Abstract
The β-xylosidase-encoding xlnD gene of Aspergillus niger 90196 was amplified by the PCR technique from first-strand cDNA synthesized on mRNA isolated from the fungus. The nucleotide sequence of the cDNA fragment was verified to contain a 2,412-bp open reading frame that encodes a 804-amino-acid propeptide. The 778-amino-acid mature protein, with a putative molecular mass of 85.1 kDa, was fused in frame with the Saccharomyces cerevisiae mating factor α1 signal peptide (MFα1s) to ensure correct posttranslational processing in yeast. The fusion protein was designated Xlo2. The recombinant β-xylosidase showed optimum activity at 60°C and pH 3.2 and optimum stability at 50°C. The Ki(app) value for d-xylose and xylobiose for the recombinant β-xylosidase was determined to be 8.33 and 6.41 mM, respectively. The XLO2 fusion gene and the XYN2 β-xylanase gene from Trichoderma reesei, located on URA3-based multicopy shuttle vectors, were successfully expressed and coexpressed in the yeast Saccharomyces cerevisiae under the control of the alcohol dehydrogenase II gene (ADH2) promoter and terminator. These recombinant S. cerevisiae strains produced 1,577 nkat/ml of β-xylanase activity when expressing only the β-xylanase and 860 nkat/ml when coexpressing the β-xylanase with the β-xylosidase. The maximum β-xylosidase activity was 5.3 nkat/ml when expressed on its own and 3.5 nkat/ml when coexpressed with the β-xylanase. Coproduction of the β-xylanase and β-xylosidase enabled S. cerevisiae to degrade birchwood xylan to d-xylose.
Plant cell walls, the major reservoir of fixed carbon in nature, contain three major polymers: cellulose (insoluble fibers of β-1,4-glucan), hemicellulose (noncellulosic polysaccharides including xylans, mannans, and glucans) and lignin (a complex polyphenolic structure) (1, 45). β-1,4-Xylans are found mainly in secondary walls of plants and can represent up to 35% of the total dry weight in certain plants. Xylan is a complex polysaccharide consisting of a backbone of β-d-1,4-linked xylopyranoside units substituted with acetyl, glucuronosyl, and arabinosyl side chains. Endo-β-xylanases (EC 3.2.1.8) act on xylans and xylo-oligosaccharides, producing mainly mixtures of xylooligosaccharides (4, 23). β-d-Xylosidases (EC 3.2.1.37) hydrolyze xylooligosaccharides, produced through the action of β-xylanases, to d-xylose. Many bacterial and fungal species are able to utilize xylans as a carbon source (18). Strains of the fungi Trichoderma and Aspergillus secrete large amounts of efficient xylan-degrading enzymes (8, 16, 51). Recently, interest in β-xylanases has increased because of their application in biobleaching (30, 44) and the food (31) and animal feed (3, 34, 47) industry.
Trichoderma reesei is a filamentous mesophilic fungus that is well known for its cellulolytic and xylanolytic enzymatic activities (12, 43). The two major inducible endo-β-xylanases secreted by this fungus are Xyn1 and Xyn2 (46). They are both relatively small protein molecules, with molecular masses of 19 and 21 kDa, respectively, but Xyn2 represents more than 50% of the total xylanolytic activity of T. reesei cultivated on xylan. Fungi of the genus Aspergillus are also efficient producers of cellulose- and xylan-degrading enzymes, regulated at the transcriptional level by the XlnR activator (49). The two endo-β-xylanases and the β-xylosidase in A. niger are encoded by xlnB, xlnC, and xlnD, respectively. The xlnD gene contains an open reading frame of 2,412 nucleotides, which encodes a protein of 804 amino acids with a predicted molecular mass of 85 kDa. The protein is N glycosylated and contains 15 potential N-glycosylation sites (48). Sequence similarity was found to β-glycosidases (β-xylosidase and β-glucosidases) of family 3, which include enzymes from both bacterial and fungal origins (20, 33, 35, 48). The condensation reaction of this β-xylosidase has been used for the synthesis of disaccharides such as β,β-1,1-xylodisaccharide, β-1,4-xylodisaccharide (xylobiose), β-1,2-xylodisaccharide, α-1,4-xylodisaccharide, and β-1,3-xylodisaccharide (17). S. cerevisiae has been successfully used for the production of related fungal β-xylosidase and β-glucosidases belonging to family 3 (7, 32, 33).
Different Candida species (C. maltosa, C. tropicalis, and C. utilis) are currently used in industry for the production of single-cell protein and ethanol from steamed hemicellulose (21). Even though these Candida strains are able to ferment d-xylose, none of them are able to tolerate the same levels of ethanol as Saccharomyces cerevisiae does, and, furthermore, they cannot ferment hexose sugars as effectively. However, the main disadvantage of S. cerevisiae is the fact that it cannot hydrolyze xylan or utilize or ferment d-xylose, the main component of xylan. While research is continuing on the development of a S. cerevisiae strain able to ferment d-xylose (9, 14, 28, 50), we are working toward the construction of strains able to break down the xylan backbone to its monomeric constituent, d-xylose.
In this paper, we describe the molecular cloning of the A. niger xlnD gene and its expression in S. cerevisiae. Expression and coexpression of xlnD and xyn2 from T. reesei in yeast was obtained with the aid of multicopy plasmids using the derepressible S. cerevisiae alcohol dehydrogenase II gene promoter (ADH2P) and terminator (ADH2T) sequences (38). The enhanced production of both the recombinant enzymes in non-selective complex medium, without the risk of losing the episomal vector, was obtained by constructing autoselective recombinant fur1 S. cerevisiae strains (29).
MATERIALS AND METHODS
Microbial strains and plasmids.
The relevant genotypes and corresponding sources of the yeast and bacterial strains that were constructed and used in this study are summarized in Table 1.
TABLE 1.
