in advanced HCC patients receiving ICIs. Analysis of the participants in the CheckMate 040 phase I/II trial revealed that the neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio were associated with survival outcomes of HCC patients after nivolumab treatment [8]. Additionally, higher baseline TGF-β concentration is reportedly correlated with poorer survival after pembrolizumab treatment [12]. A recent study shows that the combination of baseline levels of C-reactive protein and alpha-fetoprotein is associated with clinical response and survival [13]. However, it remains unknown whether these biomarkers are involved in the immune response evoked by anti-PD-1 therapy, or merely reflect the baseline tumor characteristics.
In the present study, we hypothesized that HCC treatment using anti-PD-1 blockade would induce immunological changes that could be detected in peripheral blood collected early after the therapy. To investigate this possibility, we screened various peripheral blood biomarkers from prospectively accrued patients with advanced HCC undergoing nivolumab treatment. Our results suggest that early changes in the circulating classical monocyte population were associated with clinical outcomes in these patients, and thus may serve as useful biomarkers for predicting the clinical benefit of ICI treatment in patients with advanced HCC.
Materials and methods
Study cohort and sample collection
The prospective cohort for this study comprised 48 patients with advanced HCC who were treated with nivolumab as a second or later line of treatment (clinical trial number: NCT03695952). The patients were treated with nivolumab every two weeks until tumor progression was observed. Peripheral blood samples were collected at baseline (day 0, D0) and at one or more time points after starting treatment, including cycle 1 day 8 (day 7, D7), and cycle 2 day 1 (day 14, D14) (Supplementary Table 1). Plasma was obtained by centrifugation of whole blood. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation with standard Ficoll-Paque (GE Healthcare). Plasma samples were stored at − 70 °C. PBMCs were resuspended in freezing media (RPMI 1640; Corning) supplemented with 20% fetal bovine serum (FBS) and 10% DMSO and then stored in liquid nitrogen until use. Among the participants, three patients had received immunotherapy prior to enrollment and were excluded from the analysis. Details regarding the 45 patients included in the analysis are presented in Table 1. The study protocol was approved by the Institutional Review Board of Asan Medical Center (IRB approval number 2019-1231).
Table 1.
Characteristics of the 45 patients included in the study
| Patient characteristics | Clinical benefit | P value | |
|---|---|---|---|
| DCB (n = 13) | NDB (n = 32) | ||
| Age, years (median, (range)) | 59 (50–73) | 60 (36–76) | 0.587 |
| Sex, n (%) | 0.636 | ||
| Male | 12 (92.3%) | 26 (81.2%) | |
| Female | 1 (7.7%) | 6 (18.8%) | |
| ECOG performance status, n (%) | 0.306 | ||
| 0 | 4 (30.8%) | 4 (12.5%) | |
| 1–2 | 9 (69.2%) | 28 (87.5%) | |
| Child–Pugh classification, n (%) | 0.853 | ||
| A | 10 (76.9%) | 22 (68.8%) | |
| B | 3 (23.1%) | 10 (31.2%) | |
| Etiology, n (%) | 0.955 | ||
| Hepatitis B | 10 (76.9%) | 24 (75.0%) | |
| Hepatitis C | 1 (7.7%) | 2 (6.2%) | |
| Non-B, non-C | 2 (15.4%) | 6 (18.8%) | |
| Major vessel invasion, n (%) | 0.709 | ||
| Absent | 5 (38.5%) | 16 (50.0%) | |
| Present | 8 (61.5%) | 16 (50.0%) | |
| Metastasis, n (%) | 1.000 | ||
| Any | 13 (100.0%) | 31 (96.9%) | |
| Liver | 7 (53.8%) | 26 (81.2%) | |
| Lung | 19 (59.4%) | 8 (61.5%) | |
| Bone | 11 (84.6%) | 21 (65.6%) | |
| Lymph node | 5 (38.5%) | 13 (40.6%) | |
| Others | 6 (46.2%) | 11 (34.4%) | |
| Number of metastasis, n (%) | 0.636 | ||
| 1–3 | 12 (92.3%) | 26 (81.2%) | |
| 4 ≤ | 1 (7.7%) | 6 (18.8%) | |
| Previous locoregional therapy, n (%) | 0.636 | ||
| Any | 12 (92.3%) | 26 (81.2%) | |
| Hepatectomy | 9 (69.2%) | 10 (31.2%) | |
| Transarterial chemoembolization | 21 (65.6%) | 8 (61.5%) | |
| Radiofrequency ablation | 3 (23.1%) | 4 (12.5%) | |
| Radiotherapy | 9 (69.2%) | 19 (59.4%) | |
| Number of lines of previous systemic therapy, n (%) | 1.000 | ||
| 1 | 8 (61.5%) | 19 (59.4%) | |
| 2–3 | 5 (38.5%) | 13 (40.6%) | |
| Best response to sorafenib, n (%) | 0.370 | ||
| Non-progressive disease | 8 (61.5%) | 14 (43.8%) | |
| Progressive disease | 5 (38.5%) | 16 (50.0%) | |
| Not evaluable | 0 (0.0%) | 2 (6.2%) | |
| Best response to nivolumab, n (%) | < 0.001 | ||
| Complete remission | 1 (7.7%) | 0 (0.0%) | |
| Partial remission | 8 (61.5%) | 0 (0.0%) | |
| Stable disease | 4 (30.8%) | 3 (9.4%) | |
| Progressive disease | 0 (0.0%) | 26 (81.2%) | |
| Not evaluable | 0 (0.0%) | 3 (9.4%) | |
DCB durable clinical benefit, NDB non-durable clinical benefit, ECOG Eastern Cooperative Oncology Group
Evaluation of treatment outcomes
Physical examination and laboratory tests were performed every 2 weeks. Tumor response was assessed by computed tomography every 8 weeks, according to the Response Evaluation Criteria In Solid Tumors (RECIST), version 1.1. Cases with complete response (CR), partial response (PR), or stable disease (SD) lasting over 6 months were defined as showing durable clinical benefit (DCB), and otherwise as non-durable clinical benefit (NDB). Progression-free survival (PFS) was defined as the time from the start of anti-PD-1 therapy to either disease progression (according to RECIST v1.1) or death from any cause. Overall survival (OS) was defined as the time from the start of anti-PD-1 therapy to death from any cause.
