Abstract
Alternative polyadenylation (APA) regulates gene expression by cleavage and addition of poly(A) sequence at different polyadenylation sites (PAS) in 3’UTR, thus, generating transcript isoforms with different lengths. Cleavage stimulating factor 64 (CstF64) is an APA regulator which plays a role in PAS selection and determines the length of 3’UTR. CstF64 favors the use of proximal PAS, resulting in 3’UTR shortening, which enhances the protein expression by increasing the stability of the target genes. The aim of this study is to investigate the role of CstF64 in cardiac fibrosis, a key event leading to heart failure (HF). We determined the expression of CstF64, key profibrotic genes, and their 3’UTR changes by calculating distal PAS (dPAS) usage in left ventricular (LV) tissues and cardiac fibroblasts from HF patients. CstF64 was upregulated in HF LV tissues and cardiac fibroblasts along with increased deposition of profibrotic genes such as COL1A and FN1 and significant shortening in their 3’UTR. In addition, HF cardiac fibroblasts showed increased transforming growth factor receptor β1 (TGFβR1) expression consistent with significant shortening in TGFβR1. Upon knockdown of CstF64 from HF fibroblasts, downregulation in profibrotic genes corresponding to lengthening in their 3’UTR was observed. Our finding suggests an important role of CstF64 in myofibroblast activation and promotion of cardiac fibrosis during HF through APA. Therefore, targeting CstF64 mediated RNA processing approach in human HF could provide a new therapeutic treatment strategy for limiting fibrotic remodeling.
Keywords: Alternative polyadenylation, 3’UTR, Heart failure, Fibrosis, Myofibroblast
Graphical Abstract
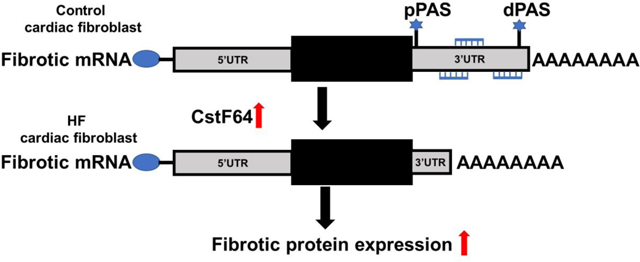
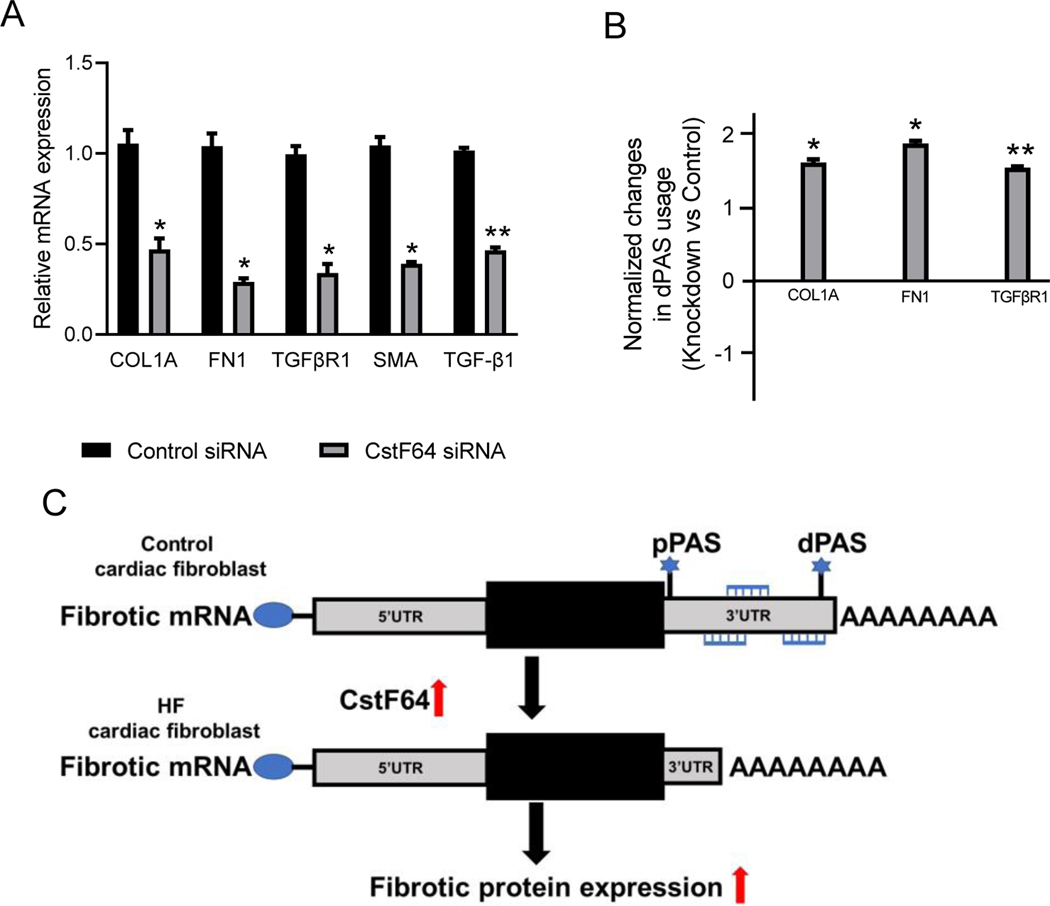
The model illustrates the effect of CstF64 upregulation on cardiac fibrosis.
