Abstract
This study applies a large proteomics panel to search for new circulating biomarkers associated with progression to kidney failure in individuals with diabetic kidney disease. Four independent cohorts encompassing 754 individuals with type 1 and type 2 diabetes and early and late diabetic kidney disease were followed to ascertain progression to kidney failure. During ten years of follow-up, 227 of 754 individuals progressed to kidney failure. Using the SOMAscan proteomics platform, we measured baseline concentration of 1129 circulating proteins. In our previous publications, we analyzed 334 of these proteins that were members of specific candidate pathways involved in diabetic kidney disease and found 35 proteins strongly associated with risk of progression to kidney failure. Here, we examined the remaining 795 proteins using an untargeted approach. Of these remaining proteins, 11 were significantly associated with progression to kidney failure. Biological processes previously reported for these proteins were related to neuron development (DLL1, MATN2, NRX1B, KLK8, RTN4R and ROR1) and were implicated in the development of kidney fibrosis (LAYN, DLL1, MAPK11, MATN2, endostatin, and ROR1) in cellular and animal studies. Specific mechanisms that underlie involvement of these proteins in progression of diabetic kidney disease must be further investigated to assess their value as targets for kidney-protective therapies. Using multivariable LASSO regression analysis, five proteins (LAYN, ESAM, DLL1, MAPK11 and endostatin) were found independently associated with risk of progression to kidney failure. Thus, our study identified proteins that may be considered as new candidate prognostic biomarkers to predict risk of progression to kidney failure in diabetic kidney disease. Furthermore, three of these proteins (DLL1, ESAM, and MAPK11) were selected as candidate biomarkers when all SOMAscan results were evaluated.
Keywords: Diabetic kidney disease, end stage kidney disease, diabetes, circulating biomarker, proteomics analysis
Graphical Abstract
INTRODUCTION
Over the last 20 years, intensive research has investigated novel biomarkers of diabetic kidney disease (DKD) and its progression to end stage kidney disease (ESKD). Proteins identified by this research may lead to a better understanding of the mechanisms responsible for DKD. This, in turn, can point to new therapeutic targets and new prognostic biomarkers (1, 2). The search for new proteins has relied primarily on antibody-based enzyme-linked immunosorbent assays (ELISAs) targeted to specified protein analytes (1). This approach has significant limitations in terms of its efficiency, reliability, and ability to find novel proteins.
The search for new proteins relevant for DKD was recently accelerated by the development and application of high throughput proteomics platforms. The SOMAscan aptamer-based proteomic platform was the first high throughput platform to be successfully used in large clinical and epidemiological studies (3–5). Unique single stranded sequences of DNA or RNA, referred to as aptamers, were demonstrated to recognize folded protein epitopes with high affinity and specificity. This property was further advanced by the Slow Off-rate Modified Aptamer (SOMAmer) utilized on the SOMAscan platform to assay concentrations of numerous proteins present in low volumes (75 μl) of biospecimens with excellent reproducibility and sensitivity.
The main goal of this study was to search for new circulating proteins that may be involved in the disease process that underlies progression of DKD to ESKD. To accomplish this goal, we measured 1,129 circulating proteins using the SOMAscan platform in multiple cohorts with type 1 diabetes (T1D) and type 2 diabetes (T2D) to identify important proteins associated with progression to ESKD. In previous studies, we used a targeted approach and examined 334 of these proteins representing specific candidate pathways for association with progression to ESKD (6–9). In this report, we used a hypothesis-free untargeted proteomic approach to examine the remaining 795 SOMAscan measured circulating proteins for association with progression to ESKD.
RESEARCH DESIGN AND METHODS
To obtain robust findings that can be generalized to all subjects with diabetes, profiles of circulating proteins were studied in four independent cohorts with T1D and T2D that were at various stages of DKD and were followed for 7–12 years to ascertain progression to ESKD. Study subjects were selected from the Joslin Kidney Study (JKS) and the Pima Indian Kidney Study. Selection of individuals in the study cohorts is described below.
Joslin Kidney Study
The Joslin Diabetes Center Committee on Human Studies approved the informed consent, recruitment and examination procedures for the JKS. The JKS is a longitudinal observational study that investigates the determinants and natural history of kidney function decline in T1D and T2D. Approximately 2,000 subjects with T1D and 1,500 subjects with T2D were recruited into the JKS from among 20,000 subjects attending the Joslin Clinic between 1991 and 2009. The protocols for recruitment, examination, and determination of clinical characteristics for these subjects were previously described (6,7). Briefly, subjects attending the Joslin Clinic aged 18–64 for T1D and 35–64 for T2D, who had had persistent Macro- or Micro- Albuminuria, were enrolled into the JKS. In addition, an equal number of subjects of the same age with Normo-Albuminuria were also enrolled. All subjects had a special examination at enrollment with blood and urine specimens obtained and Bio Banked in −85C. Similar examinations were performed biannually during subjects’ routine clinic visits or at their homes if they had stopped coming to the clinic. All JKS participants were queried every two years against the United States Renal Data System (USRDS) and the US National Death Index (NDI) rosters to ascertain individuals who developed ESKD or died.
Discovery and Replication Cohorts with late DKD:
We selected the T1D Discovery (n=219), and the T2D Replication (n=144) cohorts from participants enrolled in the JKS between 1991 and 2009 with Macro-albuminuria and impaired kidney function (eGFR 20–60 ml/min/1.73m2) at baseline. These subjects were monitored over 7–12 years for changes in kidney function and ESKD onset. Since DKD is a more homogenous disease in T1D than in T2D, we used T1D subjects with late DKD as the Discovery cohort and the T2D subjects with late DKD as the Replication cohort. In addition the T1D cohort had the largest number of subjects who progressed to ESKD; therefore, it provided the most statistical power to search for circulating proteins associated with the study outcome.
Validation Cohort with early DKD:
The T1D Validation cohort (n= 238) included JKS participants with Macro-albuminuria and eGFR 60 – 170 ml/min/1.73m2 (median eGFR 97 ml/min/1.73m2) at enrollment. They were also followed for 7–12 years to monitor kidney function changes and for ascertainment of ESKD onset.
Pima Indian Kidney Study
The study was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Disease. Pima Indians from the Gila River Indian Community in Arizona have a high prevalence of T2D and a high incidence of ESKD due to diabetes (10). This population participated in a longitudinal study of diabetes and its complications for more than 40 years. Beginning in the 1990s, informative subsets of subjects from this population were selected for more detailed longitudinal studies of DKD (11,12). The current T2D Validation cohort (n=153) included subjects from this kidney study cohort who had baseline examinations between 1994 and 2007 with baseline measured GFR of 60 – 240 ml/min (median GFR 150 ml/min) and had enough serum to be used in the proteomics analysis. More information regarding measurements of clinical characteristics including iothalamate GFR, obtaining specimens at baseline and during follow-up and ESKD ascertainment are described in a previous publication (11).
Determination of Kidney Function
In the JKS, GFR was estimated from the serum creatinine concentration using the CKD Epidemiology Collaboration equation and was expressed in ml/min/1.73m2 (13). In the Pima Indian Kidney Study, GFR was measured by the urinary clearance of iothalamate and expressed in ml/min. In both cohorts, ESKD was defined as chronic kidney disease requiring dialysis, kidney transplant, or death attributed to DKD based on the USRDS roster or a listing of kidney failure among the causes of death on a national death index (NDI) death certificate (14,15). Descriptions of other clinical measurements performed in these cohorts were reported previously (6,7).
