Abstract
Aristaless-related homeobox (ARX) is a paired-like homeodomain transcription factor playing important roles in brain development. Patients with mutations in ARX have a spectrum of neurodevelopmental disorders such as epilepsy, intellectual disability, and autism spectrum disorder, with or without structural abnormalities of the brain such as lissencephaly (smooth brain), microcephaly (small brain), and/or agenesis of corpus callosum. Mouse models have provided important clues on the pathophysiologic roles of ARX in these disorders. However, successfully isolating specific in vivo complex of ARX, with DNA and proteins, has remained as a challenge. To facilitate in vivo detection of ARX complexes, we generated a mouse line containing one epitope of FLAG-tag (1 x FLAG) targeted at the translational start site of the endogenous Arx gene using CRSPR/Cas9 strategy. Homozygous Flag-Arx mice are viable and fertile without gross abnormality, suggesting that the FLAG-tag does not perturb normal function of ARX. Using a FLAG antibody, we successfully detected ARX with immunofluorescent staining and pulled down ARX in embryonic brain tissues. This Flag-Arx mouse model will be a useful tool to isolate ARX complexes from mouse tissues for many applications.
Keywords: ARX, Flag-Arx knock-in mouse, CRSPR/cas9, brain development, detection of FLAG tagged-endogenous protein
1. INTRODUCTION
The aristaless-related homeobox (ARX) protein plays important roles in brain development and mutations are associated with a spectrum of neurodevelopmental disorders with structural and/or functional deficits (Friocourt, 2010; Kato et al., 2004; Matsuo et al., 2001; Shoubridge et al., 2010). The structural defects include lissencephaly (smooth brain), microcephaly, and agenesis of the corpus callosum, whereas the functional defects include intellectual disability, early infantile epileptic encephalopathies and other seizure types, dyskinesias (involuntary movements), and autism spectrum disorder.
ARX is expressed both in the dorsal and ventral forebrain but with distinct patterns during embryonic development (Colombo et al., 2004; Miura et al., 1997). In the dorsal forebrain (i.e., pallium), ARX is expressed in the proliferating cells of the ventricular zone (VZ), however its expression is not detected once cells exit the cell cycle and begin migrating out of the VZ (Colombo et al., 2004; Miura et al., 1997). In contrast, in the ventral forebrain (i.e., subpallium) ARX expression is weak in the VZ and stronger in the subventricular, intermediate, and mantle zones (Colombo et al., 2004; Miura et al., 1997). Interestingly, the expression of ARX in cells derived from the subpallium, unlike cells emanating from the pallial VZ, remains on even after the cells have migrated and differentiated into interneurons. Given that the cortical projection neurons are derived from the dorsal forebrain while the cortical interneurons originate from the ventral forebrain (Butt et al., 2007; Lodato and Arlotta, 2015; Ohtaka-Maruyama and Okado, 2015; Wonders and Anderson, 2006), this differential expression of ARX in the dorsal vs. ventral forebrain suggests distinct roles of ARX in these two populations of neurons, thus contributing differently to the spectrum of phenotypes (McKenzie et al., 2007; Olivetti and Noebels, 2012; Ruggieri et al., 2010; Shoubridge et al., 2010; Strømme et al., 2002). In fact, dorsal specific depletion of ARX function in mice leads to distinct phenotypes from those associated with ventral loss (Colasante et al., 2015; Marsh et al., 2009; Nasrallah et al., 2012; Simonet et al., 2015). Furthermore, we have shown that specifically ablating ARX expression in the dorsal progenitor cells, exhibit microcephaly but no seizures (Colasante et al., 2015), whereas loss of ARX in the ventral forebrain results in severe epilepsy without microcephaly (Marsh et al., 2009).
These findings prompt us to investigate the molecular basis of ARX function in distinct populations of cells. However, identification of transcriptional targets of ARX in mice via chromatin immunoprecipitation sequencing (ChIP-seq) has been challenging in part due to the performance of ARX antibodies, although ChIP-seq in Neuro2A cells transfected with Arx, or ChiP-qPCR and Chip-on-chip in mouse embryonic brains have been reported previously (Friocourt, 2011; Fulp et al., 2008; Quillé et al., 2011). In order to overcome this issue, we have generated a knock-in mouse model expressing FLAG-tagged ARX which facilitates pull-down of ARX protein in vivo via FLAG antibody. We verified that FLAG-tagged ARX reliably recapitulated most of the normal ARX expression in the embryonic mouse brains and the FLAG antibody successfully pulled down FLAG-ARX. Thus, this new mouse model will be a useful tool for the future studies involving isolation of ARX complex with DNA or with other transcription-associated factors.
2. RESULTS AND DISCUSSION
Flag-Arx knock-in mouse line was generated by introducing one epitope of the FLAG (GAC TAC AAG GAC GAC GAT GAC AAG) sequence into the first exon of Arx, immediately after the initiation codon, using the CRISPR/Cas9 system (Fig. 1A). N-or C-terminal tagging of the FLAG is often used to facilitate the endogenous detection of proteins, whose antibodies are not available or with poor performance (Ferrando et al., 2014). However, usually more than one copy of the FLAG epitope is tethered to the protein to improve detection of often low endogenous expression of the target proteins. However, the addition of multiple FLAG sequences increases the risk of disrupting the structure and function of the tagged protein. Therefore, to minimize potentially disrupting ARX function, a single copy of FLAG sequence was introduced into the Arx locus.
