Abstract
Mungbean is an important but understudied food legume compared with other major grain crops. Genetic studies through development of high-through put markers, linkage map construction and QTL analysis can accelerate and improve the efficiency of mining for genes for breeding in this crop. This study used four mungbean F5 recombinant inbred lines (RILs) from crosses of two wild types (ACC 1, ACC 87) and two cultivars (Berken, Kiloga) and DArT markers to construct individual and consensus linkage maps and to identify QTLs associated with 54 traits in mungbean. The number of polymorphic DArT markers identified among the four RIL populations varied from 1062 to 2013. The individual maps covered the lengths of 629.7–883.5 cM, comprising 672–981 DArT markers and 15–19 linkage groups (LG) with average distance between markers of 0.9–1.2 cM. The consensus map had the total length of 795.3 cM, comprising 1539 DArT markers and resolved 11 LGs with an average inter-marker distance of 0.65 cM. Sixty-two QTLs were identified for 39 traits across 10 LGs of the consensus map. Major QTLs were identified for two special traits, late flowering inherited from ACC 1 (6 QTLs, PVE of 11.2–29.9%) and perenniality inherited from ACC 87 (3 QTLs, PVE of 17.4–22.6%) in separate population analysis. Number of congruent QTLs across four mungbean populations and the consensus map was 18 for 13 traits. These results illustrated the high efficiency of DArT marker application in mungbean genetic dissection and suggested the future potential employment of identified QTLs for mungbean improvement.
Keywords: Consensus map, DArT, Improvement, QTLs, Vigna radiata
Introduction
Mungbean (Vigna radiata L.) is one of the most important leguminous food crops for human consumption because it provides sources of protein, contains most notably folate and iron, and has multitude of uses (Nair et al. 2019). Its familiar form is crunchy bean sprouts which contain high amounts of thiamine, niacin, and ascorbic acid and are widely used in oriental cuisine, salads, and healthfoods. Other diverse products produced from mungbean include fried foods and desserts. Therefore, the cultivated mungbean has become an increasingly important agricultural commodity and its production has spread worldwide in recent decades.
As with legume crops, mungbean is still considered as a village crop and its genetic improvement lags behind cereals (Wang et al. 2020). Additionally, the limited genetic variation or extremely narrow genetic base of the germplasm used in breeding, resulting in difficulty in identifying unique traits for crop development (Noble et al. 2018). To widen the germplasm base, related wild species can sometimes be employed in crop improvement. Wild species provide a potentially important source of novel traits such as pest and disease resistance, unique photothermal responses, seed traits and abiotic stress tolerance (Kim et al. 2015; Noble et al. 2018), and greater agro-morphological variability for ongoing and future breeding programs (Gayacharan et al. 2020). The putative progenitor of cultivated mungbean is the wild form, Vigna radiata spp. sublobata, which is widely distributed from western Africa to northern and eastern Australia and Papua New Guinea (Lawn and Cottrell 1988; Tomooka et al. 2002). The wild mungbean accessions have hardseededness and higher seed protein content, disease resistance and saline, calcareous or cracking clay soil tolerance, all of which are potentially useful for mungbean variety improvement (Lawn and Cottrell 1988; Lawn and Rebetzke 2006).
Genetic and genomic studies may help to improve the efficiency of mungbean breeding for high yield and quality through identifying beneficial genes. Linkage map construction and QTL analysis are important for genetic and genomic studies but have been significantly less in mungbean compared to other major crops (Lee et al. 2021). Mungbean has features making it a useful model organism among legumes, such as small genome size (about 500 Mb) (Kang et al. 2014), short life-cycle, self-pollinating, and close genetic relationship to other legumes (Kim et al. 2015). Several genetic linkage maps have been published for mungbean, e.g. Lambrides et al. (2000); Humphry et al. (2002); Zhao et al. (2010) Kajonphol et al. (2012); Isemura et al. (2012); and Wang et al. (2016). These maps were constructed from single F2 or recombinant inbred line (RIL) populations from crosses between cultivated and wild parents, such as VC3980 x TC1966 (wild from Madagascar), Berken x ACC 41 (wild from Australia), JP229096 x JP211874 (wild from Myanmar) and KUML29-1–3 × W021 (wild from Australia). The population size ranged from 58 to 250 plants. The maps differed in length (691.7–1831.8 cM), in numbers of markers (102–430), numbers of linkage groups (LGs) (11–14), and adjacent marker distances (1.78–10.2 cM). Two maps (Kajonphol et al. 2012 and Isemura et al. 2012), resolved 11 LGs which is the haploid chromosome number of mungbean. Recently, Liu et al. (2017) constructed a mungbean genetic linkage map of 11 LGs and 1010.18 cM length using 313 SSR markers and the RIL populations of two cultivars. Wang et al. (2020) seem to be the first study mapping a large number of SNP markers (21,508 SNP markers) onto 11 LGs and spanning for the length of 1060.2 cM based on the crossing population of two cultivars. Mariyammal et al. (2019) and Mathivathana et al. (2019) also applied SNP makers to construct linkage maps for the interspecific cross of Vigna radiata × Vigna umbellata.
Genes or QTLs associated with various traits encompassing insect pests, diseases and seed-related characters in mungbean were mapped. Examples were QTLs conditioning bruchid resistance (Wang et al. 2016; Mariyammal et al. 2019), powdery mildew resistance (Young et al. 1993; Chaitieng et al. 2002; Humphry et al. 2003; Kasettranan et al. 2010), hardseededness and seed size (Humphry et al. 2005), mungbean yellow mosaic virus (Alam et al. 2014; Mathivathana et al. 2019), and various agronomic characters (Kajonphol et al. 2012; Isemura et al. 2012). More recent maps are QTLs for drought (Liu et al. 2017), leaflet shapes (Wang et al. 2020) and inflorescence structure (Lee et al. 2021). None of the studies has reported either on a consensus map constructed from more than one individual segregation population or on QTL detection using the consensus map for various traits in mungbean.
Diversity Arrays Technology is a high-throughput genotyping method, which does not rely on the availability of sequence information (Vu et al. 2012). DArT markers have been widely applied to some species to investigate genetic diversity (Sansaloni et al. 2010), construct high-density DArT consensus maps and identify QTLs (Vu et al. 2015; Ates et al. 2018). Incorporation of DArT markers with other marker types has been shown to provide high-density and reliable linkage maps as well as to increase QTL detection rates. By applying the DArT marker protocols and libraries that were developed for mungbean (Vu et al. 2012) and applying successfully on soybean (Vu et al. 2015), this research investigated: (i) the application of polymorphic DArT markers to construct linkage maps in each of four individual mungbean RIL populations and the consensus map; and (ii) the detection of QTLs linked to various morphological and agronomical traits in mungbean.
Materials and methods
Plant materials and phenotypic evaluation
Four F5 RIL populations were developed from crosses of two cultivated mungbean varieties (Berken and Kiloga) and two wild parents (ACC 1 and ACC 87). The two mungbean cultivars are characterized by erect stems, determinate habit, large entire leaflets and large shiny green seed (Supplement Table 1) and were early flowering when grown in South East Queensland (Lawn 1979). In contrast, the two indigenous wild mungbean accessions are twining, viny plants with indeterminate flowering habits, small and lobed leaflets and small black seeds. The accession ACC 1 was very late flowering even when grown under short days (Rebetzke and Lawn 2006a). ACC 87 is typical of a tuberous rooted perennial form of ssp. sublobata that is endemic to the Townsville–Charters Towers region of north Queensland (Rebetzke and Lawn 2006b). ACC 87 has a thicker stem, larger leaflets, flowers and seeds than ACC 1, but nonetheless, these traits are still smaller than other cultivars.
Table 1.
Number of selected polymorphic DArT markers and level of redundancy in the four mungbean RIL populations from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
| Populations | No. selected DArT markers | Redundancy* | ||
|---|---|---|---|---|
| Cross | Notation | No. | (%) | |
| Berken x ACC 1 | Bx1 | 2013 | 345 | 17.1 |
| Kiloga x ACC 1 | Kx1 | 1995 | 299 | 15.0 |
| Berken x ACC 87 | Bx87 | 1062 | 37 | 3.5 |
| Kiloga x ACC 87 | Kx87 | 1887 | 107 | 5.7 |
*Redundancy obtained from the “Similarity of loci” calculation in Joinmap 3.0
Each cultivated parent had been crossed with each wild parent to create four hybrid populations Bx1 (Berken x ACC 1), Kx1 (Kiloga x ACC 1), Bx87 (Berken x ACC 87) and Kx87 (Kiloga x ACC 87). F1 hybrid seeds had been created and were also allowed to self in prior years, so producing F2 seeds (RJ Lawn, personal communication). F3 seeds from 84 × F2 plants of each population were available from the study on the expression and inheritance of qualitative and quantitative traits in segregating progeny generations by Nguyen et al. (2012, 2016).
