Abstract
The effect of solar UV-B radiation on the population dynamics and composition of the culturable bacterial community from peanut (Arachis hypogeae L.) was examined in field studies using plants grown under UV-B−transmitting (UV-B+) or UV-B−excluding (UV-B−) plastic filters. Our data demonstrate that solar UV-B selection alters phyllosphere bacterial community composition and that UV tolerance is a prevalent phenotype late in the season. The total bacterial population size was not affected by either UV-B treatment. However, isolates from the UV-B+ plots (n = 368) were significantly more UV tolerant than those from the UV-B− (n = 363) plots. UV sensitivity was determined as the minimal inhibitory dose of UV that resulted in an inhibition of growth compared to the growth of a nonirradiated control. The difference in minimal inhibitory doses among bacterial isolates from UV-B+ and UV-B− treatments was mainly partitioned among nonpigmented isolates, with pigmented isolates as a group being characterized as UV tolerant. A large increase in UV tolerance was observed within isolate groups collected late (89 and 96 days after planting) in the season. Identification of 200 late-season isolates indicated that the predominant UV-tolerant members of this group were Bacillus coagulans, Clavibacter michiganensis, and Curtobacterium flaccumfaciens. We selected C. michiganensis as a model UV-tolerant epiphyte to study if cell survival on UV-irradiated peanut leaves was increased relative to UV survival in vitro. The results showed an enhancement in the survival of C. michiganensis G7.1, especially following high UV-C doses (300 and 375 J m−2), that was evident between 24 and 96 h after inoculation. A dramatic increase in the in planta/in vitro survival ratio was observed over the entire 96-h experiment period for C. michiganensis T5.1.
The plant leaf surface, termed the phyllosphere, supports the growth of a diverse flora of bacteria and fungi. Phyllosphere microbial residents grow through the utilization of the limited resources available in this habitat (10, 15); survival is also predicated upon the ability of organisms to cope with varied environmental stress conditions, including fluctuating water availability, heat, osmotic stress, and exposure to solar UV radiation (UVR). The daily influx of solar UVR includes UV-A and UV-B wavelengths; high-energy UV-B wavelengths (280 to 320 nm) are particularly inhibitory to organisms and cause direct DNA damage by inciting the formation of lesions in cellular DNA, including cyclobutane pyrimidine dimers and pyrimidine(6–4)pyrimidinone photoproducts (20). These DNA lesions result in the blockage of DNA replication and RNA transcription; the accumulation of photoproducts, in the absence of efficient cellular mechanisms for their removal, can be lethal.
Results of ecological studies of the phyllosphere habitat clearly indicate that UVR has a negative impact on individual microbial species and complex microbial communities encompassing a number of trophic levels (18). For example, exposure to solar UV-B radiation was demonstrated to significantly inhibit germination of basidiospores and sporulation from infection sites of the fungal pathogen Exobasidium vexans on tea (9). Fluctuations in the composition of the bacterial community of the peanut phyllosphere were observed in another study; significantly larger percentages of isolates with higher UVR tolerance levels were consistently obtained at 1100, 1500, and 1900 than at 0700 (23). In an experiment with supplemental UV-B irradiation in experimental field plots of oak, the frequency of isolation of the yeasts Aureobasidium pullulans and Sporobolomyces roseus was significantly reduced, especially on adaxial leaf surfaces, compared to that in plots maintained under ambient conditions (16).
Differential sensitivity to UVR within and among phyllosphere microbial species is commonly observed (23, 25); however, it is currently unknown if UVR sensitivity plays an important role in species distribution in nature.
The ecological success of bacteria in UVR-exposed habitats is conferred by the ability of the organisms to effectively repair DNA damage as it is incurred or to avoid the occurrence of DNA damage through the colonization of sites shaded from UVR, if available. Avoidance of UVR in the phyllosphere is thought to be conditioned by the colonization of sites protected from UVR, either interior locations of plant leaves that are protected from UVR penetration (27) or external, physically shaded sites, such as at the base of trichomes. The abaxial leaf surface and leaves lower in the plant canopy may also represent habitat locations with reduced UVR exposure. Several reports have shown increased microbial populations on abaxial leaf surfaces (reviewed in references 5 and 23), although physical differences between leaf surfaces, such as the density of stomata or trichomes, may also affect bacterial population size.
Regardless of the avoidance strategies utilized, the relative fitness of bacteria dwelling in UVR-exposed habitats is also affected by DNA repair capabilities. In the plant pathogen and phyllosphere resident Pseudomonas syringae, the mutagenic DNA repair determinant rulAB (24) confers UVR tolerance at levels that enable strains to maintain significantly larger populations than strains lacking rulAB in the bean phyllosphere following UV-B irradiation (25). rulAB function is detectable during the time period when P. syringae cells are establishing an infection, suggesting that UVR avoidance and tolerance strategies may be simultaneously important during in planta growth of P. syringae (11). Photoreactivation also contributes to the UVR survival of P. syringae (12), although the importance of photoreactivation in the UVR survival of other phyllosphere organisms is currently unknown.
We have been investigating the effect of UVR on the phyllosphere bacterial community from field-grown peanut (Arachis hypogeae L.). Initial field studies indicated that UVR tolerance was a common phenotype and that preferential colonization of the abaxial leaf surface was an important UVR survival strategy (23). In this study, our goal was to directly determine the effects of solar UV-B radiation on population size, UVR sensitivity, and species composition of the culturable bacterial community from the peanut phyllosphere. An additional goal involved the use of a model UVR-tolerant epiphyte, Clavibacter michiganensis, to examine if UVR survival is altered during leaf colonization, possibly through the occupation of leaf sites protected from UVR.
MATERIALS AND METHODS
Experimental plots.
