ABSTRACT
Objectives:
Circulating microRNAs (miRNAs) have been discovered to play a novel role in intercellular communication and cancer biology. They are emerging candidates for noninvasive molecular biomarkers of cancer and other diseases. However, current translational researches have been limited by the lack of consensus on the optimal endogenous control of circulating miRNAs quantitation. In this study, we compared two promising miRNAs, miR-1228 and miR-16, as an endogenous control. The effects of normalizers on the relative quantification of circulating miR-31 in plasma samples of colorectal cancer (CRC) were also assessed.
Materials and Methods:
The cel-miR-39 was a spiked-in RNA used as an external control and added to plasma samples before RNA extraction. Quantitative real-time polymerase chain reaction technology was used to analyze the expression levels of circulating miRNAs in plasma samples of 4 healthy controls and 14 CRC patients. The expression stability of the candidate controls was compared by Ct analysis and NormFinder algorithms.
Results:
There was no significant difference in expression level of miR-16 and miR-1228 between healthy control group and before or after therapy of CRC patient groups. The expression of miR-1228 has smaller the range Ct values (28.25-25.64)compared with those of miR-16 (24.91-20.34). The stability value of miR-1228 (0.102) is lower than that of miR-16 (0.350). The expression of miR-1228 endogenous reference candidate has lower stability value and smaller the range Ct values compared with those in miR-16. According to the range Ct values and stability value, miR-1228 is better than miR-16 as endogenous control in CRC patients. There are significant differences in circulating miR-31 expression between healthy control and CRC patients when miR-1228 was used to standardize miR-31 expression.
Conclusions:
miR-1228 is recommended as a better endogenous control in quantification of circulating miRNAs in CRC patients.
KEYWORDS: Circulating microRNAs, Colorectal cancer, Quantification
INTRODUCTION
MicroRNAs (miRNAs) are short noncoding RNAs molecule (about 22 nucleotides) which causes either mRNA molecule degradation or translational inhibition through interactions with the 3’ untranslated regions of mRNA targets [1,2]. There are more than 2,000 mature miRNAs have been identified to regulate more than half of human mRNAs [3]. Previous studies reported that the role of miRNAs is associated with carcinogenesis, such as cell proliferation, invasion, and metastases [4,5]. The expression of miRNAs is widely altered in normal tissues, leading to carcinogenesis through abnormal expression of mRNA [6,7].
Colorectal cancer (CRC) is one of the most common cancers in the world. The development of CRC is associated with multiple epigenetic changes and genetic alterations [7]. Recent studies reported that multiple miRNAs have an aberrant expression in CRC and have been reported as diagnostic, prognostic, and predictive markers [8,9,10,11]. In addition to miRNAs playing the role of regulating gene expression in cells, historical studies have demonstrated that miRNAs could present in the cell-free body fluids such as serum, plasma, urine and saliva [10]. These extracellular miRNAs may regulate the intracellular gene expression of distant cells in some way. For example, cancer cells can control the environment of remote tissues by releasing exosomes packaged specific miRNAs to assist the metastasis, attachment and growth of cancer cells [12]. Therefore, these circulating miRNAs have the potential to be used as novel noninvasive biomarkers for clinical applications in CRC.
Comparing circulating miRNA level between healthy persons and patients with stage IV CRC cancer, the plasma levels of miR-21, miR-145, miR-203, miR155, miR-210, miR-31, and miR-345 were significantly different [8,9]. Circulating miR-17, miR-21, and miR-92 can be used as predictors to predict the likelihood of recurrence of CRC after chemotherapy [8,9]. The results of these studies indicate that in addition to routine diagnosis such as colonoscopy or tomography, these miRNAs expression in blood sample may assist in the diagnosis and treatment of clinical CRC cancer. Although many studies have described circulating miRNAs as therapeutic or diagnostic targets, the reproducibility of experimental results has been challenged. The source of the samples and the isolation and purification of the samples may cause the difference in miRNA interpretation. In addition, data normalization, especially the choice of internal control, is a major factor contributing to differences in data interpretation [13]. U6 is widely used as a normalized control for miRNA in cells, but its application as an endogenous control for normalization of circulating miRNA expression is limited due to its low expression in body fluids [14].