RILs were developed via the single seed-descent breeding method without selection via generation advancement (F3 to F5) in a glasshouse from April 2009 to March 2010. F5 RILs with population sizes of 81–84 lines were grown for the phenotypic assessment at the CSIRO Davies Laboratory in Townsville (Supplement Table 1). The phenotypic information on 54 qualitative and quantitative traits were assigned to six categories viz. qualitative morphological traits, qualitative seed traits, phenological traits, quantitative morphological traits, pod and seed traits, and yield related traits (Supplement Table 2). The measurement of 54 traits was developed previously during the phenotypic evaluation described in Nguyen et al. (2012, 2016) and Vu (2013) (Supplement Tables 3, 4 and 5) was used for the QTL analyses using DArT markers.
Young fully expanded leaves from each parental plant and each line of the four F5 RIL populations were collected and lyophilized before DNA extraction. The DArT genotyping analyses were conducted at the DArT Pty Ltd laboratory in Canberra during 2011–2012.
DNA extraction and genotyping
DNA extraction followed the standard DArT protocols (Vu et al. 2012). After DNA extraction, DNA was repurified using a DNA repurification kit [ZR–96 215 DNA clean and concentrator 216TM–5 (Zymo Research)].
Two restriction enzyme (RE) combinations, comprising the rare cutter PstI and a frequent cutter (either TaqI or BstNI), which were chosen for the creation of the mungbean and soybean DArT libraries were used for genome complexity reduction as described in Vu et al. (2012). Subsets of DNA samples from each RIL population and parents were randomly chosen for replicates. After PCR and electrophoresis gel performance, PCR products were precipitated and labeled with Cy3 and Cy5 dye (targets), and genotyping was conducted by using mungbean and soybean DArT marker libraries described in Vu et al. (2012).
Polymorphic DArT marker selection
DArT soft version 7.4 (Diversity Arrays Technology P/L, Canberra, Australia) was used to identify and score polymorphic clones with ‘1’ for the presence, ‘0’ for absence, and ‘x’ for the sample with insufficient confident score. Initially, marker selection was based on Consensus > 97% (the consensus of five different statistical methods in marker scoring) and Average Reproducibility > 97%. Markers in the range of 96.5–97% average reproducibility were only selected for average P > 80%. Then markers of discordance < 1% (overall variation of scores within the replicates), PIC > 0.2 (informativeness of a genetic marker) and average P > 65% were selected. Finally, only markers that were consistently scored across replicated parental samples and differentiated the parents in a cross were selected.
Component genetic linkage map construction
Map construction for each RIL population was performed with JoinMap 3.0. The recombination frequencies were converted into map distances (cM) using the Kosambi mapping function. Initially, a LOD (logarithm of odds) range of 2 to 10 was used to test for linkage groups. In cases, where many markers (> 200) were in a group, LOD scores were modified to a higher cutoff value. Presentation of linkage groups was drawn using Mapchart 2.3.
Genetic dissimilarities among the four RIL populations were calculated by NTSYS 2.1 based on Nei’s distance and using the total number of DArT markers that mapped in the four individual maps.
Consensus genetic linkage map construction
A consensus genetic map of mungbean was constructed based on the four individual maps. The largest individual map of Bx1 population was used as the seed map. All the linkage maps were joined into a single population consensus map by using the markers in common on each component linkage map. The consensus map construction was implemented in the DArT’s KDCompute platform. Markers were clustered into linkage groups according to the method described by Wu et al. (2008). Markers with identical genotypes are placed in redundant bins, and the resulting markers/bins within each linkage group are ordered using the traveling salesman path solver program Concorde (Applegate et al. 2006). Apparent double-crossovers are masked before reordering the linkage groups and calculating recombination fractions, with Kosambi function used to estimate genetic distances. Comparison between component maps and the consensus map was conducted and presented by MapChart 2.3.
QTL statistical analysis
The consensus map and pooled phenotypic data from the four RIL populations were used for QTL detection. QTL analysis method–Inclusive Composite Interval Mapping (ICIM)–was applied on the consensus map for 54 measured traits and performed by QTL IciMapping 3.2 (Wang et al. 2011). The ICIM with additive effects (abbreviated as ICIM–ADD) method was applied to identify putative QTL regions for traits. The scanning step was 1 cM and the LOD score threshold was determined by a permutation test with 1000 replications. The default probability in stepwise regression (PIN—the largest P-value for entering variables in the stepwise regression of phenotype on marker variables) was 0.001. Where no QTL linked to the trait were detected with a PIN of 0.001, a relaxed PIN of 0.01 was applied. QTLs of LOD > 3.0 were selected.
Congruent QTLs across four individual RILs mungbean populations and the consensus map
QTL analysis for each individual F5 mungbean population was also conducted using IciMapping 3.2 as mentioned above. Especially, for two special traits characterized by ACC 1–late flowering and by ACC 87–perenniality, QTL analysis was conducted to avoid the cases that no QTLs were detected for those traits using pooled genotype and phenotype information.
QTLs were congruent across populations and the consensus map for a trait if those QTLs shared at least a common marker regardless of its flanking position.
Results
Polymorphic DArT markers
Based on the selection criteria, there were different numbers of polymorphic DArT markers identified among the four RIL populations, varying from 1062 to 2013 markers (Table 1).
Although the number of polymorphic markers was highest in Bx1, followed by Kx1, these two populations had the highest percentage of redundancy (17% and 15%, respectively). These rates were in the range of DArT redundancy in other species such as Eucalyptus (17%) (Sansaloni et al. 2010). In contrast, the redundancy levels of Bx87 and Kx87 were much smaller (3.5 and 5.7%, respectively). Since most markers with highly similar sequences would have 99% identity of scoring, this probably reflected a level of sequence redundancy, as shown in Arabidopsis (Wittenberg et al. 2005), oat (Tinker et al. 2009), and Eucalyptus (Petroli et al. 2012). Therefore, as was concluded in the development of the DArT protocols for mungbean (Vu et al. 2012), the number of unique polymorphic DArT markers selected in these four mungbean RIL populations may be overestimated.
Component mungbean linkage maps
Data sets with 2013, 1995, 1062, and 1887 selected DArT markers, for the Bx1, Kx1, Bx87, and Kx87 populations, respectively (Table 2), were subjected to linkage map analysis. The grouping analysis with LOD > 4 resulted in only 39.3% to 63.3% of markers being grouped into 15 to 19 groups, depending on the RIL population (Table 2).
Table 2.
Mapping statistics of the DArT markers and component maps of the four mungbean F5 RIL populations from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
| Bx1 | Kx1 | Bx87 | Kx87 | ||
|---|---|---|---|---|---|
| Total mapped markers | No | 981 | 885 | 672 | 741 |
| % | 48.7 | 44.4 | 63.3 | 39.3 | |
| Segregation distortion markers | No | 416 | 298 | 321 | 327 |
| % | 42.4 | 33.7 | 47.8 | 44.1 | |
| Distortion in favour of cultivated alleles | No | 336 | 246 | 211 | 223 |
| % | 34.3 | 27.8 | 31.4 | 30.1 | |
| Distortion in favour of wild alleles | No | 80 | 52 | 110 | 104 |
| % | 8.2 | 5.9 | 16.4 | 14.0 | |
| Linkage map | |||||
| LGs | No | 15 | 19 | 17 | 17 |
| Number of markers on a LG | Min | 9 | 12 | 9 | 6 |
| Max | 184 | 114 | 95 | 87 | |
| Average | 65.4 | 46.6 | 39.5 | 43.6 | |
| LG size (cM) | Min | 19.2 | 7.1 | 12.2 | 16.6 |
| Max | 121.2 | 93.8 | 131.2 | 91 | |
| Average | 55.2 | 42.8 | 39.6 | 37.4 | |
| Inter-marker distance (cM) | Min | 0.01 | 0.01 | 0.01 | 0.01 |
| Max | 14.6 | 22.0 | 32.4 | 16.4 | |
| Average | 1.1 | 1.2 | 1.1 | 0.9 | |
| Aggregate size (cM) | 883.5 | 812.7 | 634.3 | 629.7 | |
The number of DArT markers grouped on LGs was highest in the Bx1 population (981), followed by Kx1 (885), Kx87 (741) and Bx87 (672). Segregation distortion occurred in all four RIL populations with a range of 33.7% to 47.8%. Generally, more DArT markers segregated with distortion in favour of alleles from the cultivated parents in all crosses. For instance, 336 of 981 DArT markers (34.3%) mapped on the Bx1 linkage map with aberrant segregation ratios in favour of alleles from Berken compared with 80 markers (8.2%) segregating in favour of alleles from ACC 1. Berken crosses exhibited slightly greater segregation distortion in favour of the cultivar (34.3–31.4%) than the Kiloga crosses (27.8–30.1%). Meanwhile, the ACC 87 crosses exhibited more segregation distortion favourable to the wild compared to the ACC 1 crosses (16.4–14.0% compared to 8.2–5.9%). In the ACC 1 crosses, segregation distortion seemed to be random because it occurred in all LGs in both populations, except for LG6 and LG8 of the Kx1 population. No aberrant segregation occurred on LG4, LG5 and LG6 of Bx87, and LG7 of Kx87.