Figure 1.
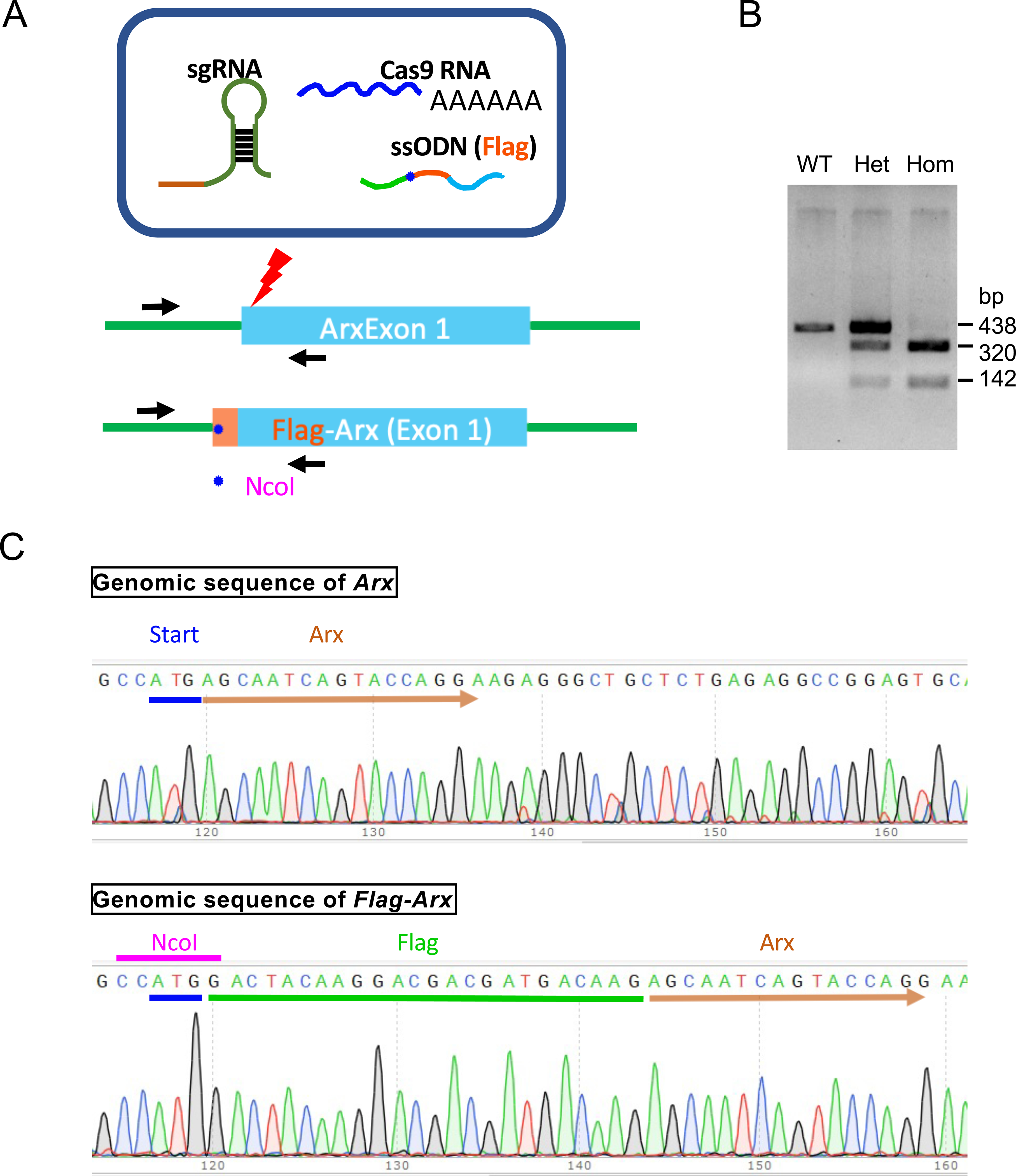
Generation of Flag-Arx knock-in (KI) mouse with CRISPR/Cas9 system. A. CRISPR/Ca9-mediated genome editing strategy to generate Flag-Arx KI mouse. sgRNA, single guidance RNA; ssODN, single strand oligonucleotide donor. NcoI, restriction enzyme site recognized by NcoI. B. Genotyping results of wild type (WT), Flag-Arx KI het (Het), and Flag-Arx KI homo (Hom). bp: base pair. C. Sequencing results of genomic DNA from wild type (Arx) and Flag-Arx KI (Flag-Arx) mice.
The genotype of each mouse was confirmed by PCR amplification followed by NcoI restriction enzyme digestion, which generates digested fragments only in Flag-Arx allele (Fig. 1B). We have also verified the N-terminal insertion of the Flag sequence into Arx locus by Sanger sequencing (Fig. 1C). Homozygous Flag-Arx knock-in mice are viable and reproduce normally. Expected phenotypes with reduced or loss of ARX include microcephaly, agenesis of corpus callosum, and epilepsy, but homozygous Flag-Arx knock-in mice did not exhibit these phenotypes in multiple litters produced over five generations bred to C57BL/6, suggesting that FLAG-tagged ARX functions normally.