Multi-color flow cytometry
After thawing, PBMCs were stained using LIVE/DEAD fixable dead cell stain kit (Invitrogen) or 7-AAD (BD Biosciences) to gate out dead cells. The cells were washed and then incubated with FcR blocking agent (Miltenyi Biotec) for 10 min at 4 °C, followed by staining with fluorochrome-conjugated antibodies against surface markers for 30 min at 4 °C. For intracellular staining, cells were fixed and permeabilized using the FoxP3 staining buffer kit (Thermo Fisher Scientific), and then stained with fluorochrome-conjugated antibodies. All staining procedures were conducted in staining buffer, phosphate-buffered saline supplemented with 1% FBS and 0.05% sodium azide. Supplementary Table 2 lists the fluorochrome-conjugated antibodies used for flow cytometry. All flow cytometric analyses were performed using an LSR II instrument (BD Biosciences), and the data were analyzed with FlowJo software (Treestar).
RNA sequencing and analysis
RNA sequencing (RNA-seq) was performed on classical monocytes (CD11b+CD3−CD19−CD14+CD16− cells) that were sorted from PBMCs collected at D0 and D7 from four patients with DCB and four patients with NDB. The cells were sorted using a FACS Aria II cell sorter (BD Biosciences), and all samples showed > 90% purity after sorting. Total RNA was extracted from the sorted cells using TRIZOL reagent (Thermo Fisher Scientific), and was assessed for quantity and quality using a Bioanalyzer 2100 (Agilent). Libraries were generated using the QuantSeq 3’ mRNA-Seq Library Prep Kit (Lexogen), and sequenced by 75-bp single-end sequencing using the NextSeq 500 (Illumina). Sequences were aligned to a human reference genome (hg19) using Bowtie2. DESeq2 was used to identify differentially expressed genes (DEGs), defined as genes with an absolute fold change of > 2 with a false discovery rate of < 0.1. Gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) were performed using the R packages fgsea and gsva, respectively.
The Cancer Genome Atlas data analysis
RNA-seq gene expression profiles (RSEM normalized) for the HCC patients (n = 370), and the matched clinical information in The Cancer Genome Atlas (TCGA) database, were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). For each patient, the R package gsva was used to calculate the GSVA enrichment score for the set of DEGs identified by comparison of the RNA-seq profile. The patients were dichotomized based on the median GSVA score to compare survival outcomes.
Statistical analysis
Continuous variables were compared using the Mann–Whitney U-test for unpaired data, and the Wilcoxon signed-rank test for paired data. The chi-squared test was used to compare the distributions of the categorical variables. Pearson’s correlation test was used to examine the correlation between continuous variables. The confidence interval of the AUC value was calculated using the Delong method. Univariate and multivariate Cox proportional hazard models were used to estimate the hazard ratio of the variables for PFS and OS. Cutoff for serum alpha-fetoprotein level was based on the previous studies [14] and other continuous variables were dichotomized according to the median values. Variables with p < 0.2 in univariate analysis were included in the multivariate model. All statistical analyses were performed using R software version 4.1.0 (https://www.r-project.org).
Results
The frequency of classical monocytes increases early after anti-PD-1 therapy in patients with DCB
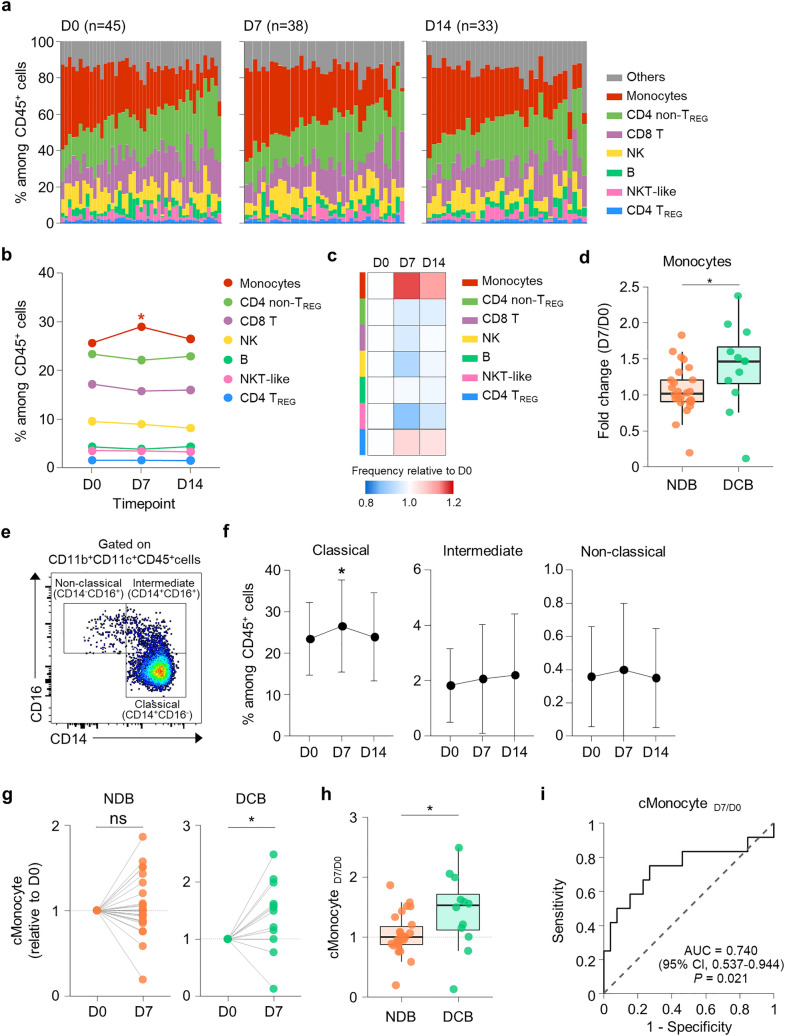
We first examined the proportions of various immune cell populations by analyzing PBMCs using multi-color flow cytometry. The markers and gating strategy for each population are shown in Supplementary Fig. 1. The relative frequencies of immune cell populations among CD45+ cells were determined at D0, D7, and D14 for each patient (Fig. 1a). The baseline (D0) frequency of each population was not associated with clinical benefit (Supplementary Fig. 2). We found that the monocyte frequency significantly increased at D7 and then subsequently decreased (Fig. 1b, c), whereas the other immune cell populations showed no significant changes. Importantly, the fold increase in the monocyte frequency at D7 over the baseline (D0) was significantly higher in patients with DCB than in patients with NDB (Fig. 1d). For other immune cell populations, the change in frequency did not significantly differ between patients with DCB versus NDB (Supplementary Fig. 3).