The DArT markers spanned the linkage maps, with the largest map size in Bx1 (883.5 cM) and the smallest map sizes in Bx87 (634.3 cM) and Kx87 (629.7 cM) (Table 2; Fig. 1; Supplement Fig. 2). The individual linkage maps contained on average similar numbers of markers per LG (in the range of 39.5–46.6), except Bx1 with 65.4, and average inter-distances of 0.9–1.2 cM.
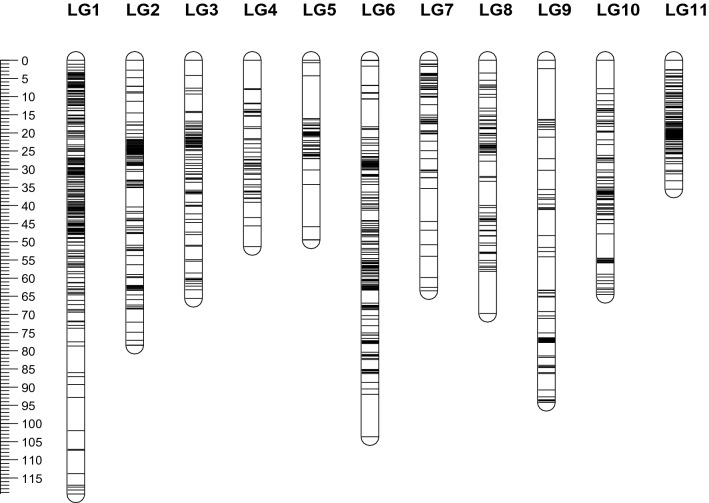
Fig. 1.
A genetic linkage map of mungbean derived from F5 RILs from the cross of Berken x ACC 1. The map includes 981 DArT markers. A centiMorgan scale is on the left
Overall, 1883 DArT markers mapped on the four individual linkage maps, and of these, 1308 (69.5%) were unique. Between 292 (Kx1 and Bx87) to 545 markers (Bx1 and Kx1) were in common (Table 3). The higher number of common markers and lower value of Nei’s genetic distance reflected lower dissimilarity between populations. The genetic distances revealed that Bx1 and Kx1 had the lowest degree of genetic dissimilarity (545 common markers; 0.54), followed by Bx87 and Kx87 (314 common markers; 0.81) whereas populations Bx1 and Bx87 were more distant (294 common markers; 1.0). These were consistent with previous observations revealing that dissimilarity between Berken and Kiloga was less than that between ACC 1 and ACC 87 (Nei’s distance of 0.19 for Berken–Kiloga and of 0.39 for ACC 1–ACC 87) (Vu et al. 2012).
Table 3.
Common markers and genetic dissimilarity between four mungbean mapping populations derived from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
| Bx1 | Kx1 | Bx87 | Kx87 | |
|---|---|---|---|---|
| Bx1 | 545 | 294 | 341 | |
| Kx1 | 0.54 | 292 | 345 | |
| Bx87 | 1.00 | 0.97 | 314 | |
| Kx87 | 0.92 | 0.85 | 0.81 |
The number of markers common between individual maps is shown above the diagonal. Pairwise Nei population genetic distances are shown below the diagonal
Consensus mungbean linkage map
The availability of sufficient numbers of common markers on individual maps facilitated the merging of the four mungbean maps into one consensus map. Using the Bx1 linkage map of 981 DArT markers as the seed map, a consensus linkage map was identified with 11 LGs, 1539 markers, and a length of 795.3 cM (Fig. 2; Table 4). The number of LGs was consistent with the haploid chromosome number of mungbean (2n = 22). Although the number of markers mapped on the consensus map was 1.2–1.5 times higher than that of component maps, its length was shorter than the Bx1 and Kx1 maps. This was because un-mapped LGs of Bx1 (3 LGs), Kx1 (3 LGs), Bx87 (2 LGs), and Kx87 (2 LGs) were excluded from the consensus map. The rates of markers from component maps mapped on the consensus map were in the range of 86.3% (580/672 in Bx87) to 93.6% (828/885) in Kx1.
Fig. 2.
A consensus genetic linkage map derived from four mungbean F5 RIL populations from the crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87). The map includes 1539 DArT markers. A centiMorgan scale is on the left
Table 4.
Mapping statistics of the mungbean consensus map and the contribution of the component maps of four mungbean F5 RIL populations derived from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
| Consensus map | Number of the markers and LGs of component maps contributing to the consensus map | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bx1 | Kx1 | Bx87 | Kx87 | |||||||||
| LG | # markers | Average inter-marker distance per LG (cM) ± SD | Maximum interval (cM) | LG length (cM) | # markers | LGa | # markers | LGa | # markers | LGa | # markers | LGa |
| LG1 | 258 | 0.47 ± 0.98 | 9.21 | 119.4 | 128 | LG1 (122), LG3 (3), LG6 (1), LG11 (1), LG14(1) | 82 | LG1 (53), LG2 (3), LG3 (24), LG7 (1), LG8(1) | 42 | LG2 (39), LG6 (3) | 40 | LG2 (1), LG6 (36), LG7 (3) |
| LG2 | 179 | 0.46 ± 0.84 | 5.37 | 78.5 | 122 | LG11 (122) | 114 | LG11 (114) | 70 | LG8 (65), LG15 (5) | 104 | LG10 (3), LG13 (66), LG15 (35) |
| LG3 | 110 | 0.66 ± 0.98 | 4.75 | 65.6 | 94 | LG12 (94) | 68 | LG12 (68) | 30 | LG12 (30) | 42 | LG9 (42) |
| LG4 | 56 | 0.95 ± 1.48 | 7.77 | 51.3 | 26 | LG4 (1), LG13 (25) | 29 | LG9 (14), LG10 (14), LG16(1) | 22 | LG15 (22) | 24 | LG10 (24) |
| LG5 | 57 | 0.90 ± 2.30 | 11.68 | 49.5 | 36 | LG15 (36) | 35 | LG17 (35) | 29 | LG14 (28) | 30 | LG8 (30) |
| LG6 | 298 | 0.41 ± 1.06 | 11.75 | 103.7 | 181 | LG3 (181) | 209 | LG2 (85), LG4 (93), LG15 (31) | 115 | LG7 (91), LG9 (24) | 139 | LG7 (33), LG11 (19), LG12 (87) |
| LG7 | 97 | 0.75 ± 1.42 | 9.05 | 63.5 | 76 | LG5 (76) | 71 | LG5 (50), LG6 (21) | 35 | LG3 (26), LG4 (9) | 51 | LG14 (51) |
| LG8 | 100 | 0.78 ± 1.53 | 11.56 | 69.7 | 68 | LG6 (68) | 73 | LG8 (54), LG18 (19) | 49 | LG11 (13), LG13 (35) | 60 | LG4 (45), LG16 (15) |
| LG9 | 114 | 0.98 ± 2.11 | 14.02 | 94.2 | 83 | LG4 (83) | 92 | LG16 (92) | 42 | LG16 (42) | 44 | LG5 (44) |
| LG10 | 118 | 0.61 ± 1.12 | 7.77 | 64.5 | 62 | LG8 (42), LG9 (20) | 55 | LG7 (55) | 53 | LG5 (32), LG10 (21) | 39 | LG3 (39) |
| LG11 | 152 | 0.24 ± 0.40 | 2.65 | 35.5 | – | – | – | – | 94 | LG1 (95) | 76 | LG2 (76) |
| Total | 1539 | 795.3 | 876 | 12 LGs | 828 | 16 LGs | 580 | 15 LGs | 649 | 15 LGs | ||
| Average | 139.9 ± 77.5 | 0.65 ± 0.24 | 8.69 ± 3.47 | 72.30 ± 24.96 | ||||||||
LG linkage group;
anumber in bracket is denoted for number of markers on the LG of respective component map, and underline indicated the LG was represented in more than one consensus LG
The number of markers per linkage group on the consensus map varied from 56 (LG4) to 258 (LG1) with an average of 139.9 markers. The maximum length of LG was observed for LG1 (119.4 cM) while the shortest was for LG11 (35.5 cM). The average inter-marker distance ranged from 0.24 cM (LG11) to 0.98 cM (LG6) with an average of 0.65 cM. Visual inspection of the consensus map identified 11 major gaps (> 7 cM) across 8 LGs (LG1, LG4, LG5, LG6, LG7, LG8, LG9, and LG10). Non-uniform distribution of markers was evident in all LGs. The largest gap between the two loci was observed as 14.0 cM on LG9.