On 18 June 1999 and 19 May 2000, peanut (A. hypogeae cv. Florunner) was hand sown at a depth of 2 to 3 cm in field plots established in a sandy loam soil adjacent to the Texas A&M University campus. The total plot size was 3.4 by 9.8 m, and individual treatment plots were 1.2 by 2.4 m with 0.3-m row spacings. Within rows, seeds were sown at a rate of one seed per 8 cm. Prior to seedling emergence, the plots were screened with wood frames covered with plastic filters designed to transmit all solar wavelengths or to exclude the UV-B spectrum. The UV-B-transparent control (UV-B+) plots were shielded with Acrolyte OP-4 plastic (Professional Plastics, Austin, Tex.) 64 mm thick. The plots in which UV-B radiation was excluded (UV-B−) had Mylar film (Cadillac Plastic and Chemical Co., Mobile, Ala.) superimposed on the Acrolyte OP-4 shields. In both years, there were two replicates of each UV-B treatment. The Acrolyte OP-4 shield transmits greater than 90% throughout the UV-A and UV-B wavelengths (Acrolyte OP-4 technical data sheet; Cyro Industries; Mt. Arlington, N.J.). The Mylar film blocks essentially all solar UVR below 310 nm (1). The plots were oriented in an east-west direction, and each plot was surrounded by additional guard rows of peanut plants to minimize the contribution of extraneous light. The filters were positioned at a distance of 45 cm from the height of the plant canopy, and all samples were taken from the center of the plots. Furrow drip irrigation was provided to the plots as necessary, ensuring that leaf moisture occurred only through dew formation.
Temperature, relative humidity, and precipitation were monitored at a site approximately 500 m from the field plots. Data readings were taken every 15 s and logged using a CR-10 Datalogger (Campbell Scientific, Logan, Utah). In 2000, solar UV-B radiation between 1100 and 1500 was monitored 26 times throughout the experiment using a UV-B detector (SED240/UVB-1/W) attached to an IL-1700 research radiometer (International Light, Newburyport, Mass.). The detector was placed at approximately 0.3 m above canopy height. UV-B radiation was measured every second, and the readings were integrated over the 4-h period, yielding a quantitative output in joules meter squared−1.
Plant sampling.
Sampling was initiated approximately 20 days after sowing. All samples were taken at 1200. Samples consisted of 10 individual leaves taken from each replicate UV-B treatment (20 leaves per UV-B treatment). Leaves of the same size and age were chosen randomly from the top of the plant canopy under the center of the filter frames. Each leaf was placed in a sterile plastic bag and transported to the laboratory on ice for immediate processing. Leaves were weighed and placed in 10 ml of prechilled buffer (0.1 M potassium phosphate [pH 7.0], 0.1% peptone), following which bacterial cells were removed by 7 min of sonication in an ultrasonic bath (model 250T; VWR Scientific, Houston, Tex.). Samples (0.1 ml) from appropriate dilutions of the sonicate were plated on King's medium B (KB) (13), amended with either 0.15 mg of cycloheximide (KBc) per ml or 300 U of nystatin (KBn) per ml to inhibit fungal growth. Bacterial colonies were counted following 72 to 96 h of incubation at 25°C. Individual counts were made for colonies appearing white or cream (nonpigmented) and for those producing yellow, orange, or pink pigments (pigmented) on KBc or KBn.
Selection of bacterial isolates.
Bacterial isolates chosen for further analyses were selected from two sampling dates (23 July and 20 August) in 1999 and six sampling dates (7 and 14 June, 12 and 19 July, and 16 and 23 August) in 2000. Isolate sampling in 1999 was limited to two dates due to a severe wind storm which blew the Mylar filters off the frames, prematurely ending the experiment. Isolates were recovered from the dilution plates used to make the bacterial counts. The method of selecting bacterial isolates involved placing the plates on a numbered grid (0 to 50); two numbers were randomly chosen, and two or three colonies in the chosen grids were selected for further testing. In this manner, 80 (1999) and 100 (2000) isolates were selected from each sampling time. Isolates were subcultured through two rounds of single-colony purification and subsequently stored at −70°C in 15% glycerol. A total of 3.8% of the isolates failed to grow during the subculturing process or could not be cultured following storage at −70°C; these isolates were removed from further analysis. A final total of 731 isolates were maintained for characterization.
UV radiation sensitivity characterization.
The sensitivity to UVR of each isolate was assayed by determining the minimal inhibitory dose of UV-C (254 nm) radiation (MIDc) necessary to inhibit the growth of cells spotted on KB agar plates. We used this method to characterize a large bacterial collection in a previous study (23). The MIDc method is a rapid, accurate, and reproducible method that effectively differentiates UVR sensitivities among closely related isolates; the MIDc values for isolates are also closely correlated with sensitivity to UV-B radiation in vitro (23). UVR assays were conducted using an XX-15 UV lamp (UVP Products, San Gabriel, Calif.) placed horizontally at a fixed height above the plates. The lamp was turned on 15 min prior to use to allow for stabilization of the UV output. The energy output of the lamp was monitored with a UV-X radiometer fitted with a UV-25 sensor (UVP Products) and determined to be 1.5 J m−2 s−1.
Identification of isolates.
A total of 394 isolates, representing all the isolates recovered on 7 and 14 June and 16 and 23 August 2000, were identified by analysis of fatty acid methyl ester (FAME) profiles. The sampling dates were chosen because they were representative of early- and late-season growth conditions. The isolates were grown on Trypticase soy agar (Difco Laboratories, Detroit, Mich.) prior to analysis. Extraction and purification of FAMEs and generation of profiles by gas chromatography were conducted using published methods (22). Isolate identification was accomplished using the Microbial Identification System software package (Microbial ID, Inc., Newark, Del.) and the TSBA library, version 3.9. FAME analyses and isolate identification were done at the Texas A&M University Plant Disease Diagnostic Laboratory.
UV survival of C. michiganensis during leaf colonization.
C. michiganensis was chosen as a model peanut epiphyte to study because this organism is highly UVR tolerant and comprises the majority of the culturable bacterial community late in the growing season. Spontaneous rifampin-resistant mutants of strains G7 and T5 (G7.1 and T5.1, respectively), isolated from peanut in a previous study (23), were selected in vitro and confirmed to maintain populations on peanut equal to those of their wild-type parents under growth chamber conditions. The sensitivity to UV-C radiation in liquid assays of strains G7.1 and T5.1 was determined to provide a baseline survival rate at various UV-C doses. Bacteria were prepared for UV-C sensitivity assays following growth overnight in 2× KB broth by washing pelleted cells in sterile saline (0.85% NaCl) solution and resuspending the cells in 15 ml of saline in a sterile glass petri dish. Following irradiation, appropriate dilutions of surviving cells were plated under safety light conditions on KB containing 75 μg of rifampin (KBrif) ml−1, and counts were determined after 72 h of incubation at 25°C.