Fig. 1.
Change in the frequency of circulating classical monocytes among immune cells predicts durable clinical benefit. a Individually plotted relative frequencies of immune cell populations among CD45+ cells at D0, D7, and D14. b Mean frequencies of immune cell populations at D0, D7, and D14. c Heat-map of relative frequency changes of immune cell populations from baseline. d Fold-change of the frequency of monocytes among CD45+ cells according to clinical benefit. e Flow cytometry plot for defining classical, intermediate, and non-classical monocytes. f Mean frequencies of classical, intermediate, and non-classical monocytes among CD45+ cells at D0, D7, and D14. g Frequencies of classical monocytes among CD45+ cells relative to D0 in patients with DCB and NDB. h Relative change in frequency of classical monocytes (cMonocyteD7/D0) according to clinical benefit. i The ROC curve for predicting DCB using cMonocyteD7/D0. The results shown in f are expressed as the mean ± SD. *p < 0.05. Statistical analyses were performed using Wilcoxon signed-rank test (b, d, f) and Mann–Whitney U-test (d, h). TREG regulatory T cells, DCB durable clinical benefit, NDB non-durable clinical benefit
Human monocytes are divided into three subpopulations according to their expressions of CD14 and CD16: classical (CD14+CD16−), intermediate (CD14+CD16+), and non-classical (CD14−CD16+) monocytes [15]. Classical monocytes were dominant among peripheral blood monocytes (Fig. 1e), and the frequency of classical monocytes among CD45+ cells significantly increased at D7 (Fig. 1f). Separate analysis of patients with DCB and NDB revealed that the frequency of classical monocytes increased at D7 only in the DCB group (Fig. 1g). Furthermore, the fold increase in the frequency of classical monocytes at D7 over the baseline (D0), abbreviated as cMonocyteD7/D0, was significantly higher in patients with DCB than in patients with NDB (Fig. 1h). The predictive power of cMonocyteD7/D0 for DCB was moderate (AUC = 0.740; Fig. 1i). Collectively, these results indicated that a favorable response to anti-PD-1 therapy was accompanied by a relative increase in the classical monocyte population.
Classical monocytes of patients with DCB and NDB exhibit distinctive transcriptional changes early after anti-PD-1 therapy
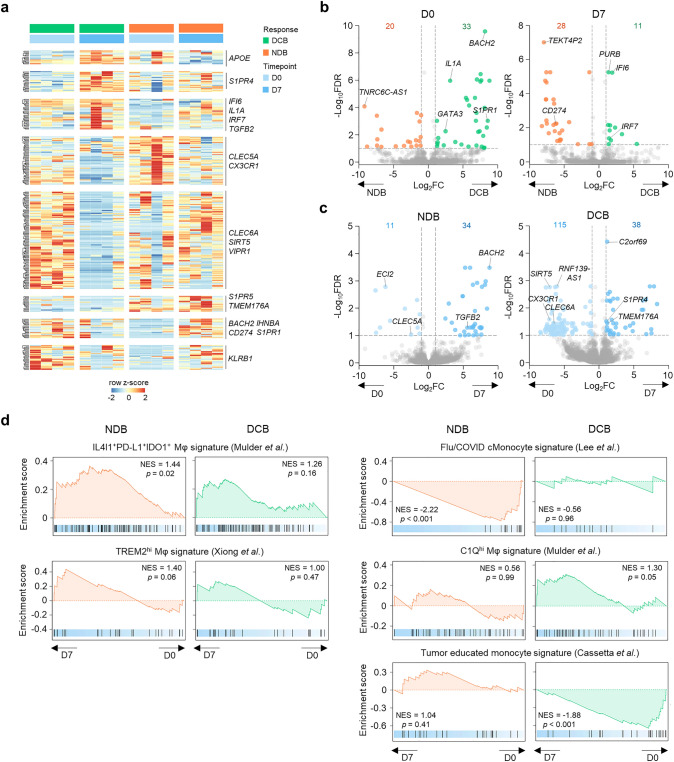
To understand the biological characteristics of classical monocytes, and their changes during anti-PD-1 therapy, we performed RNA-seq analysis of sorted classical monocytes collected at D0 and D7 from patients with DCB (n = 4) and NDB (n = 4). We observed a distinct pattern of gene expression according to the response and time-point (Fig. 2a). Comparing patients with DCB versus NDB revealed 33 upregulated and 20 downregulated genes at D0, and 28 upregulated and 11 downregulated genes at D7 (Fig. 2b; Supplementary Tables 3, 4). Notably, compared to patients with NDB, patients with DCB exhibited downregulation of CD274, encoding PD-L1, at D7. At D7, patients with DCB expressed 38 upregulated and 115 downregulated genes, and patients with NDB expressed 34 upregulated and 11 downregulated genes relative to baseline (D0) (Fig. 2c; Supplementary Tables 5, 6). To assess the clinical significance of these DEGs, we analyzed HCC patients from the TCGA database. The DEGs upregulated in NDB patients compared to DCB patients, especially at D7 (NDB > DCB at D7), were associated with poor prognosis, whereas the DEGs upregulated in DCB patients compared to NDB patients did not exhibit any association with prognosis (Supplementary Fig. 4). This result indicated that early transcriptomic changes in the peripheral blood classical monocytes from NDB patients resemble the transcriptomic features of tumor tissues from patients with poor prognosis.