Comparison of consensus and component linkage maps
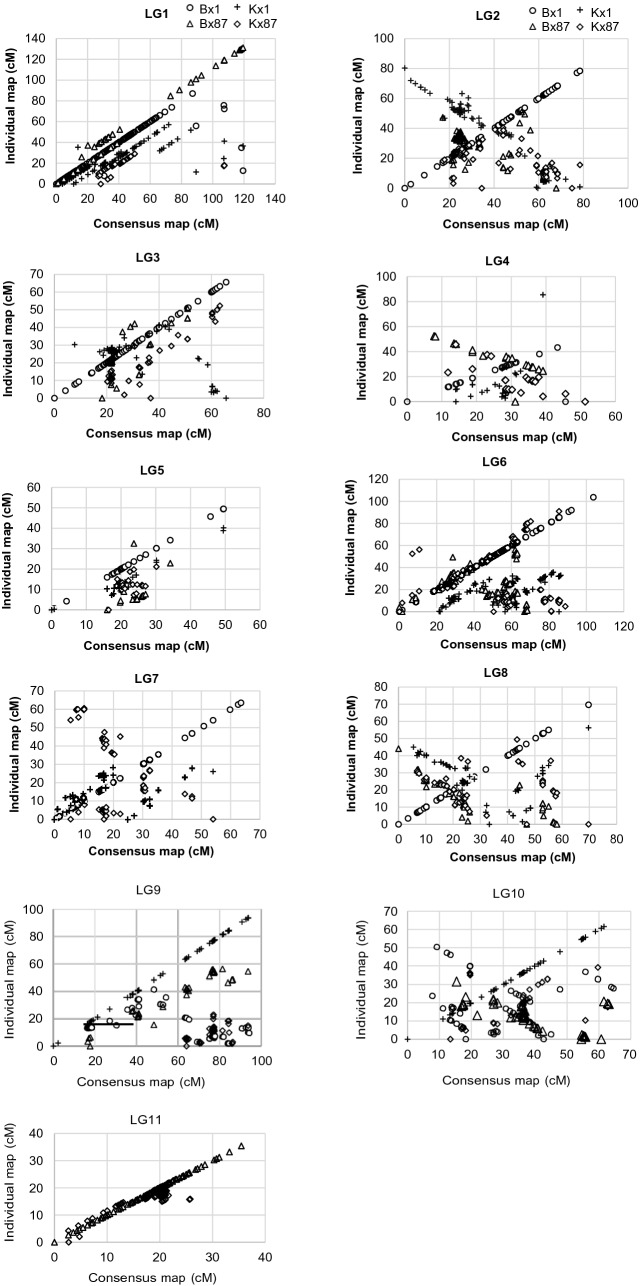
The medium to high degrees of correlation was observed for all LGs between consensus and component specific LGs (correlation coefficients r varying from 0.32 to 0.99) (Fig. 3). Some exceptional cases were observed with low correlations, such as LG8 of the consensus map had a correlation of -0.12 with LGs of the Kx87 individual map. There were also cases where LGs of the consensus map contained nearly entire one or two component specific LGs, such as LG1, LG9, LG10, and LG11 (Table 4). The conservation of marker orders was manifested in most LGs although reverse orders were observed for several component specific LGs.
Fig. 3.
Scatter plots showing the extent of correlations among the mungbean consensus genetic map and four component maps. The DArT markers integrated from four mungbean F5 RIL populations derived from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
Phenotyping and QTL detection
Overall, 81 QTLs were identified for 39 of the 54 measured traits across 10 LGs of the consensus map with positive and negative additive effects, depend on the instance (Supplement Table 6). There were some cases where one QTL conditioned for more than one trait. For example, a QTL on LG1 at the position of 9 cM conditioned for both dry pod mass and seed yield. A QTL on LG4 at the position of 24 cM conditioned for 6 traits. Taking into account such shared QTLs, the number of unique QTLs was 62. LG1 and LG2 carried more QTLs compared to others with 9 and 15 QTLs, respectively.
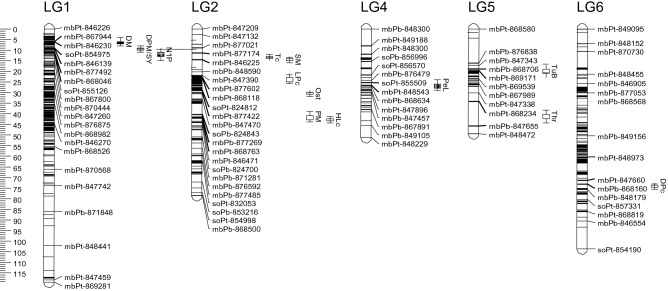
Of 62 QTLs, 19 major QTLs with PVE in the range of 10.69–41.57% were detected for qualitative pod and seed traits, a quantitative morphological trait and yield-related traits (Fig. 4). Notably, the perenniality trait, a special character of ACC 87 was conditioned by a major QTL mbPb-868706–mbPt-869171 (PVE of 15.58%) on LG5 at the position of 19 cM. This QTL had the effect of increasing the tuberisation of mungbean roots and was donated by the wild parent ACC 87.
Fig. 4.
Partial and a subset of 5 LGs of DArT linkage map showing 14 major QTLs detected in the mungbean consensus map. QTLs are in bold and black. Traits are indicated next to QTLs
Across 6 trait categories and when shared QTLs were not accounted for, qualitative morphological and seed traits had a higher number of detected QTLs than other categories with 39 and 19 QTLs, respectively. While the suggested single and double gene action models, based on phenotypic segregation data, were broadly consistent between the F2 and F5 generations for most traits (Nguyen et al. 2012; Vu 2013), the numbers of associated QTLs, which were identified for qualitative traits were variable. Several traits were identified with 1 or 2 major QTL models which supported the phenotypic observations, such as leaflet lobing, dry pod colour (single gene in ACC 87 crosses), powdery mildew (single gene in ACC1 crosses), perenniality (single gene in Bx87 cross), and hilum colour (single gene) (Table 5).
Table 5.
Summary and comparison of gene action models for qualitative traits suggested by phenotypic data at F2 (Nguyen et al. 2012) and F5 generations (Vu 2013) with models suggested by QTL detection
| Traits | F2 phenotypic model* (Nguyen et al. 2012) | F5 phenotypic model* (Vu 2013) | Number of detected QTLs on the consensus map† | Suggested QTL models |
|---|---|---|---|---|
| Bx1/Kx1/Bx87/Kx87 | ||||
| Qualitatively inherited traits | ||||
| Hypocotyl pigment | - | 2/2/-/- | 5 | Generally, minor and additive QTLs |
| Stem colour | - | 2/2/2/- | 1 | |
| Leaf rachis colour | - | 1/1/2/2 | 2 | |
| Leaf petiole colour | - | 1/2/-/- | 7 | |
| Plant hair density | - | 1 | 2 | |
| Plant hair colour | - | 1/1/2/2 | 1 | |
| Growth habit | 2 | 2/2/2/- | 4 | |
| Twining | 2/2/1/1 | 2/2/1/1 | 3 | |
| Leaflet lobbing | 1 | 1/-/-/- | 5 | |
| Flower colour | - | 1/1/2/2 | 3 | |
| Inflorescence structure | - | 2/2/1/- | 1 | |
| Dry pod colour | - | 2/2/1/1 | 3 | A major QTL and minor additive QTLs; |
| Pod dehiscence | 2/2/1/1 | 2 | 2 | Additive minor QTLs |
| Powdery mildew | 1/1/2/2 | 2 | 1 | A major QTL |
| Thrips | - | 2 | 3 | Two major QTLs |
| Perenniality | -/-/2/2 | -/-/1/2 | 1 | A major QTL |
| Qualitative seed traits | ||||
| Testa colour | 2 | 2 | 2 | A major QTL |
| Seed mottling | - | 2 | 3 | A major QTL |
| Seed coat ridging | 2/2/1/1 | -/-/1/2 | 1 | A minor QTL |
| Lustre | 2/2/2/1 | -/-/1/2 | - | No QTLs |
| Depth of texture layer | - | 2/2/1/2 | 3 | Additive QTLs; |
| Hilum colour | 1 | 1 | 2 | A major QTL |
| Texture layer colour | - | 1/1/2/2 | 2 | Additive QTLs |
| Overall visual seed traits | - | - | 1 | A major QTL |
Numbers in -/-/-/- respectively denotes number of genes operating in Bx1/Kx1/Bx87/Kx87 crosses
*: 1. single gene model; 2. two genes model; -. No suggested model; † bold numbers indicate where major QTLs were detected for corresponding traits;
Congruent QTLs across four mungbean RIL populations and the consensus map
All detected QTLs for four mungbean populations and the consensus map are listed in Supplement Table 7. The total number of QTLs detected for each RILs population was in the range of 42–86 for 30–41 traits (Table 6). The total numbers of detected QTLs were fewer in the ACC 87 crosses compared to ACC 1 crosses. The higher number of selected DArT markers and lower levels of marker segregation distortion observed in the ACC 1 crosses may have contributed to these differences (Table 2).