SOMAscan proteomics platform to measure concentration of circulating proteins
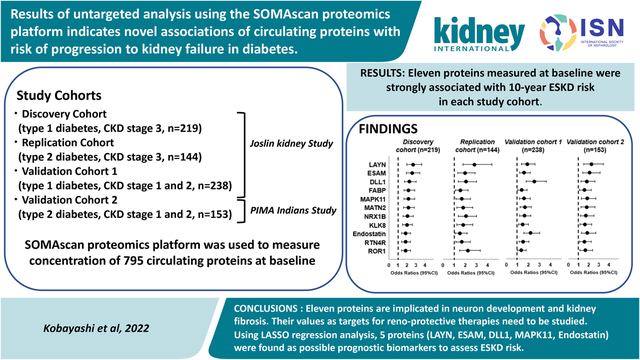
Concentrations of circulating proteins were measured using the SOMAscan proteomics platform as described previously (SomaLogic, Inc.; Boulder, CO, USA). Due to the high cost of measurements of all proteins (N=1,129) present on SOMAscan platform, we performed measurements in two stages (see Fig. 1). In the 1st stage, all 1,129 proteins on the SOMAscan platform were measured in 113 (56 ESKD cases) individuals selected randomly from among the 219 included in the T1D Discovery (late DKD) Joslin Cohort. Proteins potentially associated with risk of ESKD in these individuals were identified by univariate logistic regression analysis. In these analyses, 103 proteins with Odds Ratio (OR) < 0.7 (P<0.01) were identified as potentially protective, and 129 proteins with OR >1.60 (P<0.01) were identified as potential risks. The above thresholds were determined by considerations on the statistical power of the study. All other proteins (n=897) were considered unassociated with risk of ESKD.
Figure 1. Two stage study design for SOMAscan measurements.
* Parenthesis shows number of proteins which we measured only in ½ Discovery cohort
The aim of the 2nd stage was to measure half of the proteins present on the SOMAscan platform, i.e. 564 proteins, in the rest of the cohorts. In the smaller panel, we included all 232 potentially ESKD associated proteins and 332 out of 897 proteins not associated with risk of ESKD that were selected randomly from among all SOMAscan proteins. Proteins on the smaller panel were measured in baseline samples in the rest of the Discovery Cohort and in the 3 other cohorts. Twelve anchored samples were included in all batches to allow for universal calibration. The measurements in baseline plasma specimens from the Joslin Cohorts and in baseline serum specimens from the Pima Indian Cohort were performed at the lab of the SomaLogic, Inc.; Boulder, CO, USA. The measurements in the 1st and 2nd stage were performed using the same 1,129 protein SOMAscan platform, except that in the 2nd stage only 564 proteins were read.
The SOMAscan data were analyzed as outlined in Figure 1. In the first 4 publications, we analyzed circulating proteins that were members of specific pathways implicated in the etiology of DKD. These included 194 inflammatory proteins (6), 42 axon guidance pathway proteins (7), 73 candidate proteins identified as protective factors against progression to ESKD (8), and 25 TGF-β pathway related proteins (9). In total, 334 proteins were examined. In the present study, we use an untargeted approach without an a priori hypothesis to search for circulating proteins associated with risk of ESKD among the remaining 254 proteins measured in both the 1st and 2nd stages and 541 proteins measured only in the 1st stage (Figure 1); in total 795 proteins.
Statistical Analyses
Clinical characteristics and summary of outcomes were expressed as counts and percentages (proportions) for categorical variables, means (standard deviations) for normally distributed continuous variables, and as medians (25th–75th percentiles) for variables with skewed distributions.
Effects of baseline proteins on 10-year risk of ESKD were estimated using a logistic regression model and were expressed in terms of OR and their confidence interval (CI). To evaluate the results of the study in the whole discovery cohort, a Bonferroni correction for n=795 independent tests was used (254 proteins examined in both stages and 541 examined only in 1st stage), which yielded a threshold of P < 6.3×10−5. Nominal significance P < 0.05 was used to evaluate the findings obtained in the other study cohorts. Multivariable logistic regression analyses were performed to examine the association of specific candidate proteins with risk of ESKD in all study cohorts. In these analyses, the protein levels were modeled as 1-quartile change for easier interpretability and uniform scaling of the OR, and the models were adjusted for sex, duration of diabetes, HbA1c, SBP, and baseline GFR, ACR, and stratified by subject cohort.
To identify the best overall set of predictors of progression to ESKD, the least absolute shrinkage and selection operator (LASSO) regression analysis was used for variable selection. LASSO shrinks coefficients for weaker predictors toward zero. The degree of shrinkage is determined by an optimal parameter lambda. Optimal lambda, a penalty factor for penalized maximum likelihood estimation, was calculated by 10-fold cross-validation at its minimum level. Prognostic performances of logistic regression models for the selected variables by LASSO regression were compared with continuous net reclassification improvement (NRI) and Uno’s C-statistic in the data set comprising the four cohorts. We used receiver operating characteristic (ROC) curve analysis with the Youden index to determine the optimal cutoff value of the models that maximized the sum of sensitivity and specificity. Statistical analyses were performed with SAS version 9.4 (SAS Institute, Cary, NC), and R (version 4.0.3. and glmnet package for LASSO selection).
RESULTS
Characteristics of study cohorts
By design, the 4 independent study cohorts representing both T1D and T2D and early and late CKD stages differed at baseline with regard to age, diabetes duration, glycemic control, systolic blood pressure, ACR and GFR. In the Joslin cohorts, 88% of subjects were of European ancestry, whereas all subjects in the Pima cohort were American Indians. During 10 years of follow-up, 227 of 754 individuals progressed to ESKD. Baseline clinical characteristics of individuals who progressed and did not progress to ESKD during 10-year follow-up are shown in Table 1. Those who progressed to ESKD had higher HbA1c, systolic blood pressure, ACR and lower GFR than those who did not progress. These differences were observed in all 4 study cohorts.
Table 1. |.
Clinical characteristics of study cohorts
| Joslin | Joslin | Joslin | Pima Indians | |||||
|---|---|---|---|---|---|---|---|---|
| T1D Discovery cohort | T2D Replication cohort | T1D Validation cohort 1 | T2D Validation cohort 2 | |||||
| Characteristics at baseline | Non-ESKD | ESKD | Non-ESKD | ESKD | Non-ESKD | ESKD | Non-ESKD | ESKD |
| N | 111 | 108 | 109 | 35 | 188 | 50 | 119 | 34 |
| Male/Female | 49/62 | 57/51 | 72/37 | 22/13 | 101/87 | 30/20 | 31/88 | 12/22 |
| Age (years) | 47 ± 10 | 42 ± 8 * | 60 ± 5 | 59 ± 7 | 39 ± 10 | 38 ± 9 | 46 ± 10 | 43 ± 9 |
| Diabetes duration (years) | 32 ± 10 | 28 ± 9 * | 15 ± 9 | 19 ± 9 * | 26 ± 9 | 26 ± 10 | 15 ± 7 | 17 ± 5 |
| HbA1c (DCCT, %) | 8.4 ± 1.5 | 9.3 ± 1.7 * | 7.5 ± 1.6 | 7.7 ± 1.4 | 8.8 ± 1.7 | 9.9 ± 1.5 | 8.9 ± 2.2 | 10.9 ± 2.1 * |
| SBP (mmHg) | 132 ± 20 | 138 ± 19 * | 139 ± 18 | 143 ± 21 | 130 ± 16 | 135 ± 18 | 123 ± 12 | 125 ± 12 |
| ACR (mg/g) | 404 (46, 871) | 1427 (709, 2637) *** | 163 (49, 614) | 1679 (643, 3447) *** | 481 (130, 986) | 1287 (679, 2281) *** | 3.5 (1.0, 11) | 232 (17, 1927)*** |
| eGFR (ml/min/1.73m 2 ) ┼ | 48 ± 9 | 38 ± 10*** | 51 ± 10 | 44 ± 11** | 100 ± 21 | 90 ± 21** | 153 ± 46 | 143 ± 49 |
T1D, Type 1 Diabetes; T2D, Type 2 diabetes; ESKD, End Stage Kidney Disease, i.e., dialysis or kidney transplant; HbA1c, hemoglobin A1c; SBP, Systolic blood pressure; ACR, urine albumin to creatinine ratio; eGFR, estimated glomerular filtration.
For Pima cohort, GFR (ml/min) was measured directly using urinary clearance of iothalamate.
Data are expressed as mean ± standard deviation or median (25th and 75th percentiles).
Mann-Whitney U test for continuous variables and Fisher exact test for sex proportion were performed for comparison between patients with non-ESKD and ESKD within 10-years in each cohort (*P<0.05, **P<0.01, ***P<0.001).