Next, we examined the expression of FLAG-ARX by immunofluorescent labeling using both anti-FLAG and anti-ARX antibodies. The detection of the single FLAG tag, as predicted, required antigen retrieval and fluorescent amplification methods (Fig. 2 and see Materials and Methods). We successfully detected FLAG-ARX with anti-FLAG antibody in E14.5 embryonic brains. Furthermore, the specificity of the FLAG antibody labeling was confirmed by demonstrating co-localization with anti-ARX labeling. Detection of the FLAG tagged ARX was observed in all expected brain regions including the subventricular zone (SVZ) and outer SVZ of the ganglionic eminence (ventral telencephalon), as well as in the marginal zone (MZ) and cortical plate of the dorsal telencephalon (Figure 2). It is known that ARX expression is much lower in the cells of the cortical ventricular zone (VZ) when compared to GE cells. This was the one populations of cells we could not detect with FLAG antibody labeling, while a weak staining with anti-ARX antibody was detected (Figure 2) as previously reported (Colombo et al., 2004; Miura et al., 1997).
Figure 2.
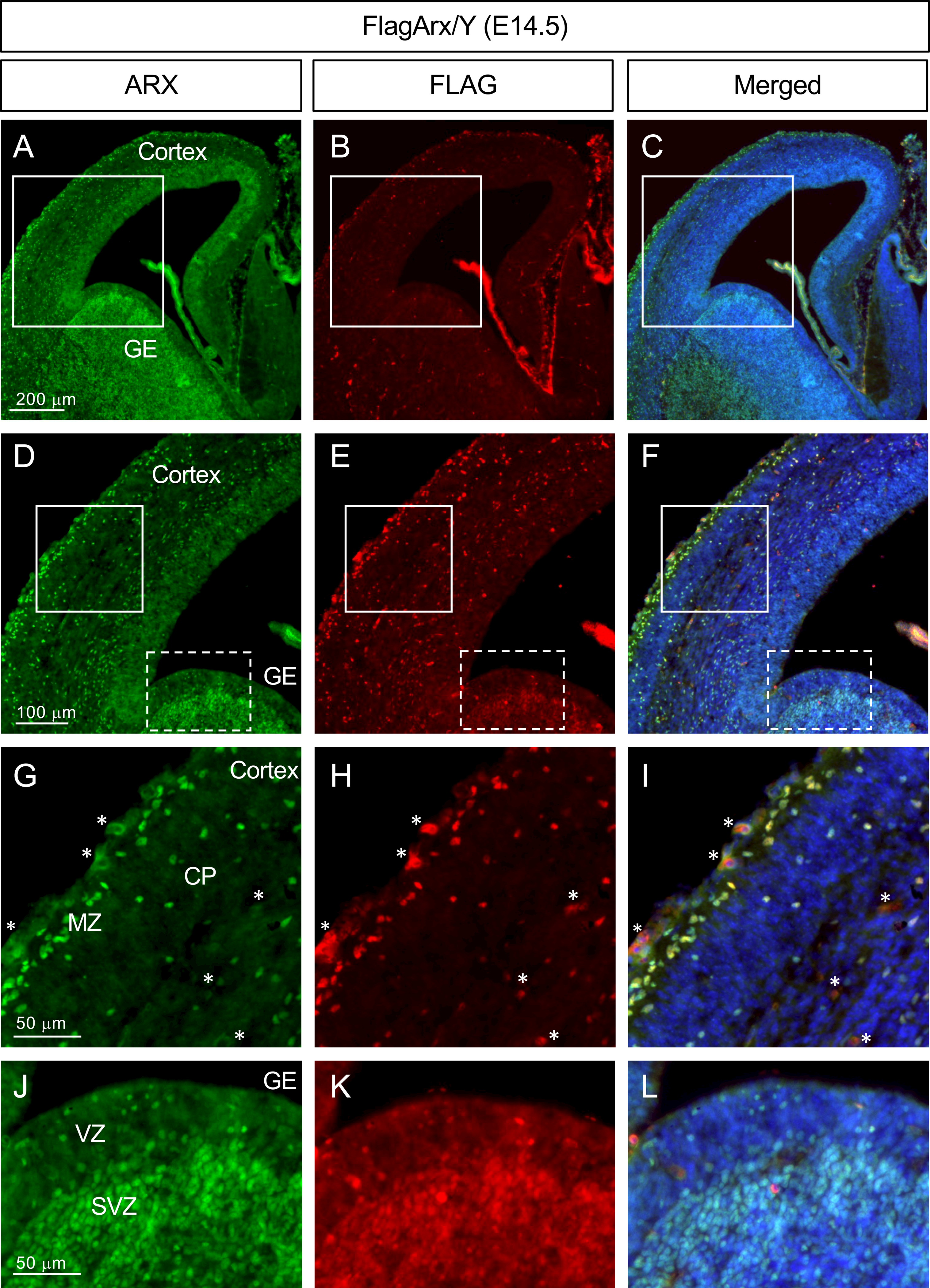
FLAG-tagged ARX expression detected by anti-FLAG immunofluorescent staining. Representative images of the embryonic cortex and ganglionic eminence (E14.5, coronal section) of the Flag-Arx KI mouse stained with anti-ARX (A, D, G, J; green)- or anti-FLAG (B, E, H, K; red) antibody (C, F, I, L: merged images). DAPI (blue) was used for nuclear staining. D,E,F: magnified images of the boxed area in A, B, C; G,H,I: magnified images of the solid boxed areas in D, E, F; J, K, L: magnified images of the dotted boxed areas in D, E, F. Abbreviations: CP, cortical plate; GE, ganglionic eminence; MZ, marginal zone; SVZ, subventricular zone; VZ, ventricular zone. Asterisks (*) in G, H and I are examples of non-specific blood vessel staining.