Fig. 2.
Transcriptomic analysis of classical monocytes from patients with DCB and NDB. a Expression pattern of differentially expressed genes according to clinical benefit and time-point. b, c Volcano plots comparing patients with DCB and NDB at the same time-point (b), and before and after anti-PD-1 therapy in the same patients (c). d Gene set enrichment analyses of monocyte/macrophage-related gene sets at D7 compared to D0 in patients with DCB and NDB. DCB durable clinical benefit, NDB non-durable clinical benefit, FDR false-discovery rate, NES normalized enrichment score
We next focused on the transcriptomic changes from D0 to D7 in patients with DCB and NDB by performing GSEA using monocyte/macrophage-related gene sets (Fig. 2d). A gene set of IL4I1+PD-L1+IDO1+ macrophages [16] (which are abundant in various tumors and thought to play a suppressive role) was significantly enriched at D7 in patients with NDB, but not in patients with DCB. A gene set of TREM2hi macrophages [17] (an abundance of which is associated with resistance to anti-PD-1 therapy) also tended to be enriched at D7 in patients with NDB, although this change was not statistically significant. These data indicated that the classical monocytes of patients with NDB acquire transcriptomic features of immunosuppressive monocytes/macrophages that are related to anti-PD-1 resistance. Additionally, at D7, patients with NDB exhibited significant downregulation of a gene set of monocytes from patients with influenza or COVID-19 [18]. On the other hand, at D7, patients with DCB exhibited downregulation of a gene set of tumor-educated monocyte signature [19], which is upregulated in monocytes from breast cancer patients. Taken together, these findings indicate that the classical monocytes of patients with DCB and NDB undergo distinctive transcriptional changes early after anti-PD-1 therapy.
The frequency of PD-L1+ classical monocytes increases early after anti-PD-1 therapy in patients with NDB
Having observed the upregulation of CD274, and signature enrichment of IL4I1+PD-L1+IDO1+ macrophages, in patients with NDB following treatment, we next directly examined PD-L1 expression on classical monocytes. The frequency of PD-L1+ classical monocytes among classical monocytes did not significantly change after treatment among all patients (Fig. 3a), but it significantly increased at D7 in patients with NDB (Fig. 3b, c). Additionally, the fold increase in the frequency of PD-L1+ classical monocytes at D7 over D0 (abbreviated as cMonocyte-PDL1D7/D0) tended to be higher in patients with NDB than with DCB, although this difference was not statistically significant (Fig. 3d). Notably, cMonocyte-PDL1D7/D0, was predictive of NDB (AUC = 0.689; Fig. 3e). We further examined the phenotypes of PD-L1+ classical monocytes collected at D7, and found higher expression of the T-cell inhibitory ligands VISTA and CD155, compared to in their PD-L1− counterparts (Fig. 3f, g). Additionally, cMonocyte-PDL1D7/D0 was significantly associated with upregulation of the IL-6- or IL-10-stimulated gene signature [20, 21] (Fig. 3h, i), indicating that the numerical increase of PD-L1+ classical monocytes may be caused by IL-6 and IL-10. Taken together, our results showed that in patients with NDB, anti-PD-1 therapy increases the frequency of PD-L1+ classical monocytes that co-express other inhibitory ligands.
Fig. 3.
Expression of PD-L1 on circulating classical monocytes during anti-PD-1 therapy. a Mean frequencies of PD-L1+ cells among classical monocytes at D0, D7, and D14. b Representative flow cytometry plots of PD-L1 expression on classical monocytes from patients with DCB and NDB at D0 and D7. c Frequencies of PD-L1+ classical monocytes among classical monocytes relative to D0 in patients with DCB and NDB. d Relative change in frequency of PD-L1+ classical monocytes (cMonocyte-PDL1D7/D0) according to clinical benefit. e The ROC curve for predicting DCB using PD-L1+ cMonocyteD7/D0. f Representative flow cytometry plots of VISTA and CD155 expression on PD-L1− and PD-L1+ classical monocytes at D7. g Expression of VISTA and CD155 on PD-L1− and PD-L1+ classical monocytes at D7. h, i Correlation between PD-L1+ cMonocyteD7/D0 and changes in GSVA scores of an IL-6-stimulated macrophage-upregulated gene set (h) and an IL-10-treated peripheral blood mononuclear cell-upregulated gene set (i). The results shown in a are expressed as the mean ± SD. *p < 0.05, ****p < 0.0001. Statistical analyses were performed using the Mann–Whitney U-test (d), Wilcoxon signed-rank test (c, g), and Pearson’s correlation test (h, i). DCB durable clinical benefit, NDB non-durable clinical benefit, GSVA gene set variation analysis