Microbial strains and plasmids used in this study
| Strain or plasmida | Relevant genotype | Source or reference |
|---|---|---|
| Yeast and fungal strains | ||
| S. cerevisiae Y294 | leu2-3,112 ura3-52 his3 trp1-299 | ATCC 201160 |
| S. cerevisiae Y294 | ||
| (fur1::LEU2 pDLG1) | bla URA3 ADH2PT | 24 |
| (fur1::LEU2 pDLG5) | bla URA3 ADH2P-xyn2-ADH2T | 24 |
| (fur1::LEU2 pDLG55) | bla URA3 ADH2P-MFα1S-xlnD-ADH2T | This work |
| (fur1::LEU2 pDLG56) | bla URA3 ADH2P-xyn2-ADH2TADH2P-MFα1S-xlnD-ADH2T | This work |
| T. reesei QM6a | ATCC 13631 | |
| A. niger | ATCC 90196 | |
| Bacterial strain | ||
| E. coli XL1-Blue MRF′ | Δ (mcrA)193 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | ZAP-cDNA synthesis kit (Stratagene) |
| Plasmids | ||
| pGEM-T Easy | bla | Promega |
| pRR1 | bla URA3 ADH2P-MFα1S-ADH2T | 37 |
| pDF1 | bla fur1::LEU2 | 24 |
S. cerevisiae Y294 (fur1::LEU2 pDLG1) is designated Y294 (VECT); S. cerevisiae Y294 (fur1::LEU2 pDLG5) is designated Y294 (XYN2); S. cerevisiae Y294 (fur1::LEU2 pDLG55) is designated Y294 (XLO2); S. cerevisiae Y294 (fur1::LEU2 pDLG56) is designated Y294 (XYN2 XLO2).
Media and culture conditions.
Escherichia coli was cultivated on Luria-Bertani medium (39), supplemented with ampicillin (100 μg/ml) for plasmid selection. S. cerevisiae Y294 was cultivated on either YPD medium (1% yeast extract, 2% peptone, 2% glucose) or selective synthetic complete (SC) medium (containing 1 or 2% glucose, yeast nitrogen base without amino acids [Difco], 20 mM succinate [pH 6], and all the required growth factors except uracil [SC−Ura]]. Solid media contained 2% agar. A. niger was also cultivated in SC medium with all the necessary growth factors but with 0.3% oat spelt xylan as the sole carbon source for induction of the xylanolytic enzymes. Bacteria were routinely cultured at 37°C and yeast and A. niger were cultured at 30°C in 300-ml Erlenmeyer flasks, containing 100 ml of medium, on a rotary shaker at 150 rpm. Approximately 2 × 106 cells were used as inoculum in yeast cultures for enzymatic assays.
RNA isolation, first-strand cDNA preparation, and PCR amplification.
Total cellular RNA and mRNA from A. niger were prepared as described previously (24). The A. niger xlnD gene was amplified from a first-strand cDNA copy prepared from mRNA with the aid of two oligonucleotides: ASNXLND-left (5′-GATCATCGATCAACCATGGCGCACTCA-3′) and ASNXLND-right (5′-CATGCTCGAGGTAATAGGCTGACTCTCATCCC-3′). These primers were based on the sequence of the mature region of the A. niger xlnD gene (accession number Z84377). DNA was amplified in 50-μl reaction mixtures (10 pmol of each primer, AMV/Tfl reaction buffer, 1 mM MgSO4, 200 μM each deoxynucleoside triphosphate, 1 μl of mRNA [100 ng/μl], 5 U of avian myeloblastosis virus reverse transcriptase [Promega, Access RT-PCR system], and 5 U of Tfl DNA polymerase [Promega, Access RT-PCR system]) under mineral oil with a Biometra Trio Thermoblock TB1 (Biometra Biomedizinische Analytik, Göttingen, Germany). The reaction mixture was incubated at 48°C for 45 min to allow first-strand cDNA synthesis to take place. Subsequently, denaturation, annealing, and polymerization were carried out for 30 s at 94°C, 1 min at 53°C, and 2 min 30 s at 68°C, respectively, for 33 cycles. The amplified DNA fragment was ligated to pGEM-T-Easy using the pGEM-T-Easy vector system (Promega), as specified by the manufacturer.
DNA manipulations and plasmid constructions.
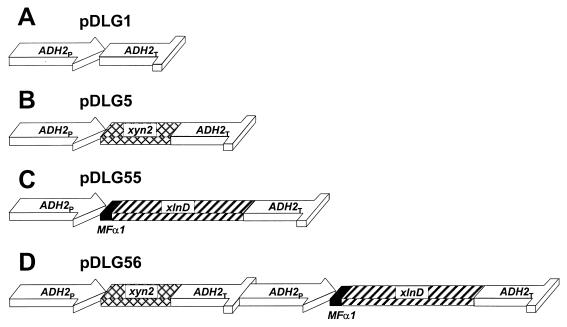
Standard protocols were followed for DNA manipulation (39). Restriction endonuclease-digested DNA was eluted from agarose gels by the method of Tautz and Renz (41). Restriction endonucleases, T4 DNA ligase, the Klenow fragment of E. coli DNA polymerase I, and DNA linkers were purchased from Roche Molecular Biochemicals and used as recommended by the manufacturer. The construction of pDLG1 and pDLG5 (24), as well as pRR1 (37), was described previously. The xlnD gene cloned into plasmid pGEM-T-Easy was sequenced, and the derived sequence was used to design PCR primer DAANXLND-left (5′-GATCATCGATACACCAGCTATGTCGATTAC-3′). The xlnD gene was amplified from the pGEM-T-Easy vector without its native signal sequence with the aid of primers DAANXLND-left and ASNXLND-right and cloned as a 2.4-kb ClaI-SalI fragment into the ClaI and XhoI sites of pRLR1, to create the XLO2 fusion gene in plasmid pDLG55. The PCR was done in a 50-μl reaction mixture (10 pmol of each primer, Pfu reaction buffer, 1 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 5 μl of pGEM-T-Easy with xlnD DNA [10 ng/μl], and 2.5 U of cloned Pfu DNA polymerase [Stratagene]) with a Perkin-Elmer GeneAmp PCR System 2400 apparatus (The Perkin-Elmer Corp., Norwalk, Conn.). Denaturation, annealing, and polymerization were carried out for 1 min at 94°C, 1 min at 50°C, and 5 min at 72°C, respectively, for 28 cycles. Plasmid pDLG5 was digested with HindIII, the overhanging ends were filled in with DNA polymerase I (Klenow fragment), and a BamHI linker was inserted at this site to create plasmid pDLG7. Plasmid pDLG56 was constructed by digesting plasmid pDLG7 with BamHI, isolating the 2.7-kb ADH2P-xyn2-ADH2T fragment, and inserting it into the corresponding site of pDLG55. Plasmid pDF1 (24) was used to construct autoselective S. cerevisiae strains. The relevant expression cassettes transformed to S. cerevisiae are illustrated in Fig. 1.