Circulating proteins associated with risk of ESKD – untargeted approach
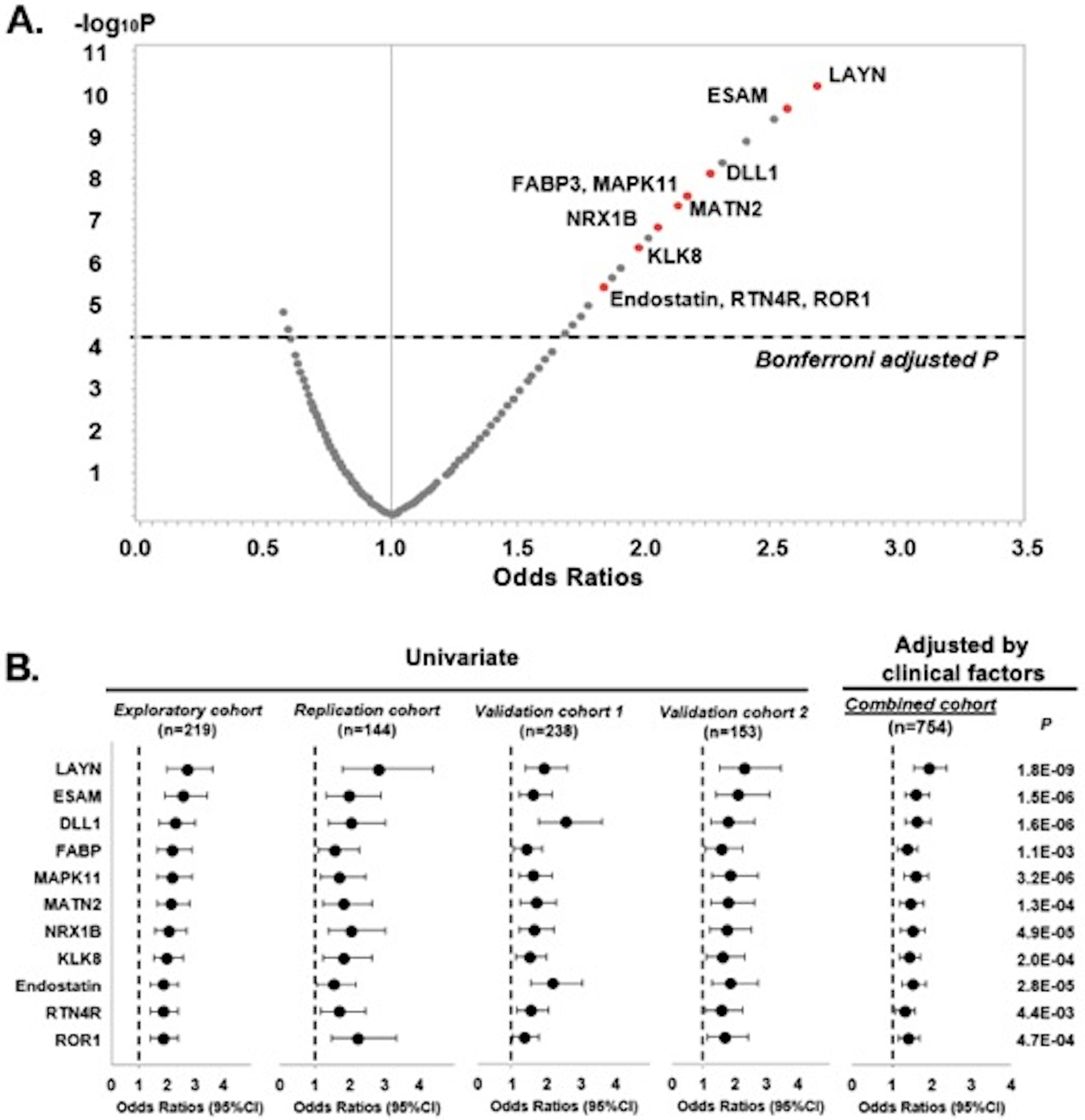
A volcano plot of Odds Ratios (OR) for progression to ESKD according to concentration of these proteins and P values is shown in Figure 2A. Thirty-two proteins showed highly statistically significant (P < 6.3×10−5, threshold for Bonferroni correction for multiple comparisons) association with risk of progression to ESKD. Out of these 32 proteins, 11 were robustly associated with progression to ESKD among subjects in each of the study cohorts, who had measurements done during the 2nd stage. The remaining twenty-one proteins were associated with risk of progression to ESKD, but only in some of the study cohorts. These proteins are not reported in further analyses. The results of univariate logistic regression analysis for the 11 proteins in each of the study cohorts are presented in Supplementary Table S1 and for the other 21 in Supplementary Table S2.
Figure 2. Results of logistic regression analysis for circulating proteins associated with risk of 10-year ESKD.
A. Volcano plot of effect sizes (Odds Ratio per 1-quartile increase in concentration of circulating proteins) and strengths of associations (p value- y axis) with risk of 10-year ESKD. Proteins that are considered in 2nd stage are presented in the figure.
A total of 32 potential candidate proteins were significantly associated with risk of 10-year ESKD. Red indicates 11 proteins that were confirmed in all study cohorts.
B. Odds Ratios and 95% CIs for risk of 10-year ESKD are presented per 1-quartile increase in protein level for each of the 11 candidate proteins in each cohort (univariate logistic analysis) and in combined cohort (multivariable logistic analysis adjusted by sex, duration of diabetes, HbA1c, systolic blood pressure, eGFR, and ACR). See also Supplementary Table 1 and Table 2
A graphical presentation of the findings from Supplementary Table S1 is shown in Figure 2B. Although subjects in the Discovery, Replication and Validation cohorts had different types of diabetes and different stages of CKD, the associations between the 11 candidate proteins and progression to ESKD, expressed as ORs, were remarkably similar among these cohorts. Therefore, these cohorts were combined for further analyses. In the combined cohort, multivariable logistic regression analysis was performed for each protein. The models were adjusted for sex, duration of diabetes, HbA1c, SBP, baseline GFR, and ACR (Figure 2B and Supplementary Table S1). All 11 candidate proteins were strongly associated with progression to ESKD.
Functional and biological characteristics of candidate proteins
Since the current study used an untargeted approach for selecting mechanisms/pathways of DKD progression, we used bioinformatics analysis to search for pathways and functions in which the candidate 11 proteins might be involved. Gene Ontology (GO) gene sets from the Broad Institute’s MSigDB (http://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=GO) were analyzed using R package, clusterProfiler (16). This analysis identified 9 neuron development related processes, including neuron differentiation, neurogenesis, regulation of nervous system development, axon development, neuron projection regeneration, response to axon injury, negative regulation of nervous system development and presynapse, which were enriched with 6 of 11 candidate proteins (ROR1, KLK8, DLL1, RTN4R, NRX1B, and MATN2) with P < 1.0×10−2 for enrichment (Table 2). The pathway of organic acid binding was enriched with 3 of 11 candidate proteins (LAYN, FABP3, and RTN4R). Three proteins (ESAM, MAPK11 and endostatin) were not enriched in any of the pathways in the GO Database.
Table 2. |.
Biological Processes in Gene Ontology enriched by genes targeted by 11 candidate proteins (genes)
| Biological processes | Gene Ratio | Background Ratio | P | Targets |
|---|---|---|---|---|
| Neuron Differentiation | 6/11 | 137/819 | 4.4E-3 | ROR1/KLK8/DLL1/RTN4R/NRX1B/MATN2 |
| Neurogenesis | 6/11 | 158/819 | 9.3E-3 | ROR1/KLK8/DLL1/RTN4R/NRX1B/MATN2 |
| Regulation of nervous system development | 5/11 | 90/819 | 3.9E-3 | ROR1/KLK8/DLL1/RTN4R/NRX1B |
| Axon development | 4/11 | 68/819 | 9.2E-3 | KLK8/RTN4R/NRX1B/MATN2 |
| Neuron projection regeneration | 3/11 | 16/819 | 9.2E-4 | KLK8/RTN4R/MATN2 |
| Negative regulation of neuron differentiation | 3/11 | 21/819 | 2.1E-3 | KLK8/DLL1/RTN4R |
| Response to axon injury | 3/11 | 21/819 | 2.1E-3 | KLK8/RTN4R/MATN2 |
| Organic acid binding | 3/11 | 32/819 | 7.2E-3 | LAYN/FABP3/RTN4R |
| Negative regulation of nervous system development | 3/11 | 33/819 | 7.9E-3 | KLK8/DLL1/RTN4R |
| Presynapse | 3/11 | 33/819 | 7.9E-3 | ROR1/RTN4R/NRX1B |
Biological processes enriched with at least 3 candidate proteins and P < 1.0×10−2 are presented in the table.