Monocyte index predicts clinical benefit of anti-PD-1 therapy
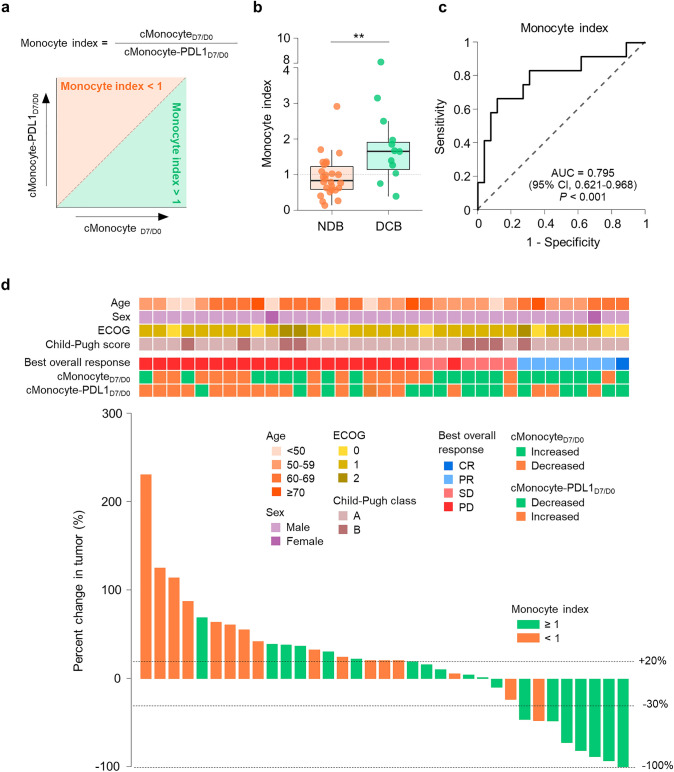
Since we identified two distinct monocyte-related parameters—cMonocyteD7/D0 and cMonocyte-PDL1D7/D0—that predicted the clinical benefit of anti-PD-1 therapy, we combined these two parameters to improve the prediction power. The combined biomarker, termed the “monocyte index”, was calculated by dividing cMonocyteD7/D0 by cMonocyte-PDL1D7/D0 (Fig. 4a). A monocyte index of greater than 1 indicates that the numerical increase of classical monocytes outpaces the upregulation of PD-L1. Patients with DCB showed a higher monocyte index than patients with NDB (Fig. 4b). Other peripheral blood biomarkers—including neutrophil-to-lymphocyte ratio, monocyte-to-lymphocyte ratio, and alpha-fetoprotein—were not associated with clinical benefit (Supplementary Fig. 5). The monocyte index predicted DCB with an AUC value of 0.795 (Fig. 4c). A monocyte index cut-off value of 1.37 maximized the sum of sensitivity and specificity; however, we performed further analyses using a cut-off value of 1 for the convenience of the application. For predicting DCB, the monocyte index (≥ 1 vs. < 1) had a positive prediction value of 0.526, and a negative prediction value of 0.895. The accuracy was 0.711, sensitivity was 0.833, and specificity was 0.654. Additionally, the monocyte index was strongly associated with changes in tumor size (p = 0.009, Pearson’s correlation test; Fig. 4d).
Fig. 4.
Monocyte index as a biomarker of clinical benefit. a Definition of the monocyte index. b Monocyte index according to clinical benefit. c The ROC curve for predicting durable clinical benefit using the monocyte index. d Waterfall plot of tumor size change and clinical parameters and biomarkers derived from circulating classical monocytes. **p < 0.01. Statistical analyses were performed using the Mann–Whitney U-test (b). DCB durable clinical benefit, NDB non-durable clinical benefit, ECOG Eastern Cooperative Oncology Group, CR complete response, PR partial response, SD stable disease, PD progressive disease
When we analyzed the classical monocytes from peripheral blood at D14, the frequencies of classical monocytes among CD45+ cells were not changed in either DCB patients nor NDB patients, and the fold change in the frequency (cMonocyteD14/D0) was not significantly different between DCB patients and NDB patients (Supplementary Fig. 6a, b). The frequency of PD-L1+ classical monocytes at D14 was not significantly different from D0 in patients with DCB; however, the frequency of PD-L1+ classical monocytes was significantly higher at D14 compared to D0 in patients with NDB, although the fold change of PD-L1+ classical monocytes (cMonocyte-PDL1D14/D0) was not significantly different according to clinical benefit (Supplementary Fig. 6c, d). Notably, a monocyte index, calculated by dividing cMonocyteD14/D0 by cMonocyte-PDL1D14/D0, was significantly higher in DCB patients compared to NDB patients (Supplementary Fig. 6e). The AUC value of the monocyte index for prediction of DCB was 0.715 (Supplementary Fig. 6f). When we multiplied the two monocyte indices derived from days 7 and 14, the combined index showed improved predictive performance (AUC = 0.811; Supplementary Fig. 6 g).
Monocyte index is an independent prognostic factor of survival outcomes
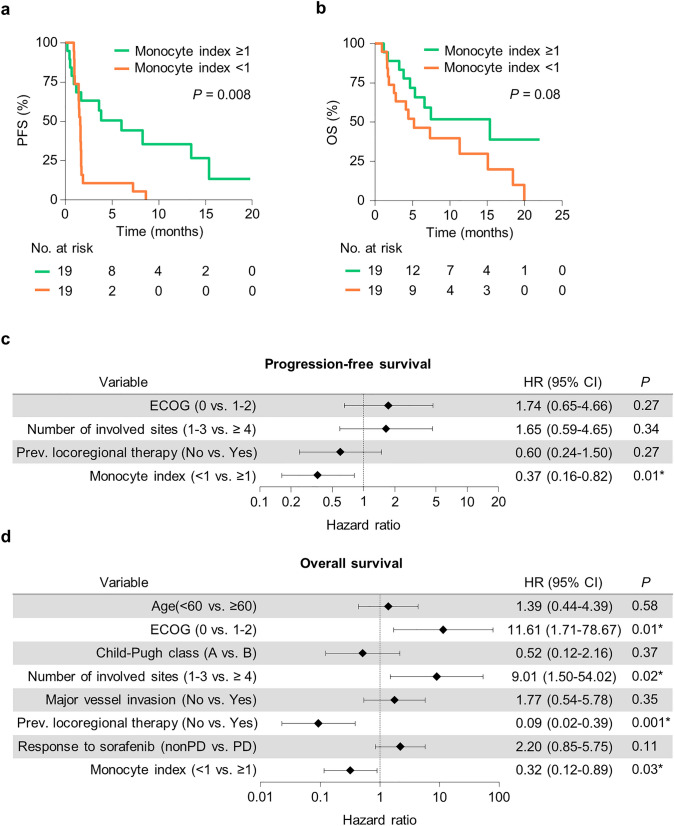
To determine whether the monocyte index is an independent prognostic factor of survival, we first analyzed PFS and OS according to the monocyte index. Patients with a monocyte index ≥ 1 showed better outcomes than patients with a monocyte index < 1 (Fig. 5a, b). In multivariate analysis, the monocyte index was an independent prognostic factor for both PFS (Fig. 5c; Supplementary Table 7) and OS (Fig. 5d; Supplementary Table 8). Additionally, prolonged OS was associated with good performance status, small number of metastases (< 4), and prior locoregional therapy. Similarly, higher monocyte index based on parameters from D14 was associated with better PFS and OS (Supplementary Fig. 6 h, i), although this was not significant in multivariate analysis.