FIG. 1.
Schematic representation of the expression cassettes used, indicating the xylanase (XYN2) and β-xylosidase (XLND) genes as well as the ADH2 promoter (ADH2P) and terminator (ADH2T) and the mating factor α secretion signal (MFα1S). XYN2 is indicated by cross-hatched boxes, xlnD is indicated by hatched boxes, the ADH2 promoter and terminator sequences are indicated by open boxes, and the MFα1S secretion signal is indicated by solid boxes.
Subcloning and sequencing of XLO2.
Plasmid pGEM-T-Easy containing the β-xylosidase gene was used to construct six deletion subclones for sequencing. The XLO2 nucleotide sequence was determined by amplifying DNA fragments with the Big Dye Terminator cycle-sequencing reader reaction with AmpliTaq DNA polymerase F5 (Applied Biosystems kit) using fluorescently labeled nucleotides, and the reaction mixtures were subjected to electrophoresis on an Applied Biosystems automatic DNA sequencer (model ABI Prism 377). Sequence data were analyzed by using the PC/GENE software package (IntelliGenetics, Inc., Mountain View, Calif.).
DNA transformation and PCR confirmation of gene replacement.
E. coli and S. cerevisiae transformations were carried out by standard techniques described by Sambrook et al. (39) and the lithium acetate dimethylsulfoxide method described by Hill et al. (13), respectively. Plasmid pDF1 digested with NsiI and NcoI (24) was used to construct autoselective S. cerevisiae fur1 strains. Replacement of the wild-type FUR1 gene with the LEU2 disrupted allele on plasmid pDF1 was confirmed by PCR using primers FUR1-left (5′-TCCGTCTGGCATATCCTA-3′) and FUR1-right (5′-TTGGCTAGAGGACATGTA-3′). These primers annealed near the NsiI and NcoI sites of the FUR1 gene (24). Total cellular DNA was isolated from S. cerevisiae strains by the method described by Hoffman and Winston (15). DNA was amplified in 25-μl reaction mixtures (10 pmol of each primer, Taq reaction buffer, 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 4 μl of genomic DNA [100 ng/μl] and 1 U of Taq DNA polymerase [Roche Molecular Biochemicals]) with a GeneAmp PCR System 2400 apparatus. Denaturation, annealing, and polymerization were carried out for 30 s at 94°C, 30 s at 57°C, and 3 min at 72°C, respectively, for 30 cycles. PCR with the wild-type strain produced a DNA fragment of 1.33 kb, while successful gene replacement in the recombinant strains produced a DNA fragment of 3.27 kb.
Xylanase and β-xylosidase activity determination.
β-Xylanase- and β-xylosidase-producing cultures were grown in YPD for 160 h, and the enzyme activities were determined. All enzyme activity determinations were done in triplicate in 50 mM sodium citrate buffer at pH 5 and 50°C for 5 min, unless stated otherwise. The β-xylanase activity was determined by the method described by Bailey et al. (2). The β-xylosidase activity was quantitated using the chromophoric substrate p-nitrophenyl-β-d-xyloside (PNPX) (25). The chromophoric substrate was used at a final concentration of 5 mM unless stated otherwise. The culture supernatant was used as source of β-xylanase and supernatant with intact yeast cells was used as source of β-xylosidase for the growth curves. All activities were expressed in katals per milliliter; 1 katal is the amount of enzyme needed to produce 1 mol of reducing sugar (or d-xylose equivalent) from birchwood xylan (or chromophoric substrate) per s (2).
The pH and temperature optima, thermostability, and inhibition studies were performed with the extracellular fraction of an S. cerevisiae Y294 (XLO2) culture. The thermostability of the recombinant β-xylosidase was tested by heating enzyme samples for different times at various temperatures and subsequently determining the activity at 50°C for 5 min. For the determination of the optimum pH of the β-xylosidase, the buffers used were 50 mM citrate (pH 3.0 to 6.2) and 50 mM potassium phosphate (pH 6.2 to 8.0). Inhibition of β-xylosidase activity on PNPX in the presence of xylobiose, d-xylose, cellobiose, and d-glucose was studied by determining the β-xylosidase activity using PNPX at a final concentration of 2 mM at different inhibitor concentrations (0 to 20 mM).
Analysis of xylobiose, xylotriose, and xylan degradation.
Xylobiose and xylotriose hydrolysis by strain Y294 (XLO2) was carried out in 800-μl reaction mixtures at 60°C. Xylobiose or xylotriose (100 μl of a 50 mM solution in water), 400 μl of 100 mM citrate buffer (pH 3.4), and 200 μl of water were thermally equilibrated before the reactions were started by adding 100 μl of a 50-h-old culture of Y294 (XLO2), grown in YPD, to the mixture. The total activity on PNPX of the enzyme mix used in the above-mentioned reactions was 3.93 nkat/ml. Aliquots (80 μl) of the reaction mixtures were obtained, and the reactions were stopped at different time intervals by incubating at 100°C for 10 min. Thin-layer liquid chromatography on silica gel plates (Silica gel 60; Merck) in a solvent mixture of n-propanol, ethanol, water (7:1:2) was used to separate the hydrolysis products. After removal of the solvent, the spots of sugar were visualized by dipping in a solution of ethanol and sulfuric acid (95:5) followed by incubation at 180°C for about 2 min.