Table 6.
QTL detection and congruent QTLs resolved in the four mungbean RIL populations derived from crosses between cultivars (Berken and Kiloga) and wild accessions (ACC 1 and ACC 87) and the consensus map
| Common marker* | Populations | Consensus map | PVE % range | ||||
|---|---|---|---|---|---|---|---|
| Bx1 | Kx1 | Bx87 | Kx87 | ||||
| # QTLs detected | 59 | 86 | 42 | 65 | 81 | ||
| # traits for which QTLs were detected | 30 | 41 | 32 | 36 | 39 | ||
| Congruent QLTs | |||||||
| # Congruent markers | 18 | 9 | 4 | 4 | 7 | 14 | 3.8–54.7 |
| # traits for which congruent markers were detected | 13 | 9 | 4 | 3 | 5 | 12 | |
| Leaf rachis colour | mbPb-877288 | ✓/ + | ✓/ + | 7.3–18.0 | |||
| Growth habit | mbPb-868471 | ✓/ + | ✓/ + | 3.8–13.5 | |||
| Twining | mbPt-876465 | ✓/ + | ✓/ + | 5.6–10.2 | |||
| Leaflet lobing | mbPt-876514 | ✓/ + | ✓/ + | 5.5–23.4 | |||
| Dry pod colour | mbPb-868160 | ✓/ + | ✓/ + | 15.1–22.8 | |||
| Powdery mildew | mbPb-877269 | ✓/ + | ✓/- | 8.9–11.6 | |||
| Perenniality | mbPb-868706 | ✓/ + | ✓/ + | 15.6–17.5 | |||
| Testa colour | |||||||
| mbPb-847032 | ✓/ + | ✓/ + | 24.2–29.1 | ||||
| mbPt-846225 | ✓/ + | ✓/ + | 11.1–45.6 | ||||
| Seed mottling | |||||||
| mbPb-847032 | ✓/ + | ✓/ + | 22.4–33.6 | ||||
| mbPt-846225 | ✓/ + | ✓/ + | 14.4–54.7 | ||||
| Texture layer depth | mbPb-877325 | ✓/ + | ✓/ + | 8.2–22.5 | |||
| Texture layer colour | mbPb-871281 | ✓/ + | ✓/ + | 9.8–13.2 | |||
| mbPt-868828 | ✓/ + | ✓/ + | ✓/ + | 7.7–37.3 | |||
| mbPb-877325 | ✓/ + | ✓/ + | ✓/ + | 9.8–21.9 | |||
| Overall visual seed traits | mbPb-877269 | ✓/ + | ✓/ + | 12.1–13.8 | |||
| soPt-824812 | ✓/ + | ✓/ + | 16.6–30.8 | ||||
| Growth duration | mbPb-868706 | ✓/ + | ✓/ + | 18.2–25.4 | |||
*Loci nearby the corresponding putative additive QTL are common among populations
✓: indicating the loci are present; + and − : indicating the additive effect direction
The number of congruent QTLs resolved in the four mungbean RIL populations and the consensus map based on common markers was 18 for 13 traits and with PVE range of 3.8–54.7% (Table 6). The number of pairwise congruent QTLs was highest in the consensus map (14), followed by the Bx1 (9). Generally, congruent QTLs occurred in qualitative morphological and seed traits, except for common marker mbPb-868706 of growth duration in Bx87 and Kx87. Most of these QTLs showed positive additive effects and variation in PVE ranges, such as the QTLs for leaflet lobing, testa colour, and seed mottling.
Especially, most of these congruent QTLs governed seed appearance traits, such as testa colour and seed mottling, texture layer colour and depth, and overall seed appearance. All of these QTLs came from the wild and had positive additive effects. For example, the two QTL/markers mbPb-847032 and mbPt-846225 were linked to both testa colour and seed mottling and were pairwise common across Bx1, Kx1, Kx87, and the consensus map. The similar observation was for QTL mbPt-868828 and mbPb-877325. The QTL mbPb-868706 linked to both perenniality and growth duration, was present in both Bx87 and Kx87 and had the effect of increasing the tuberisation of mungbean roots.
QTL detection for late flowering and perenniality
The cultivars contributed alleles conditioning earlier flowering and shorter durations of flowering, pod growth, and total plant growth, while the wild parents contributed alleles conditioning for longer durations. While no QTLs conditioning late flowering were detected on the consensus map, separate QTL analyses in Bx1 and Kx1 detected 3 QTLs only in the Kx1 population with PVE range of 8.1 to 28.6% (Table 7). Two major QTLs were identified with PVE > 20% on LG7 and LG11 of Kx1 and effects of later flowering enhancement. The presence of major and positive additive effect QTLs originating from ACC 1 suggested relatively few major genes may control late flowering in ACC 1 crosses. Besides, 4 major QTLs for time to flowering were also exhibited in ACC 87 crosses with PVE range of 11.2–29.9%. The phenotyping of the ACC 87 crosses indicated a small number of very late flowering plants (Vu 2013).
Table 7.
Detected QTLs on the mungbean component maps by ICIM-ADD method at PIN of 0.001 for time to flowering and perenniality in mungbean RIL populations derived from crosses between two cultivars (Berken and Kiloga) and two wild accessions (ACC 1 and ACC 87)
| Time to flowering | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kx1 | Bx87 | Kx87 | ||||||||||||
| LG | Pos (cM) | Marker interval | LOD | PVE (%) | LG | Pos (cM) | Marker interval | LOD | PVE (%) | LG | Pos (cM) | Marker interval | LOD | PVE (%) |
| 7 | 40.0 | mbPt-846324/mbPt-868542 | 10.6 | 27.2 | 9 | 23.0 | soPb-824730/soPb-825660 | 5.1 | 21.0 | 4 | 1.0 | mbPb-847400/mbPt-868384 | 3.7 | 11.6 |
| 11 | 36.0 | soPb-824755/mbPt-867926 | 11.6 | 28.6 | 16 | 15.0 | mbPt-847782/mbPt-847390 | 3.0 | 11.2 | 13 | 31.0 | mbPt-868828/mbPt-868260 | 8.6 | 29.9 |
| 19 | 27.0 | mbPb-846990/soPb-832109 | 3.7 | − 8.1 | ||||||||||
| Perenniality trait | ||||||||||||||
| Bx87 | Kx87 | |||||||||||||
| LG | Pos (cM) | Marker interval | LOD | PVE (%) | LG | Pos (cM) | Marker interval | LOD | PVE (%) | |||||
| 17 | 18.0 | mbPt-870410/mbPt-877415 | 5.3 | − 22.6 | 8 | 18.0 | mbPb-868706/mbPb-871035 | 5.3 | 17.5 | |||||
| 8 | 19.0 | mbPb-871035/mbPb-847338 | 4.0 | 17.4 | ||||||||||
Bold number indicated major QTL with PVE % ≥ 10%
For the novel perenniality trait, 3 major QTLs linked to tuberisation were detected in Bx87 and Kx87 with various PVE from 17.4 to 22.6% (Table 6). In Bx87, although a QTL on LG17 accounted for 22.6% of the variation, it originated with Berken and had the effect of decreasing trait expression. In Kx87, 2 QTLs of close position on LG8 equally contributed around 17% to the total variation in tuberisation. The marker mbPb-868706 was also detected with the QTL on LG5 of the consensus map.