1. Introduction
Alternative polyadenylation (APA) regulates gene expression post-transcriptionally and produces different lengths of 3′ untranslated regions (3’UTR) of messenger RNA (mRNA), as well as an altered reading frame. APA has a major impact on diverse cell processes that include mRNA metabolism, protein localization, differentiation, development, senescence, and cell proliferation [1]. Mammals have around 70% of their mRNA-encoding genes with more than one polyadenylation site (PAS), most of which are found in 3’UTR regions [2]. The differential use of proximal or distal PAS can result in APA isoforms with shorter or longer transcriptions or 3’UTRs. This can affect the stability of mRNAs in various ways, such as altering the availability of microRNA (miRNA) binding sites, RNA binding protein (RBP) sites, and lncRNA binding sites, which are usually found at the 3’UTR; or altering the repression of mRNA by AU-rich elements (ARE) [3, 4]. As a result, mRNAs with shorter 3’UTRs are more stable since they escape regulatory mechanisms and tend to be overexpressed. Similarly, mRNAs with lengthened 3’UTRs are typically underexpressed.
The choice of PAS is determined by the interaction between cis-elements in pre-mRNA and existing levels of trans-acting protein complexes involved in cleavage and polyadenylation. There are nearly 20 core trans-acting proteins which include four protein complexes - cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor Im (CFIm), and cleavage factor IIm (CFIIm); and multiple other single proteins [5]. The majority of these protein complexes favor the use of distal PAS, whereas CstF64, one of the three components of the CstF complex, has been demonstrated to use proximal PAS, resulting in transcripts with shortened 3’UTRs [1]. Through its interaction with the conserved G/U-rich downstream sequence element (DSE), the CstF complex contributes to the selection of PAS [6]. CstF64 is the RNA binding component of the CstF complex that has been associated with upregulation in many cancer types including those where 3’UTR shortening also occurred [7]. As a result of the dysregulation of APA core proteins, global 3’UTR shortening events have been reported in several human diseases, including cardiovascular disease [8]. However, studies of the role of APA and, specifically, CstF64 in heart failure (HF) and its involvement in the progression of cardiac fibrosis have not been previously investigated.
Cardiac fibrosis is the primary cause of HF, and it is often associated with other cardiovascular diseases [9]. When cardiac fibroblasts are activated and differentiated into myofibroblasts, they deposit extracellular matrix proteins (ECM), especially collagen type I, which causes matrix stiffness, reduced tissue compliance, and impaired heart muscle function, all factors accelerating the progression of HF [10]. Despite a significant increase in treatment options for other organs, there are no FDA-approved drugs to directly target cardiac fibrosis, indicating a crucial need for innovative therapeutic approaches. The molecular mechanisms driving the deposition of ECM proteins and the trigger of profibrotic genes are not completely understood, particularly with RNA regulation. In this study, we investigated the role of CstF64 in cardiac fibrosis using tissues and fibroblast cells derived from the left ventricle (LV) of HF patients collected during transplantation.
2. Materials and Methods
2.1. Human heart tissues collection
The tissue samples were obtained from the LV of human HF patients during transplantation at the DeBakey Heart & Vascular Center, Houston Methodist Hospital as described previously [11]. Control tissue samples were obtained from the LV of healthy donor patients that were not used for transplantation. The tissues were immediately flash frozen in liquid nitrogen and stored at −80 °C for protein and RNA isolation until use. Tissues were immediately fixed in 2% paraformaldehyde and the standard protocol was followed to embed them in paraffin. For cell culture, fresh tissues were immediately stored in cold phosphate-buffered saline (PBS). This protocol is approved by the Houston Methodist Hospital Institutional Review Board [IRB (2) 0511–0100].
2.2. Cardiac fibroblast isolation and culture
Human LV tissues were collected in cold phosphate-buffered saline (PBS) and cut into small pieces about 1–2 millimeters in size. The LV tissues were digested with Liberase TM 0.56 U/ml (Roche, USA) in DMEM (Invitrogen, USA) at 37°C for approximately 1 hour [12]. The cell suspension was washed with DMEM 4–5 times and seeded into 6-well plates in a Fibroblast Growth Medium −3 (FGM-3) (Promocell, USA). Human cardiac ventricular fibroblast (Promocell, USA) derived from healthy hearts was cultured in FGM-3 and used as a control.