Xylan degradation by the recombinant yeast strains was analyzed by inoculating the relevant strains (100 μl of a 24-h-old culture) into YPD medium buffered at pH 5 with 0.1 mM citrate buffer containing 5% birchwood xylan. Aliquots were removed at different time intervals, extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1), and analyzed by thin-layer liquid chromatography. The amount of xylose produced was quantified by a high-performance liquid chromatography system (model DX500; Dionex, Sunnyvale, Calif.) using an anion-exchange column (Carbopac PA-100, 4 × 250, and Carbopac PA-100, guard) and a pulsed amperometric detector (ED40). A gradient of sodium acetate (20 to 100 mM) in 60 mM NaOH was used. Data were analyzed using the Dionex Peaknet software package.
Nucleotide sequence accession number.
The XLO2 sequence was deposited at GenBank (accession number AF108944).
RESULTS
Cloning and expression of the A. niger xlnD gene and T. reesei xyn2 gene in yeast.
The A. niger xlnD gene was amplified from first-strand cDNA prepared from A. niger by using sequence-specific PCR primers. The PCR product (lacking the native 26-amino-acid signal-encoding region) was inserted into plasmid pRLR1 in frame with the yeast mating factor α secretion signal (MFα1s) under the control of the derepressible ADH2 gene promoter and terminator, creating plasmid pDLG55 (Fig. 1C). Correct processing by S. cerevisiae would lead to the production of an 804-amino-acid Xlo2 protein with a putative molecular mass of 85.1 kDa. Plasmid pDLG55 was subsequently transformed into S. cerevisiae Y294, and the production of functional β-xylosidase was confirmed by determining the enzymatic hydrolysis of PNPX to p-nitrophenol and d-xylose. The ADH2P-xyn2-ADH2T gene cassette was isolated from plasmid pDLG7 (Fig. 1B) and cloned into plasmid pDLG55 (creating plasmid pDLG56 [Fig. 1D]), which contained both the T. reesei xyn2 and A. niger XLO2 genes. Plasmid pDF1 was used to disrupt the FUR1 gene of S. cerevisiae strains containing plasmids pDLG1, pDLG5, pDLG55, and pDLG56, creating strains Y294 (VECT), Y294 (XYN2), Y294 (XLO2), and Y294 (XYN2 XLO2), respectively.
Effects of pH and temperature on β-xylosidase activity.
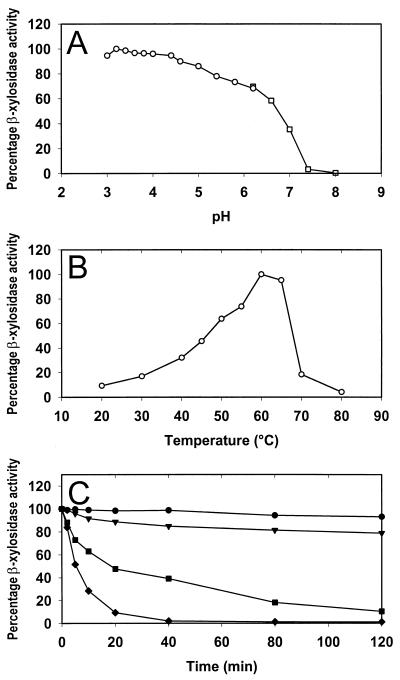
The recombinant β-xylosidase activity peaked between pH 3 and 5, with the highest activity (5.4 nkat/ml) measured at pH 3.2 in 50 mM citrate buffer (Fig. 2A). The optimum temperature for this enzyme was at 60°C (Fig. 2B). Although the highest β-xylosidase activity was measured at 60°C, the enzyme was unstable at this temperature (Fig. 2C). The β-xylosidase activity decreased by almost 60% after a 20-min incubation at 60°C, and less than 10% activity could be measured after a 2-h incubation. However, the recombinant enzyme was relatively stable at 55°C, with more than 80% of the activity remaining after 2 h at this temperature.
FIG. 2.
(A and B) Effect of pH at 50°C (A) and temperature at pH 5.0 (B) on the activity of Xlo2. The buffers used in the enzyme reactions were 50 mM citrate buffer (pH 3 to 6.2) (○) and 50 mM phosphate buffer (pH 6.2 to 9) (□). (C) The temperature stability of Xlo2 at 50°C (●), 55°C (▾), 60°C (■), and 65°C (⧫), was determined by preincubating the enzyme at these temperatures in the absence of the substrate for 0, 2, 5, 10, 20, 40, 90, and 120 min before determining the β-xylosidase activity on PNPX. The β-xylosidase activity prior to the preincubations (time 0 min) was taken as 100%.
Xlo2 activity on xylobiose and xylotriose.
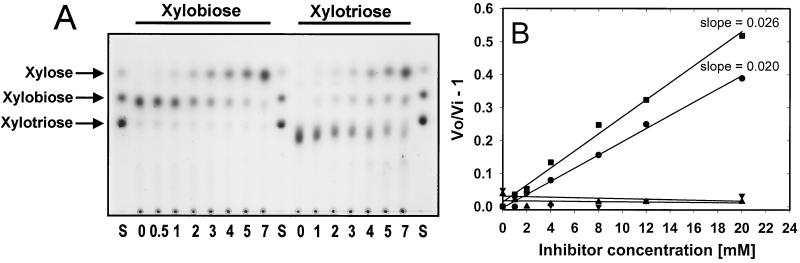
The hydrolysis of xylobiose and xylotriose by the recombinant A. niger β-xylosidase produced in yeast was determined by incubating the enzyme in the presence of these substrates at 50°C. The β-xylosidase hydrolyzed xylotriose less efficiently than it hydrolyzed xylobiose; however, hydrolysis of both substrates to d-xylose was almost complete after 7 h (Fig. 3A).
FIG. 3.
(A) Thin-layer chromatogram of the hydrolysis of xylobiose and xylotriose by S. cerevisiae (XLO2). Reaction mixtures were incubated at 50°C, and samples were taken after 0 and 30 min and 1, 2, 3, 4, 5, and 7 h. d-Xylose, xylobiose, and xylotriose were used as standards (S). (B) A plot of (vo/vi) − 1 versus the xylobiose concentration to investigate competitive inhibition of β-xylosidase activity on PNPX in the presence of xylobiose (■), d-xylose (●), cellobiose (▾), or d-glucose (▴).