ACC 87 contributed alleles for tuberisation while Kiloga and Berken provided alleles for fibrous roots. The classical Mendelian analysis of tuberisation expression in the F2 and BC progeny (Nguyen et al. 2012) and the Kx87 F5 RIL population (Vu 2013), suggested two complementary dominant genes. The QTL results here supported that conclusion to the extent that QTLs from both parental sides were present in both the Bx87 and Kx87 F5 RILs and additive genes contributing to the degree of expression of tuberisation from weak to strong in the ACC 87 crosses.
Discussion
Linkage map construction in mungbean
As a result of the use of wild accessions to create the hybrid mungbean populations, the number of polymorphic DArT markers was quite high (Table 1). Number of polymorphic DArT markers identified and mapped in the four mungbean RIL populations (1062–2013 markers) were around twice those in three soybean RIL populations (550–1272 markers) (Vu et al. 2015) when using the same DArT array developed by Vu et al. (2012). This was also consistent with polymorphism levels evaluated during array development and reflected the large genetic distances between the two cultivated and two wild mungbean accessions.
DArT marker segregation distortion was also observed. The levels of segregation distortion were in the range observed for other markers in mungbean mapping projects such as 36.7% for RFLP markers (Humphry et al. 2005), 38.3% for SSR, RAPD and STS markers (Zhao et al. 2010), and 20.4% for SSR (Liu et al. 2017). In addition, the segregation distortion was also within the reported ranges for other marker types in other species, such as for DArT markers in Triticale mapping (8.3–34.3%) (Alheit et al. 2011) and soybean (17.4–41.6%) (Vu et al. 2015). In many cases, the markers segregating with distortion occurred in linked blocks and may simply reflect a lack of recombination in the distorted regions of the genome for these populations. More severe segregation distortion occurred in populations involving wild and landrace parents: i.e. segregation distortion levels were more severe in the four mungbean populations involving the wild accessions ACC 1 and ACC 87. This phenomenon has been reported in other studies in which mapping populations were derived from wide genetic crosses (Lambrides et al. 2000; Wang et al. 2012). The high levels of polymorphisms together with severe segregation distortion in the four mungbean RIL populations, even though the RILs were intra-specific crosses, could suggest that the Australian forms of sublobata are distinct from cultivated mungbean (Lawn and Cottrell 1988; Lambrides et al. 2000). Moreover, because segregation distortion was lower in the crosses involving ACC 87, it also suggests that Australian forms are diverse within themselves. This is consistent with the observations that ACC 1 and ACC 87 are distant at the molecular level (Vu et al. 2012) and morphologically different (Nguyen et al. 2012, 2016) and with phenotypic variation reported for other accessions in V. radiata spp. subolata collected in Australia (Lawn and Cottrell 1988; James et al. 1999; Rebetzke and Lawn 2006a, b).
The DArT mungbean component and consensus linkage map lengths were variable but comparable with published linkage maps using RAPD, RFLP, SSR, STS, EST-SSR and SNP markers (e.g. Lambrides et al. 2000; Humphry et al. 2002; Zhao et al. 2010; Isemura et al. 2012; Kajonphol et al. 2012; Wang et al. 2016, 2020; Liu et al. 2017). The average distances between adjacent markers in published maps were at least three times greater than in the present DArT marker maps (3–10.2 cM compared to 0.9–1.2 cM, respectively), except for the linkage maps constructed by Isemura et al. (2012) (1.78 cM) and by Wang et al. (2020) (0.54 cM). Most linkage maps have resolved 12–14 LGs (i.e. more than 11, the haploid chromosome number of mungbean), the exceptions being the maps by Kajonphol et al. (2012), Isemura et al. (2012), Liu et al. (2017) and Wang et al. (2020) composed of, respectively, 150, 430, 331 and 21,508 markers in 11 LGs. The differences in design, size, marker density, as well as technical errors of genotyping could result in genetic map variation among multiple population maps (Wang et al. 2011). In addition, genomic structural variation such as rearrangements, translocations and deletions can contribute to variation observed in lengths, number of LGs and marker orders among multiple population maps (Khan et al. 2012) i.e. among the four mungbean F5 RIL populations in this study or with other populations in previous studies.
DArT markers were not evenly distributed along the chromosomes with large ranges in the number of markers per LG (e.g. 9–184 in Bx1), LG sizes (e.g. 12.2–131.2 cM in Bx87), and inter-marker distances (e.g. 0.01 to 22 cM in Kx1). Cluster formation of markers in different regions of the chromosomes was also observed (Fig. 1; Supplement Fig. 1). The uneven patterns of DArT marker distribution and clustering in different chromosome regions observed in this study were also observed in Triticale and soybean linkage maps when only DArT markers were applied (Alheit et al. 2011; Vu et al. 2015). Similar occurrences were for other marker types, such as RFLP and SSRs in mungbean (Fatokun et al.1992; Kajonphol et al. 2012). Additionally, map resolution can be improved by incorporation of DArT markers with other marker types as shown in Triticale (Tyrka et al. 2011) and lentil (Ates et al. 2018). Thus, these DArT linkage maps for mungbean would be improved, in terms of both their resolution and the identification of LGs corresponding to the respective chromosome numbers, by incorporating other markers used in mungbean projects, such as RFLPs and RAPDs (Lambrides et al. 2000; Humphry et al. 2005), STS and SSRs (Zhao et al. 2010; Kajonphol et al. 2012; Isemura et al. 2012). This approach would also allow the comparison for congruency of marker orders and mapping positions.
The consensus mungbean map covered a large number of markers from the component maps (> 85%; Table 4) and resolved for 11 LGs. However, the severe segregation distortion in the mungbean RILs possibly caused some degree of re-arrangement of marker orders on some LGs of the consensus map (Fig. 3). Alheit et al. (2011) illustrated other possible consequences of segregation distortion which not only affect genetic map distances and ordering of loci but can even result in complete chromosomes being absent from genetic maps. To this time, there have not been any reports on the construction of a mungbean consensus map from more than two hybrid populations. Given that in the present study, from 1883 DArT markers mapped across the four mungbean RIL populations, an ample set of 1539 DArT markers were constructed for 11 LGs of the consensus mungbean map.
Validation of QTLs through congruent QTL analysis
QTL detection based on a single population usually results in only a limited number of QTLs and the results are often not conclusive. Therefore, research on QTL effects in more than one genetic background can provide more authentic and reliable QTL information. Several QTL studies have used this approach. For instance, five independent and two related mapping populations were involved in comprehensive QTL scanning in rice (Uga et al. 2010; Li et al. 2012). Additionally, pooled analysis of data from multiple quantitative trait locus mapping populations or meta-quantitative trait loci are also methods for evaluating the overall evidence for the existence of a QTL from different studies (Guo et al. 2006; Delfino et al. 2019; Shariatipour et al. 2021). Some other studies using Near- isogenic lines developed by repeated backcrossing with the parent for validation of detected QTLs (Jang et al. 2020). Thus, in the mungbean, this is the first study to apply DArT markers, with four related populations derived from crosses between cultivars and wild accessions used to construct the consensus map and identify QTLs for 54 traits (using pooled analysis) and analyze the congruent QTLs. Based on common markers observed among the individual populations and with the consensus map, congruent QTLs i.e. QTLs which are common across at least two populations or with the consensus map were identified (Table 6; Supplement Table 7).
Many QTL analysis studies have revealed the phenomenon where different QTLs could be detected for traits, especially quantitative traits, regardless of whether genetically related background populations were used, or sometimes when the traits were evaluated on the same population but in different environments (Hossain et al. 2010; Negeri et al. 2011; Ding et al. 2011; Li et al. 2012). This phenomenon was also observed in this study where four related mungbean populations were assessed and the consensus map was constructed. For example, no common QTLs were detected for hypocotyl pigment, stem colour, leaf petiole colour, and petiole length across component populations and the consensus map (Supplement Table 7). This may perhaps relate to the genetic distance between the populations (Table 2) or simply indicate that minor additive QTLs also accounted for trait expression. In addition, different sets of polymorphic DArT markers selected for each RIL population which were used for QTL analysis obviously would result in different QTLs being detected for the same traits. The inability to identify sequencing redundancy of DArT markers may also have resulted in an underestimate of the presence of common markers among populations, the consensus map, and traits. However, in this study, congruent QTLs detected were reasonable, especially for qualitative morphological and seed traits, suggesting their validation (Table 6).