Fig. 5.
Monocyte index as a prognostic factor for survival outcomes. a, b Kaplan–Meier curves for progression-free survival (PFS) (a) and overall survival (OS) (b) according to the monocyte index. c, d Forest plots for the hazard ratio of PFS (c) and OS (d) from multivariate analysis. The results shown in c and d are hazard ratio and 95% confidence interval. Statistical analysis was performed using a Cox proportional hazard model (a–d). PFS progression-free survival, OS overall survival, ECOG Eastern Cooperative Oncology Group, PD progressive disease
Discussion
Clinical trials of ICI treatment for advanced HCC have led to novel treatment options to improve clinical outcomes for these patients. Based on the results of prospective clinical trials [2–5, 8], the PD-1-blocking antibodies nivolumab and pembrolizumab can be considered for use in patients who have not responded to first-line treatment. Moreover, combination of the anti-PD-L1 antibody atezolizumab with the anti-VEGF antibody bevacizumab remarkably increases the survival of advanced HCC patients compared to sorafenib treatment [22], expanding the role of ICIs in the management of unresectable HCC. However, less than 20% of patients show clinical response to nivolumab or pembrolizumab [2–6]. Thus, there is a need for biomarkers predicting the clinical outcomes of anti-PD-1 therapy. In the present study, we discovered a peripheral blood biomarker based on circulating classical monocytes, which predicts the clinical benefit of anti-PD-1 therapy in HCC patients.
Numerous peripheral blood biomarkers have been found to predict the clinical response to ICIs in other types of cancer, especially melanoma and non-small cell lung cancer (NSCLC) [23]. The majority of these biomarkers are related to the characteristics of peripheral blood T cells. The activation and differentiation of peripheral CD8+ T cells are associated with a better response to anti-PD-1 therapy [24–26]. Additionally, recent studies have demonstrated that the clonal diversity and dynamics of peripheral T cells are associated with the treatment response [27–30]. These studies not only demonstrate that peripheral blood biomarkers predict the response, but also emphasize the importance of a systemic immune response upon ICI treatment, suggesting the potential underlying mechanisms of the therapy. The immunological features of HCC are distinct from those in other solid tumors, and HCC mostly arises from chronic inflammation [31]. Furthermore, recent studies suggest that a tumor within the liver exerts a distinct impact on anti-tumor immune responses [32, 33]. Thus, there is a need for HCC-specific peripheral blood biomarkers.
Our present analysis characterized early changes of circulating classical monocytes upon anti-PD-1 therapy for advanced HCC and demonstrated that the parameters were correlated with DCB. Previously, several studies have reported that monocytes are associated with response to ICIs. An analysis of PBMCs from melanoma patients revealed that the baseline frequency of classical monocytes was positively correlated with responsiveness to anti-PD-1 therapy [34]. Another study of melanoma discovered that a higher baseline frequency of S100A9+ monocytes is associated with poor response to PD-1 blockade [35]. Other studies examining PD-L1 expression on monocytes in various cancer types have consistently showed that higher baseline PD-L1 expression is correlated with worse clinical outcomes after ICI treatment [36–39]. In the present study, we developed a combined biomarker, termed the monocyte index, by integrating numeric changes in total and PD-L1+ classical monocytes early after anti-PD-1 therapy.
A previous study of melanoma demonstrated that pre-treatment PD-L1 levels on classical monocytes are upregulated in responders compared to non-responders, probably due to elevated IFN-γ [34]. However, in our current study, we observed the upregulation of PD-L1 on classical monocytes after anti-PD-1 treatment in patients with NDB. In line with these present findings, other recent reports have shown a negative relationship between PD-L1 expression on monocytes and prognosis [36–39]. Interestingly, we also found that upregulation of an IL-6- or IL-10-stimulated gene signature was significantly associated with cMonocyte-PDL1D7/D0. This suggests that IL-6 and/or IL-10 contribute to the numerical increase of PD-L1+ classical monocytes early after anti-PD-1 treatment. Previous reports demonstrate that IL-6 and IL-10 can induce PD-L1 expression on monocytes and macrophages [38, 40, 41]. PD-L1+ monocytes express T-cell inhibitory ligands, such as VISTA and CD155, indicating their immunosuppressive role. Indeed, previous studies show that peritumoral PD-L1+ monocytes exert immunosuppressive roles in HCC [42, 43].
Many studies have shown that the majority of TAMs are recruited and differentiated from circulating inflammatory monocytes [44–47]. Peripheral monocytes can also fuel tumor-infiltrating myeloid-derived suppressor cells (MDSCs), which suppress anti-tumor immunity. Hence, changes in circulating monocytes may impact the biological properties of TAMs or MDSCs. In the current study, GSEA revealed that in patients with NDB, classical monocytes acquire transcriptomic features of anti-PD-1 resistance-related, immunosuppressive monocyte/macrophages. It will be interesting to investigate whether the peripheral PD-L1+ monocytes that were increased early after anti-PD-1 treatment directly migrate to tumors and exert immunosuppressive roles.