2.3. Cell transfection
Human cardiac fibroblasts derived from the LV of the HF patients were grown in antibiotic-free media, transfected with 50 ng/ml siRNA (Santa Cruz Biotechnology, USA) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, USA). Seventy-two hours post-transfection, cells were harvested for western blot, qRT-PCR, or fixed for immunofluorescence staining.
2.4. Western blot analysis
Cardiac tissues and cultured cardiac fibroblasts were lysed in RIPA buffer (Boston BioProducts, USA) with protease inhibitor (Thermo Scientific, USA) and phosphatase inhibitor (Thermo Scientific, USA) as described previously [13]. The total protein concentration in the samples was measured by the bicinchoninic acid method (Thermofisher Scientific, USA). Equal amounts of proteins were resolved by electrophoresis and transferred to PVDF membrane. The blots were blocked with 1% casein blocker (Bio-Rad, USA) and incubated with primary antibodies following incubation with horseradish peroxidase conjugated secondary antibodies (Santa Cruz Biotechnology, USA). Clarity™ western ECL substrate (Bio-Rad, USA) was used to detect the bands. The photographs of stained immunoblots were taken with a BIO-RAD ChemiDoc imaging system (Bio-Rad, USA).
2.5. Immunofluorescence staining and quantification
Control and HF fibroblasts were grown to 70–80% confluency and fixed with 4% formaldehyde for 20 minutes at room temperature. Cells were stained with collagen type 1 alpha (COL1A), transforming growth factor-beta 1 (TGF-β1), fibronectin 1 (FN1), CstF64 (Proteintech, USA), and alpha-smooth muscle actin (α-SMA) (Sigma Aldrich, USA) antibodies. Images were acquired using EVOS™ M5000 microscope (Thermofisher Scientific, USA). The area stained by each marker was quantified using ImageJ (NIH) software following guidelines from NIH.
2.6. Fibrosis assessment
Tissues sections from LV of HF and control patients were stained using a Masson’s trichrome kit (Sigma-Aldrich, USA) following manufacturer’s guidelines and images were taken using an Olympus AX70 microscope (Olympus, Japan). Fibrosis was quantified using the methods described previously [11].
2.7. RNA isolation and RT-qPCR for dPAS usage
LV tissues and cultured cardiac fibroblasts were homogenized in QIAzol Lysis Reagent (Qiagen, USA) and isolated using the RNeasy Mini Kit (Qiagen, USA). cDNA was constructed using qScript® cDNA SuperMix (Quantabio, USA). Real-time PCR was performed using CFX96™ Real Time System (Bio-Rad, USA). Two pairs of primers were designed for each gene to calculate dPAS usage. The first pair was designed to target an open reading frame representing the total transcripts and the second pair was designed to target a region slightly proximal of distal PAS, which represented the mRNAs with longer 3’UTR (Fig 1E). The dPAS usage was calculated as ΔCT=CTdistal − CTtotal described previously [14, 15]. Data were presented as differences normalized to control by calculating ΔΔCT= ΔCTaverage target − ΔCTaverage of control. A negative ΔΔCT indicates 3’UTR shortening, and a positive value indicates 3’UTR lengthening compared to control.
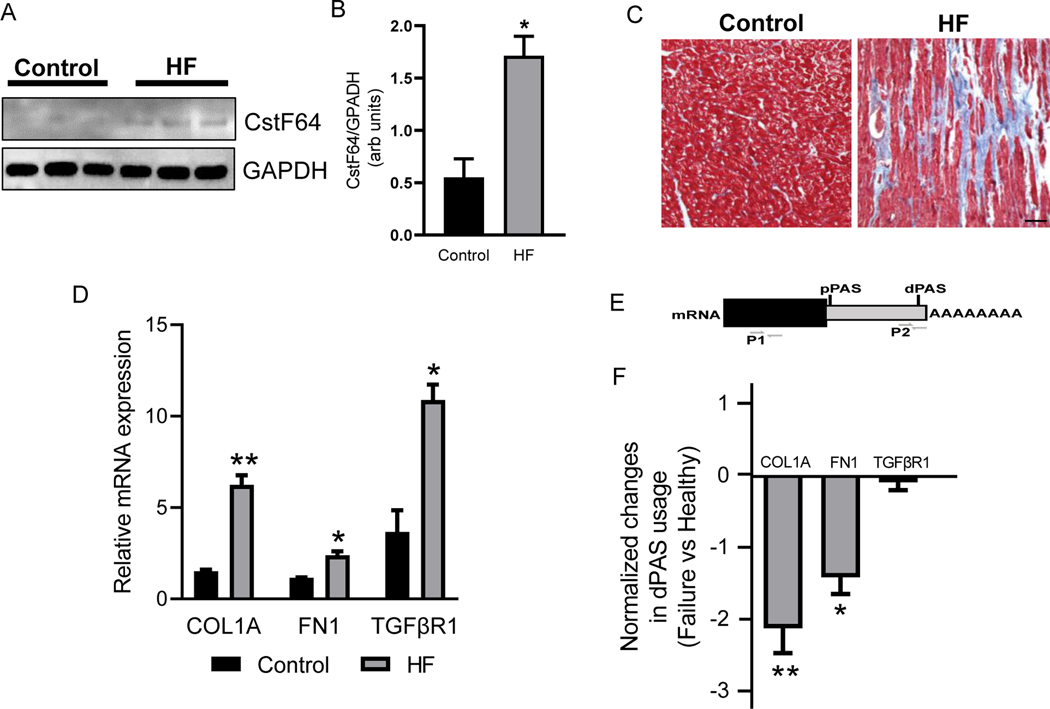
Figure 1. CstF64 expression and 3’UTR changes in profibrotic genes in the left ventricle (LV) tissues of heart failure (HF) patients.