The normalization of miRNA quantitation by using a suitable reference gene is an essential component of reliable quantitative polymerase chain reaction (qPCR) assay [15]. However, the optimal reference miRNA for normalization of circulating miRNAs expression was not clear. Although there are a few reports on the issues, there are no universal consensus on endogenous miRNAs for this purpose in various cancers. Huang et al. used miR-16 as the internal control for plasma miRNA quantification in CRC [16]. Hu et al. reported miR-1228 as the most stable endogenous control for quantification of circulating miRNA in cancer patients through microarray approaches and critically appraised the optimal endogenous control after comparing it with the other control (miR-16, miR-223, let-7a, and RNU6B) commonly used in the literature [15]. Danese et al. used miRTarBase to estimate target genes of selected miRNAs. The results showed that the miR-16 regulated multiple oncogenes and tumor suppressor genes including BCL1, KRAS, BMI-1, MYB, and PT53. In addition, the report also pointed out that miR-1228 does not interact with any known oncogenes or tumor suppressor genes [17,18].
MiR-16 was often used as endogenous reference genes for various diseases, including chronic kidney disease, papillary thyroid carcinoma, breast cancer, and prostate cancer [19]. MiR-1228 was reported to be a stable endogenous control and recently it was verified [15,20]. To clarifying the ambiguous issue on endogenous miRNA control, we set up experiments to compare miR-1228 with miR-16 which is frequently adopted as endogenous miRNA control in the literature as optimal reference miRNAs in estimate circulating miRNA expression in CRC patients. In this study, we selected these two miRNAs and exogenous cel-miR-39 as the research targets, and compared their stability values and range of Ct values in CRC plasma samples to explore whether changes occurred before and after CRC treatment, as suitable reference miRNA candidates.
MATERIALS AND METHODS
Samples collection and processing
The plasma samples were collected from three groups of participants (HC: Healthy controls, BT: Before therapy of CRC patients, AT: After therapy of CRC patients). The samples were obtained from 4 HC and 14 CRC patients including before and AT in Taipei Buddhist Tzu Chi hospital. An amount of 5 ml of whole blood from each participant was collected in EDTA tube. Blood samples were obtained by centrifugation at 1,200 g for 10 min at 4°C. The supernatant was transferred into new microcentrifuge tubes and centrifuged at 12,000 g for 10 min at 4°C to completely remove cellular components. The supernatant plasma was stored at −80°C until use. The study protocol was approved by Taipei Tzu Chi Hospital Institutional Review Board (IRB No. 05-XD37-067 and 05-XD38-068). Inform consent was obtained from all participants prior to collection of blood samples.
RNA isolation and reverse transcription polymerase chain reaction
Total miRNAs were isolated from 300 μL of plasma sample using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) and eluting it into 30 μL RNase-free water. For normalization of sample-to-sample variation during RNA isolation procedures, 0.25 pmole of synthetic cel-miR-39 was added into each sample. Small RNAs from plasma samples were reverse transcribed using the TaqMan miRNAs Reverse Transcription kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instruction. Reverse transcription primers were following:
hsa-miR-16-5p, 5’-GTCGTATCCAGTGCAGGGTCCGA GGTATTCGCACTGGATACGACCGCCAA-3’, hsa-miR-31-5p,5’-GTCGTATCCAGTGCAGGGTCCGA GGTATTCGCACTGGATACGACAGCTAT-3’, hsa-miR-1228-3p,5’-GTCGTATCCAGTGCGTGTCGTG GAGTCGGCAATTGCACTGGATACGACGGGGGG-3’, cel-miR-39-3p,5’-GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACCAAGCT-3’.
Quantitative real-time polymerase chain reaction
For miRNAs quantification, a real-time quantitative PCR of miRNAs was performed using Luna Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA) on the ABI7900 PCR machine. The reaction was performed in 96 well plates under the following condition: 95°C for 2 min, 40 cycles of 95°C for 15 sec, 60°C for 1 min. The primers of quantitative PCR were following:
hsa-miR-16-5p forward primer, 5’-CGGCG TAGCAGCACGTAAATA-3’, hsa-miR-16-5p reverse primer, 5’-CCAGTGCAGGGTCCGAGGTA-3’, hsa-miR-31-5p forward primer,5’-CGGCGAGGCAAGATGCTGGC-3’, hsa-miR-31-5p reverse primer, 5’-CCAGTGCAGGGTCCG AGGTA-3’, hsa-miR-1228-3p forward primer, 5’-CGGC GTCACACCTGCCTCG-3’, hsa-miR-1228-3p reverse primer, 5’-CAGTGCGTGTC GTGGAGTC-3’, cel-miR-39-3p forward primer, 5’-CGGCGTCACCGGGTGTAAATC-3’, cel-miR-39-3p reverse primer, 5’-CAGTGCGTGTCGTGGAGTC-3’, The quantitative end point of real time PCR is the threshold cycle (Ct). miRNAs expression by relative quantification using the 2-ΔΔCt method to determine fold changes in expression.