ESAM, MAPK11, and endostatin did not enrich any biological processes in the GO database.
In addition to the GO analysis, we manually reviewed the literature to assess if the 11 candidate proteins were reported in kidney disease pathology in cellular or animal studies. The results of the GO analysis and the manual literature review are shown in Table 3. Importantly, 6 of 11 candidate proteins, LAYN, DLL1, MAPK11, MATN2, endostatin, and ROR1, were associated with kidney fibrosis in the manual review (17–23). Another 2 of 11 proteins, ESAM and NRX1B, were associated with adhesion of cells in the glomerulus (24–26). The remaining 3 proteins, FABP3, KLK8, and RTN4R, were not previously reported to be associated with kidney diseases.
Table 3. |.
Candidate proteins classified according to biological processes and relevance for mechanisms involved in kidney diseases.
| Target Full Name | Target | Biological Process in Gene Ontology Database | Cellular Component in Gene Ontology Database | Relevance for kidney diseases |
|---|---|---|---|---|
| Layilin | LAYN | Organic acid binding | No information | Profibrotic (17) |
| Endothelial cell-selective adhesion molecule | ESAM | Cell-cell junction assembly | Bicellular tight junction | Adhesion molecule of glomerulus (24,25) |
| Delta-like protein 1 | DLL1 | Neuron development related processes | Cytoplasmic vesicle | Profibrotic (18,19) |
| Fatty acid-binding protein, heart | FABP3 | Organic acid binding | Extracellular exosome | N/A |
| Mitogen-activated protein kinase 11 | MAPK11 | Negative regulation of muscle tissue | Nucleoplasm | Profibrotic (20) |
| Matrilin-2 | MATN2 | Neuron development related processes | No information | Profibrotic (21) |
| Neurexin-1-beta | NRX1B | Neuron development related processes | Component of membrane | Adhesion molecule of glomerulus (26) |
| Kallikrein-8 | KLK8 | Neuron development related processes | Secretory and cytoplasmic vesicle | N/A |
| Endostatin | Endostatin | Cell morphogenesis | Extracellular exosome | Profibrotic (22) |
| Reticulon-4 receptor | RTN4R | Neuron development related processes | Membrane raft | N/A |
| Tyrosine-protein kinase transmembrane receptor | ROR1 | Neuron development related processes | Actin filament bundle | Profibrotic (23) |
Candidate proteins as predictors of progression to ESKD
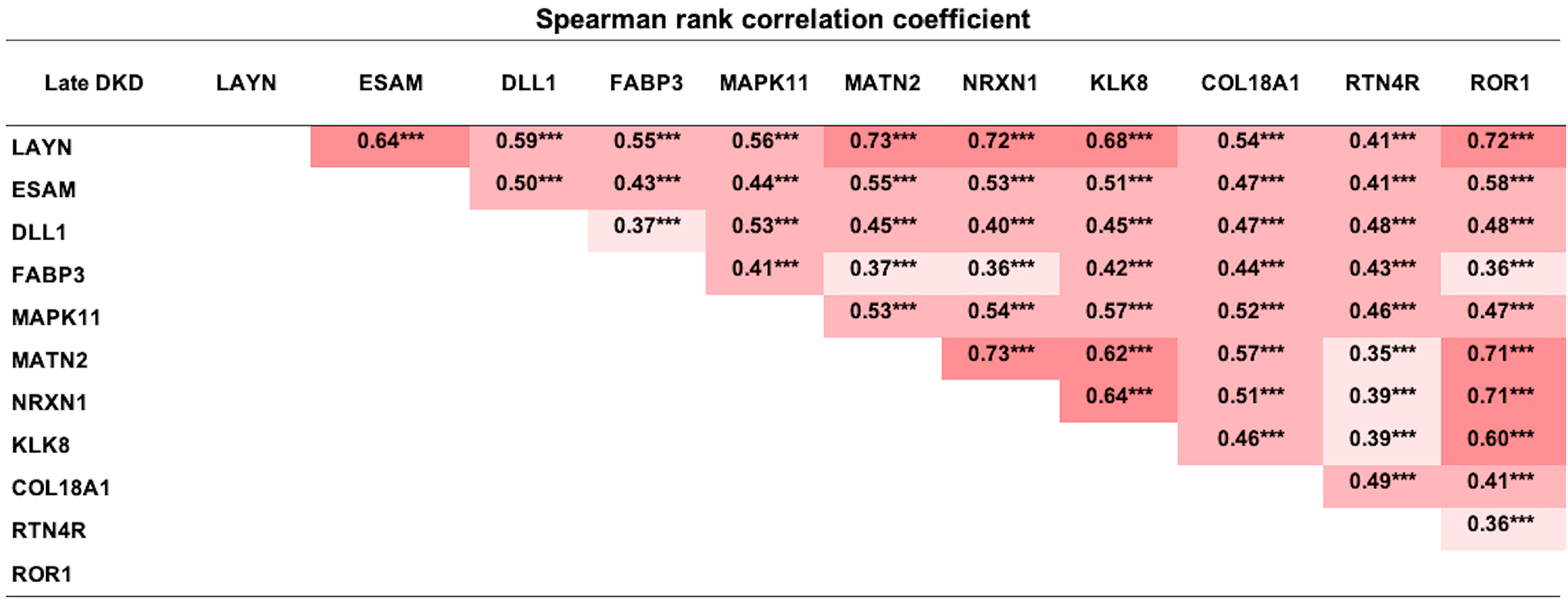
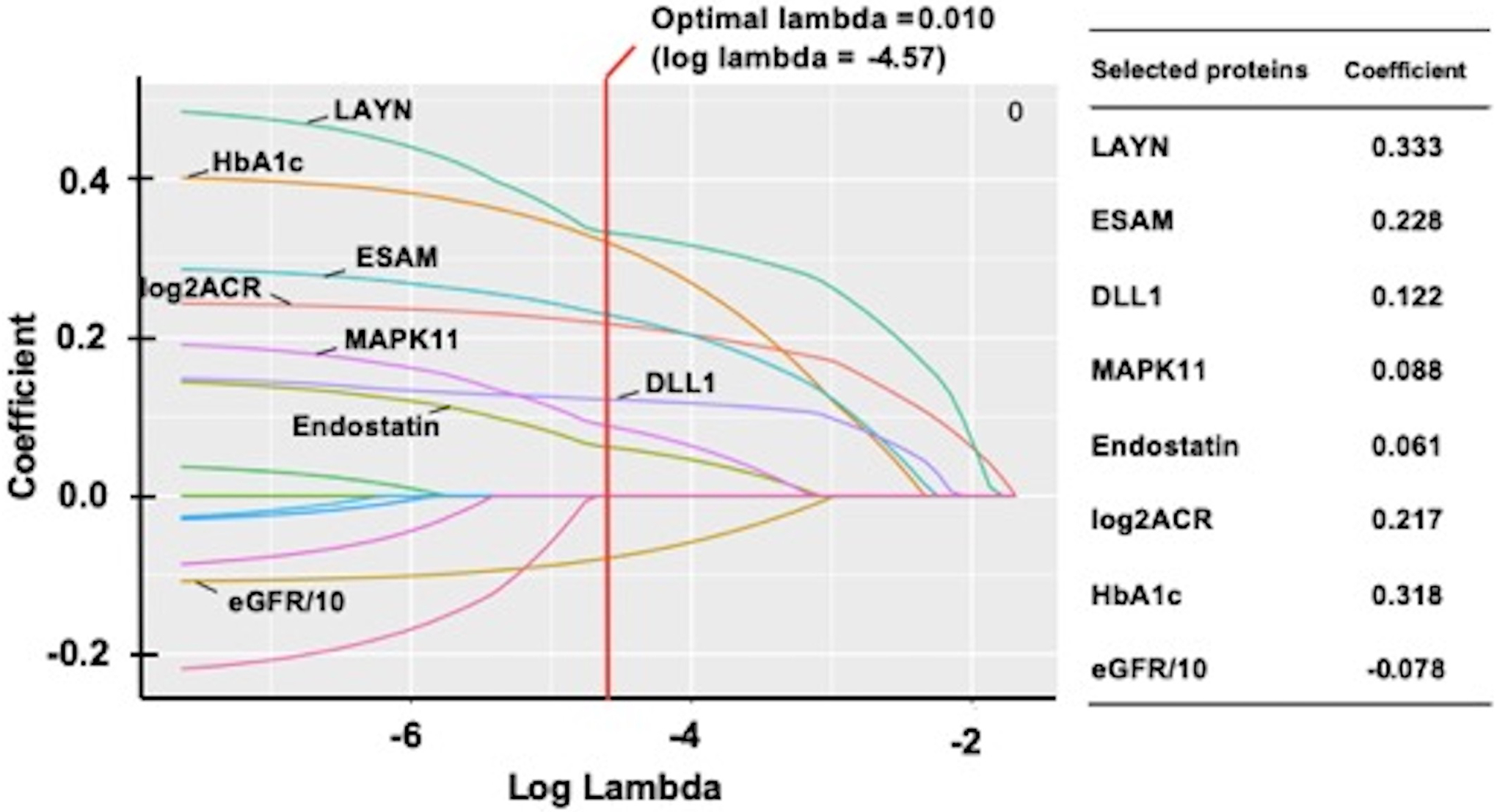
The relative concentrations of the 11 candidate proteins were inter-correlated. The strongest correlation was observed between circulating LAYN and the neuron development related proteins (Figure 3). Therefore, to identify which of the candidate proteins were the best independent predictors of 10-year risk of ESKD, LASSO logistic regression analysis was performed. The model included the 11 candidate proteins and clinical factors including ACR, HbA1c, and baseline GFR. As shown in Figure 4, LAYN, ESAM, DLL1, MAPK11, endostatin, log2ACR, HbA1c, and GFR/10 were selected as a set of biomarkers that can predict 10-year risk of ESKD. To examine the performance of this set of biomarkers as predictors of progression to ESKD, multivariable logistic regression models were employed in the combined cohorts (n=754). The 1st model included only clinical variables that were selected by logistic regression analysis with backward elimination. In the model, sex, duration of diabetes, HbA1c, SBP, eGFR, and log2ACR were included and, only three variables were selected as significant contributors