(A) Western blot showing expression of CstF64 and internal control GAPDH proteins in LV tissues of healthy and HF patients. (B) Densitometry analysis of the expression of CstF64 from western blot normalized to GAPDH. (C) Representative photomicrographs of healthy and HF LV tissue cross-section stained with Masson’s Trichrome. Scale bar, 50μm. (D) Plot showing mRNA expression of COL1A, FN1, and TGFβR1 in HF and healthy LV tissues by RT-qPCR. (E) An illustration depicting primers used to detect total (P1) and long variants (P2) transcript. (F) Plot showing significant shortening of COL1A and FN1 in HF LV tissues compared to healthy donors by dPAS using RT-qPCR. Data are presented as mean ± SEM. Student’s t-test was used to analyze the data; n = 3, * p < 0.05 and ** p < 0.01.
2.8. Statistical analyses
Results are presented as mean ± standard error mean (SEM). Unpaired Student’s t-test was performed to test statistical differences between two groups. All the statistical analyses were carried using the Graph Pad Prism software V9.0 (Graph Pad Software Inc., USA).
3. Results
3.1. LV of HF patients show increased expression of CstF64
The HF LV tissues showed significantly increased expression of CstF64 compared to the healthy controls by western blot (Fig. 1A, B). To assess the fibrosis state in the corresponding HF tissues, we performed Masson’s trichome staining, which showed significantly increased (29.38 ± 1.77%, *** p < 0.001, n = 3) fibrosis compared to the healthy controls (Fig. 1C). Similarly, the expression of key profibrotic genes, such as COL1A, FN1, and TGFβR1, were also significantly increased in the HF (Fig. 1D). So, we asked if the difference in fibrosis was because of APA events by the promotion of 3’UTR shortening in profibrotic genes. To find if COL1A, FN1, and TGFβR1 are regulated by APA, we used RT-qPCR to calculate the percentage of mRNA that are using the distal Poly-A site (dPAS). Using this method, we showed that COL1A and FN1 had a greater number of shortened transcripts in HF compared to the healthy controls (Fig. 1F). The shortening in 3’UTR allows escaping from regulatory RNAs and proteins hence, increasing the protein expression. Here, we also correlated the fibrosis state in HF with the increase in the number of 3’UTR shortened transcripts of fibrosis genes.