The plant experiments were conducted in growth chambers at high relative humidities to facilitate leaf surface colonization. Cells of strains G7.1 and T5.1 grown on KBrif for 48 h were used as inocula for plant experiments. The cells were resuspended in 0.1 M potassium phosphate buffer (pH 7.0), and suspensions were adjusted turbidimetrically to approximately 108 CFU ml−1. Peanut plants were grown in a soilless medium under controlled conditions in a growth chamber (24°C, 75% relative humidity, 240-μmol-m−2-s−1 light intensity, and a 12-h photoperiod). Care was taken during watering not to wet leaves in order to prevent nonspecific leaf surface colonization. An inoculum of strain G7.1 or T5.1 was applied to the peanut leaves with a hand-held sprayer until the leaf surfaces were uniformly wet. Immediately and at 24, 48, 72, and 96 h after inoculation, four leaves per treatment were randomly excised, placed in sterile plastic trays, and irradiated on the abaxial and adaxial leaf surfaces with UV-C radiation by using the XX-15 lamp as described above. The UV-C doses ranged from 50 to 375 J m−2, depending on the sensitivity of the strains used. An additional four leaves were excised, and these leaves were not irradiated as a control. After irradiation, leaves were processed by sonication (to effectively quantify cells located in external leaf sites) and dilution plating as described above. Percent survival values were determined by comparing counts of cells recovered from UV-C-irradiated and nonirradiated leaves. A ratio was then derived by dividing the in planta percent survival value by the corresponding percent survival value determined in vitro. Three independent experiments were conducted for each strain.
RESULTS
Weather conditions during the field experiments.
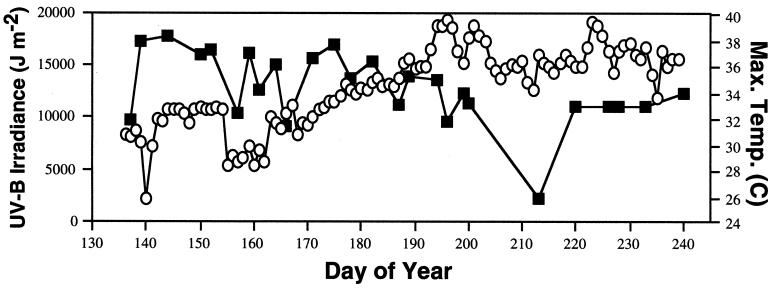
The climatic conditions observed in the two sampling seasons were typical for this region of Texas. In 2000, 4-h (1100 to 1500) solar UV-B measurements ranged from 9.1 to 17.8 kJ m−2, with only one reading (2.2 kJ m−2 on day 213) falling below the typical range (Fig. 1). A decrease in average daily relative humidity values from 79.1% (days 137 to 186 in 2000) to 60.9% (days 194 to 243 in 2000), accompanied by long periods without measurable rainfall (data not shown), contributed to the overall dry conditions observed, especially later in the sampling seasons. A steady increase in daily maximum temperatures during the sampling seasons was also noted in both years (data shown for 2000 in Fig. 1).
FIG. 1.
Integrated 4-h (1100 to 1500) solar UV-B irradiance readings (squares) and maximum (Max.) daily temperature measurements (in degrees Celcius) (circles) taken at College Station, Tex., in 2000 during the time period of the field experiments.
Population dynamics and UVR sensitivity characterization for peanut phyllosphere isolates.
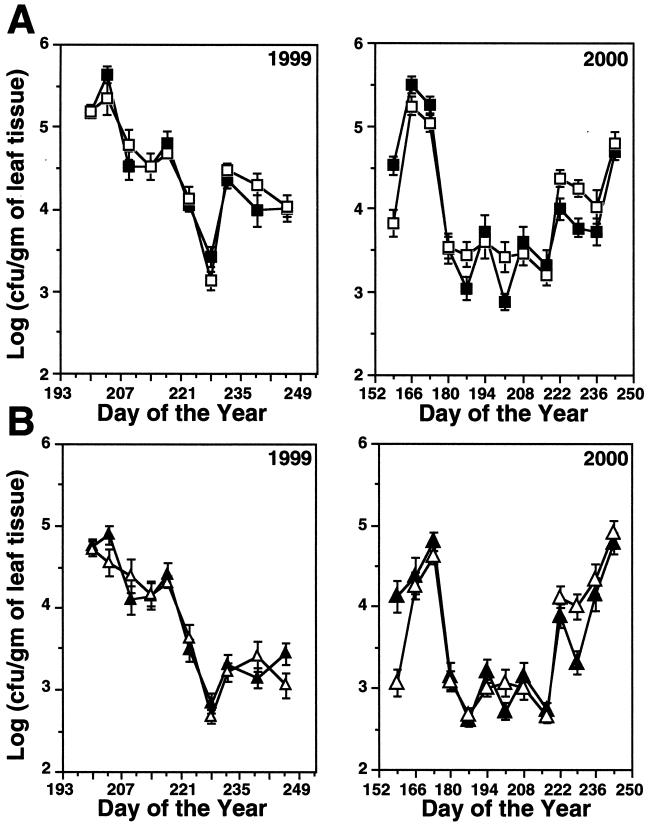
Total culturable bacterial populations ranged from approximately 103 to 7 × 106 CFU/g, and populations from the UV-B+ and UV-B− treatments were similar at most sampling points in both seasons (Fig. 2A). The dynamics of the pigmented portion of the culturable bacterial community essentially mirrored those of the total population (Fig. 2B). UVR sensitivity analyses were performed on a total of 731 isolates, including 368 and 363 isolates from the UV-B+ and UV-B− treatments, respectively. An MIDc value was determined for each isolate, and mean MIDc values for the total, pigmented, and nonpigmented collections were calculated and compared statistically using the Student t test. The results indicated that, for the total collection, the mean MIDc for isolates from the UV-B+ treatment was significantly higher (P < 0.05) than the mean MIDc for isolates from the UV-B− treatment (Table 1). This difference in mean MIDc between UV-B treatments was partitioned mainly among the nonpigmented isolates and was highly significant (P < 0.01), while the difference in mean MIDc between treatments for pigmented isolates was not significant (P > 0.5) (Table 1). The percentage of UVR-sensitive isolates (MIDc, ≤50 J m−2) recovered was higher for the UV-B− plots than for the UV-B+ plots (39.2 versus 28.6%). Also, the percentage of isolates with higher UVR tolerance (MIDc, ≥200 J m−2) was slightly lower in the UV-B− plots than in the UV-B+ plots (14.3 versus 20.2%).