to 10-year ESKD risk: eGFR, log2ACR and HbA1c. The 2nd model included the set of biomarkers selected by LASSO regression analysis including 3 clinical variables. Prognostic performance of these models was evaluated by C-statistics and continuous NRI, and ROC curve analyses were performed for each model to identify optimal cutoff with sensitivity and specificity (Supplementary figure S1). The C-statistic for the 1st model (clinical model) was 0.847; it increased to 0.869 for 2nd model. The improvement from the 1st model to the 2nd model was statistically significant by the Uno’s concordance statistic (2nd model vs 1st model: P=0.006). As assessed by the continuous NRI, adding LAYN, ESAM, DLL1, MAPK11, and endostatin (2nd model) to the 1st model resulted in significant improvement of reclassification (NRI: 0.55; P < 0.0001), with 31% of events correctly reclassified (P < 0.0001) and 23% of nonevents correctly reclassified (P < 0.0001).
Figure 3. Spearman rank correlation analysis for 11 candidate proteins in type 1 and type 2 diabetic subjects with CKD stage 3 in Joslin cohorts.
***P<0.001
Figure 4. Paths of regression coefficient for proteins and clinical factors selected as predictors of risk of 10-year ESKD shrinking towards zero using penalized LASSO logistic regression.
A total of 11 candidate proteins and clinical factors are included in the least absolute shrinkage and selection operator (LASSO) regression model and the coefficient of 8 selected variables are shown. Optimal lambda, a penalty factor for penalized maximum likelihood estimation, was calculated by 10-fold cross-validation at its minimum level. Each curve corresponds to a protein selected as a result of shrinkage for selection, and draws shrinkage during estimation of regression coefficient. LASSO penalizes the sum of the absolute values of regression coefficients, and a predictor with a coefficient of zero was excluded from the model and was not presented in the figure.
C-statistics of logistic regression for clinical model (HbA1c, log2ACR, and eGFR/10): 0.847. C-statistics of logistic regression for selected variables by LASSO regression analysis (HbA1c, log2ACR, eGFR/10, LAYN. ESAM. DLL1, MAPK11, endostatin): 0.869.
Clinical model vs New model: Difference in C-statistics, 0.022 (P=0.006); NRI, 0.55 (P<0.0001).
Summary of findings from SOMAscan analyses in Joslin Kidney Study and Pima Indian Kidney Study
To assess the importance of the current findings, we compared ORs of the 11 new proteins on risk of progression to ESKD with ORs for the other 35 proteins that we previously reported. Table 4 shows the list of the proteins reported as significant in each of our publications. After adjustments for baseline HbA1c, GFR, log2ACR, and cohort indicator, effects of some of the proteins became insignificant. This was seen for the protective proteins. In the original report, the study outcome was slow or no decline in eGFR during 10 years of follow-up. The results shown in Table 4 were obtained from logistic regression analysis where progression to ESKD was the outcome. Excluding the 8 protective proteins, all other proteins had highly statistically significant association with risk of progression to ESKD, with OR per 1-quartile increase in protein level varied between 1.27 and 2.27. It is important to notice that the proteins identified in the current study showed statistical significance and odds ratio on risk of ESKD that was similar to proteins in our other publications.
Table 4. |.