Statistical analysis
For the statistical analysis, we used the Graph Pad Prism software package (GraphPad software Inc., La Jolla, CA, USA). The Mann–Whitney test was used to analyze the difference in relative expression levels of miRNA between patients and controls. The quantitation cycle value (Ct) was used to estimate the level of gene expression. The expression variation range of Ct value (DCt) was used as the first parameter to assess the expression stability of the candidate reference miRNA. A narrow range of Ct value indicates a gene with more stable expression. The NormFinder algorithm was then used to assess the expression stability of the candidate reference genes [15]. It is based on an analysis of variance mathematical model, estimates both the inter-group and intra-group variations and finally calculates the stability of a candidate reference gene. A lower stability value indicates a more stably expressed gene. The sample with minimum Ct or maximum expression was used as a calibrator with a set value of 1.
RESULTS
Demographic and clinical features of enrolled patients and healthy subjects
A total of 14 CRC patients (mean age 63.9 ± 11.4 years; range 41–73) and 4 healthy controls (mean age (49.5 ± 8.4 years; range 41–46) were recruited in this study with equal numbers of male and female patients but 100% male in healthy subjects [Table 1]. The TNM stage distribution was Stage I (7.1%), stage II (35.7%), Stage III (35.7%), and Stage IV (21.4%). The majority of tumor differentiation was moderate differentiated (85.7%) with poor differentiated (14.3%). Tumor was located in proximal colon (37.5%), distal colon (28.6%), and rectum (35.7%). The histologic subtypes of tumor included adenocarcinoma (85.7%), mucinous adenocarcinoma (7.1%), and signet-ring carcinoma (7.1%).
Table 1.
Demographic and clinical features of enrolled patients and healthy subjects
| Characteristics | n (%) |
|---|---|
| Healthy controls | |
| Age (years), mean±SD | 49.5±8.4 |
| Range | 41-61 |
| Sex | |
| Male | 4 (100.0) |
| Female | 0 |
| CRC patients | |
| Age (years), mean±SD | 63.9±11.4 |
| Range | 41-73 |
| Sex | |
| Male | 7 (50.0) |
| Female | 7 (50.0) |
| Tumor TNM stage | |
| I | 1 (7.1) |
| II | 5 (35.7) |
| III | 5 (35.7) |
| IV | 3 (21.4) |
| Tumor differentiation | |
| Moderate | 12 (85.7) |
| Poor | 2 (14.3) |
| Tumor location | |
| Proximal colon | 5 (35.7) |
| Distal colon | 4 (28.6) |
| Rectum | 5 (35.7) |
| Histology subtype | |
| Adenocarcinoma | 12 (85.7) |
| Mucous adenocarcinoma | 1 (7.1) |
| Signet-ring carcinoma | 1 (7.1) |
SD: Standard deviation, CRC: Colorectal cancer, TNM: Tumor-node-metastasis
Expression levels of candidate reference microRNAs
We selected the three candidate reference miRNAs including two endogenous reference candidates (miR-16, miR-1228) and one extrinsic control (cel-miR-39) to analyze their expression using quantitative real-time PCR from plasma samples including 4 HC, 14 BT of CRC patients (BT) and 5 AT of CRC patients (AT). The median circulating levels of miR-1228 and miR-16 in the early CRC stage (Stage I and II) and advanced stage (Stage III and IV) were 26.81 (range 26.99-25.79) versus 21.46 (23.59-20.38), and 26.20 (28.25-25.64) versus 23.12 (24.91-20.52), respectively. The circulating level of miR-1228 was not influenced by tumor stage but miR-16 level was higher in advanced stage. The spike-in cel-miR-39 displayed the lowest DCt value (1.32) among all plasma samples (n = 23). The DCt value of miR-1228 was 2.61 less than that of miR-16 which was 4.57 in all plasma samples. In addition, the Ct value of miR-1228 was also lower than that of miR-16 in plasma samples of HC, BT and AT [Table 2].
Table 2.