FIG. 2.
Population dynamics of total bacteria (A) and pigmented bacteria (B) recovered from the phyllosphere of field-grown peanut in UV-B− (▪ in A and ▴ in B) and UV-B+ (□ in A and ▵ in B) plots. Each sampling point represents a population mean for 20 individual leaf samples. The standard error of the mean is also shown.
TABLE 1.
Statistical comparison, based on UV-B treatment, of the MIDcs for the peanut phyllosphere bacterial isolates (1999 and 2000) and the bacterial isolates subdivided by the presence or absence of pigmentation
| Isolate type | Resulta with the following treatment:
|
|||
|---|---|---|---|---|
| UV-B−
|
UV-B+
|
|||
| No. of isolates | MIDc | No. of isolates | MIDc | |
| Total | 363 | 100.6 (3.6) | 368 | 112.7 (3.8) A |
| Pigmented | 184 | 136.7 (5.1) | 200 | 139.5 (5.0) NS |
| Nonpigmented | 179 | 63.5 (3.2) | 168 | 81.1 (4.6) B |
MIDcs are reported as the (standard error of the mean) in joules per square meter. Significant differences between means within a row were calculated using the Student t test and are as follows: A, P < 0.05; B, P < 0.01; NS, not significant, P > 0.5.
In 2000, 50 isolates were recovered from each UV-B treatment plot at six specific times (a total of 600 isolates) to determine if there was any seasonal variation in UVR sensitivity among isolates. The sampling times were designated early (7 and 14 June), middle (12 and 19 July), and late (16 and 23 August) seasons. The isolate MIDcs from each UV-B treatment and seasonal sampling time were compiled, and a one-way analysis of variance was computed. Differences among the mean MIDcs were assessed using the Student-Newman-Keuls test. Two main points were noted: (i) significantly higher (P < 0.05) mean MIDcs for the UV-B+ treatments were observed only in the total and nonpigmented group late-season samples (Table 2); and (ii) an overall trend toward the isolation of organisms with higher MIDcs as the season progressed was consistently observed for both UV-B treatments and for both pigmented and nonpigmented isolates (Table 2).
TABLE 2.
Statistical comparison of the MIDcs, based on UV-B treatment and seasonal time of isolation, for the peanut phyllosphere bacterial isolates (2000) and the bacterial isolates subdivided by the presence or absence of pigmentation
| Treatmenta | Resultb for the following isolates:
|
|||||
|---|---|---|---|---|---|---|
| Total
|
Pigmented
|
Nonpigmented
|
||||
| No. | MIDc | No. | MIDc | No. | MIDc | |
| UV-B+, late | 99 | 173.2 (7.2) A | 72 | 183.0 (7.1) A | 27 | 147.2 (17.7) A |
| UV-B−, late | 97 | 136.1 (7.7) B | 67 | 168.3 (7.4) A | 30 | 64.2 (10.4) C |
| UV-B+, middle | 98 | 102.6 (6.4) C | 42 | 132.7 (11.4) B | 56 | 79.9 (5.6) B |
| UV-B−, middle | 99 | 108.6 (6.6) C | 54 | 129.2 (9.1) B | 45 | 83.9 (8.2) B |
| UV-B+, early | 100 | 67.8 (3.9) D | 48 | 81.8 (5.3) C | 52 | 54.8 (5.1) C |
| UV-B−, early | 100 | 61.8 (3.6) D | 28 | 81.3 (9.4) C | 72 | 54.2 (3.0) C |
Early-season isolates were collected on 7 and 14 June, middle season isolates were collected on 12 and 19 July, and late-season isolates were collected on 16 and 23 August 2000.
MIDs are reported as the mean (standard error of the mean) in joules per square meter. Within the MIDc columns, means not followed by the same letter were significantly different at P < 0.05, as determined by an analysis of variance and the Student-Newman-Keuls test.
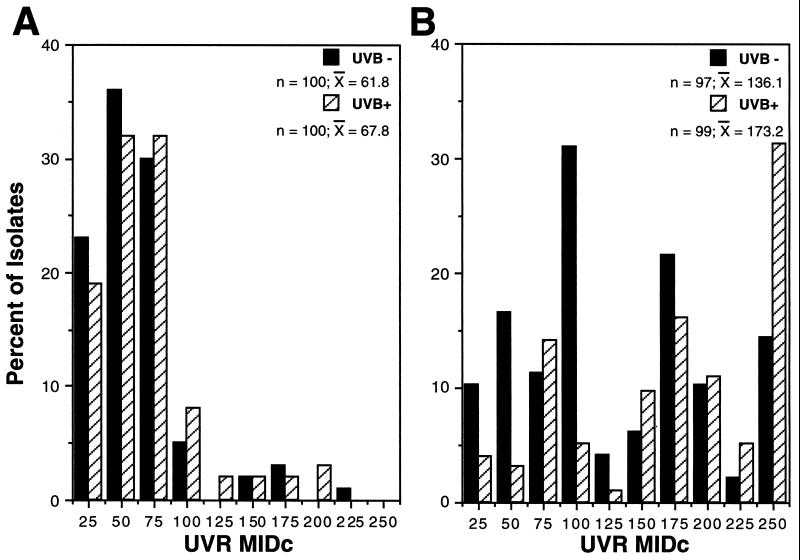
The distribution of MIDc phenotypes among the early-season isolates was characterized by a predominance of UVR-sensitive isolates (Fig. 3A). Only 15% of this collection had an MIDc of >100 J m−2, the dose level used to distinguish UVR tolerance in our previous study (23). The late-season MIDcs were more widely distributed, with a twofold higher recovery of highly tolerant isolates (MIDc, 250 J m−2) from UV-B+ plots (Fig. 3B). We used the chi-square analysis to compare MIDc distributions between UV-B treatments and seasonal times of isolation. The distributions of MIDcs for early-season isolates based on UV-B treatment (Fig. 3A) were not significantly different (χ2 = 8.59; P = 0.48). However, the MIDc distributions for late-season isolates (Fig. 3B) were highly significantly different (χ2 = 23.28; P = 0.006) depending on UV-B treatment, indicating the recovery of more UV-tolerant isolates from the UV-B+ plots. Comparisons of early- versus late-season isolates from UV-B+ plots (χ2 = 99.02; P = 0.51 × 10−18) and UV-B− plots (χ2 = 65.9; P = 0.97 × 10−12) also showed a highly significant effect, confirming the trend toward the isolation of UV-tolerant individuals late in the season.