List of SOMAscan measured circulating proteins strongly associated with progression to ESKD in Joslin Kidney Study and Pima Indian Kidney Study
| Proteins reported | Multivariable logistic regression analysis* | LASSO results¶ | |||
|---|---|---|---|---|---|
| Gene name | Protein name | OR (95%CI) | P | ||
| Inflammatory proteins (17/194) (6) | |||||
| TNFRSF1A | TNF sR-I | 2.27 (1.82, 2.84) | 5.4E-13 | X | V |
| TNFRSF1B | TNF sR-II | 1.94 (1.58, 2.39) | 3.7E-10 | V | |
| IL15RA | IL-15 Ra | 1.88 (1.55, 2.30) | 4.2E-10 | ||
| TNFRSF19 | TAJ | 1.84 (1.50, 2.25) | 3.3E-9 | ||
| RELT | RELT | 1.75 (1.43, 2.15) | 7.9E-8 | V | |
| EDA2R | XEDAR | 1.69 (1.40, 2.04) | 5.6E-8 | X | |
| TNFSF15 | TNFSF15 | 1.61 (1.34, 1.94) | 5.6E-7 | X | |
| CCL15 | MIP-5 | 1.59 (1.32, 1.92) | 1.4E-6 | X | |
| CCL14 | HCC-1 | 1.55 (1.28, 1.88) | 4.4E-6 | ||
| TNFRSF21 | DR6 | 1.55 (1.28, 1.88) | 9.5E-6 | V | |
| CSF1 | CSF-1 | 1.51 (1.26, 1.82) | 1.2E-5 | ||
| HAVCR2 | TIMD3 | 1.47 (1.21, 1.78) | 7.4E-5 | V | |
| IL17F | IL-17F | 1.45 (1.18, 1.77) | 3.4E-4 | ||
| CD300C | CLM6 | 1.40 (1.17, 1.68) | 2.3E-4 | ||
| CD55 | DAF | 1.37 (1.15, 1.64) | 5.7E-4 | ||
| IL18R1 | IL-18 Ra | 1.35 (1.13, 1.62) | 8.7E-4 | ||
| IL1R1 | IL-1 sRI | 1.27 (1.06, 1.52) | 8.8E-3 | ||
| Axon guidance proteins (6/42) (7) | |||||
| EPHA2 | Epithelial cell kinase | 2.01 (1.62, 2.49) | 1.7E-10 | X | V |
| UNC5C | UNC5H3 | 1.81 (1.47, 2.22) | 1.4E-8 | V | |
| EFNA4 | Ephrin-A4 | 1.79 (1.46, 2.20) | 3.2E-8 | ||
| EPHB6 | EphB6 | 1.55 (1.27, 1.89) | 1.9E-5 | V | |
| EFNA5 | Ephrin-A5 | 1.50 (1.24, 1.82) | 3.5E-5 | ||
| EPHB2 | EPHB2 | 1.39 (1.16, 1.67) | 4.3E-4 | ||
| TGF-β signaling proteins (4/25) (9) | |||||
| NBL1 | DAN | 2.27 (1.81, 2.84) | 1.2E-12 | X | V |
| FSTL3 | FSTL3 | 1.75 (1.43, 2.15) | 9.2E-8 | V | |
| RGMB | RGMB | 1.35 (1.11, 1.64) | 2.6E-3 | ||
| TGFBR3 | TGF-b R III | 1.16 (0.96, 1.40) | 1.1E-1 | ||
| Protective proteins (8/73) (8) | |||||
| TNFSF12 | TWEAK | 0.71 (0.60, 0.85) | 2.3E-4 | X | |
| FGF20 | FGF-20 | 0.80 (0.68, 0.95) | 1.1E-2 | X | V |
| ANGPT1 | Angiopoietin-1 | 0.88 (0.74, 1.05) | 1.5E-1 | X | |
| SPARC | ON | 0.95 (0.80, 1.12) | 5.2E-1 | ||
| APP | amyloid precursor protein | 0.93 (0.79, 1.11) | 4.2E-1 | ||
| PF4 | PF-4 | 0.97 (0.81, 1.15) | 6.9E-1 | ||
| DNAJC19 | DnaJ homolog | 1.03 (0.87, 1.23) | 7.3E-1 | ||
| CCL5 | RANTES | 0.98 (0.83, 1.16) | 8.3E-1 | ||
| Untargeted proteins (11/795) Current publication | |||||
| LAYN | Layilin | 1.85 (1.50, 2.27) | 6.3E-9 | ||
| DLL1 | DLL1 | 1.61 (1.33, 1.96) | 1.8E-6 | X | |
| ESAM | ESAM | 1.60 (1.33, 1.94) | 1.2E-6 | X | |
| MAPK11 | MK11 | 1.58 (1.30, 1.91) | 3.3E-6 | X | |
| COL18A1 | Endostatin | 1.50 (1.23, 1.82) | 4.3E-5 | ||
| NRXN1 | NRX1B | 1.45 (1.19, 1.75) | 1.7E-4 | ||
| KLK8 | kallikrein 8 | 1.43 (1.19, 1.73) | 1.7E-4 | ||
| MATN2 | MATN2 | 1.38 (1.15, 1.67) | 7.0E-4 | ||
| FABP3 | FABP | 1.36 (1.13, 1.64) | 1.1E-3 | ||
| ROR1 | ROR1 | 1.36 (1.13, 1.63) | 1.2E-3 | V | |
| RTN4R | Nogo Receptor | 1.31 (1.09, 1.58) | 4.3E-3 | ||
| Clinical variables | |||||
| HbA1c | 1.38 (1.25, 1.52) | 8.9E-11 ** | X | ||
| GFR/10 | 0.83 (0.77, 0.90) | 5.2E-6 ** | X | ||
| log2ACR | 1.63 (1.49, 1.79) | 4.8E-25 ** | X | ||
Multivariable logistic regression analyses for each circulating protein adjusted by HbA1c, GFR, log2ACR, and cohort indicator were performed in the combined cohort (n=754). Effect size (Odds Ratio and 95% CI) for risk of 10-years ESKD are presented per 1-quartile increase in protein level.
Odds Ratios of univariate logistic regression analysis for each clinical variable.
List of proteins selected by LASSO regression model.
Significant proteins associated with eGFR slope after adjustment for baseline eGFR and ACR reported by Ngo et al. (27)
Many of the 46 ESKD associated circulating proteins were highly inter-correlated. To identify those least correlated and valuable for the development of prognostic indexes, we performed a LASSO logistic regression analysis. A total of 11 such proteins and 3 clinical variables were selected as a set of independent predictors of ESKD. Importantly, 3 of the 11 proteins, including DLL1, ESAM, and MAPK11, were identified in the current study, see Table 4. All these proteins can be considered candidate biomarkers to develop a prognostic index to identify individuals with diabetes at high risk of ESKD. Due to too small study cohorts, we have not pursued such a goal. In Table 4, we compared our findings with those reported in general population cohorts by Ngo et al. Out of 46 ESKD associated circulating proteins reported in our studies, 12 proteins were positively associated with eGFR decline in studies reported by Ngo et al. (27). This agreement further supports the generalizability of our findings, considering the significant differences between the studies. As an outcome, we used progression to ESKD, the gold standard; they used eGFR slope, a surrogate for ESKD. We used only patients with diabetes with early and late DKD; they studied a general population with a small proportion of subjects with impaired kidney function and a small proportion of subjects with diabetes.
Discussion
Using a global proteomic approach, we found 11 previously unreported circulating proteins associated with risk of progression to ESKD in diabetes. These associations were very robust and were validated in patients from four cohorts with type 1 and type 2 diabetes, with different CKD stages, and in different races. Although the mechanisms accounting for these associations are presently unknown, progression to ESKD is ultimately associated with tissue remodeling and fibrosis in the kidney, and these proteins may in some way contribute to or be associated with these pathways.
Applying bioinformatics analysis and using Gene Ontology Databases, we identified statistically significant association of 6 candidate proteins (DLL1, MATN2, NRX1B, KLK8, RTN4R and ROR1) with neuron related pathways. This finding compliments our recent discovery that 6 axon guidance pathway proteins (EFNA4, EFNA5, EPHA2, EPHB2 EPHB6 and UNC5C) significantly contributed to progression to ESKD (7). Taken together, the results of these two studies point to the important role of neuron development and repair related proteins in mechanisms that underlie progression to ESKD, i.e., tissue remodeling and development of kidney fibrosis. Two possible mechanisms linking these proteins with risk of ESKD can be postulated. The first is indirect and posits that variations in circulating concentration of the candidate proteins contribute to the development of neuropathy and specifically to autonomic neuropathy. Alterations in kidney structure and function in response to autonomic neuropathy may contribute to kidney injury and progressive kidney decline (28). Several publications have shown an association between autonomic neuropathy and risk of kidney function decline in diabetes (29,30). The second mechanism may be a direct effect of variations in circulating concentrations of the candidate proteins on podocytes. Podocytes share many similarities with neurons, which are polarized and form microtubule dependent processes with branching morphology with actin-based projections (dendrites and foot processes) (31). Along with these morphological characteristics, several reports highlight the functional and biochemical similarities between podocytes and neurons, and an increasing number of proteins, known as neuron-specific proteins, are now identified in podocytes (31). The staining intensity of NRX1B in podocyte is lowered, and their staining pattern is shifted to a more discontinuous patchy pattern, in disease models showing severe proteinuria (26). In addition, ROR1 is one of the important components of the planar cell polarity pathway that regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes (32). Further studies are needed to better understand the biological mechanisms linking elevated levels of the circulating candidate proteins with podocyte damage.
Independent from the bioinformatics analysis, a manual literature review of the candidate proteins identified 6 proteins (DLL1, MATN2, ROR1, LAYN, MAPK11, endostatin) implicated in kidney fibrosis in in vivo or in vitro studies (see references for Table 3). Importantly the first 3 proteins were also members of multiple neuron related pathways, whereas the last 3 pro-fibrotic proteins were not, suggesting their association with different mechanism of progressive kidney function decline. The role of the two other candidate proteins, ESAM and FABP3, could neither be classified as neuron related pathway proteins nor as pro-fibrotic proteins.