QTLs associated with morphological and agronomical traits and implication for mungbean improvement
The four RIL populations were generated from genetically distant parents and segregated for multiple traits (Nguyen et al. 2012, 2016; Vu 2013). As expected, this proved advantageous and allowed QTLs controlling different traits to be detected on a single map, and therefore QTL detection in mungbean was relatively straightforward for both qualitative and quantitative traits. Most quantitative traits also had moderate to high broad-sense heritability indicating moderate to large additive genetic effects. Therefore, not surprisingly, many QTLs were detected for the majority of the evaluated traits in the four populations (62 QTLs detected for 39 traits across 10 LGs and 19 major QTLs–Fig. 4; Supplement Table 6).
QTLs for morphological traits
Many QTLs associated with both qualitative and quantitative morphological traits were identified on the consensus map. In general, additive QTL models were suggested for most traits with phenotypic variation explanation (PVE) effects ranging from minor (2.98%) to highly significant (41.57%). More often, QTLs had the effect of enhancing the wild-type phenotype such as increasing the degree of twining and leaflet lobing.
QTLs for powdery mildew and thrip resistance
Putative resistance to thrips in mungbean appears to be complex because 3 additive QTLs from both parental sides were identified. In contrast, only a major QTL conditioned powdery mildew resistance. Generally, the major QTLs for both resistance to thrips and powdery mildew had the effect of enhancing plant resistance and were donated by the wild parents. These QTLs were not co-located with other QTLs governing wild phenotypes such as prostrate growth habit, twining, and seed phenotypes. This raises the possibility of pyramiding different resistance genes to provide a broader spectrum of resistance to develop mungbean varieties resistant to powdery mildew disease and thrip insects.
High genetic variability in reaction to powdery mildew in mungbean landraces was reported by Yohe and Poehlman (1972). Several studies based on phenotypic data have suggested different gene models conferring powdery mildew resistance in mungbean, from single gene action (Khajudparn et al. 2007; Nguyen et al. 2012) to two genes with both additive and dominant gene actions (Kasettranan 2009; Nguyen et al. 2012). At a molecular level, one to three major loci conditioning resistance were identified in earlier studies on mapping powdery mildew resistance (Young et al. 1993; Chaitieng et al. 2002; Humphry et al. 2003; Kasettranan et al. 2010).
In contrast to powdery mildew, little is known about resistance to thrips in plants in general and in mungbean in particular (Nair et al. 2019). Multi-genic resistance mechanisms were suggested with additive, dominance and epistatic gene effects (Omo-Ikerodah et al. 2009). Two QTLs in common bean (Frei et al. 2005) and 2, 3, and 6 QTLs in cowpea (Omo-Ikerodah et al. 2008; Muchero et al. 2010) associated with thrips resistance were reported. Although some studies screened for thrips resistance of mungbean (Khattak et al. 2004b; Sandhya et al. 2008), no other molecular markers appear to be available for thrips resistance in mungbean.
QTLs for seed appearance traits
Testa colour, hilum colour, seed mottling, and seed coat ridging are visual traits and can be easily screened for selection. However, they are all maternal tissue traits, so that the detection of associated QTLs can still be useful for marker-assisted selection (MAS). Maternally inherited traits require generation advancement before they become evident, making MAS useful since it allows for selection before the seeds are formed, perhaps even before sowing (Mogensen 1996). Most of the seed trait QTLs detected in the present study governed wild seed phenotypes. In particular, 4 major QTLs were identified as potentially useful for early screening out of lines that carry undesirable wild seed appearance phenotypes.
QTLs for pod and seed traits
Pod dehiscence (shattering) before harvest is generally an undesirable trait since it is one of the main reasons for seed loss and low yield in mungbean. Thus, breeding for dehiscence–resistant varieties is an important objective in mungbean genetic improvement programs. In this study, 2 QTLs on LG8 linked to pod dehiscence but with the minor effect of accelerating pod dehiscence (PVE of 5.55 and 4.34%) (Supplement Table 6). They did not co-localize with undesirable traits, except for the twining habit. Since only DArT markers were employed, the comparison with other pod shattering QTLs in previous studies was not possible (Isemura et al. 2012).
Hardseededness is considered a possible trait in mungbean breeding programs where the aim is to develop weather-resistance (Lawn et al. 1988). Both wild parents showed extremely strong hardseededness, while the cultivated lines were almost completely soft seeded. This study detected only a minor QTL on LG3 for hardseededness (PVE of 4.82%). Previous observations showed few genes control this trait (Lawn et al. 1988), or a single common QTL that explained 11% to 23% of the hardseededness in field and glasshouse conditions was identified by Humphry et al. (2005). Isemura et al. (2012) reported 4 QTLs conditioning the loss of seed dormancy or increase of water absorption.
A number of QTLs linked to pod and seed traits have also been reported in other mungbean studies e.g. Fatokun et al. (1992), Humphry et al. (2005), Kajonphol et al. (2012), and Isemura et al. (2012). Humphry et al. (2005) showed co-localization for QTLs of hardseededness and seed weight and alleles associated with hardseededness and small seed weight were usually inherited together. Although this study detected a minor QTL for hardseedeness, this QTL co-localized with QTLs conditioning testa colour and texture layer depth. In addition, significant negative phenotypic correlations between hardseededness and seed size (data not shown) were observed in all four RIL populations. Those linkages are likely to make attempts to breed large and hard-seeded mungbean varieties difficult to achieve.
QTLs for yield related traits
In addition to traits that were significantly correlated with seed yield, such as number of seeds per pod, and total number of pod clusters, crop improvement aiming for high yield could be based on selection for yield attributes such as total pod mass and standing dry biomass. This study detected numerous QTLs linking to pod dry mass, seed yield, and standing dry biomass. In particular, the co-localization of the seed yield, pod dry mass, and standing dry biomass QTLs either at the same position or on the same linkage groups of the consensus map (LG1 and LG4) were consistent with significant phenotypic correlations among those traits (Vu 2013).
QTLs for phenology
Two minor QTLs, respectively controlling duration of flowering and days to maturity were detected on the same LG4 of the consensus map. Isemura et al. (2012) and Kajonphol et al. (2012) reported 3 and 4 QTLs associated with earliness. Although flowering time is one of the keystones in plant adaptation, there is little information on the genetics of flowering in mungbean, especially late flowering (Weller and Ortega 2015). Depending on abiotic (photoperiod, temperatures, and nutrients) and biotic (competition, pollinators) conditions, different flowering time strategies can be adopted between and even within a wild plant species. In the Kx1 crosses, 2 major QTLs associated with late flowering–the special character of the wild mungbean ACC 1–were revealed. The ACC 87 crosses also produced a few very late flowering genotypes, indicating that lateness genes might exist in that accession, but they are likely masked by other genes for earliness. This was in line with QTLs of the effect of increasing time to flowering in ACC 87 populations (Table 6).
The time to flowering is an agronomically important trait in many legume species such as pea, soybean, and chickpea (Weller and Ortega 2015). The inheritance of time to flowering is known as a quantitative and complex trait (Khattak et al. 2004a; Nguyen et al. 2016) and influenced by environmental factors, especially day length and temperature (Khattak et al. 2001; Rehman et al. 2010). Various hypotheses on gene action for flowering in mungbean have been suggested, including both additive and non-additive gene action controlling the earliness of mungbean (Rehman et al. 2010), or both additive and dominant gene action (Tah 2009).
Therefore, detection of these first QTLs is a start for a better understanding of the control of flowering in mungbean. Transferring late flowering from the wild to cultivated mungbean can be useful in broadening latitudinal adaptation, extending the crop duration for higher yield potential, or delaying pod maturity until unfavourable weather conditions have ceased (Nair et al. 2019).
QTLs for perenniality
While phenotyping in the F5 generation in both Bx87 and Kx87 crosses suggested two different gene models for inheritance of perenniality, the QTL analysis indicated a major QTL on the consensus map contributing to trait expression. This QTL also conditioned the growth habit with insignificant effects. As such, it should prove relatively straightforward for breeders to transfer the trait across into cultivated mungbean backgrounds. Together with the study by Nguyen et al. (2012), the present study is among the first to explore the inheritance of perenniality in the mungbean.