Competitive inhibition of the hydrolysis of 2 mM PNPX by the recombinant β-xylosidase was determined in the presence of 0 to 20 mM xylobiose, d-xylose, cellobiose, and d-glucose. The Km value for cell wall-bound recombinant Xlo2 β-xylosidase on PNPX was determined to be 0.4 mM (5). To determine the approximate Km value of the recombinant Xlo2 β-xylosidase on its natural substrate, xylobiose, the apparent inhibition constant, Ki(app), for xylobiose as competitive inhibitor for β-xylosidase on PNPX can be determined from the equation (27)
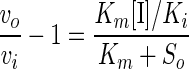 |
When the PNPX concentration (2 mM) and Km for PNPX (0.4 mM) are substituted into the equation, it can be simplified to
 |
The plot of (vo/vi) − 1 versus the xylobiose concentration (Fig. 3B) was linear and thus allows the determination of the Ki(app) for Xlo2 as 0.167/slope. The slope was determined by linear regression in Sigmaplot for Windows (version 4.0), and the Ki(app) values for xylose and xylosidase were determined as 8.33 and 6.41 mM, respectively. Glucose and cellobiose did not act as competitive inhibitors of PNPX in the 0 to 20 mM range (Fig. 3B).
Production of β-xylosidase and β-xylanase by recombinant yeast strains.
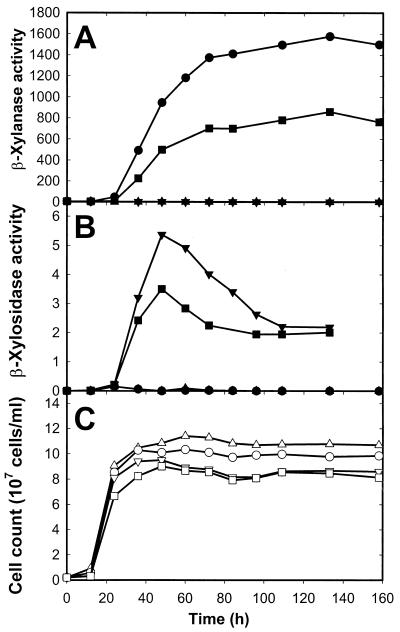
The production of β-xylanase and β-xylosidase, as well as the cell growth of the different recombinant yeast strains, was monitored over a 160-h period (Fig. 4). Relatively high levels of β-xylanase activity were recorded in Y294 (XYN2) and Y294 (XYN2 XLO2) at ca. 72 h of growth (Fig. 4A). After 72 h, the activity still increased gradually to reach a maximum of 1,577 and 860 nkat/ml in Y294 (XYN2) and Y294 (XYN2 XLO2), respectively. The β-xylosidase activity in Y294 (XLO2) and Y294 (XYN2 XLO2) increased rapidly to reach its maximum of 5.3 and 3.5 nkat/ml, respectively, after only 48 h of growth (Fig. 4B). The activity subsequently decreased and stabilized at a constant level of about 2 nkat/ml in both strains. In all three recombinant strains expressing the T. reesei β-xylanase and/or the A. niger β-xylosidase gene, reduced cell yield rates were obtained compared with the parental strain containing plasmid pDLG1 (Fig. 4C).
FIG. 4.
Time course of β-xylanase (A), β-xylosidase (B), and cell mass (C) produced by S. cerevisiae Y294 (VECT) (▴, ▵), Y294 (XYN2) (●, ○), Y294 (XLO2) (▾, ▿), and Y294 (XYN2 XLO2) (■, □) in shake flask cultures. The β-xylanase activities were assayed by the method of Bailey et al. (2) using the culture supernatant as the source of enzyme, and the β-xylosidase activities were assayed as described by La Grange et al. (25), using cell cultures as the source of enzyme. Enzyme activities were expressed in katals per milliliter and are indicated by solid symbols. The enzyme activities represent the average of three independent cultures. The maximum deviation for the β-xylanase and β-xylosidase activities did not exceed 11 and 12%, respectively. Yeast cell counts were determined with a haemocytometer and are indicated by open symbols.
Xylan degradation by S. cerevisiae.
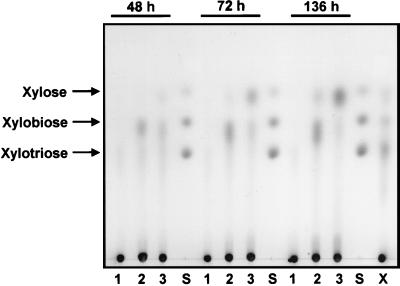
Y294 (VECT), Y294 (XYN2), and Y294 (XYN2 XLO2) were individually inoculated in YPD medium plus 5% birchwood xylan to analyze the ability of the recombinant S. cerevisiae strains to degrade xylan. Samples were taken after 48, 72, and 136 h of growth at 30°C. Both Y294 (XYN2) and Y294 (XYN2 XLO2) were able to degrade xylan; however, Y294 (XYN2) produced primarily xylobiose while Y294 (XYN2 XLO2) was able to degrade xylan to its monomeric constituent, d-xylose (Fig. 5). HPLC analysis showed that Y294 (XYN2 XLO2) released more than 20 g of d-xylose per liter from 50 g of birchwood xylan per liter after 136 h of growth (Table 2) while Y294 (XYN2) released only ca. 7 g of d-xylose per liter. d-Xylose is the major end product released from birchwood by Y294 (XYN2 XLO2), whereas Y294 (XYN2) predominantly released xylobiose with xylotriose as minor product. Although the T. reesei Xyn2 β-xylanase released d-xylose, xylobiose, and xylotriose from birchwood xylan, the A. niger Xlo2 β-xylosidase synergistically enhanced the release of d-xylose as dominant end product.
FIG. 5.
Xylan degradation by Y294 (VECT) (lanes 1), Y294 (XYN2) (lanes 2), and Y294 (XYN2 XLO2) (lanes 3). Samples were taken after 48, 72, and 136 h of growth at 30°C. d-Xylose, xylobiose, and xylotriose were used as standards (S). The right-hand lane (labeled X) contains the standard (d-xylose, xylobiose, and xylotriose), as well as the 136-h sample of Y294 (VECT), to monitor the effects of the medium components on the migration of d-xylose, xylobiose, and xylotriose.