We found that the monocyte index predicted not only the clinical benefit but also survival outcomes. Notably, the prognostic power of the index for OS was not statistically significant in the univariate analysis but was significant in the multivariate analysis. This phenomenon was likely due to the higher proportion of patients with a large number of metastases (≥ 4) in the higher monocyte index group (21.1% vs. 10.5%). Further studies are needed to examine the clinical parameters that affect the circulating classical monocytes during anti-PD-1 therapy.
Despite these novel findings, a limitation of our study is the relatively small number of patients included in our analysis without internal or external validation, which may limit the statistical power and interpretation of our data. Given the heterogeneity of classical monocytes among patients of the same response group, as shown through RNA-seq analysis, the usefulness of the monocyte index should be investigated in a larger and, preferably, independent cohort. Furthermore, the positive prediction value of the monocyte index was relatively low because the cut-off value of 1 used in the analysis is substantially different from the optimal cut-off value. The cut-off value must be optimized to support utilization of the monocyte index in clinical practice. Additionally, the mechanisms by which the changes in circulating classical monocytes are associated with clinical benefit were not thoroughly investigated, and further study with tumor tissue samples of anti-PD-1-treated patients is necessary.
The current analysis provides a monocyte-based peripheral blood biomarker that can predict the clinical benefit and survival outcomes of advanced HCC patients in their first week of anti-PD-1 treatment. To our knowledge, this is the first study to suggest that changes in monocytes following ICI treatment can be used as biomarkers for clinical benefit, in any type of cancer. Future studies are needed to validate our findings in larger cohorts and to investigate the mechanisms for how the changes of circulating classical monocytes impact the anti-tumor response in the tumor microenvironment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We appreciate our laboratory members for their support and critical opinions.
Abbreviations
- CR
Complete response
- DCB
Durable clinical benefit
- DEG
Differentially expressed gene
- FBS
Fetal bovine serum
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variation analysis
- HCC
Hepatocellular carcinoma
- ICI
Immune checkpoint inhibitor
- MDSC
Myeloid-derived suppressor cell
- NDB
Non-durable clinical benefit
- OS
Overall survival
- PBMC
Peripheral blood mononuclear cell
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- PFS
Progression-free survival
- PR
Partial response
- RECIST
Response Evaluation Criteria In Solid Tumors
- RNA-seq
RNA sequencing
- SD
Stable disease
- TAM
Tumor-associated macrophage
- TCGA
The Cancer Genome Atlas
Author contributions
SHJ and YJL designed and performed experiments, analyzed and interpreted the data, and wrote the manuscript. H-DK designed the experiments, analyzed and interpreted the data, and wrote the manuscript. HN assisted with flow cytometry analysis. B-YR collected the human samples. S-HP gave suggestions for the manuscript. CY collected the human samples, designed the experiments, and gave suggestions for the manuscript. E-CS interpreted the data and wrote the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by a National Research Foundation Grant NRF-2018M3A9D3079498, which is funded by the Ministry of Science and ICT, and funded in part by grants from Asan Institute for Life Sciences at Asan Medical Center in Seoul, Korea (2018-0634 and 2022IP0095).
Availability of data and material
The clinical information and flow cytometry data analyzed in the current study are available from the corresponding author on reasonable request due to patient privacy concern. The raw and processed RNA-seq data can be accessed from Gene Expression Omnibus database (Accession Number GSE202145).
Declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Informed consent was obtained from all enrolled patients.
Consent for publication
Patients signed informed consent regarding publishing their data.
Ethical approval
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki. The samples were collected according to the protocols approved by the Institutional Review Boards at Asan Medical Center (IRB No. 2019-1231).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors Seung Hyuck Jeon, Yong Joon Lee and Hyung-Don Kim contributed equally to this work.
Contributor Information
Changhoon Yoo, Email: cyoo.amc@gmail.com.
Eui-Cheol Shin, Email: ecshin@kaist.ac.kr.