At E16.5 more interneurons have migrated from the GE to the cortex, and, as noted above, these cells continue to express ARX. Anti-FLAG and anti-ARX immunofluorescent labeling identified the migrating cells in the marginal zone and those that had reached the cortical plate (Figure 3).
Figure 3.
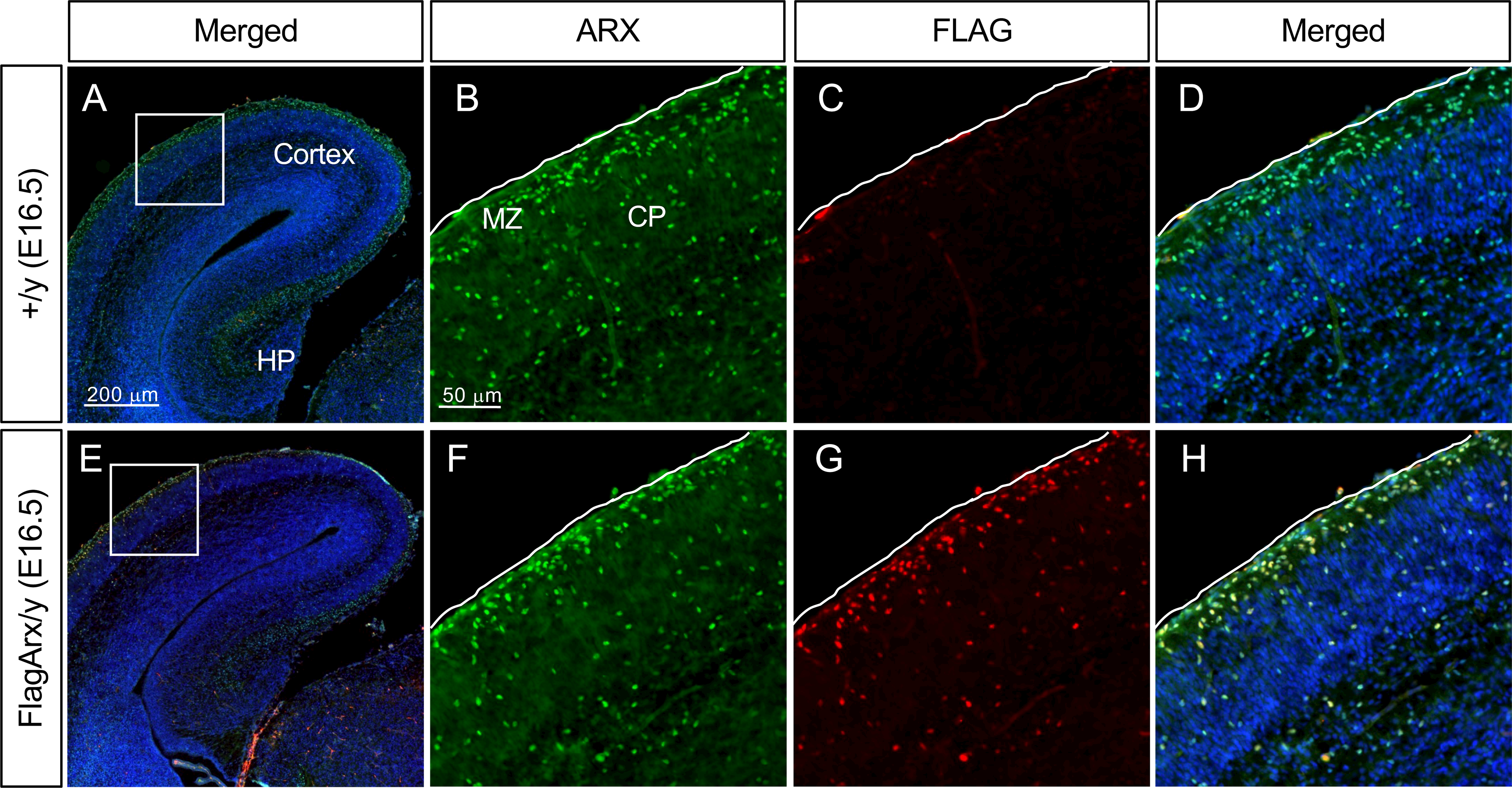
FLAG-tagged ARX recapitulates normal ARX expression. A-D, Representative images of the wild type (+/y) embryonic cortex stained with ARX (B) or FLAG (C) antibody (E16.5). B, C, D are magnified images of the boxed area in A (merged). E-H, Representative images of the Flag-Arx KI (FlagArx/y) embryonic cortex stained with ARX (F) or FLAG (G) antibody (E16.5). F, G, H are magnified images of the boxed area in E (merged). DAPI (blue): nuclear staining. CP, cortical plate; HP, hippocampus; MZ, marginal zone. Pial surface is outline with white line in each image.