TABLE 2.
Release of d-xylose, xylobiose, and xylotriose from birchwood xylan (50 g/liter) after incubation with recombinant S. cerevisiae strains
| Strain | Amt (g/liter) of:
|
|||||
|---|---|---|---|---|---|---|
| Xylotriose
|
Xylobiose
|
d-Xylose
|
||||
| 72 h | 136 h | 72 h | 136 h | 72 h | 136 h | |
| S. cerevisiae Y294 (VECT) | NDa | ND | 0.93 | 1.4 | 1.1 | 1.3 |
| S. cerevisiae Y294 (XYN2) | 1.6 | 4.3 | 21.0 | 11.4 | 4.0 | 9.2 |
| S. cerevisiae Y294 (XYN2 XLO2) | ND | ND | 6.4 | 7.7 | 13.2 | 23.4 |
ND, not detected.
DISCUSSION
mRNA was isolated from the xylanolytic fungus A. niger ATCC 90196, and the xlnD gene encoding the β-xylosidase XlnD was amplified with the aid of sequence-specific PCR primers. The DNA sequence was verified and compared with the DNA sequence published by Van Peij et al. (48) and with other DNA sequences available in the GenBank database. The nucleotide sequence of the cDNA fragment is 94 and 98% identical to the DNA and amino acid sequences, respectively, of the A. niger xlnD sequence reported by Van Peij et al. (48) (accession number Z84377). The native xlnD contains a predicted signal peptide of 26 or 27 amino acids, and the mature protein has a predicted molecular mass of 88 kDa. It also exhibits significant levels of similarity at the amino acid level to the A. nidulans (66% identity), A. oryzae (64% identity), and T. reesei (63% identity) β-xylosidases (20, 33, 35). It is noteworthy that the Bxl1 of T. reesei also exhibited α-l-arabinofuranosidase and α-l-arabinopyranosidase activities (33).
Translation in yeast can be modulated at the level of initiation by four aspects of mRNA structure: (i) the position of the initiation codon, i.e., whether it is the first AUG; (ii) the primary sequence or context surrounding the AUG codon; (iii) the secondary structure both upstream and downstream of the AUG codon; and (iv) the leader length (22). By placing xlnD under the transcriptional control of a strong yeast promoter (ADH2P), none of the above should pose a problem. The native secretion signal was replaced with the MFα1S secretion signal to facilitate secretion (36). XlnD and Xyn2 have S. cerevisiae codon bias index values of 0.25 and 0.33, respectively, which are acceptable for efficient translation in S. cerevisiae (40).
The xlnD and xyn2 genes were expressed from episomal plasmids, and the resulting recombinant yeast strains were kept under selective conditions to ensure vector stability. However, the use of a selective synthetic medium was not ideal for the production of high levels of heterologous proteins. We therefore genetically altered the recombinant yeast strains to allow autoselection for the episomal plasmids (24). The FUR1 gene of S. cerevisiae encodes uracil phosphoribosyltransferase, which catalyzes the conversion of uracil into uridine 5′-phosphate in the pyrimidine salvage pathway (19). By disrupting the FUR1 gene, S. cerevisiae Y294 was forced to utilize the complementing URA3 gene product to synthesize uridine 5′-phosphate de novo, even in complex (YPD) medium. The URA3 gene was provided as the yeast selectable marker on the YEp352-based vectors used for the expression of XYN2 and/or XLO2 genes. In this study the ADH2 promoter and terminator were used for the expression of both the xyn2 and XLO2 genes. However, in the construction of a recombinant industrial yeast strain, the promoter and terminator of one of these genes need to be changed to ensure genetic stability.
The highest total β-xylanase activities obtained in YPD medium in shake flask cultures of Y294 (XYN2) and Y294 (XYN2 XLO2) were 1,577 and 860 nkat/ml, respectively (Fig. 4). The maximum β-xylosidase activities in Y294 (XLO2) and Y294 (XYN2 XLO2) were 5.3 and 3.5 nkat/ml, respectively, after only 48 h of growth (Fig. 4). Aspergillus β-xylosidases are usually cell wall bound (35), and intact yeast cells in the growth medium were used to determine these activities; however, this does not include intracellular activity, which accounts for almost 40% of the total β-xylosidase measured (data not shown). Production of the recombinant β-xylanase and β-xylosidase caused a reduction in cell yields (Fig. 4), probably because of the increased metabolic burden imposed on the cells through the high-level expression of the heterologous β-xylanase and β-xylosidase protein (10). The recombinant β-xylosidase produced by Y294 (XLO2) and Y294 (XYN2 XLO2) is active on both xylobiose and xylotriose. As expected, the enzyme is less active on xylotriose than on xylobiose; however, both these substrates were degraded to their monomeric constituent, d-xylose (Fig. 3A). Xylobiose and d-xylose acted as competitive inhibitors of recombinant Xlo2 when PNPX was used as the substrate, but glucose and cellobiose did not (Fig. 3B). The XlnD β-xylosidase was most probably sensitive to product (d-xylose) inhibition, as has frequently been found for β-glucosidases belonging to the same hydrolase family (family 3) (11, 52). The Ki(app) values for both xylobiose and d-xylose were higher than reported for native A. niger XlnD (42). However, (hyper)glycosylation of the β-xylosidase, which frequently occurs during heterologous protein expression in S. cerevisiae (24), could affect its Ki value, as observed for the β-xylosidase of Arxula adeninivorans (6).