FIG. 3.
Percentage of total bacterial isolates from the UV-B− and UV-B+ plots, along with the corresponding MIDc, recovered on 7 and 14 June (A) or 19 and 26 August (B) 2000.
Identification of early- and late-season isolates.
We subjected the early- and late-season isolates from both UV-B treatments to FAME analysis for identification purposes. Members of 25 named genera were identified. Only 1.3% of samples did not match any entries in the MIS Aerobic Bacteria library; these bacteria were unnamed and were subsequently listed as “No match.” Data regarding seasonal time of isolation and MIDc were tabulated for each bacterium identified by FAME analysis. As in our previous study (23), the majority of strains identified were gram positive; in this study, Bacillus spp. accounted for 39% of the total (Table 3). A close examination of the Bacillus sp. data indicated that only one species, Bacillus coagulans, showed a UVR tolerance phenotype (Table 3). It should also be noted that B. coagulans represented 80% of the Bacillus strains isolated in the late season from the UV-B+ treatment plots (Table 3). In contrast, B. coagulans represented only 24% of the strains isolated in the late season from the UV-B− treatment plots, as other, more UVR-sensitive species, especially Bacillus megaterium, were also recovered from these plots (Table 3).
TABLE 3.
Frequency distribution of MIDcs for peanut phyllosphere isolates, based on UV-B treatment, as identified by FAME analysis
| Treatment | Strain | No. of strainsa
|
|||||
|---|---|---|---|---|---|---|---|
| Total | Early | Late | For which: the MIDc was as follows:
|
||||
| 25–75 | 100–150 | 175–250 | |||||
| UV-B+ | Bacillus spp.b | 9 | 8 | 1 | 8 | 1 | 0 |
| Bacillus coagulans | 24 | 4 | 20 | 1 | 2 | 21 | |
| Bacillus megaterium | 24 | 21 | 3 | 23 | 1 | 0 | |
| Bacillus pumilus | 12 | 11 | 1 | 12 | 0 | 0 | |
| Clavibacter michiganensis | 35 | 0 | 35 | 0 | 3 | 32 | |
| Curtobacterium flaccumfaciens | 14 | 1 | 13 | 0 | 2 | 12 | |
| Pantoea agglomerans | 23 | 17 | 6 | 19 | 4 | 0 | |
| Pantoea ananas | 15 | 13 | 2 | 12 | 3 | 0 | |
| Pseudomonas putida | 13 | 13 | 0 | 10 | 3 | 0 | |
| Other gram negativec | 14 | 7 | 7 | 11 | 3 | 0 | |
| Other gram positived | 14 | 5 | 9 | 7 | 5 | 2 | |
| UV-B− | Bacillus spp.e | 13 | 6 | 7 | 13 | 0 | 0 |
| Bacillus coagulans | 10 | 4 | 6 | 0 | 4 | 6 | |
| Bacillus megaterium | 47 | 37 | 10 | 46 | 1 | 0 | |
| Bacillus pumilus | 15 | 12 | 3 | 15 | 0 | 0 | |
| Clavibacter michiganensis | 46 | 1 | 45 | 2 | 6 | 38 | |
| Curtobacterium flaccumfaciens | 8 | 1 | 7 | 0 | 3 | 5 | |
| Pantoea agglomerans | 11 | 5 | 6 | 9 | 2 | 0 | |
| Pantoea ananas | 11 | 6 | 5 | 11 | 0 | 0 | |
| Pseudomonas putida | 9 | 8 | 1 | 8 | 1 | 0 | |
| Other gram negativef | 15 | 9 | 6 | 15 | 0 | 0 | |
| Other gram positiveg | 11 | 10 | 1 | 8 | 1 | 2 | |
Early-season isolates were collected on 7 and 14 June, and late-season isolates were collected on 16 and 23 August 2000. MIDcs are given in joules per square meter.
Includes one or two strains from the following Bacillus species: B. cereus, B. circulans, B. licheniformis, B. pathothenticus, B. sphaericus, B. subtilus, and B. thuringiensis.
Includes one to three strains from each of the following genera: Acetobacter, Acinetobacter, Burkholderia, Comamonas, Escherichia, Gluconobacter, Kluyvera, Pseudomonas, and Salmonella.
Includes one to four strains from each of the following genera: Aureobacterium, Brevibacterium, Cellulomonas, and Micrococcus.
Includes one or two strains from the following Bacillus species: B. cereus, B. licheniformis, and B. subtilus.
Includes one to four strains from each of the following genera: Agrobacterium, Bradyrhizobium, Burkholderia, Cedecea, Comamonas, Escherichia, Klebsiella, Kluyvera, Pseudomonas, and Salmonella.
Includes one to three strains from each of the following genera: Brevibacillus, Cellulomonas, Enterococcus, Micrococcus, Paenibacillus, Rhodococcus, and Staphylococcus.
Further examination of trends in species recovery among the UV-B treatments showed that the UVR-sensitive genus Pantoea was recovered in equal numbers in both early and late seasons from the UV-B− plots (Table 3). However, the recovery of this genus from the UV-B+ plots was skewed toward the early season. Two UVR-tolerant species recovered, C. michiganensis and Curtobacterium flaccumfaciens, were almost exclusively recovered late in the season and in almost equal numbers from the UV-B+ and UV-B− treatment plots (Table 3). We tabulated the mean MIDcs for the most commonly isolated species and genera and used the Student t test to determine the influences of UV-B treatment. The results indicated that the mean MIDcs for C. michiganensis and Pantoea ananas from the UV-B+ treatment were significantly higher than those for the corresponding strains from the UV-B− treatment (Table 4). Relatively large, but nonsignificant differences were observed for B. coagulans and C. flaccumfaciens; significant differences in mean MIDcs based on UV-B treatment were not observed for B. megaterium, Bacillus pumilus, Pantoea agglomerans, and Pseudomonas putida (Table 4). A more general comparison of all identified gram-negative or gram-positive strains showed significantly higher MIDcs for isolates from the UV-B+ treatment and also indicated that the MIDcs for gram-positive organisms were almost double those for gram-negative organisms, regardless of UV-B treatment (Table 4).