To determine if the FLAG tagged ARX could be detected in biochemical assays, Western blot analysis of embryonic brain lysates were performed. At E12.5 few cells have migrated from the GE to the dorsal forebrain (Faux et al., 2012). Thus, lysates from isolated dorsal or ventral E12.5 forebrain were interrogated with anti-ARX (Figure 4A) and anti-FLAG (Figure 4B) antibodies. Both antibodies detected comparable levels of protein in both the dorsal and ventral forebrain, with slightly lower levels dorsally as expected (Figure 4A, B). Knowing that the vast majority of ARX positive cells in the dorsal telencephalon at this embryonic stage are derived from and located in the VZ, this result supports the conclusion that FLAG antibody can detect FLAG-ARX present in the cortical VZ by Western blot analysis, where we were not able to detect FLAG-ARX by immunofluorescence (data not shown).
Figure 4.
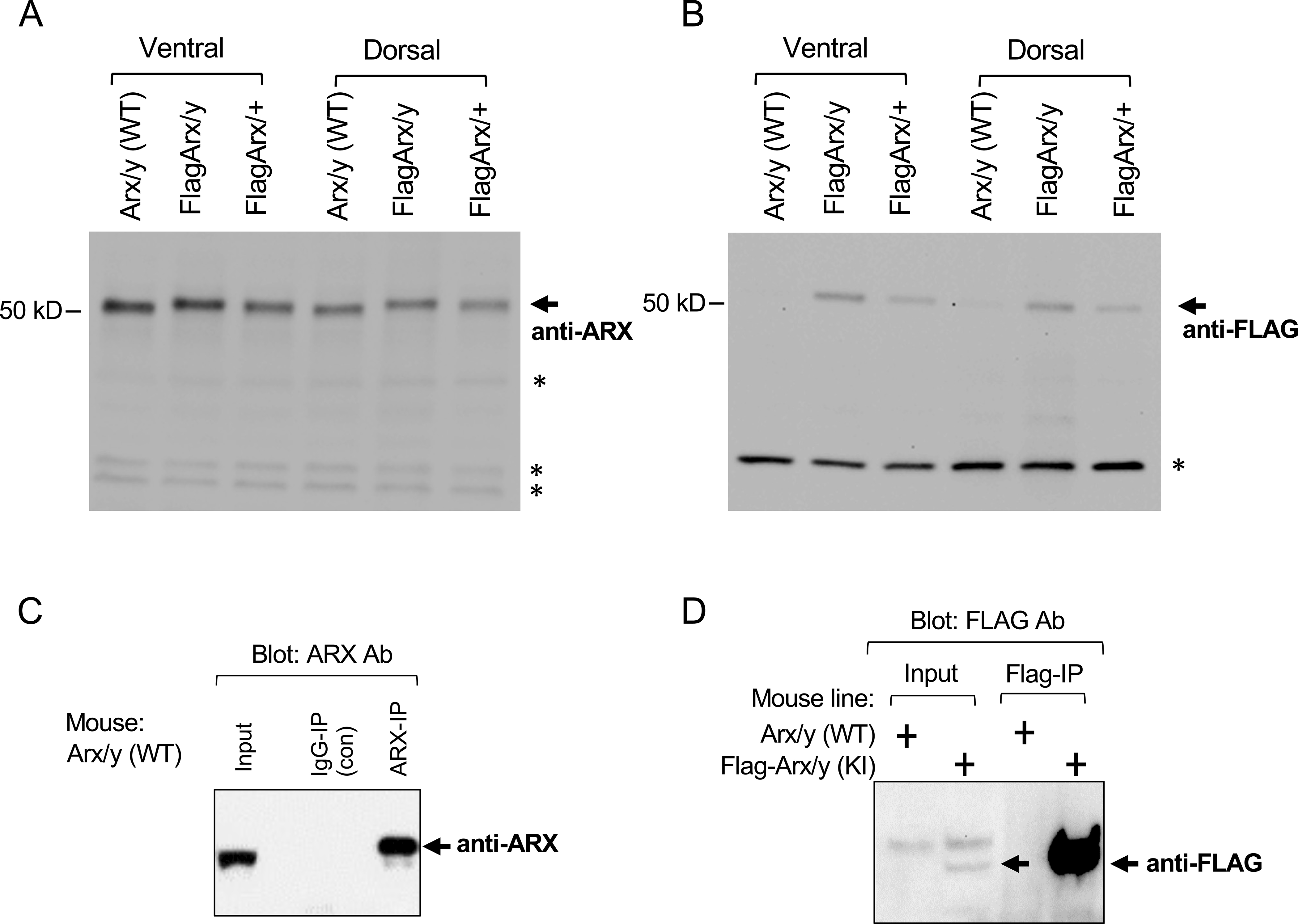
Biochemical analysis of Flag-Arx KI embryonic brain. A-B, Western blot analysis of the embryonic brain lysates (E12.5) probed with anti-ARX (A) or anti-FLAG (B) antibody. Dorsal and ventral telencephalon lysates from the WT male (Arx/y), Flag-Arx KI male (FlagArx/y), and Flag-Arx KI het female (FlagArx/+), were probed with sheep anti-ARX or mouse anti-FLAG antibody. Arrow indicates expected size of ARX or FLAG-ARX band. Asterisks indicate non-specific bands. C-D, Immunoprecipitation of ARX using wild type (C, D) and Flag-Arx KI embryonic brain lysates (D) (E14.5). For immunoprecipitation of ARX with anti-ARX antibody using WT brain lysates (IP with rabbit anti-ARX; Western with rabbit anti-ARX 1–221 from ref. Kitamura et al.), 2% of input and 30% of IP eluate were loaded (~3% of ARX was precipitated) (C). For immunoprecipitation of ARX with anti-FLAG antibody using WT and Flag-Arx KI brain lysates (IP with mouse anti-FLAG-M2; Western with rabbit anti-FLAG), 2% of input and 30% of IP eluate were loaded (~30% of ARX was precipitated) (D).
