Abstract
Background
The evidence linking vitamin D (VitD) levels and spontaneous intracerebral hemorrhage (ICH) remains inconclusive. We tested the hypothesis that lower genetically determined VitD levels are associated with higher risk of ICH.
Methods and Results
We conducted a 2 sample Mendelian Randomization (MR) study using publicly available summary statistics from published genome‐wide association studies of VitD levels (417 580 study participants) and ICH (1545 ICH cases and 1481 matched controls). We used the inverse‐variance weighted approach to generate causal estimates and the MR Pleiotropy Residual Sum and Outlier and MR‐Egger approaches to assess for horizontal pleiotropy. To account for known differences in their underlying mechanism, we implemented stratified analysis based on the location of the hemorrhage within the brain (lobar or nonlobar). Our primary analysis indicated that each SD decrease in genetically instrumented VitD levels was associated with a 60% increased risk of ICH (odds ratio [OR], 1.60; [95% CI, 1.05–2.43]; P=0.029). We found no evidence of horizontal pleiotropy (MR‐Egger intercept and MR Pleiotropy Residual Sum and Outlier global test with P>0.05). Stratified analyses indicated that the association was stronger for nonlobar ICH (OR, 1.87; [95% CI, 1.18–2.97]; P=0.007) compared with lobar ICH (OR, 1.43; [95% CI, 0.86–2.38]; P=0.17).
Conclusions
Lower levels of genetically proxied VitD levels are associated with higher ICH risk. These results provide evidence for a causal role of VitD metabolism in ICH.
Keywords: intracerebral hemorrhage, mendelian randomization, vitamin D
Subject Categories: Genetics, Precision Medicine
Vitamin D (VitD) metabolism is an appealing target for preventive interventions in cerebrovascular disease, as it can be easily and safely intervened upon. 1 A number of studies have explored the role of VitD in ischemic stroke. 2 , 3 Two observational studies demonstrated that depleted levels of VitD were associated with increased mortality and worse outcome in this condition. 2 , 3 Additionally, Zhou et al found that lower VitD levels were related to a higher risk of ischemic stroke in a large meta‐analysis. 4 Genetic studies have also examined the relationship between VitD levels and ischemic stroke. A Mendelian Randomization (MR) study evaluating the association between genetically determined VitD and ischemic stroke did not find an association between these 2 traits. 5 Despite the significant amount of research outlined above, the role of VitD metabolism in intracerebral hemorrhage (ICH) remains relatively unexplored. While ICH shares some biological features with ischemic stroke, the overall pathophysiology of these 2 types of cerebrovascular diseases is significantly different. We therefore aimed to use MR analyses to evaluate the role of circulating levels of VitD in ICH, hypothesizing that genetically reduced levels of VitD lead to higher risk of ICH.
METHODS
Because of the sensitive nature of the data collected for this study, requests to access the data set from qualified researchers trained in human subject confidentiality protocols may be sent to dbGap. Institutional Review Board approval was not required for this study.
Study Design
We conducted a 2‐sample MR study, a study design where the genetic information for the exposure (VitD levels) and the outcome (risk of ICH) come from different studies, using summary statistics from large genetic studies of these traits. Importantly, there was no overlap between both study populations.
Instruments
We used independent (r 2>0.1) single nucleotide polymorphisms (SNPs) associated with VitD levels at genome‐wide levels (P<5×10−8) 6 , 7 from the largest GWAS (Genome‐Wide Association Study) on VitD available to date. 7 Palindromic SNPs were excluded from all analyses. For the final list of VitD associated instruments, we abstracted effect estimates and standard errors from the largest GWAS of ICH conducted to date, 8 using proxy variants (r 2>0.9) where available. The average F‐statistic for the genetic instrument was 216 and the median was 39. The I2 for the genetic instrument was 0.99. In combination, these metrics suggest that the genetic instrument had sufficient strength. We did not adjust for ancestry in the MR analysis because both GWAS (for the exposure and for the outcome) conducted principal component analyses, a robust method to identify genetic ancestry, to identify study participants from European ancestry and exclude population outliers.
Statistical Analysis
Our primary MR analysis used the inverse‐variance weighted method. We tested for horizontal pleiotropy (the possibility that the effect of the instrument on the outcome of interest is exerted through a pathway other than VitD levels) using the MR‐Egger intercept and Mendelian Randomization Pleiotropy Residual Sum and Outlier (MR‐PRESSO) global test. In sensitivity analyses, we implemented the weighted median method and calculated the MR‐PRESSO outlier corrected estimate.