3.2. Isolated cardiac fibroblasts from HF patients show dysregulation of CstF64 and 3’UTR shortening of key profibrotic genes
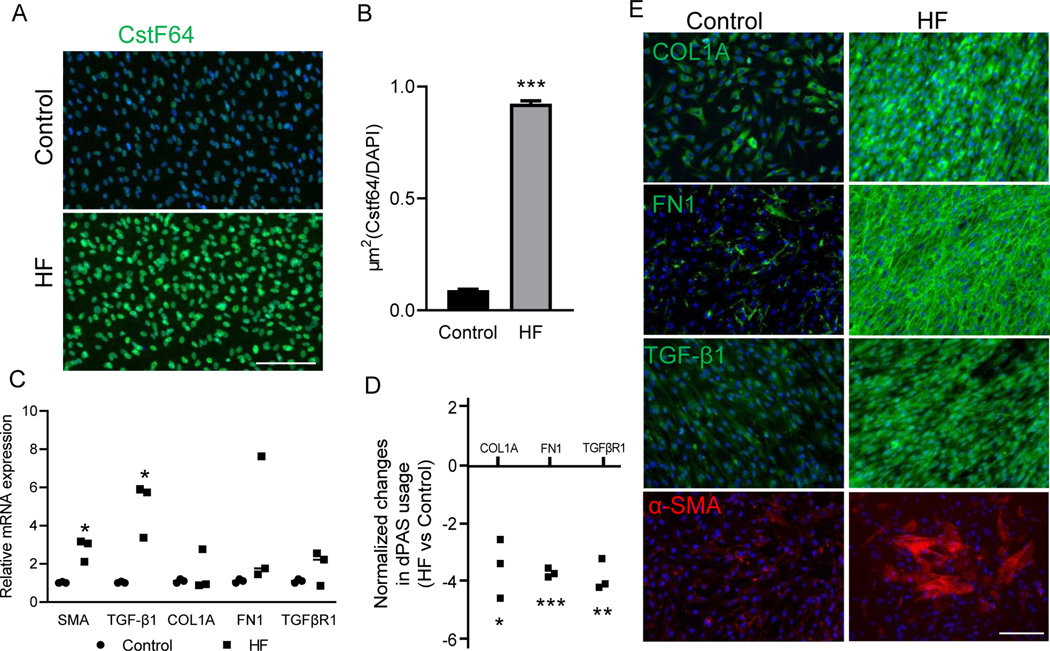
The immunofluorescence staining showed a significantly higher expression of CstF64 in cardiac fibroblasts from HF patients than the controls (Fig. 2A, B). Using RT-qPCR, we investigated the transcript levels of some of the key profibrotic markers, including α-SMA, the marker of myofibroblast differentiation and activation. The transcript levels of α-SMA and TGF-β1 were significantly higher in cardiac fibroblasts from HF patients. We also found that not all the HF cardiac fibroblasts showed significant differences in COL1A, FN1, and TGFβR1 expression compared to the healthy controls (Fig. 2C). On the other hand, the investigation of dPAS usage showed that there is a significant 3’UTR shortening of COL1A, FN1, TGFβR1 in cardiac fibroblasts from HF compared to healthy controls (Fig. 2D). Then, we investigated the protein expression of COL1A and FN1 along with other key genes in fibrosis, TGF-β1, and α-SMA, in the HF fibroblasts to relate them to 3’UTR shortening in these genes. The immunofluorescence staining of HF fibroblasts showed significantly increased deposition of ECM genes such as COL1A and FN1 compared to the healthy controls (Fig. 2E). In addition, a significant number of activated myofibroblasts marked by α-SMA and upregulated TGF-β1 expression were seen in HF fibroblasts compared to healthy controls (Fig. 2E). This confirmed the presence of α-SMA positive-myofibroblasts in HF, which were excessively depositing ECM proteins such as COL1A and FN1 as well as resulting in increased TGF-β expression. These data suggested that irrespective of the total transcript levels in HF and control fibroblasts, 3’UTR shortening is sufficient to enhance the protein expression of those genes. Hence, our results indicated dysregulation of one of the key machinery of APA associated with an increase in 3’UTR shortening and protein levels of profibrotic markers.
Figure 2. CstF64 expression and 3’UTR changes in profibrotic genes within isolated fibroblasts from the HF patients’ LV.
(A) Immunofluorescence staining of CstF64 expression in isolated fibroblasts from LV of HF patients. Scale bar, 150μm. (B) Bar plot showing quantification of CstF64 expression from immunofluorescence staining normalized to DAPI in the control and HF fibroblasts. (C) Plot showing mRNA expression of α-SMA, TGF-β1, COL1A, FN1, and TGRβR1 in heart failure and control fibroblast by RT-qPCR. (D) Plot showing significant shortening of COL1A, FN1, and TGFβR1 in HF fibroblasts compared to control by dPAS usage RT-qPCR. (E) Immunofluorescence staining of COL1A, FN1, TGF-β1, and α-SMA proteins in isolated fibroblasts from LV of HF patients. Scale bar, 150μm. Data are presented as mean ± SEM. Student’s t-test was used to analyze the data; n = 3, * p < 0.05; ** p < 0.01; ***p < 0.001.