In univariate analysis, all 11 candidate proteins showed strong associations with risk of ESKD. However, in multivariable LASSO regression analysis only 5 of these proteins, LAYN, ESAM, DLL1, MAPK11, endostatin, were selected as independent predictors of progression to ESKD. Among these proteins only DLL1 belonged to the neuron related candidate proteins. The absence of the other neuron related proteins among the predictors most likely resulted from the fact that their effect was represented/accounted for by LAYN. Concentration of LAYN was highly correlated with concentrations of the neuron related proteins. The other predictors identified in the LASSO regression analysis were less correlated with neuron related proteins and with LAYN, so their association with risk of ESKD might reflect their involvement in other biological pathways or different down-stream targets. Clinically relevant features of the 5 proteins selected as predictors of ESKD are described below.
LAYN was the strongest predictor of future progression to ESKD but, unfortunately, there is limited literature regarding this protein. LAYN, a transmembrane protein with a C-type lectin-like domain, has been reported as a receptor of hyaluronic acid (33). Studies in mice showed that LAYN was expressed in various organs, including the liver, heart, kidney, and spleen (34). In an in vitro study, LAYN was involved in the TNF-α-induced epithelial mesenchymal transition (EMT) of kidney tubular epithelial cells, implying that it may be involved in the generation of kidney interstitial fibrosis (17). LAYN signaling was also involved in the enhancement of inflammation (35). Although our study showed strong correlations of LAYN with other neuron related proteins, no literature suggests possible mechanisms to account for such associations. The source of circulating LAYN and its regulation are unknown.
ESAM is a member of the immunoglobulin superfamily that is selectively expressed by vascular endothelial cells (25). It was reported that ESAM was highly expressed in glomerular endothelial cells, and its level was significantly reduced in streptozotocin-induced diabetic mice. Importantly, urinary ACR in ESAM−/− mice was significantly higher than in ESAM+/+ mice, suggesting that ESAM regulates vascular permeability in the glomerulus (24). However, our study showed that an elevated circulating level of ESAM is associated with high risk of ESKD. We cannot reconcile these discrepant findings.
DLL1 is a transmembrane ligand protein that binds the extracellular domain of NOTCH 1–3 receptors. DLL1 plays a role in central nervous system development at a different level by regulating neuronal differentiation of neural precursor cells (36). In addition, DLL1 maintains quiescence of neural stem cells and plays a role as a fate determinant that segregates asymmetrically to one daughter cell during neural stem cells mitosis (37). Studies implicate the involvement of the NOTCH signaling pathway, with DLL1 as its ligand, in the development of kidney fibrosis (18). Activation of this pathway in kidney tubular cells was both necessary and sufficient for development of kidney fibrosis. In human kidney tissue (various primary diseases), degree of glomerulosclerosis, severity of tubulointerstitial fibrosis and eGFR correlated with expression of cleaved NOTCH1 receptor evaluated by immunostaining in podocytes and in tubulointerstitium (19). Our study is the first to show the importance of elevated DLL1 in progressive kidney function decline and progression to ESKD. It is important to note that circulating level of NOTCH1 receptor was not associated with progression to ESKD in our study (Data are not shown).
MAPK11 is one of the four p38 MAPKs that play an important role in the cascade of cellular responses evoked by extracellular stimuli, such as proinflammatory cytokines or physical stress, leading to direct activation of transcription factors. Importantly, p38 MAPK plays an important role in glucose-induced EMT in tubular epithelial cells (20).
Endostatin is a fragment of collagen XVIII that is highly expressed in kidney glomeruli and peritubular capillaries (38). Importantly, endostatin is involved in fibrosis of the aging kidney (22). It is formed during extracellular matrix remodeling and has anti-angiogenic effects that may lead to impaired kidney repair after injury. Elevated levels of circulating endostatin level may reflect endothelial dysregulation. In a recent nested case-control study in the Action to Control Cardiovascular Disease (ACCORD) trial, plasma endostatin predicted kidney outcomes in patients with T2D during 4.6 years of follow-up (39). Our current study expands this result to patients with T1D and to Pima Indians.
This is our fifth report that examines the association of circulating proteins measured on the SOMAscan platform version 1 with the risk of progression to ESKD among individuals with DKD. The four previous reports (6–9) targeted 334 circulating proteins that were included in 4 candidate pathways hypothesized to be important in the development and progression of DKD. Thirty-five of those proteins were found to be significantly associated with progression to ESKD. This study focused on the remaining 795 proteins that were measured on the SOMAscan platform. As discussed above, the 11 circulating proteins that were found to be associated with progression to ESKD are important markers/indicators of disease processes that underly progressive kidney decline. However, since many of these proteins were inter-correlated, we used LASSO logistic regression to select proteins that were least correlated. We found 5 such proteins and they can be of value as candidate biomarkers for the development of a prediction index to identify subjects at high risk of ESKD. In this study, however, we did not attempt to develop such an index due to too small study cohorts.
The strength of all our reports, including this one, was the inclusion of multiple different cohorts of patients with diabetes, clearly demonstrating that the abnormalities in circulating proteins preceded the onset of ESKD in all cohorts. A limitation of both the current and previous findings is that they are derived from follow-up observational studies. Therefore, we cannot determine whether the circulating proteins are simply markers of risk of progression to ESKD or if they are causally related to this outcome. The latter needs to be established through animal studies and clinical trials. Additionally, because our study cohorts were small and not population-based, we were not able to assess prognostic performance of the candidate circulating proteins with certainty, and clinical relevance of the biomarkers are still uncertain. Further studies with large cohorts and long follow-up are needed.
Supplementary Material
Supplementary Figure S1. Receiver Operating Characteristic (ROC) curve analysis for each model Optimal cutoff points are calculated by Youden Index, which was calculated as sensitivity + specificity – 1. First model included log2ACR, HbA1c, and GFR/10. Second model included log2ACR, HbA1c, GFR/10, LAYN, ESAM, DLL1, MAPK11, and endostatin.
Supplementary Table S1. Results of logistic regression analysis for 11 candidate proteins associated with 10-year follow-up risk of ESKD in each study cohort
Supplementary Table S2. List of 21 potential candidate proteins association with 10-year follow-up risk of ESKD replicated only partially in other study cohorts
Acknowledgments
Funding:
We acknowledge grants support from: the National Institutes of Health (NIH) (DK041526, DK110350 and DK126799) to A.S.K, the Novo Nordisk Foundation grant NNF14OC0013659 (PROTON) to A.S.K. The Uehara Memorial Foundation (Postdoctoral Fellowship), and the Japan Society for the Promotion of Science (Overseas Research Fellowship) to H.K. The Mary K. Iacocca Fellowship, the Sunstar Foundation, Japan (Hiroo Kaneda Scholarship) and the Foundation for Growth Science from Japan to E.S. This research was also supported by the American Diabetes Association (Clinical Science Award 1-08-CR-42) to R.G.N., by the Intramural Research Program of the NIH NIDDK to R.G.N., H.C.L. P.J.S. and by NIH DERC grant (P30 DK036836) to Joslin Diabetes Center. The datasets that support the findings of this study are available from the corresponding authors upon request in a deidentified manner.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Material:
Supplementary information is available on Kidney International’s web site.
Competing interests: A.S.K. and M.A.N. are co-inventor of the “TNF-R1 and TNF-R2 patent for predicting risk of ESRD”. This patent was licensed by the Joslin Diabetes Center to the Renalytix AI PLC. J.M.W is an employee of Eli Lilly and Company and holds equity in Eli Lilly and Company. K.L.D. is an employee of Eli Lilly and Company and has ownership interest in Eli Lilly and Company and Pfizer. The other authors of this report declare no competing conflicts of interest.