We also examined whether any of the instruments were potentially associated with known confounders of the association between VitD and ICH. To this end, we performed sensitivity analyses using Cook's distance, defining outlier instruments as those with a Cook's distance >4/number of SNPs. We also looked for evidence of SNP outliers with RadialMR. In addition, we performed sensitivity analyses using multivariable MR to adjust for the genetic contribution of VitD going through other pathways associated with ICH. Finally, we completed leave‐one‐out plots (Figures S1 through S3). We recalibrated the effect estimates to express the change in ICH risk associated with an SD decrease in genetically instrumented VitD levels. We declared statistical significance at P<0.05 (2‐tailed) when testing the single primary hypothesis that lower genetically determined VitD levels are associated with higher risk of ICH. We used R (version 3.6.0) 9 and its TwoSampleMR and MR‐PRESSO packages.
Results
From a GWAS of VitD that analyzed data on 417 580 people of European ancestry, we identified 108 SNPs that met our criteria to select instruments for this trait (Table 1). Of these, 88 SNPs (Table S1) had available summary results in a GWAS of ICH that included 1545 ICH cases (664 lobar and 881 nonlobar ICH cases) and 1481 matched controls of European ancestry (Table 1). Our primary analysis using the inverse‐variance weighted MR method indicated that each SD decrease in genetically instrumented VitD levels was associated with an 60% increased risk of ICH (odds ratio [OR], 1.60; [95% CI, 1.05–2.43]; P=0.029) (Figure). To rule out heterogeneity in estimates across SNPs, we calculated Cochran Q statistics, which did not reveal heterogeneity. For all ICH, Q was 105.01, with 87 degrees of freedom and P value of 0.09. For lobar ICH, Q was 89.43 with 87 degrees of freedom and P value of 0.41. For nonlobar ICH, Q was 88.77, with 88 degrees of freedom and P value of 0.46.
Table 1.
Characteristics of GWAS of Vitamin D and Intracerebral Hemorrhage
| Characteristic | GWAS of vitamin D | GWAS of intracerebral hemorrhage |
|---|---|---|
| Sample size | 417 580 | 3026 |
| Mean age, y | 47–66 [0.34–14.12] | 67 [SD, 10] |
| Female sex | 51% | 45% |
| Genotyping platform | UK Biobank Axiom Array |
Affymetrix 6.0 Illumina HumanHap610 |
| No. of SNPs evaluated | 8 806 780 | 5 258 103 |
| No. of genome‐wide significant loci | 143 | 2 |
GWAS indicates Genome‐Wide Association Study; and SNP, single nucleotide polymorphism.
Figure Figure. . Mendelian randomization plot.
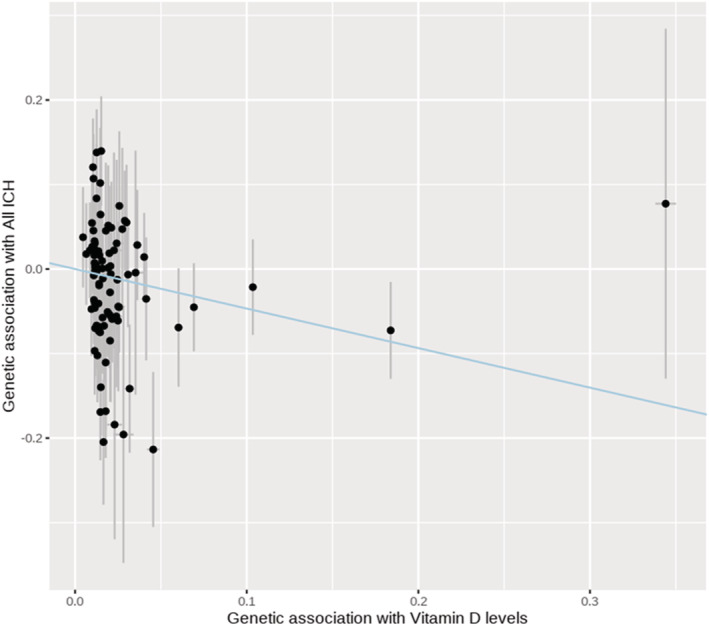
The plot presents the effect estimates of association tests between the single nucleotide polymorphisms and vitamin D levels (X axis) and risk of intracerebral hemorrhage (Y axis). The blue summary line corresponds to the slope of the inverse‐variance weighted method. ICH indicates intracerebral hemorrhage.