Finally, we performed immunoprecipitation of ARX using embryonic brain lysates from the Flag-Arx knock-in mouse or wild type control (E14.5) (Figure 4C, D). The anti-FLAG antibody specifically pulled down ARX in the brain lysates prepared from Flag-Arx KI, but not from WT mouse (Figure 4D). Immunoprecipitation of ARX with an anti-ARX antibody was also performed as a control (Figure 4C).
In summary, we have generated a Flag-Arx KI mouse line and successfully detected FLAG-ARX with immunostaining as well as with Western blot using FLAG antibody. Furthermore, FLAG antibody successfully pulled down ARX in vivo from the brain lysates isolated from Flag-Arx KI mouse, with higher efficiency than the anti-ARX antibody. We anticipate this mouse line will be a useful tool for other analyses requiring ARX complex isolation (e.g., ChIP-seq) as the function of this gene is further elucidated in understanding a range of human neurodevelopmental conditions.
3. MATERIALS AND METHODS
3.1. Generation of Flag-Arx knock-in mouse
All animal experiments were performed in accordance with the relevant guidelines and regulations approved by the Brigham and Women’s Hospital Institutional Animal Care and Use Committee. Flag-Arx knock-in mouse line was generated using CRISPR/Cas9 gene editing system. Cas9 mRNA was purchased from TriLink BioTechnologies (San Diego, CA), and all the oligonucleotides were synthesized at IDT integrated DNA Technologies (Coralville, IA). Targeting sequence for Cas9 within Arx genome locus was selected using an online tool (https://zlab.bio/guide-design-resources). The single guide RNA (sgRNA) was transcribed in vitro according to the manufacturer’s instruction (HiScribe™ T7 High Yield RNA Synthesis Kit, NEB). The donor template DNA bearing FLAG tag sequence was prepared by annealing complementary Ultramer oligonucleotides. Cas9 mRNA/sgRNA/donor template were injected into C57BL/6 mouse eggs at the DF/HCC transgenic mouse core. In-frame insertion of FLAG tag at the N-terminus of ARX was confirmed by sequencing (Macrogen, USA). The sgRNA (5’-AAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACGGTACTGATTGCTCATGGCTC-3’) was transcribed using the following oligos:
Template oligo for in vitro transcription of sgRNA, 5’-AAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACGGTACTGATTGCTCATGGCTCCCTATAGTGAGTCGTATTA-3’,
T7 primer, 5’- TAATACGACTCACTATAGGG-3’.
Ultramer for donor template:
Top strand, 5’-CCCCAACACACACCCTTTTCATCCAACCCTCGAGGAGAGAGGAGCAGAGGTCCGCTCTCTGAGCCCAAGGAAAAAGCACCAGCCATGGACTACAAGGACGACGATGACAAGAGCAATCAGTACCAGGAAGAGGGCTGCTCTGAGAGGCCGGAGTGCAAGAGTAAATCTCCAACTTTGCTCTCCTCCTACTGCATC-3’,
Bottom strand, 5’-GATGCAGTAGGAGGAGAGCAAAGTTGGAGATTTACTCTTGCACTCCGGCCTCTCAGAGCAGCCCTCTTCCTGGTACTGATTGCTCTTGTCATCGTCGTCCTTGTAGTCCATGGCTGGTGCTTTTTCCTTGGGCTCAGAGAGCGGACCTCTGCTCCTCTCTCCTCGAGGGTTGGATGAAAAGGGTGTGTGTTGGGG-3’
For genotyping, mouse genomic DNA was isolated from tail biopsies by quick NaOH extraction (Truett et al., 2000), and DNA fragment (Flag-Arx,462 bp; wild type, 438 bp) and was amplified by PCR using the following oligos: Flag-Arx-F, GGGACGGAAAGGAACAAAGATC, Flag-Arx-R, GGGACTTGTCAAGTTGGAGAC. PCR-amplified DNA was digested with NcoI restriction enzyme which generates 142 bp and 320 bp DNA fragments only from the Flag-tag inserted allele.