The recombinant β-xylanase has a pH optimum between 4 and 6 (24), while the β-xylosidase has an optimum of between 3 and 5 (Fig. 2A). In the recombinant yeast Y294 (XYN2 XLO2), both enzymes should function optimally at pH 5. At the optimum temperature of growth for S. cerevisiae (30°C), both these enzymes are only about 30% active. When Y294 (XYN2 XLO2) was cultivated in YPD medium containing 5% birchwood xylan, breakdown of xylan to d-xylose was visible after 72 h (Fig. 5; Table 2). When the residual amounts of xylobiose (1.4 g/liter) and d-xylose (1.3 g/liter) released by nonspecific activities present in S. cerevisiae Y294 (VECT) were subtracted Y294 (XYN2) released ca. 4.3 g of xylotriose per liter, 10.0 g of xylobiose per liter, and 6.8 g of d-xylose per liter with the aid of recombinant Xyn2 β-xylanase, which amounts to about 42% conversion of birchwood to these end products. Y294 (XYN2 XLO2), producing both the Xyn2 β-xylanase and Xlo2 β-xylosidase, released ca. 6.6 g of xylobiose per liter and 22.0 g of d-xylose per liter, which amounts to about 57% conversion of birchwood to these end products. The simultaneous production of both the T. reesei β-xylanase and A. niger Xlo2 β-xylosidase thus synergistically enhanced the hydrolysis of birchwood xylan to d-xylose as the dominant end product. These results are in constrast with the results obtained with the recombinant S. cerevisiae strain Y294 (XYN2 XLO1) coproducing the T. reesei β-xylanase and B. pumilus Xlo1 β-xylosidase (26). The inability of Y294 (XYN2 XLO1) to yield d-xylose as the major product from 5% birchwood xylan could most probably be ascribed to the very low Xlo1 activity (0.5 nkat/ml) produced.
Considering the eventual use of recombinant S. cerevisiae strains to convert xylan to cell mass or ethanol through simultaneous saccharification and fermentation, the ability of recombinant S. cerevisiae Y294 (XYN2 XLO1) to degrade xylan in culture was assessed. Actively growing cells were used to ensure maximum levels of enzyme activity throughout the experiment. The described xylan-degrading S. cerevisiae strain, together with the current development of S. cerevisiae strains capable of fermenting d-xylose (9, 14), thus paves the way to efficient xylan degradation and utilization by yeast.
ACKNOWLEDGMENT
We thank the Foundation for Research Development, South Africa, for financial support.
REFERENCES
- 1.Aristidou A, Penttilä M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 2.Bailey M J, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23:257–270. [Google Scholar]
- 3.Beauchemin K A, Rode L M, Sewalt V J H. Fibrolytic enzymes increase fiber digestibility and growth rate of steers fed dry forages. Can J Anim Sci. 1995;75:641–644. [Google Scholar]
- 4.Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286–290. [Google Scholar]
- 5.Biely P, Hirsch J, la Grange D C, van Zyl W H, Prior B A. A chromogenic substrate for a β-xylosidase-coupled assay of α-glucuronidase. Anal Biochem. 2000;286:289–294. doi: 10.1006/abio.2000.4810. [DOI] [PubMed] [Google Scholar]
- 6.Buttner R, Bode R. Purification and characterization of β-xylosidase activities from the yeast Arxula adeninivorans. J Basic Microbiol. 1992;32:159–166. doi: 10.1002/jobm.3620320304. [DOI] [PubMed] [Google Scholar]
- 7.Cummings C, Fowler T. Secretion of Trichoderma reesei β-glucosidase by Saccharomyces cerevisiae. Curr Genet. 1996;29:227–233. [PubMed] [Google Scholar]
- 8.De Vries R P, Kester H C M, Poulsen C H, Benen J A E, Visser J. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr Res. 2000;327:401–410. doi: 10.1016/s0008-6215(00)00066-5. [DOI] [PubMed] [Google Scholar]
- 9.Eliasson A, Christensson C, Wahlbom C F, Hahn-Hägerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–3386. doi: 10.1128/aem.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Görgens J F, Van Zyl W H, Knoetze J H, Hahn-Hägerdal B. The metabolic effect of the PGK1 and ADH2 promoter systems for heterologous xylanase production by Saccharomyces cerevisiae in defined medium. Biotechnol Bioeng. 2001;73:238–245. doi: 10.1002/bit.1056. [DOI] [PubMed] [Google Scholar]
- 11.Gueguen Y, Chemardin P, Arnaud A, Galzy P. Purification and characterization of an intracellular β-glucosidase from Botrytis cinerea. Enzyme Microb Technol. 1995;78:900–906. [Google Scholar]
- 12.Herrmann M C, Vrsanska M, Jurickova M, Hirsch J, Biely P, Kubicek C P. The β-d-xylosidase of Trichoderma reesei is a multifunctional β-d-xylan xylohydrolase. Biochem J. 1997;321:375–381. doi: 10.1042/bj3210375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill J, Ian K A, Donald G, Griffiths D E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho N W Y, Chen Z, Brainard A P. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 16.Hrmova M, Biely P, Vrsanska M. Cellulose- and xylan-degrading enzymes of Aspergillus terreus and Aspergillus niger. Enzyme Microb Technol. 1989;11:610–616. [Google Scholar]
- 17.Iizuka Y, Shinoyama H, Kamiyama Y, Yasui T. The condensation reaction of Aspergillus niger crude β-xylosidase using xylose. Biosci Biotechnol Biochem. 1992;56:331–332. [Google Scholar]
- 18.Jeffries T W. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- 19.Kern L, De Montigny J, Jund R, Lacroute F. The FUR1 gene of Saccharomyces cerevisiae: cloning, structure and expression of wild-type and mutant alleles. Gene. 1990;88:149–157. doi: 10.1016/0378-1119(90)90026-n. [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto N, Yoshino S, Ohmiya K, Tsukagoshi N. Sequence analysis, overexpression, and antisense inhibition of a β-xylosidase gene, xylA, from Aspergillus oryzae KBN616. Appl Environ Microbiol. 1999;65:20–24. doi: 10.1128/aem.65.1.20-24.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein R D, Favreau M A. The Candida species: biochemistry, molecular biology and industrial applications. In: Hui Y H, Khachatourians G G, editors. Food biotechnology microorganisms. New York, N.Y: VCH Publishers, Inc.; 1995. pp. 297–371. [Google Scholar]