TABLE 4.
MIDc analysis of strains from eight identified species or genera, based on UV-B treatment
| Species | Resulta with the following treatment:
|
|||
|---|---|---|---|---|
| UV-B−
|
UV-B+
|
|||
| No. of isolates | MIDc | No. of isolates | MIDc | |
| Bacillus megaterium | 47 | 51.6 (3.0) | 24 | 53.1 (4.3) NS |
| Bacillus pumilus | 15 | 38.3 (4.1) | 12 | 35.4 (4.8) NS |
| Bacillus coagulans | 10 | 180.0 (17.0) | 24 | 208.3 (10.1) NS |
| Clavibacter michiganensis | 46 | 187.0 (6.5) | 35 | 207.1 (6.2) A |
| Curtobacterium flaccumfaciens | 7 | 192.9 (18.2) | 13 | 209.6 (13.5) NS |
| Pantoea agglomerans | 11 | 70.5 (7.4) | 23 | 75.0 (5.0) NS |
| Pantoea ananas | 11 | 63.6 (6.2) | 15 | 80.0 (2.7) A |
| Pseudomonas putida | 9 | 66.7 (5.9) | 13 | 73.1 (10.8) NS |
| All gram negativeb | 46 | 60.9 (3.2) | 65 | 73.1 (3.4) B |
| All gram positivec | 150 | 109.3 (6.2) | 132 | 142.6 (7.4) C |
MIDcs are reported as the mean (standard error of the mean) in joules per square meter. Significant differences between mean UV-B+ and UV-B− MIDcs within a row were calculated using the Student t test and are as follows: A, P ≤ 0.05; B, P ≤ 0.01; C, P ≤ 0.001; NS, not significant, P > 0.05.
Includes 65 (UV-B+) and 46 (UV-B−) strains identified by FAME analysis.
Includes 132 (UV-B+) and 150 (UV-B−) strains identified by FAME analysis.
UV survival of C. michiganensis G7.1 and T5.1 during leaf colonization.
Comparisons of populations from UV-C-irradiated (five different doses) and nonirradiated peanut leaves inoculated with either strain G7.1 or strain T5.1 were carried out to generate in planta percent survival values (Table 5). In vitro survival values at the corresponding UV-C doses (data not shown) were then used to derive an in planta/in vitro survival ratio (Table 5). Ratios of greater than 1 indicated that UV-C survival was increased for the in planta populations. Immediately after inoculation, the survival of G7.1 was similar to that determined in vitro (Table 5), indicating that the cell concentrations used in the experiment did not result in survival enhancement from cell shading in planta. Between the time of inoculation of strain G7.1 and 24 h after inoculation, an average cell survival increase of 4.5-fold relative to the survival of the nonirradiated controls was observed for all of the UVR treatments (data not shown). Also, the rate of cell survival of strain G7.1 at the higher UV-C doses (300 and 375 J m−2) beginning 24 h after inoculation was consistently increased in planta, with in planta/in vitro survival ratios ranging from 2.1 to 18.7 (Table 5). The survival of strain T5.1 on leaves irradiated with the highest UV-C doses (150, 200, and 250 J m−2) was 15- to 3,140-fold greater than that observed in vitro. This enhancement of strain survival was evident immediately after inoculation and continued throughout the entire experiment (Table 5).
TABLE 5.
Survival of C. michiganense G7.1 and T5.1 on peanut leaves irradiated with UV-C
| Strain | Time p.i. (h)a | UV-C dose (J m−2) | % Survivalb | In planta/in vitro survival ratioc |
|---|---|---|---|---|
| G7.1 | 0 | 75 | 31.0 | 0.5 |
| 150 | 12.3 | 0.4 | ||
| 225 | 5.2 | 0.6 | ||
| 300 | 4.0 | 1.4 | ||
| 375 | 2.6 | 5.0 | ||
| 24 | 75 | 52.9 | 0.9 | |
| 150 | 34.4 | 1.1 | ||
| 225 | 62.6 | 7.0 | ||
| 300 | 8.5 | 3.1 | ||
| 375 | 9.7 | 18.7 | ||
| 48 | 75 | 85.8 | 1.4 | |
| 150 | 33.7 | 1.1 | ||
| 225 | 24.2 | 2.7 | ||
| 300 | 15.2 | 5.5 | ||
| 375 | 8.2 | 15.8 | ||
| 72 | 75 | 61.4 | 1.0 | |
| 150 | 23.8 | 0.8 | ||
| 225 | 14.2 | 1.6 | ||
| 300 | 5.8 | 2.1 | ||
| 375 | 3.9 | 7.5 | ||
| 96 | 75 | 52.9 | 0.9 | |
| 150 | 30.4 | 1.0 | ||
| 225 | 9.2 | 1.0 | ||
| 300 | 10.1 | 3.6 | ||
| 375 | 7.7 | 14.8 | ||
| T5.1 | 0 | 50 | 49.1 | 0.9 |
| 100 | 20.5 | 2.0 | ||
| 150 | 11.5 | 15.7 | ||
| 200 | 8.5 | 202.4 | ||
| 250 | 2.7 | 540.0 | ||
| 24 | 50 | 41.1 | 0.8 | |
| 100 | 20.1 | 2.0 | ||
| 150 | 12.3 | 16.8 | ||
| 200 | 8.2 | 195.2 | ||
| 250 | 15.7 | 3,140.0 | ||
| 48 | 50 | 43.9 | 0.8 | |
| 100 | 31.1 | 3.0 | ||
| 150 | 11.2 | 15.3 | ||
| 200 | 8.4 | 200.0 | ||
| 250 | 11.1 | 2,220.0 | ||
| 72 | 50 | 100.0 | 1.9 | |
| 100 | 41.1 | 4.0 | ||
| 150 | 47.6 | 64.9 | ||
| 200 | 27.9 | 664.3 | ||
| 250 | 13.6 | 2,720.0 | ||
| 96 | 50 | 67.8 | 1.3 | |
| 100 | 21.6 | 2.1 | ||
| 150 | 19.0 | 25.9 | ||
| 200 | 8.9 | 211.9 | ||
| 250 | 1.5 | 300.0 |
p.i., postinoculation. After spray inoculation of bacteria, plants were incubated in a growth chamber under high-humidity conditions.