3.3. Depletion of CstF64 from HF fibroblasts downregulates profibrotic genes
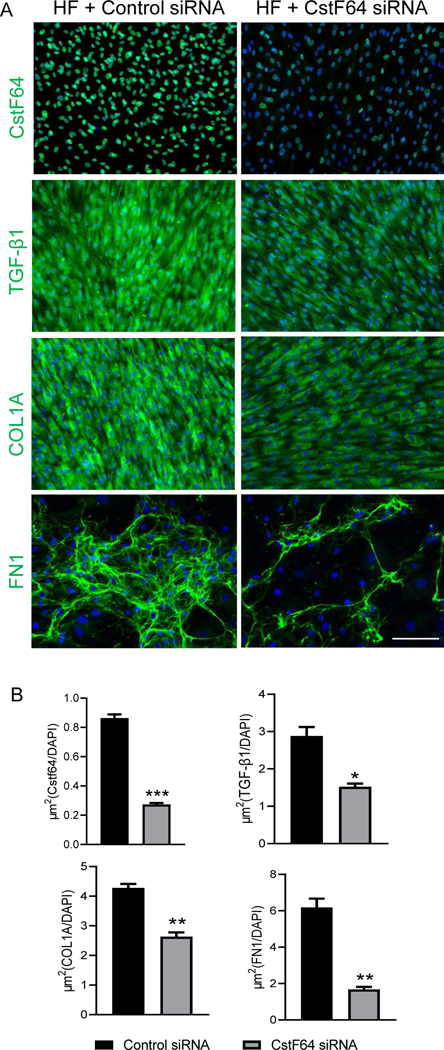
We established that CstF64 is upregulated in HF fibroblasts and these fibroblasts also show 3’UTR shortening in some of the profibrotic genes that correlate with the protein expression. The association between increased CstF64 and shortening of 3’UTR of profibrotic genes led us to hypothesize that CstF64 directly targets these genes in HF fibroblasts and promotes myofibroblast activation. Therefore, we knocked down the expression of CstF64 in the HF fibroblasts using siRNA. The immunofluorescence staining of the HF fibroblasts transfected with CstF64 siRNA showed efficient knockdown of CstF64 after 72 hours compared to cells transfected with control siRNA (Fig. 3A, B). With the efficient knockdown of CstF64, we also noticed the decrease in the expression of some of the key ECM genes such as FN1 and COL1A; and TGF-β1, a key cytokine implicated in fibrosis (Fig. 3A, B).
Figure 3. CstF64 knockdown attenuates fibrotic protein expression in HF fibroblasts.
(A) Immunofluorescence staining of CstF64, COL1A, TGF-β1 and FN1 in an HF fibroblast 72 hours post-transfection post-transfection with control and CstF64 siRNA. Scale bar, 150μm. (B) Bar plots showing quantification of CstF64, TGF-β1, COL1A, and FN1 protein expression normalized to DAPI corresponding to the immunofluorescence staining in Figure 3A. Data are represented as mean ± SEM. Student’s t-test was used to analyze the data; n = 3, * p < 0.05; ** p < 0.01; ***p < 0.001.
3.4. CstF64 knockdown changes the 3’UTR length of profibrotic genes in HF fibroblast
Next, we investigated the transcript levels of fibrosis genes after the knockdown by using RT-qPCR. The key fibrosis associated genes such as COL1A, FN1, and α-SMA, including the major cytokine TGF-β1 and its receptor TGFβR1, were significantly downregulated by at least 2-fold after the knockdown of CstF64 (Fig. 4A). Lastly, we investigated if the knockdown of CstF64 changed the 3’UTR length of these target genes. We ran RT-qPCR to check the dPAS usage, and the data showed that the usage in dPAS increased after the knockdown of CstF64 (Fig. 4B). Compared to the control siRNA, there was an increase in the length of 3’UTR of COL1A, FN1, and TGFβR1 genes which also correlated with the downregulation of these genes observed at the protein level (Fig. 3A). Thus, we showed that CstF64 directly targets the 3’UTR region of COL1A, FN1, and TGFβR1 genes to regulate their protein expression in HF fibroblasts.
Figure 4. CstF64 knockdown promotes the 3’UTR lengthening of profibrotic genes expression in HF fibroblasts.
(A) Bar plot showing relative mRNA expression of COL1A, FN1, TGRβR1, TGF-β1, and α-SMA in an HF fibroblast 72 hours post-transfection with CstF64 and control siRNA by RT-qPCR. (B) Plot showing significant lengthening of COL1A, FN1, and TGFβR1 in HF fibroblasts 72 hours post-transfection with CstF64 siRNA compared to control siRNA by dPAS using RT-qPCR. Data are represented as mean ± SEM. Student’s t-test was used to analyze the data; n = 3, * p < 0.05; ** p < 0.01. (C) The model illustrates the effect of CstF64 upregulation on cardiac fibrosis.
4. Discussion
The present study has demonstrated that the APA machinery gene, CstF64, plays a significant role in the regulation of major profibrotic genes in LV cardiac tissues and isolated fibroblasts from patients with HF. A novel profibrotic role for CstF64 is revealed by the shortening of 3’UTR length of profibrotic genes, which ultimately leads to the activation of myofibroblast differentiation. The expression of CstF64 was found to be upregulated in many cancer types which also exhibited global 3’UTR shortening [7]. Upregulation of CstF64 has been shown to favor the use of weaker proximal PAS [16, 17]. In contrast, depletion of CSTF64 in HeLa cells resulted in global 3’UTR lengthening [18]. However, the role of CstF64 in 3’UTR shortening of fibrosis-associated genes in HF fibroblasts has not been investigated. First, we showed a significant increase in CstF64 expression in LV tissues from HF patients along with shortened 3’UTR in COL1A and FN1 genes and a consistent increase in collagen deposition in the LV failing tissues. This study confirmed an association between an increase in CstF64 and an increase in 3’UTR shortening events in the HF (Fig 4C), similarly to what has been reported in various types of cancer [7].