There was no evidence of significant horizontal pleiotropy, as evidenced by the nonsignificant results of the MR‐Egger intercept and MR‐PRESSO Global tests (both P>0.05, Table 2). The direction and size of the effect were consistent in secondary analyses aimed at reducing the impact of any pleiotropy (Table 2). Summary statistics for analyses stratified by location were based on the analysis of 664 lobar and 881 nonlobar ICH cases (Table 1). In these stratified analyses according to location (Tables S2 and S3), the results remained significant for nonlobar ICH (OR, 1.87; [95% CI, 1.18–2.97]; P=0.007) but not for lobar ICH (OR, 1.43; [95% CI, 0.86–2.38]; P=0.17) (Table 2). For nonlobar ICH, there was no evidence of significant horizontal pleiotropy (P>0.05 for both the MR‐Egger intercept and MR‐PRESSO Global tests) and the direction and size of the effect were consistent in secondary analyses aimed at reducing pleiotropy (Table 2). The direction of effect for the MR‐Egger estimate was consistent with other methods. For all ICH, the MR‐Egger estimate was 1.42 (95% CI, 0.82–2.46; P=0.21). For nonlobar ICH, the MR‐Egger estimate was 1.51 (95% CI, 0.83–2.73; P=0.19). For lobar ICH, the MR‐Egger estimate was 1.21 (95% CI, 0.62–2.36; P=0.57). After performing Cook’s distance, we found 2 outliers for all ICH, 1 outlier for lobar ICH and 3 outliers for nonlobar ICH. The Cook’s distance plots can be found in Figures S4 through S6. After removing these outliers, the association remained significant for both all ICH (OR, 1.95; [95% CI, 1.08–3.55]; P=0.027) and nonlobar ICH (OR, 2.37; [95% CI, 1.20–4.67]; P=0.013).
Table 2.
Results of Different Mendelian Randomization Analyses
| Mendelian randomization method | All intracerebral hemorrhage | Lobar intracerebral hemorrhage | Nonlobar intracerebral hemorrhage | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Association tests | ||||||
| Inverse‐variance weighted | 1.60 (1.05–2.43) | 0.029 | 1.43 (0.86–2.38) | 0.17 | 1.87 (1.18–2.97) | 0.007 |
| Weighted median | 1.42 (0.81–2.50) | 0.22 | 1.36 (0.66–2.79) | 0.40 | 1.46 (0.76–2.80) | 0.26 |
| MR‐Egger estimate | 1.42 (0.82–2.46) | 0.21 | 1.21 (0.62–2.36) | 0.57 | 1.51 (0.83–2.73) | 0.19 |
| Weighted mode | 1.37 (0.89–2.11) | 0.15 | 1.11 (0.62–1.98) | 0.73 | 1.45 (0.84–2.52) | 0.19 |
| Tests for horizontal pleiotropy* | ||||||
| MR‐Egger intercept | 1.01 (0.99–1.02) | 0.52 | 1.01 (0.99–1.03) | 0.45 | 1.01 (0.99–1.03) | 0.26 |
| MR‐PRESSO global test | … | 0.11 | … | 0.44 | … | 0.48 |
IVW indicates inverse‐variance weighted; MR, Mendelian Randomization; MR‐PRESSO, Mendelian Randomization Pleiotropy Residual Sum and Outlier; and OR, odds ratio.
Horizontal pleiotropy is absent if P>0.05.
In additional sensitivity analyses using multivariable MR to adjust for the genetic contribution of VitD to other pathways associated with ICH, we found that the association between VitD and both all and nonlobar ICH remained significant (Table S4). Finally, we have conducted a sensitivity analysis using an R 2 threshold of 0.001 for clumping, that resulted in the selection of 64 SNPs. Results were similar and consistent with the original analyses for all ICH (OR, 1.55; [95% C,I 1.01–2.40]; P=0.047), lobar ICH (OR, 1.47; [95% CI, 0.86–2.54]; P=0.16), and nonlobar ICH (OR, 1.78; [95% CI, 1.08–2.92]; P=0.02). Moreover, when testing for potential outliers with RadialMR, we did not detect any significant outliers for all ICH, lobar ICH, or nonlobar ICH using a Bonferroni‐corrected threshold. Using a nominal threshold of 0.05, there were 9 outliers for all ICH, 6 outliers for lobar ICH, and 4 outliers for nonlobar ICH. The inverse‐variance weighted MR remained significant for both all ICH (P=0.0467) and nonlobar ICH (P=0.016) after removing these outliers.