(Mean total population from UV-C-irradiated leaves/mean total population from nonirradiated leaves) × 100. Cell populations from leaves were removed by sonication and enumerated by dilution plating.
Ratio derived from the percent survival determined in planta divided by the corresponding percent survival determined in vitro at the same UV-C dose.
DISCUSSION
The exclusion of solar UV-B radiation from the phyllosphere of field-grown peanut did not affect the population size of the culturable bacteria from this habitat. However, the phyllosphere bacterial community, defined in terms of UVR sensitivity and species composition, differed among UV-B treatment plots, and these parameters were especially apparent in the nonpigmented members of the community. These observations suggest that solar UV-B radiation is an important environmental stress for nonpigmented phyllosphere bacteria. Examinations of the bacterial counts showed that the cell numbers were lower than those observed in a previous experiment (23), suggesting that the plastic filters may have affected the canopy microclimate, compared to a no-filter situation. However, the effects of the filters should have been equivalent for both treatments and thus should not have affected comparisons, as others have shown that UV-B exclusion treatments do not affect leaf or soil temperatures (3).
An overall examination of the pigmented bacterial isolates indicated that these organisms possessed similar MIDcs regardless of UV-B treatment, and the overall mean MIDc was 1.5 to 2 times higher than that of their nonpigmented counterparts. The occurrence of higher MIDcs in pigmented isolates correlates with prior observations (23). Although differences in MIDc based on UV-B treatment were observed within subsamples of the pigmented community (e.g., C. michiganensis), it is possible that many indigenous phyllosphere pigmented organisms already are tolerant of the existing solar UV-B radiation flux and are ecologically competitive even in the absence of UV-B stress.
A seasonal trend toward increased UVR tolerance within both pigmented and nonpigmented isolates, as reflected by higher MIDcs and alterations in the frequency distribution of MIDcs (defined by χ2 analysis), was observed for both UV-B treatments. The general trend toward elevated MIDcs during the season implicates factors in addition to solar UV-B in the manipulation of the phyllosphere community. The weather data from 2000 indicate a steady seasonal increase in average daily temperature (Fig. 1), a decrease in average daily relative humidity, long periods without measurable rainfall, and fairly constant daily UV-B flux (Fig. 1). Thus, it appears that the hot and dry weather conditions occurring during this study also contributed to the change in community composition observed late in the growing season and that UVR tolerance may be linked with other stress tolerance phenotypes in organisms that are commonly isolated at this time.
Many of the organisms identified by FAME showed preferences in seasonal times of isolation, and some were clearly influenced by solar UV-B. For example, Bacillus spp., mostly nonpigmented, UV-sensitive organisms, were commonly recovered from both UV-B treatments early in the season (sampling times, 18 and 25 days after planting). The usual habitat of some of these organisms, particularly B. megaterium, is thought to be soil (21). Indeed, the UV sensitivity of B. megaterium correlates with occupancy of the soil habitat. Although the phyllosphere and soil bacterial communities are distinctly different (5), it is possible that organisms such as B. megaterium transiently colonize peanut as the seed germinates and initially grows through soil. It is also clear that B. megaterium can inhabit the phyllosphere, as this organism has been detected in previous phyllosphere studies and has effectively colonized leaves in experimental studies (7, 8, 23). The survival of Bacillus spp. as spores could also contribute to increased UVR tolerance (17), thus enhancing epiphytic fitness. However, only the UV-tolerant B. coagulans was isolated with regularity from the late-season UV-B+ treatment (80% of Bacillus isolates), demonstrating the selective effect of solar UV-B on Bacillus spp.
The FAME data also show a clear trend toward the late-season isolation of other UV-tolerant organisms, such as Clavibacter and Curtobacterium spp. These data provide evidence for an ecological succession of organisms which was especially apparent in the UV-B+ treatment and thus would be predicted to occur under natural conditions. Seasonal studies of sugar beet and wheat phyllosphere bacteria from England did not show trends reflective of the replacement of organisms depending upon the time of year (14, 26). It is possible that the more stressful climatic conditions in Texas played a role in the differences observed in these studies. In France, Bacillus, Lactobacillus, and Pseudomonas spp. were preferentially isolated from evergreen oak leaves of various ages during different seasons (19). The ability to tolerate higher temperatures was suggested to favor Bacillus sp. colonization versus Pseudomonas sp. colonization during the summer months (19). Likewise, the ability to tolerate hotter, drier conditions was thought to favor the colonization of bean leaves in Wisconsin by pink-pigmented facultative methylotrophs instead of P. syringae (10). In Argentina, a reduction in the diversity of Bacillus spp. from the soybean phyllosphere occurred throughout the season, with only one species (B. pumilus) being recovered late in the season (2). The role of heat and dessiccation stresses in the seasonal succession observed in phyllosphere bacterial communities remains an open question. Our data indicate that solar UV-B also contributes to these seasonal ecological trends and suggest the importance of a UV tolerance phenotype for late-season phyllosphere inhabitants in Texas.
Beattie and Lindow (5) discussed the general ecological strategies of tolerance and avoidance of environmental stresses as characteristics of bacterial saprophytes and pathogens, respectively, in the phyllosphere. The ability of pathogens to become internalized within leaf tissue and thereby become more protected from abiotic stresses is an effective means of avoiding dessiccation and UVR stresses (4, 6, 27). Saprophytes, meanwhile, are thought to survive by tolerating environmental stresses, as these organisms are noninvasive for plant tissue. Our results indicate that the UVR survival of these organisms can be enhanced (especially at higher UV-C doses) on leaves relative to the in vitro UVR survival; however, the contributing factors are still unclear. Possibilities include colonization of external shaded sites, increased cell aggregation over time, or other alterations in cell physiology. Nevertheless, our data suggest the potential for saprophytic organisms to utilize both strategies of avoidance and tolerance of UVR stress in the phyllosphere.