A total of five founder mice were obtained as a result of Cas9 mRNA/sgRNA/donor template injection into C57BL/6 mouse eggs. Among these, only one had the correct Flag sequence targeted to the Arx locus, immediately after the initiation codon, and this line was used for all analyses in this study after backcrossing to C57BL/6 for five generations. Two of the other lines lost initiation codon, consequently producing 5’ deletion (40 amino acid) in Arx, and the other two were wild type. The characterized mouse line will be available to the research community upon acceptance of the manuscript.
3.2. Immunofluorescence staining and imaging
Embryonic brains were fixed with 4% paraformaldehyde overnight and processed for cryosection as described previously (Lim et al., 2019). Coronal sections (14 μm) were used for immunostaining. Briefly, sections were subjected to antigen retrieval (boiled for 10–15 min in antigen unmasking solution, citrate-based, Vector Lab) (Lim et al., 2019; Shi et al., 2020), blocked with R.T.U. animal free-blocker (Vetor Lab) for 30 min, and incubated with primary antibody (FLAG-M2, 1:1000, Sigma-Aldrich; anti-ARX, 1: 100, sheep polyclonal, R&D Systems) overnight at 4°C. Next, the sections used for FLAG-M2 primary antibody were incubated with EnVision Flex+ mouse linker (raised in rabbit) (DAKO, neat) for 30min, goat anti-rabbit IgG secondary antibody conjugated with polymer-HRP, neat) for 30min, and finally Tyramide-594 (ThermoFisher) with H2O2 in amplification buffer provided by the manufacturer, for 5 min. The sections used for ARX primary antibody were incubated with biotinylated secondary antibody (goat anti-sheep IgG, ThermoFisher, 1:200) for 1hr and then with streptavidin-Alexa-488 (1:1000, ThermoFisher) for 30 min. DAPI was used for nuclei staining. Images were acquired on Zeiss Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-Flash4.0 camera using Zeiss Zen Pro software.
3.3. Immunoprecipitation and Western blot analysis
For E12.5 brains, the dorsal and ventral embryonic telencephalons were dissected out separately and used for Western blot analysis as described previously (anti-ARX antibody, 1:1000, sheep polyclonal, R&D; anti-FLAG antibody, FLAG-M2, mouse monoclonal, 1:1000, Sigma Aldrich) (Shi et al., 2020). For ARX immunoprecipitation, either the dorsal forebrains (Figure 4C) or the whole embryonic brains (Figure 4D) were isolated from C57BL/6 mouse (Figure 4C) or Flag-Arx KI (Flag-Arx/y) and WT (Arx/y) male mouse (Figure 4D) at E14.5. The brains were lysed in RIPA buffer (25 mM Tris-Cl, pH 7.4, 10% glycerol, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholic acid, 0.1% SDS,1 mM DTT, 1 mM EDTA, protease inhibitors; Roche Biochem) and sonicated on ice (Branson digital sonifier 250, 5 cycles of sonication at 2 sec “On” and 2 sec “Off’ at 10% output). The lysates were spun at 13,000 × g for 10 min at 4°C and the cleared lysates were incubated with an anti-ARX antibody (rabbit polyclonal, generated by ARX polypeptide, in Golden lab) for 1 hr at 44°C then protein G-conjugated beads (Invitrogen), or anti-FLAG M2 magnetic beads (Sigma Aldrich) for 1 hr at 4°C. The beads were washed with RIPA buffer five times and bound proteins were eluted using 2x sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer by boiling for 5 min. For Western blotting, 2% input and 30% eluate were run on 4–15% SDS-PAGE gel (Bio-Rad, Mini-PROTEAN® Precast Gels) and blotted on PVDF membrane (Bio-Rad,Trans-Blot® Turbo™ Mini PVDF Transfer Packs) using Trans-Blot® Turbo™ Transfer System (Bio-Rad). The blot was probed with anti-ARX antibody (rabbit polyclonal, against 158–181 amino acid, 1:1000, a gift from Dr. Kitamura)(Kitamura et al., 2002) or rabbit anti-FLAG antibody (1:1,000; Cell signaling Technology) and developed using ECL kit (SuperSignal Chemiluminescent Substrate, Thermo Scientific). The image was scanned using ChemiDoc MP imaging system (Bio-Rad).
Acknowledgments
The authors thank Dr. Kitamura (National Center of Neurology and Psychiatry, Japan) for providing anti-ARX antibody, and Xiuyu Shi and Brenna Stallings for maintaining the mouse line.