Discussion
We report the results of a 2‐sample MR study that used summary level results to evaluate the relationship between genetically determined VitD levels and risk of ICH. We found that genetically reduced levels of VitD associated with a modest increase in the risk of ICH, without evidence of significant horizontal pleiotropy and consistent direction of effects in secondary, more conservative analyses. However, only substantial pleiotropic effects can be ruled out because of the limited statistical power of methods such as MR‐Egger. Stratification by location indicated that these associations were predominantly driven by nonlobar ICH, where associations were stronger despite a significant reduction in sample size.
Prior studies investigated the link between VitD deficit and ischemic stroke. A systematic review and meta‐analysis demonstrated a 62% higher risk of stroke among individuals in the lowest versus highest categories of VitD concentrations. 4 Importantly, a randomized controlled trial evaluating cardiovascular disease broadly failed to show a protective effect of VitD supplementation on stroke risk. 10 From a population genetics perspective, a prior MR study that evaluated the relationship between genetically instrumented VitD levels and risk of ischemic stroke failed to find a protective association. 11
Our study adds important new evidence to the field of VitD metabolism and ICH, an area of research that remains relatively unexplored. A prior MR study examined causal associations between VitD levels and several cardiovascular traits, including myocardial infarction, ischemic heart disease, ischemic stroke, subarachnoid hemorrhage, and ICH among Chinese. 5 Importantly, the effect size for the association between genetically determined VitD levels and ICH risk observed in this study 5 was similar (hazard ratio, 1.09 [95% CI, 1.01–1.18] per 25 nmol/L higher plasma 25(OH)D) to that obtained in our study. Another study implemented MR analyses using data from the China Kadoorie Biobank, the Copenhagen City Heart Study, and the Copenhagen General Population Study, and did not find any significant associations. However, the study only considered VitD‐related variants in the CYP2R1 and DCHR7 genes. In contrast, the present study used a more powerful approach using several dozen genetic risk variants known to modify VitD levels. In addition, by focusing on a single outcome, we increase our discovery power by reducing multiple testing.
There are several possible pathophysiological mechanisms that could mediate the inverse association between VitD and ICH. First, VitD plays an important role in the occurrence of various cardiometabolic traits that predispose to ICH, including diabetes, 12 metabolic syndrome, 13 and renin‐angiotensin system activation. 14 Another potential mechanism involves an upregulation of inflammatory pathways, as VitD inhibits the production of various inflammation factors. 15
In conclusion, our results point to an inverse association between genetically determined VitD levels and risk of ICH. This association was driven by nonlobar (also known as deep) hemorrhages. Further research is needed to confirm these findings, extend these results to non‐Europeans, and identify the putative mechanisms behind this association.
Sources of Funding
Acosta is supported by the American Heart Association (AHA) Bugher Fellowship in Hemorrhagic Stroke Research. Leasure is supported by the AHA Medical Student Research Fellowship. Gill is supported by the Yale Claude D. Pepper Older Americans Independence Center and by the National Institute on Aging (P30AG021342). Sheth is supported by the National Institutes of Health and the AHA, and reports grants from Hyperfine, Biogen, and Bard unrelated to this work. Falcone is supported by the National Institutes of Health and the AHA.
Disclosures
Sheth received compensation from Sense, Cerevasc, Certus, Rhaeos, Zoll Medical Corporation, CSL Behring, Astrocyte, and a patent for Alva Health. The remaining authors have no disclosures to report.
Supporting information
Tables S1–S4
Figures S1–S6
The abstract of this work was presented at the Annual Meeting of the American Academy of Neurology, April 17 to 22, 2021.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.024141
For Sources of Funding and Disclosures, see page 5.