References
- 1.Fitzmaurice C, Akinyemiju T, Abera S, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 4.Yau T, Park J-W, Finn RS, et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 2022;23:77–90. doi: 10.1016/s1470-2045(21)00604-5. [DOI] [PubMed] [Google Scholar]
- 5.Finn RS, Ryoo BY, Merle P, et al. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: a randomized, double-blind, phase III trial. J Clin Oncol. 2020;38:193–202. doi: 10.1200/JCO.19.01307. [DOI] [PubMed] [Google Scholar]
- 6.Kudo M, Matilla A, Santoro A, et al. CheckMate 040 cohort 5: a phase I/II study of nivolumab in patients with advanced hepatocellular carcinoma and Child–Pugh B cirrhosis. J Hepatol. 2021;75:600–609. doi: 10.1016/j.jhep.2021.04.047. [DOI] [PubMed] [Google Scholar]
- 7.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73:1460–1469. doi: 10.1016/j.jhep.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu CL, Ou DL, Bai LY, et al. Exploring markers of exhausted CD8 T cells to predict response to immune checkpoint inhibitor therapy for hepatocellular carcinoma. Liver Cancer. 2021;10:346–359. doi: 10.1159/000515305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita M, Nishida N, Sakai K, et al. Immunological microenvironment predicts the survival of the patients with hepatocellular carcinoma treated with anti-PD-1 antibody. Liver Cancer. 2021;10:380–393. doi: 10.1159/000516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng HHM, Lee RY, Goh S, et al. Immunohistochemical scoring of CD38 in the tumor microenvironment predicts responsiveness to anti-PD-1/PD-L1 immunotherapy in hepatocellular carcinoma. J Immunother Cancer. 2020;8:987. doi: 10.1136/jitc-2020-000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feun LG, Li YY, Wu C, et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer. 2019;125:3603–3614. doi: 10.1002/cncr.32339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheiner B, Pomej K, Kirstein MM, et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy—development and validation of the CRAFITY score. J Hepatol. 2022;76:353–363. doi: 10.1016/j.jhep.2021.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Kelley RK, Meyer T, Rimassa L, et al. Serum alpha-fetoprotein levels and clinical outcomes in the phase III CELESTIAL study of cabozantinib versus placebo in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2020;26:4795–4804. doi: 10.1158/1078-0432.CCR-19-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cros J, Cagnard N, Woollard K, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder K, Patel AA, Kong WT, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54:1883.e5–1900.e5. doi: 10.1016/j.immuni.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Xiong D, Wang Y, You M. A gene expression signature of TREM2hi macrophages and γδ T cells predicts immunotherapy response. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-18546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, Park S, Jeong HW, et al. Immunophenotyping of covid-19 and influenza highlights the role of type i interferons in development of severe covid-19. Sci Immunol. 2020;5:1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassetta L, Fragkogianni S, Sims AH, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35:588.e10–602.e10. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang R, Pauleau AL, Parganas E, et al. SOCS3 regulates the plasticity of gp130 signaling. Nat Immunol. 2003;4:546–550. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- 21.Teles RMB, Graeber TG, Krutzik SR, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science (80-) 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/nejmoa1915745. [DOI] [PubMed] [Google Scholar]
- 23.Kim KH, Kim CG, Shin EC. Peripheral blood immune cell-based biomarkers in anti-PD-1/PD-L1 therapy. Immune Netw. 2020;20:e8. doi: 10.4110/in.2020.20.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Schenkel JM, Pauken KE, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KH, Cho J, Ku BM, et al. The first-week proliferative response of peripheral blood PD-1+CD8+ T cells predicts the response to anti-PD-1 therapy in solid tumors. Clin Cancer Res. 2019;25:2144–2154. doi: 10.1158/1078-0432.CCR-18-1449. [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi T, Hoki T, Oba T, et al. T-cell CX3CR1 expression as a dynamic blood-based biomarker of response to immune checkpoint inhibitors. Nat Commun. 2021;12:1402. doi: 10.1038/s41467-021-21619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cader FZ, Hu X, Goh WL, et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat Med. 2020;26:1468–1479. doi: 10.1038/s41591-020-1006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairfax BP, Taylor CA, Watson RA, et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med. 2020;26:193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Duan J, Bai H, et al. TCR repertoire diversity of peripheral PD-1+CD8+ T cells predicts clinical outcomes after immunotherapy in patients with non-small cell lung cancer. Cancer Immunol Res. 2020;8:146–154. doi: 10.1158/2326-6066.CIR-19-0398. [DOI] [PubMed] [Google Scholar]
- 30.Watson RA, Tong O, Cooper R, et al. Immune checkpoint blockade sensitivity and progression-free survival associates with baseline CD8+ T cell clone size and cytotoxicity. Sci Immunol. 2021 doi: 10.1126/SCIIMMUNOL.ABJ8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringelhan M, Pfister D, O’Connor T, et al. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 32.Lee JC, Mehdizadeh S, Smith J, et al. Regulatory T cell control of systemic immunity and immunotherapy response in liver metastasis. Sci Immunol. 2020;5:759. doi: 10.1126/sciimmunol.aba0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Green MD, Li S, et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med. 2021;27:152–164. doi: 10.1038/s41591-020-1131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg C, Nowicka M, Guglietta S, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 35.Rad Pour S, Pico De Coaña Y, Demorentin XM, et al. Predicting anti-PD-1 responders in malignant melanoma from the frequency of S100A9+ monocytes in the blood. J Immunother Cancer. 2021;9:e002171. doi: 10.1136/jitc-2020-002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasuoka H, Asai A, Ohama H, et al. Increased both PD-L1 and PD-L2 expressions on monocytes of patients with hepatocellular carcinoma was associated with a poor prognosis. Sci Rep. 2020;10:10377. doi: 10.1038/s41598-020-67497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riemann D, Schütte W, Turzer S, et al. High PD-L1/CD274 expression of monocytes and blood dendritic cells is a risk factor in lung cancer patients undergoing treatment with PD1 inhibitor therapy. Cancers (Basel) 2020;12:1–13. doi: 10.3390/cancers12102966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Liu Y, Yan Z, et al. IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J Immunother Cancer. 2020;8:285. doi: 10.1136/jitc-2019-000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ando K, Hamada K, Shida M, et al. A high number of PD-L1+ CD14+ monocytes in peripheral blood is correlated with shorter survival in patients receiving immune checkpoint inhibitors. Cancer Immunol Immunother. 2021;70:337–348. doi: 10.1007/s00262-020-02686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Garcia M, Porichis F, de Jong OG, et al. Expression of PD-L1 and PD-L2 on human macrophages is up-regulated by HIV-1 and differentially modulated by IL-10. J Leukoc Biol. 2011;89:507–515. doi: 10.1189/jlb.0610327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copland A, Sparrow A, Hart P, et al. Bacillus Calmette–Guérin induces PD-L1 expression on antigen-presenting cells via autocrine and paracrine interleukin-STAT3 circuits. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-40145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang DM, Zhao Q, Peng C, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327–1337. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen DP, Ning WR, Jiang ZZ, et al. Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol. 2019;71:333–343. doi: 10.1016/j.jhep.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Movahedi K, Laoui D, Gysemans C, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 45.Qian BZ, Li J, Zhang H, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science (80-) 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Feng X, Herting CJ, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77:2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical information and flow cytometry data analyzed in the current study are available from the corresponding author on reasonable request due to patient privacy concern. The raw and processed RNA-seq data can be accessed from Gene Expression Omnibus database (Accession Number GSE202145).