References
- 1.Colhoun HM, Marcovecchio ML. Biomarkers of diabetic kidney disease. Diabetologia. 2018; 61: 996–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coca SG, Nadkarni GN, Huang Y, et al. Plasma Biomarkers and Kidney Function Decline in Early and Established Diabetic Kidney Disease. J Am Soc Nephrol. 2017; 28: 2786–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold L, Ayers D, Bertino J, Bock C, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010; 5: e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016; 315: 2532–2541. [DOI] [PubMed] [Google Scholar]
- 5.Ngo D, Sinha S, Shen D, et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation. 2016; 134: 270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niewczas MA, Pavkov ME, Skupien J, et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med. 2019; 25: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satake E, Saulnier PJ, Kobayashi H, et al. Comprehensive search for novel circulating miRNAs and axon guidance pathway proteins associated with risk of end stage kidney disease in diabetes. J Am Soc Nephrol. 2021; 32: 2331–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Md Dom ZI, Satake E, Skupien J, et al. Circulating Proteins Protect Against Renal Decline and Progression to End Stage Renal Disease in Diabetes: Results of Global Proteomics Analysis. Science Translat Med. 2021; 13: eabd2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi H, Looker HC, Satake E, et al. Neuroblastoma suppressor of tumorigenicity 1 (NBL1) a novel circulating protein associated with progression to end-stage kidney disease in diabetes. 2021, (submitted). [DOI] [PMC free article] [PubMed]
- 10.Nelson RG, Newman JM, Knowler WC, et al. Incidence of end-stage renal disease in type 2 (non-insulin-dependent) diabetes mellitus in Pima Indians. Diabetologia. 1988; 31: 730–736. [DOI] [PubMed] [Google Scholar]
- 11.Nelson RG, Bennett PH, Beck GJ, et al. Development and progression of renal disease in Pima Indians with non-insulin-dependent diabetes mellitus. Diabetic Renal Disease Study Group. N Engl J Med. 1996; 335: 1636–1642. [DOI] [PubMed] [Google Scholar]
- 12.Weil EJ, Fufaa G, Jones LI, et al. Effect of losartan on prevention and progression of early diabetic nephropathy in American Indians with type 2 diabetes. Diabetes. 2013; 62: 3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levey AS, Stevens LA, Schmid CH, et al. ; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009; 150: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention, National Center for Health Statistics: Data Access—National Death Index. Available at: http://www.cdc.gov/nchs/ndi.htm. Queried September 1, 2013.
- 15.U.S. Renal Data System 2013 Annual Data Report.: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD: (2013). [Google Scholar]
- 16.Guangchuang Yu, Wang Li-Gen, Han Yanyan, et al. clusterProfiler: An r Package for Comparing Biological Themes Among Gene Clusters. Omics. 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi T, Arito M, Suematsu N, et al. Roles of layilin in TNF-alpha-induced epithelial-mesenchymal transformation of renal tubular epithelial cells. Biochem Biophys Res Commun. 2015; 467: 63–69. [DOI] [PubMed] [Google Scholar]
- 18.Bielesz B, Sirin Y, Si H, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010; 120: 4040–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murea M, Park JK, Sharma S, et al. Expression of Notch pathway proteins correlates with albuminuria, glomerulosclerosis, and renal function. Kidney Int. 2010; 78: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv ZM, Wang Q, Wan Q, et al. The role of the p38 MAPK signaling pathway in high glucose-induced epithelial-mesenchymal transition of cultured human renal tubular epithelial cells. PLoS One. 2011; 6: e22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Zhang M, Huang H, et al. High glucose-induced Matrilin-2 expression in mouse mesangial cells was mediated by transforming growth factor beta 1 (TGF-β1). Biochem Biophys Res Commun. 2016; 474: 303–308. [DOI] [PubMed] [Google Scholar]
- 22.Lin CH, Chen J, Zhang Z, et al. Endostatin and transglutaminase 2 are involved in fibrosis of the aging kidney. Kidney Int. 2016; 89: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y, Liang Y, Zhu X, et al. The signaling protein Wnt5a promotes TGF beta1-mediated macrophage polarization and kidney fibrosis by inducing the transcriptional regulators Yap/Taz. J Biol Chem. 2018; 293: 19290–19302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Ishida T, Cangara HM, et al. Endothelial cell-selective adhesion molecule regulates albuminuria in diabetic nephropathy. Microvasc Res. 2009; 77: 348–355. [DOI] [PubMed] [Google Scholar]
- 25.Kacso IM, Kacso G. Endothelial cell-selective adhesion molecule in diabetic nephropathy. Eur J Clin Invest. 2012; 42: 1227–1234. [DOI] [PubMed] [Google Scholar]
- 26.Saito A, Miyauchi N, Hashimoto T, et al. Neurexin-1, a presynaptic adhesion molecule, localizes at the slit diaphragm of the glomerular podocytes in kidneys. Am J Physiol Regul Integr Comp Physiol. 2011; 300: R340–8. [DOI] [PubMed] [Google Scholar]
- 27.Ngo D, Wen D, Gao Y, et al. Circulating testican-2 is a podocyte-derived marker of kidney health. Proc Natl Acad Sci U S A. 2020; 117: 25026–25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petras D, Koutroutsos K, Kordalis A, et al. The role of sympathetic nervous system in the progression of chronic kidney disease in the era of catheter based sympathetic renal denervation. Curr Clin Pharmacol. 2013; 8: 197–205. [DOI] [PubMed] [Google Scholar]
- 29.Orlov S, Cherney DZ, Pop-Busui R, et al. Cardiac autonomic neuropathy and early progressive renal decline in patients with nonmacroalbuminuric type 1 diabetes. Clin J Am Soc Nephrol. 2015; 10: 1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahrani AA, Dubb K, Raymond NT, et al. Cardiac autonomic neuropathy predicts renal function decline in patients with type 2 diabetes: a cohort study. Diabetologia. 2014; 57: 1249–1256. [DOI] [PubMed] [Google Scholar]
- 31.Boyer O, Mollet G, Dorval G. Neurological involvement in monogenic podocytopathies. Pediatr Nephrol. 2021. Mar 31, doi: 10.1007/s00467-020-04903-x. [DOI] [PubMed] [Google Scholar]
- 32.Babayeva S, Zilber Y, Torban E. Planar cell polarity pathway regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes. Am J Physiol Renal Physiol. 2011; 300: F549–560. [DOI] [PubMed] [Google Scholar]
- 33.Bono P, Rubin K, Higgins JM, et al. Layilin, a novel integral membrane protein, is a hyaluronan receptor. Mol Biol Cell. 2001; 12: 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borowsky ML, Hynes RO. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J Cell Biol. 1998; 143: 429e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asano K, Arito M, Kurokawa MS, et al. Secretion of inflammatory factors from chondrocytes by layilin signaling. Biochem Biophys Res Commun. 2014; 452: 85–90. [DOI] [PubMed] [Google Scholar]
- 36.Campos LS, Duarte AJ, Branco T, et al. mDll1 and mDll3 expression in the developing mouse brain: role in the establishment of the early cortex. J Neurosci Res. 2001; 64: 590–598. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi D, Furutachi S, Kawai H, et al. Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat Commun. 2013; 4: 1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomono Y, Naito I, Ando K et al. Epitope-defined monoclonal antibodies against multiplexin collagens demonstrate that type XV and XVIII collagens are expressed in specialized basement membranes. Cell Struct. 2002; 27: 9–20. [DOI] [PubMed] [Google Scholar]
- 39.Chauhan K, Verghese DA, Rao V, et al. Plasma endostatin predicts kidney outcomes in patients with type 2 diabetes. Kidney Int. 2019; 95: 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Receiver Operating Characteristic (ROC) curve analysis for each model Optimal cutoff points are calculated by Youden Index, which was calculated as sensitivity + specificity – 1. First model included log2ACR, HbA1c, and GFR/10. Second model included log2ACR, HbA1c, GFR/10, LAYN, ESAM, DLL1, MAPK11, and endostatin.
Supplementary Table S1. Results of logistic regression analysis for 11 candidate proteins associated with 10-year follow-up risk of ESKD in each study cohort
Supplementary Table S2. List of 21 potential candidate proteins association with 10-year follow-up risk of ESKD replicated only partially in other study cohorts