The activation and differentiation of cardiac fibroblasts into ECM producing α-SMA- positive myofibroblasts is central to cardiac fibrosis [19]. The expression of CstF64 was found to be higher in cardiac fibroblasts isolated from HF patients than in healthy controls. Aside from that, HF cardiac fibroblasts were α-SMA-positive, showed elevated levels of TGF- β signaling, and displayed an excess of genes known to be involved in the ECM, including COL1A and FN1. Our study provides evidence for ECM genes including COL1A and FN1; and TGFβR1, a key receptor in TGF-β signaling, have shortened 3’UTR in HF fibroblasts. Although the total transcript levels of genes involved in fibrosis were not significantly higher in HF fibroblasts, the significantly shorter 3’UTRs of these genes could explain the increase in the expression of those genes. Inhibitory miRNAs and RBPs negatively regulate transcript stability by binding mainly to 3’UTRs, thereby controlling protein expression [3]. As a result of such expression of APA isoforms with shorter 3’UTR and without binding sites for miRNA regulation, nearly 10% of targeted miRNAs are altered, resulting in dysregulated protein synthesis [20]. It is also possible that similar mechanisms may be involved in HF fibroblasts to alter genes associated with fibrosis expression by affecting the regulation by their target miRNAs. This suggested APA events are also playing a role in myofibroblast activation and ECM deposition in addition to the canonical TGF-β signaling pathways.
Depletion of CSTF64 resulted in global 3’UTR lengthening and inhibits the target genes [18]. A significant finding here was that depleting CstF64 alone in cardiac fibroblasts isolated from HF patients was sufficient to induce 3’UTR lengthening in COL1A, FN1, and TGFβR1 genes; and reduce gene or protein expression of ECM genes such as COL1A and FN1. In addition, The knockdown of CstF64 in HF cardiac fibroblasts also showed downregulation of key marker of fibroblast to myofibroblast differentiation, α-SMA, and a key cytokine implicated in fibrosis – TGF-β1 and its receptor TGFβR1 [21]. These results showed that CstF64 depletion lengthened these key genes associated with fibrosis, proving that these genes are directly targeted by APA. Thus, the downregulation of profibrotic genes in HF cardiac fibroblasts induced by CstF64 knockdown suggests that CstF64 may be a new therapeutic target for the treatment of cardiac fibrosis.
One of the limitations of the current study is the grasp of the extent of global APA changes occurring after the depletion of CstF64 from HF fibroblasts; a better understanding of this process could bring potential treatment for cardiac fibrosis. High-throughput sequencing such as Poly(A)-ClickSeq RNA sequencing to sequence the 3’UTR of mRNAs and PolyA-miner algorithm to measure the global patterns of APA are necessary to address those limitations [22].
In conclusion, our study showed the dysregulation of the key component of APA regulators, CstF64, in tissues and isolated fibroblasts from failing human hearts that correlated with 3’UTR shortening and increased expression of key profibrotic genes. The knockdown of CstF64 from the cardiac fibroblasts from HF patients mitigated the excess deposition of some ECM genes and downregulated the TGF-β1 cytokine and its receptor (components of a central profibrotic signaling pathway) consistently with lengthening of 3’UTR in those genes. Our study provides new insights into the role of CstF64 via APA–dependent regulation of cardiac fibrosis.
Highlights.
Alternative polyadenylation (APA) generates different isoforms of mRNA with varying lengths of 3’UTR and plays a role in diverse cellular processes.
CstF64, an APA regulator, is upregulated in left ventricular samples from human heart failure patients.
CstF64 causes 3’UTR shortening of profibrotic genes.
Knockdown of CstF64 from heart failure fibroblasts downregulates profibrotic genes by increasing the length of their 3’UTR.
Acknowledgments
This study was supported by grants from the American Heart Association (19TPA34880039 and 18IPA34170497 R.A.T), 5R01HL138510 to HKQ, and the Roswell and Ann Vaughan Fund to A.G. and R01AG059599 to K.A.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests
There are no conflicts of interest.