Reference miRNAs expression in plasma samples
| miRNAs | HC (n=4) | BT (n=14) | AT (n=5) | All (n=23) | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| Median (range) | △Ct | Median (range) | △Ct | Median (range) | △Ct | Median (range) | △Ct | |
| miR-16 | 21.77 (22.62-20.34) | 2.28 | 22.15 (24.91-20.38) | 4.53 | 22.16 (24.20-21.05) | 3.15 | 22.00 (24.91-20.34) | 4.57 |
| miR-1228 | 26.83 (27.08-26.52) | 0.56 | 26.36 (28.25-25.64) | 2.61 | 26.84 (27.57-26.63) | 0.94 | 26.80 (28.25-25.64) | 2.61 |
| cel-miR-39 | 14.58 (14.65-14.34) | 0.31 | 14.95 (15.66-14.36) | 1.30 | 15.01 (15.64-14.68) | 0.96 | 14.94 (15.66-14.34) | 1.32 |
HC: Healthy controls, BT: Before therapy of CRC patients, AT: After therapy of CRC patients,△Ct: Ctmaximum -Ctminimum, CRC: Colorectal cancer
Expression stability of candidate reference microRNAs
The variable stability of the three candidate reference miRNAs was analyzed with the NormFinder algorithm. The stability value of the spike-in cel-miR-39 (0.069) was the lowest value in all plasma samples. The stability value of miR-1228 was 0.102 less than that of miR-16, which was 0.350 in all plasma samples. The results indicating miR-1228 was more stable than miR-16 as endogenous reference control in the three group of plasma samples [Table 3].
Table 3.
Ranking of reference miRNAs using NormFinder algorithm
| Rank | HC (n=4) | BT (n=14) | AT (n=5) | All (n=23) | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| miRNA name | Stability value | miRNA name | Stability value | miRNA name | Stability value | miRNA name | Stability value | |
| 1 | cel-miR-39 | 0.042 | cel-miR-39 | 0.247 | miR-1228 | 0.101 | cel-miR-39 | 0.069 |
| 2 | miR-1228 | 0.234 | miR-1228 | 0.534 | cel-miR-39 | 0.266 | miR-1228 | 0.102 |
| 3 | miR-16 | 0.720 | miR-16 | 0.940 | miR-16 | 0.981 | miR-16 | 0.350 |
HC: Healthy controls, BT: Before therapy of CRC patients, AT: After therapy of CRC patients, CRC: Colorectal cancer
Effect of the selected reference genes on relative expression of circulating miR-31
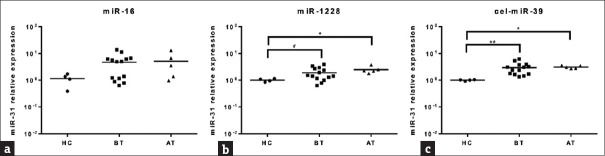
The expression level of circulating miR-31 was elevated in CRC and used as a diagnostic marker for CRC [15]. Therefore, to ensure that the three candidate reference miRNAs as a stable control, the influences of normalizers on the relative quantification of circulating miR-31 were also assessed. We found there was a significant difference in the level of circulating miR-31 between healthy controls and CRC patients when miR-1228 or cel-miR-39 was used as normalizers. However, when miR-16 was used as a normalizer, there was no significant differences between HC and CRC patients [Figure 1].
Figure 1.
The expression of miR-31 was analyzed by quantitative real-time polymerase chain reaction. Relative miR-31 expression levels were normalized to (a) miR-16, (b) miR-1228 and (c) cel-miR-39. HC: Healthy controls, BT: Before therapy of CRC patients, AT: After therapy of CRC patients, **: P < 0.01, *: P < 0.05, #: P < 0.1
DISCUSSION
Recent studies have demonstrated that miRNAs were involved in tumorigenesis, including cell proliferation, apoptosis, invasion and metastases [4,21]. The aberrant expression of miRNAs contributed to multiple epigenetic changes and genetic alterations via posttranscriptional regulation of gene expression [2,6]. In our study, there were no significant difference in the expression level of miR-16 and miR-1228 between HC group and before or AT of CRC patient groups. The expression of miR-1228 endogenous reference candidate has lower stability value and smaller the range Ct values compared with those in miR-16. In our study results in CRC, the miR-1228 also showed less variation as compared with miR-16. Recently, Duran-Sanchon et al. observed that miR-16 was significantly influenced by hemolysis, but miR-1228 was not influenced by hemolysis and therefore suggested that miR-1228 should be the optimal endogenous control for circulating miRNA analysis in CRC [20].
The report showed that circulating miR-31 expression is higher in CRC patients and miR-16 was selected an endogenous reference miRNA in the study 10 22]. In our experiments, we also observed significant differences in circulating miR-31expression between HC and CRC patients when miR-1228 was used to standardize miR-31 expression. However, the significance of difference was gone when miR-16 was used as endogenous control. Therefore, our results demonstrate that the use of different reference miRNA can lead to misjudgments in clinical blood samples, regardless of whether the experiment is derived from individual differences or experimental operating techniques.