References
- 1. Ma L, Wang S, Chen H, Cui L, Liu X, Yang H, Li G, Liu S, Qi T, Tian H. Diminished 25‐OH vitamin D(3) levels and vitamin D receptor variants are associated with susceptibility to type 2 diabetes with coronary artery diseases. J Clin Lab Anal. 2020;34:e23137. doi: 10.1002/jcla.23137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wajda J, Świat M, Owczarek AJ, Brzozowska A, Olszanecka‐Glinianowicz M, Chudek J. Severity of vitamin D deficiency predicts mortality in ischemic stroke patients. Dis Markers. 2019;2019:3652894. doi: 10.1155/2019/3652894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen H, Liu Y, Huang G, Zhu J, Feng W, He J. Association between vitamin D status and cognitive impairment in acute ischemic stroke patients: A prospective cohort study. Clin Interv Aging. 2018;13:2503–2509. doi: 10.2147/cia.S187142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou R, Wang M, Huang H, Li W, Hu Y, Wu T. Lower vitamin D status is associated with an increased risk of ischemic stroke: A systematic review and meta‐analysis. Nutrients. 2018;10:1–12. doi: 10.3390/nu10030277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang T, Afzal S, Yu C, Guo Y, Bian Z, Yang L, Millwood IY, Walters RG, Chen Y, Chen N, et al. Vitamin D and cause‐specific vascular disease and mortality: A mendelian randomisation study involving 99,012 Chinese and 106,911 European adults. BMC Med. 2019;17:160. doi: 10.1186/s12916-019-1401-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang X, O’Reilly PF, Aschard H, Hsu YH, Richards JB, Dupuis J, Ingelsson E, Karasik D, Pilz S, Berry D, et al. Genome‐wide association study in 79,366 European‐ancestry individuals informs the genetic architecture of 25‐hydroxyvitamin D levels. Nat Commun. 2018;9:260. doi: 10.1038/s41467-017-02662-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Revez JA, Lin T, Qiao Z, Xue A, Holtz Y, Zhu Z, Zeng J, Wang H, Sidorenko J, Kemper KE, et al. Genome‐wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat Commun. 2020;11:1647. doi: 10.1038/s41467-020-15421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, Anderson CD, Brouwers HB, Valant V, Battey TWK, et al. Meta‐analysis of genome‐wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. doi: 10.1016/j.ajhg.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Team RC . R: A Language and Environment for Statistical Computing. Viena, Austria: R Foundation for Statistical Computing; 2018. Available from: http://www.r‐project.org/. [Google Scholar]
- 10. Scragg R, Stewart AW, Waayer D, Lawes CMM, Toop L, Sluyter J, Murphy J, Khaw KT, Camargo CA Jr. Effect of monthly high‐dose vitamin D supplementation on cardiovascular disease in the vitamin D assessment study: A randomized clinical trial. JAMA Cardiol. 2017;2:608–616. doi: 10.1001/jamacardio.2017.0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsson SC, Traylor M, Mishra A, Howson JMM, Michaëlsson K, Markus HS. Serum 25‐hydroxyvitamin D concentrations and ischemic stroke and its subtypes. Stroke. 2018;49:2508–2511. doi: 10.1161/strokeaha.118.022242 [DOI] [PubMed] [Google Scholar]
- 12. Dadrass A, Mohamadzadeh Salamat K, Hamidi K, Azizbeigi K. Anti‐inflammatory effects of vitamin D and resistance training in men with type 2 diabetes mellitus and vitamin D deficiency: A randomized, double‐blinded, placebo‐controlled clinical trial. J Diabetes Metab Disord. 2019;18:323–331. doi: 10.1007/s40200-019-00416-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park JE, Pichiah PBT, Cha YS. Vitamin D and metabolic diseases: Growing roles of vitamin D. J Obes Metab Syndr. 2018;27:223–232. doi: 10.7570/jomes.2018.27.4.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui C, Xu P, Li G, Qiao Y, Han W, Geng C, Liao D, Yang M, Chen D, Jiang P. Vitamin D receptor activation regulates microglia polarization and oxidative stress in spontaneously hypertensive rats and angiotensin II‐exposed microglial cells: Role of renin‐angiotensin system. Redox Biol. 2019;26:101295. doi: 10.1016/j.redox.2019.101295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krivoy A, Satz J, Hornfeld SH, Bar L, Gaughran F, Shoval G, Hochman E, Weizman A, Taler M. Low levels of serum vitamin D in clozapine‐treated schizophrenia patients are associated with high levels of the proinflammatory cytokine IL‐6. Int Clin Psychopharmacol. 2020;35:208–213. doi: 10.1097/yic.0000000000000303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S4
Figures S1–S6