- 22.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 23.Kulkarni N, Shendye A, Rao M. Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev. 1999;23:411–456. doi: 10.1111/j.1574-6976.1999.tb00407.x. [DOI] [PubMed] [Google Scholar]
- 24.La Grange D C, Pretorius I S, Van Zyl W H. Expression of a Trichoderma reesei β-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:1036–1044. doi: 10.1128/aem.62.3.1036-1044.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Grange D C, Pretorius I S, Van Zyl W H. Cloning of the Bacillus pumilus β-xylosidase gene (xynB) and its expression in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;47:262–266. doi: 10.1007/s002530050924. [DOI] [PubMed] [Google Scholar]
- 26.La Grange D C, Claeyssens M, Pretorius I S, Van Zyl W H. Coexpression of the Bacillus pumilus β-xylosidase gene (xynB) with the Trichoderma reesei β-xylanase (xyn2) gene in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2000;54:195–200. doi: 10.1007/s002530000372. [DOI] [PubMed] [Google Scholar]
- 27.Leggio L, Pickersgill R W. Xylanase-oligosaccharide interactions studied by a competitive enzyme assay. Enzyme Microb Technol. 1999;25:701–709. [Google Scholar]
- 28.Lidén G, Walfridsson M, Ansell R, Anderlund M, Adler L, Hahn-Hägerdal B. A glycerol-3-phosphate dehydrogenase-deficient mutant of Saccharomyces cerevisiae expressing the heterologous XYL1 gene. Appl Environ Microbiol. 1996;62:3894–3896. doi: 10.1128/aem.62.10.3894-3896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loison G, Nguyen-Juilleret M, Alouani S, Marquet M. Plasmid-transformed ura3 fur1 double-mutants of S. cerevisiae: an autoselection system applicable to the production of foreign proteins. Bio/Technology. 1986;4:433–437. [Google Scholar]
- 30.Luonteri E, Tenkanen M, Siika-aho M, Buchert J, Viikari L. α-Arabinosidases of Aspergillus terreus and their potentials in pulp and paper applications. In: Srebotnik E, Messner K, editors. Biotechnology in the pulp and paper industry. Vienna, Austria: Facultas-Universitätsverlag; 1996. pp. 119–122. [Google Scholar]
- 31.Maat J, Roza M, Verbakel J, Stam H, Santos da Silva M J, Bosse M, Egmond M R, Hagemans M L D, Gorcom R F M V, Hessing J G M, van der Hondel C A M J J, Rotterdam C V. Xylanases and their application in bakery. In: Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 349–360. [Google Scholar]
- 32.Machida M, Ohtsuki I, Fukui S, Yamashita I. Nucleotide sequences of Saccharomycopsis fibuligera genes for extracellular β-glucosidases as expressed in Saccharomyces cerevisiae. Appl Environ Microbiol. 1988;54:3147–3155. doi: 10.1128/aem.54.12.3147-3155.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolles-Clark E, Tenkanen M, Nakari-Setälä T, Penttilä M. Cloning of the genes encoding α-l-arabinofuranosidase and β-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:3840–3846. doi: 10.1128/aem.62.10.3840-3846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newbold C J. Microbial feed additives for ruminants. In: Wallace R J, Chesson A C, editors. Biotechnology in animal feeds and animal feeding. New York, N.Y: VCH Publishers; 1995. pp. 259–278. [Google Scholar]
- 35.Pérez-González J A, Van Peij N N M E, Bezoen A, MacCabe A P, Ramón D, De Graaff L H. Molecular cloning and transcriptional regulation of the Aspergillus nidulans xlnD gene encoding a β-xylosidase. Appl Environ Microbiol. 1998;64:1412–1419. doi: 10.1128/aem.64.4.1412-1419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanos M A, Scorer C A, Clare J J. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 37.Roth R L. Molecular cloning, manipulation and expression of the laccase gene (lacA) of Pleurotus osteatus in Saccharomyces cerevisiae. M.S. thesis. Stellenbosch, South Africa: University of Stellenbosch; 1997. [Google Scholar]
- 38.Russell D W, Smith M, Williamson V M, Young E T. Nucleotide sequence of the yeast alcohol dehyrogenase II gene. J Biol Chem. 1983;258:2674–2682. [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Sharp P M, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- 41.Tautz D, Renz N. An optimized freeze squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983;132:503–517. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- 42.Tavobilov I M, Rodionova N A, Bezborodov A M. Properties of the exo-1,4-β-xylosidase of Aspergillus niger 15. Prikl Biokhim Mikrobiol. 1983;19:232–239. . (In Russian.) [PubMed] [Google Scholar]
- 43.Teeri T T, Koivula A, Linder M, Wohlfahrt G, Divne C, Jones T A. Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose? Biochem Soc Trans. 1998;26:173–178. doi: 10.1042/bst0260173. [DOI] [PubMed] [Google Scholar]
- 44.Tenkanen M, Buchert J, Puls J, Poutanen K, Viikari L. Two main xylanases of Trichoderma reesei and their use in pulp processing. In: Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 547–550. [Google Scholar]
- 45.Thomson J A. Molecular biology of xylan degradation. FEMS Microbiol Rev. 1993;104:65–82. doi: 10.1111/j.1574-6968.1993.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 46.Törönnen A, Mach R L, Messner R, Gonzalez R, Kalkkinen N, Harkki A, Kubicek C P. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Bio/Technology. 1992;10:1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- 47.Van Paridon P A, Boonman J C P, Selten G C M, Geerse C, Barug D, De Bot P H M, Hemke G. The application of fungal endoxylanase in poultry diets. In: Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 371–378. [Google Scholar]
- 48.Van Peij N N M E, Brinkmann J, Vrsanská M, Visser J, De Graaff L H. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 49.van Peij N N, Gielkens M M, De Vries R P, Visser J, de Graaff L H. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walfridsson M, Bao X, Anderland M, Lilius G, Bülow L, Hahn-Hägerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol. 1996;62:4648–4651. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong K K Y, Saddler J N. Trichoderma xylanases, their properties and application. Crit Rev Biotechnol. 1992;12:413–435. [Google Scholar]
- 52.Woodward J, Wiseman A. Fungal and other β-glucosidases—their properties and applications. Enzyme Microb Technol. 1882;15:62–65. [Google Scholar]