In the selection of suitable miRNA reference genes in CRC research, in addition to considering that the reference genes do not affect the tumor formation and progression, it is also necessary to consider whether these reference genes are affected by oncogene/tumor suppressor genes in the process of cancerization. Most miRNAs regulate the expression of target genes, and miRNAs themselves may also be regulated by other noncoding RNAs such as long noncoding RNAs or circular RNAs [23]. Recent studies have found that miR-1228 can be regulated by circRNA_100395 and participates in the carcinogenesis of ovarian cancer and lung cancer [24,25]. However, the performance of circRNA_100395 in the large intestine tissue has not yet been explored. Similarly, miR-16 can be regulated by long noncoding RNA AGAP2-AS1, SNHG12, and UCA1 to participate in the carcinogenesis of lung cancer, gastric cancer, and bladder cancer [26,27,28]. Therefore, the above research results suggest that the reference genes used in the research of different types of cancer should be individualized.
Although some studies have explored the applicability of miR-1228 and miR-16 as reference genes for CRC, the changes of these reference genes before and after surgical treatment of CRC patients have not been explored in the literature [17,20]. Our research results indicate that miR-1228 was not affected by surgical treatment and should be considered as an optimal reference gene for treatment studies. Besides, the circulating level of miR-1228 was not influenced by tumor stage but miR-16 level was higher in advanced stage. Although we recruited all the patients with CRC stage ranging from stage I to stage IV, all the cases in healthy control were male. Besides, due to the small sample size and the limitation of the interpretation of research data in a single research center, further validations from larger cohorts are needed to verify our research results in the future.
CONCLUSION
We recommended miR-1228 as a better endogenous control to quantify circulating miRNA expression in CRC patients. The limitation of our study is the small sample size. Our significant findings should be validated in large-scale clinical studies.
Financial support and sponsorship
This research was funded by grants from the Taipei Tzu Chi Hospital through the Buddhist Tzu Chi Medical Foundation (TCRD-TPE-106-C2-1, TCRD-TPE-106-C2-2, TCRD-TPE-106-C2-3, TCRD-TPE-110-35).
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–40. [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conev NV, Donev IS, Konsoulova-Kirova AA, Chervenkov TG, Kashlov JK, Ivanov KD. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci Trends. 2015;9:393–401. doi: 10.5582/bst.2015.01170. [DOI] [PubMed] [Google Scholar]
- 9.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–25.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nassar FJ, Msheik ZS, Itani MM, Helou RE, Hadla R, Kreidieh F, et al. Circulating miRNA as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics (Basel) 2021;11:341. doi: 10.3390/diagnostics11020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br J Cancer. 2017;116:762–74. doi: 10.1038/bjc.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating MicroRNAs in cancer: Potential and challenge. Front Genet. 2019;10:626. doi: 10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R, Ramasubramanian B, Kanji S, Chakraborty AR, Haque SJ, Chakravarti A. Circulating microRNAs in cancer: Hope or hype? Cancer Lett. 2016;381:113–21. doi: 10.1016/j.canlet.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135:1187–94. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 17.Danese E, Minicozzi AM, Benati M, Paviati E, Lima-Oliveira G, Gusella M, et al. Reference miRNAs for colorectal cancer: Analysis and verification of current data. Sci Rep. 2017;7:8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–47. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duran-Sanchon S, Vila-Navarro E, Marcuello M, Lozano JJ, Muñoz J, Cubiella J, et al. Validation of miR-1228-3p as housekeeping for MicroRNA analysis in liquid biopsies from colorectal cancer patients. Biomolecules. 2019;10:16. doi: 10.3390/biom10010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W, et al. MicroRNAs involved in carcinogenesis, prognosis, therapeutic resistance and applications in human triple-negative breast cancer. Cells. 2019;8:1492. doi: 10.3390/cells8121492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota N, Taniguchi F, Nyuya A, Umeda Y, Mori Y, Fujiwara T, et al. Upregulation of microRNA-31 is associated with poor prognosis in patients with advanced colorectal cancer. Oncol Lett. 2020;19:2685–94. doi: 10.3892/ol.2020.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080–90. doi: 10.1080/15384101.2018.1515553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C, et al. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J Cancer. 2020;11:599–609. doi: 10.7150/jca.35041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:194. doi: 10.1186/s13046-019-1188-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Zhao G, Wang S, Liang X, Wang C, Peng B. Oncogenic role of long non-coding RNA SNHG12 in gastric cancer cells by targeting miR-16. Exp Ther Med. 2019;18:199–208. doi: 10.3892/etm.2019.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J Clin Oncol. 2015;45:1055–63. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]