Funding information:
National Institute of Neurological Disorders and Stroke, Grant number: R01NS100007
REFERENCES
- Butt SJB, Cobos I, Golden J, Kessaris N, Pachnis V, Anderson S, 2007. Transcriptional Regulation of Cortical Interneuron Development. J Neurosci 27, 11847–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G, Simonet JC, Calogero R, Crispi S, Sessa A, Cho G, Golden JA, Broccoli V, 2015. ARX Regulates Cortical Intermediate Progenitor Cell Expansion and Upper Layer Neuron Formation Through Repression of Cdkn1c. Cereb Cortex 25, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Galli R, Cossu G, Gécz J, Broccoli V, 2004. Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev Dynam 231, 631–639. [DOI] [PubMed] [Google Scholar]
- Faux C, Rakic S, Andrews W, Britto JM, 2012. Neurons on the Move: Migration and Lamination of Cortical Interneurons. Neurosignals 20, 168–189. [DOI] [PubMed] [Google Scholar]
- Ferrando RE, Newton K, Chu F, Webster JD, French DM, 2014. Immunohistochemical Detection of FLAG-Tagged Endogenous Proteins in Knock-In Mice. J Histochem Cytochem 63, 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G and Parnavelas JG, 2010. Mutations in ARX result in several defects involving GABAergic neurons. Front. Cell. Neurosci. 4:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G and Parnavelas JG, 2011. Identification of Arx targets unveils new candidates for controlling cortical interneuron migration and differentiation. Front. Cell. Neurosci. 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA, 2008. Identification of Arx transcriptional targets in the developing basal forebrain. Hum Mol Genet 17, 3740–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Das S, Petras K, Kitamura K, Morohashi K, Abuelo DN, Barr M, Bonneau D, Brady AF, Carpenter NJ, Cipero KL, Frisone F, Fukuda T, Guerrini R, Iida E, Itoh M, Lewanda AF, Nanba Y, Oka A, Proud VK, Saugier-Veber P, Schelley SL, Selicorni A, Shaner R, Silengo M, Stewart F, Sugiyama N, Toyama J, Toutain A, Vargas AL, Yanazawa M, Zackai EH, Dobyns WB, 2004. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum. Mutat. 23, 147–159. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo S, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi K, 2002. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet 32, 359–369. [DOI] [PubMed] [Google Scholar]
- Lim Y, Cho I-T, Shi X, Grinspan JB, Cho G, Golden JA, 2019. Arx Expression Suppresses Ventralization of the Developing Dorsal Forebrain. Sci Rep-uk 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Arlotta P, 2015. Generating Neuronal Diversity in the Mammalian Cerebral Cortex. Annual Review of Cell and Developmental Biology 31, 699–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, Brooks-Kayal A, Golden JA, 2009. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain 132, 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo A, Matsuzaka T, Tsuru A, Moriuchi H, Nakashita Y, Tanaka S, Baba C, Tomimasu K, 2001. Epidemiological and clinical studies of West syndrome in Nagasaki Prefecture, Japan. Brain and Development 23, 575–579. [DOI] [PubMed] [Google Scholar]
- McKenzie O, Ponte I, Mangelsdorf M, Finnis M, Colasante G, Shoubridge C, Stifani S, Gecz J, Broccoli V, 2007. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience 146, 236–247. [DOI] [PubMed] [Google Scholar]
- Miura H, Yanazawa M, Kato K, Kitamura K, 1997. Expression of a novel aristaless related homeobox gene “Arx” in the vertebrate telencephalon, diencephalon and floor plate. Mech Dev 65, 99–109. [DOI] [PubMed] [Google Scholar]
- Nasrallah MP, Cho G, Simonet JC, Putt ME, Kitamura K, Golden JA, 2012. Differential effects of a polyalanine tract expansion in Arx on neural development and gene expression. Hum Mol Genet 21, 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka-Maruyama C, Okado H, 2015. Molecular Pathways Underlying Projection Neuron Production and Migration during Cerebral Cortical Development. Front. Neurosci. 9, 4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti PR, Noebels JL, 2012. Interneuron, interrupted: molecular pathogenesis of ARX mutations and X-linked infantile spasms. Current Opinion in Neurobiology 22, 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillé M-L, Carat S, Quéméner-Redon S, Hirchaud E, Baron D, Benech C, Guihot J, Placet M, Mignen O, Férec C, Houlgatte R, Friocourt G, 2011. High-Throughput Analysis of Promoter Occupancy Reveals New Targets for Arx, a Gene Mutated in Mental Retardation and Interneuronopathies. PLoS ONE 6, e25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri M, Pavone P, Scapagnini G, Romeo L, Lombardo I, Volti GL, Corsello G, Pavone L, 2010. The aristaless (Arx) gene: one gene for many “interneuronopathies”. Front Biosci (Elite Ed) 2, 701–710. [DOI] [PubMed] [Google Scholar]
- Shi X, Lim Y, Myers AK, Stallings BL, Mccoy A, Zeiger J, Scheck J, Cho G, Marsh ED, Mirzaa GM, Tao T, Golden JA, 2020. PIK3R2/Pik3r2 Activating Mutations Result in Brain Overgrowth and EEG Changes. Ann Neurol 88, 1077–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge C, Fullston T, Gécz J, 2010. ARX spectrum disorders: making inroads into the molecular pathology. Hum. Mutat. 31, 889–900. [DOI] [PubMed] [Google Scholar]
- Simonet JC, Sunnen CN, Wu J, Golden JA, Marsh ED, 2015. Conditional Loss of Arx From the Developing Dorsal Telencephalon Results in Behavioral Phenotypes Resembling Mild Human ARX Mutations. Cerebral Cortex 25, 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SME, Bruyere H, Lütcherath V, Gedeon ÁK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SGM, Fryns J-P, Sutherland GR, Mulley JC, Gécz J, 2002. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30, 441–445. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML, 2000. Preparation of PCR-Quality Mouse Genomic DNA with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 29, 52–54. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA, 2006. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696. [DOI] [PubMed] [Google Scholar]