In summary, three important observations were noted in this study, namely, the impact of solar UV-B radiation on the composition of the phyllosphere community, the seasonal trend resulting in the isolation of organisms with higher UVR tolerance later in the season, and the observation of enhanced UVR survival of leaf-associated populations of C. michiganensis. Further work on the genetic nature of UVR tolerance in species such as B. coagulans and C. michiganensis must be done together with ecological studies to understand the relative effects of solar UVR and other environmental stresses on the seasonal dynamics of these organisms.
ACKNOWLEDGMENTS
We thank M. Hall for the weather data, F. Pate for constructing the UV-B shield frames and platform stand for the IL-1700 radiometer, and L. Barnes and R. Henson for performing the FAME analyses. We also thank two anonymous reviewers whose comments strengthened the manuscript.
This work was supported by the Texas Agricultural Experiment Station.
REFERENCES
- 1.Aas P, Lyons M M, Pledger R, Mitchell D L, Jeffrey W H. Inhibition of bacterial activities by solar radiation in nearshore waters and the Gulf of Mexico. Aquat Microb Ecol. 1996;11:229–238. [Google Scholar]
- 2.Arias R S, Sagardoy M A, Van Vuurde J W L. Spatio-temporal distribution of naturally occurring Bacillus spp. and other bacteria on the phylloplane of soybean under field conditions. J Basic Microbiol. 1999;39:283–292. doi: 10.1002/(sici)1521-4028(199912)39:5/6<283::aid-jobm283>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 3.Ballare C L, Scopel A L, Stapelton A E, Yanovsky M J. Solar ultraviolet-B radiation affects seedling emergence, DNA integrity, plant morphology, growth rate, and attractiveness to herbivore insects in Datura ferox. Plant Physiol. 1996;112:161–170. doi: 10.1104/pp.112.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beattie G A, Lindow S E. Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on leaves. Appl Environ Microbiol. 1994;60:3790–3798. doi: 10.1128/aem.60.10.3790-3798.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie G A, Lindow S E. The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol. 1995;33:145–172. doi: 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- 6.Beattie G A, Lindow S E. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology. 1999;89:353–359. doi: 10.1094/PHYTO.1999.89.5.353. [DOI] [PubMed] [Google Scholar]
- 7.Bora R S, Murty M G, Shenbagarathai R, Sekar V. Introduction of a lepidopteran-specific insecticidal crystal protein gene of Bacillus thuringiensis subsp. kurstaki by conjugal transfer into a Bacillus megaterium strain that persists in the cotton phyllosphere. Appl Environ Microbiol. 1994;60:214–222. doi: 10.1128/aem.60.1.214-222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ercolani G L. Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb Ecol. 1991;21:35–48. doi: 10.1007/BF02539143. [DOI] [PubMed] [Google Scholar]
- 9.Gunasekera T S, Paul N D, Ayres P G. The effects of ultraviolet-B (UV-B: 290–320 nm) radiation on blister blight disease of tea (Camellia sinensis) Plant Pathol. 1997;46:179–185. [Google Scholar]
- 10.Hirano S S, Upper C D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J-J, Sundin G W. Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J Bacteriol. 2000;182:6137–6144. doi: 10.1128/jb.182.21.6137-6144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J-J, Sundin G W. Construction and analysis of photolyase mutants of Pseudomonas aeruginosa and Pseudomonas syringae: contribution of photoreactivation, nucleotide excision repair, and mutagenic DNA repair to cell survival and mutability following exposure to UV-B radiation. Appl Environ Microbiol. 2001;67:1405–1411. doi: 10.1128/AEM.67.4.1405-1411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 14.Legard D E, McQuilken M P, Whipps J M, Fenlon J S, Fermor T R, Thompson I P, Bailey M J, Lynch J M. Studies of seasonal changes in the microbial populations on the phyllosphere of spring wheat as a prelude to the release of a genetically modified microorganism. Agric Ecosyst Environ. 1994;50:87–101. [Google Scholar]
- 15.Mercier J, Lindow S E. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol. 1999;66:369–374. doi: 10.1128/aem.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newsham K K, Low M N R, McLeod A R, Greenslade P D, Emmett B A. Ultraviolet-B radiation influences the abundance and distribution of phylloplane fungi on pedunculate oak (Quercus robur) New Phytol. 1997;138:287–297. [Google Scholar]
- 17.Nicholson W L, Munakata N, Horneck G, Melosh H J, Setlow P. Resistance of Bacillus endopsores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul N D, Rasanayagam S, Moody S A, Hatcher P E, Ayres P G. The role of interactions between trophic levels in determining the effects of UV-B on terrestrial ecosystems. Plant Ecol. 1997;128:296–308. [Google Scholar]
- 19.Perissol C, Roux M, Le Petit J. Succession of bacteria attached to evergreen oak leaf surfaces. Eur J Soil Biol. 1993;29:167–176. [Google Scholar]
- 20.Pfeifer G P. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 21.Slepecky R A, Hemphill H E. The genus Bacillus—nonmedical. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 1663–1696. [Google Scholar]
- 22.Stead D E. Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol. 1992;42:281–295. [Google Scholar]
- 23.Sundin G W, Jacobs J L. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.) Microb Ecol. 1999;38:27–38. doi: 10.1007/s002489900152. [DOI] [PubMed] [Google Scholar]
- 24.Sundin G W, Kidambi S P, Ullrich M, Bender C L. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene. 1996;177:77–81. doi: 10.1016/0378-1119(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 25.Sundin G W, Murillo J. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290–320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ Microbiol. 1999;1:75–87. doi: 10.1046/j.1462-2920.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 26.Thompson I P, Bailey M J, Fenlon J S, Fermor T R, Lilley A K, Lynch J M, McCormack P J, McQuilken M P, Purdy K J, Rainey P B, Whipps J M. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris) Plant Soil. 1993;150:177–191. [Google Scholar]
- 27.Wilson M, Hirano S S, Lindow S E. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl Environ Microbiol. 1999;65:1435–1443. doi: 10.1128/aem.65.4.1435-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]