References
- [1].Tian B, Manley JL, Alternative polyadenylation of mRNA precursors, Nat Rev Mol Cell Biol, 18 (2017) 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T, A quantitative atlas of polyadenylation in five mammals, Genome Res, 22 (2012) 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell, 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Garneau NL, Wilusz J, Wilusz CJ, The highways and byways of mRNA decay, Nat Rev Mol Cell Biol, 8 (2007) 113–126. [DOI] [PubMed] [Google Scholar]
- [5].Di Giammartino DC, Nishida K, Manley JL, Mechanisms and consequences of alternative polyadenylation, Mol Cell, 43 (2011) 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang W, Hsu PL, Yang F, Song JE, Varani G, Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3’-end processing signals, Nucleic Acids Res, 46 (2018) 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W, Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’-UTR landscape across seven tumour types, Nat Commun, 5 (2014) 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dharmalingam P, Mahalingam R, Yalamanchili HK, Weng T, Karmouty-Quintana H, Guha A, Thandavarayan RA, Emerging roles of alternative cleavage and polyadenylation (APA) in human disease, J Cell Physiol, (2021). [DOI] [PubMed] [Google Scholar]
- [9].Murtha LA, Schuliga MJ, Mabotuwana NS, Hardy SA, Waters DW, Burgess JK, Knight DA, Boyle AJ, The Processes and Mechanisms of Cardiac and Pulmonary Fibrosis, Front Physiol, 8 (2017) 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Centurion OA, Alderete JF, Torales JM, Garcia LB, Scavenius KE, Mino LM, Myocardial Fibrosis as a Pathway of Prediction of Ventricular Arrhythmias and Sudden Cardiac Death in Patients With Nonischemic Dilated Cardiomyopathy, Crit Pathw Cardiol, 18 (2019) 89–97. [DOI] [PubMed] [Google Scholar]
- [11].Wang G, Cruz AS, Youker K, Marcos-Abdala HG, Thandavarayan RA, Cooke JP, Torre-Amione G, Chen K, Bhimaraj A, Role of Endothelial and Mesenchymal Cell Transitions in Heart Failure and Recovery Thereafter, Front Genet, 11 (2020) 609262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cieslik KA, Trial J, Carlson S, Taffet GE, Entman ML, Aberrant differentiation of fibroblast progenitors contributes to fibrosis in the aged murine heart: role of elevated circulating insulin levels, FASEB J, 27 (2013) 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Thandavarayan RA, Watanabe K, Ma M, Gurusamy N, Veeraveedu PT, Konishi T, Zhang S, Muslin AJ, Kodama M, Aizawa Y, Dominant-negative p38alpha mitogen-activated protein kinase prevents cardiac apoptosis and remodeling after streptozotocin-induced diabetes mellitus, Am J Physiol Heart Circ Physiol, 297 (2009) H911–919. [DOI] [PubMed] [Google Scholar]
- [14].Weng T, Ko J, Masamha CP, Xia Z, Xiang Y, Chen NY, Molina JG, Collum S, Mertens TC, Luo F, Philip K, Davies J, Huang J, Wilson C, Thandavarayan RA, Bruckner BA, Jyothula SS, Volcik KA, Li L, Han L, Li W, Assassi S, Karmouty-Quintana H, Wagner EJ, Blackburn MR, Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation, J Clin Invest, 129 (2019) 1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ, CFIm25 links alternative polyadenylation to glioblastoma tumour suppression, Nature, 510 (2014) 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Takagaki Y, Seipelt RL, Peterson ML, Manley JL, The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation, Cell, 87 (1996) 941–952. [DOI] [PubMed] [Google Scholar]
- [17].Shell SA, Hesse C, Morris SM Jr., Milcarek C, Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection, J Biol Chem, 280 (2005) 39950–39961. [DOI] [PubMed] [Google Scholar]
- [18].Yao C, Choi EA, Weng L, Xie X, Wan J, Xing Y, Moresco JJ, Tu PG, Yates JR 3rd, Shi Y, Overlapping and distinct functions of CstF64 and CstF64tau in mammalian mRNA 3’ processing, RNA, 19 (2013) 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gibb AA, Lazaropoulos MP, Elrod JW, Myofibroblasts and Fibrosis: Mitochondrial and Metabolic Control of Cellular Differentiation, Circ Res, 127 (2020) 427–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP, Global analyses of the effect of different cellular contexts on microRNA targeting, Mol Cell, 53 (2014) 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Meng XM, Nikolic-Paterson DJ, Lan HY, TGF-beta: the master regulator of fibrosis, Nat Rev Nephrol, 12 (2016) 325–338. [DOI] [PubMed] [Google Scholar]
- [22].Yalamanchili HK, Alcott CE, Ji P, Wagner EJ, Zoghbi HY, Liu Z, PolyA-miner: accurate assessment of differential alternative poly-adenylation from 3’Seq data using vector projections and non-negative matrix factorization, Nucleic Acids Res, 48 (2020) e69. [DOI] [PMC free article] [PubMed] [Google Scholar]