QTLs identified in this study may be potentially useful in mungbean genetic improvement include those linked to powdery mildew and thrips resistance, seed appearance traits, perenniality, pod dehiscence, hardseedeedness, late flowering, and dry biomass. The application or transfer of these genes from the wild across cultivated mungbean backgrounds will be more straightforward for those QTLs that are not located in regions of the genome where severe distortion in favour of cultivated alleles was detected, have large PVE and are not co-localized with undesirable traits. For the putative resistance to powdery mildew and thrips, the resilience of the traits needs to be re-checked in different environments and years. As mentioned above, breeding for large and hard-seeded mungbean could still be challenging as many of the detected QTLs for enhancing hardseededness were co-localized with QTLs decreasing seed size.
Before attempts are made to use these QTLs, such as routine MAS in breeding programs, it would be desirable that further studies are required to verify the observations made in this study. QTL verification is defined as the repeated detection of the same marker alleles at a similar position on the genetic map of a chromosome, or of a QTL controlling a trait under more than one set of experimental conditions. As with mungbean, some previous mapped genes and QTLs have been corrected or re-confirmed in Isemura et al. (2012) for the bruchid resistance gene, 100-seed weight, black mottle on the seed coat, or in Alam et al. (2014) for mungbean yellow mosaic disease. Although all DArT QTLs investigated in this study could not be used for comparison due to the different types of markers, major and important QTLs, as well as identified congruent QTLs can be selected for sequencing and blast with available draft mungbean reference genome (Kang et al. 2014) as done with soybean (Vu et al. 2015) for confirmation. This will then make QTLs useful for the genetic improvement of the mungbean.
Currently, together with the QTL approach, breeders have increasingly moved towards Genomic Selection (GS) which uses genome-wide molecular markers to predict breeding values and make selections of individuals before phenotyping. Additionally, the second platform of DArT technology, DArTseq makers based on the benefit of the complexity reduction method optimized for species and Genotype by Sequence (GBS) has been successfully applied for genetic diversity studies, linkage map construction, and QTL analysis (Pascual et al. 2020; Siekmann et al. 2021). Thus, the complexity reduction method optimized for mungbean by Vu et al. (2012) and the effectiveness of DArT mungbean markers applied in this study suggest the potential for the development of DArTseq platform and SNP markers for mungbean in the future. These will probably saturate the mungbean linkage map and make it comparable to other current linkage maps constructed of SNP markers (Mariyammal et al. 2019; Mathivathana et al. 2019; Wang et al. 2020; Lee et al. 2021).
Conclusions
The present study is the first to develop and apply DArT markers in mungbean, to construct a mungbean consensus marker map, and to identify QTLs associated with various traits in cultivated and wild mungbean. Overall, DArT development was successful with high polymorphism. DArT application in linkage map construction was effective and successful in mungbean, with map sizes comparable to published maps. The linkage maps constructed from DArT markers had close inter-marker distances, which were highly suitable for QTL scanning. DArT technology, with its advantages in terms of speed of marker discovery, analysis and high-throughput, therefore has great potential for use in constructing high-resolution genetic maps in mungbean in combination with other marker systems. Various QTLs were also successfully identified for various traits in the mungbean with major effects due to large genetic variation for the traits of interest in the populations.
Thus, this study provides an improved understanding of the expression and inheritance of a range of cultivated and wild morphological and agronomical traits in mungbean at the molecular level. It also confirms two Australian wild mungbean accessions to be part of the primary gene pool of cultivated mungbean and a potential source of useful genetic variation for mungbean improvement. Although most wild traits would be undesirable, some wild traits are potentially useful, such as resistance to powdery mildew and thrips, late flowering, hardseededness, and perenniality. Potentially useful QTLs identified for many mungbean traits can contribute to more effective and efficient breeding programs with the potential to save time and resources. Further studies should be suggested for sequences and blast of DArT markers to the available mungbean reference genome.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The first author acknowledges the support from Australian Endeavour Scholarship, James Cook University, Townsville and Alan and Elizabeth Finkel Foundation to conduct this research. Special thanks to staff at Diversity Arrays Technology Pty Ltd, Canberra for technical support, and to CSIRO Agriculture–ATSIP, James Cook University, Townsville and CSIRO–St Lucia, Queensland Biosciences Precinct for research facility access.
Author contributions
All authors contributed to the study conception and design. Conceptualization: TTHV, AK, RJL; Methodology: TTHV, AK, RJL, JC; Data curation: TTHV, JC; Writing–original draft preparation: TTHV, RJL; Writing-review and editing: TTHV, AKRJL, JC. All authors commented on the previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Declarations
Conflict of interest
Not applicable.
Availability of data and material
The datasets analysed in the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01201-8.
References
- Alam AM, Somta P, Srinives P. Identification and confirmation of quantitative trait loci controlling resistance to mungbean yellow mosaic disease in mungbean [Vigna radiata (L.) Wilczek] Mol Breed. 2014;34:1497–1506. doi: 10.1007/s11032-014-0133-0. [DOI] [Google Scholar]
- Alheit KV, Reif J, Maurer HP, et al. Detection of segregation distortion loci in Triticale (x Triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics. 2011;12:380–393. doi: 10.1186/1471-2164-12-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate D, Bixby R, Chvátal V, Cook WJ (2006) Concorde TSP Solver. http://www.tsp.gatech.edu/concorde
- Ates D, Aldemir S, Alsaleh A, et al. A consensus linkage map of lentil based on DArT markers from three RIL mapping populations. PLoS ONE. 2018;13:e0191375. doi: 10.1371/journal.pone.0191375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitieng B, Kaga A, Han OK, et al. Mapping a new source of resistance to powdery mildew in mungbean. Plant Breed. 2002;121:521–525. doi: 10.1046/j.1439-0523.2002.00751.x. [DOI] [Google Scholar]
- Delfino P, Zenoni S, Imanifard Z, Tornielli GB, Bellin D. Selection of candidate genes controlling veraison time in grapevine through integration of meta-QTL and transcriptomic data. BMC Genomics. 2019;20:1–19. doi: 10.1186/s12864-019-6124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding A-M, Li J, Cui F, Zhao C-H, Ma H-Y, Wang H-G. Mapping QTLs for yield related traits using two associated RIL populations of wheat. Acta Agron Sin. 2011;37:1511–1524. doi: 10.1016/S1875-2780(11)60041-2. [DOI] [Google Scholar]
- Fatokun CA, Menancio-Hautea DI, Danesh D, Young ND. Evidence for orthologous seed weight genes in cowpea and mungbean based on RFLP mapping. Genetics. 1992;132:841–846. doi: 10.1093/genetics/132.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei A, Blair MW, Cardona C, et al. QTL mapping of resistance to Thrips palmi Karny in common bean. Crop Sci. 2005;45:379–387. doi: 10.2135/cropsci2005.0379. [DOI] [Google Scholar]
- Gayacharan TK, Meena SK, et al. Understanding genetic variability in the mungbean (Vigna radiata L.) genepool. Ann Appl Biol. 2020;177:346–357. doi: 10.1111/aab.12624. [DOI] [Google Scholar]
- Guo B, Sleper DA, Sun J, Nguyen HT, Arelli PR, Shannon JG. Pooled analysis of data from multiple quantitative trait locus mapping populations. Theor Appl Genet. 2006;113:39–48. doi: 10.1007/s00122-006-0268-4. [DOI] [PubMed] [Google Scholar]
- Hossain S, Ford R, McNeil D, Pittock C, Panozzo JF. Development of a selection tool for seed shape and QTL analysis of seed shape with other morphological traits for selective breeding in chickpea (Cicer arietinum L.) Aust J Crop Sci. 2010;4:278–286. [Google Scholar]
- Humphry ME, Konduri V, Lambrides CJ, et al. Development of a mungbean (Vigna radiata) RFLP linkage map and its comparison with lablab (Lablab purpureus) revelas a high level of colinearity between the two genomes. Theor Appl Genet. 2002;105:160–166. doi: 10.1007/s00122-002-0909-1. [DOI] [PubMed] [Google Scholar]
- Humphry ME, Magner T, McIntyre CL, et al. Identification of a major locus conferring resistance to powdery mildew (Erysiphe polygoni DC) in mungbean (Vigna radiata L. Wilczek) by QTL analysis. Genome. 2003;46:738–744. doi: 10.1139/g03-057. [DOI] [PubMed] [Google Scholar]
- Humphry ME, Lambrides CJ, Chapman SC, et al. Relationships between hard-seededness and seed weight in mungbean (Vigna radiata) assessed by QTL analysis. Plant Breed. 2005;124:292–298. doi: 10.1111/j.1439-0523.2005.01084.x. [DOI] [Google Scholar]
- Isemura T, Kaga A, Tabata S, et al. Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata) PLoS ONE. 2012;7:e41304. doi: 10.1371/journal.pone.0041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AT, Lawn RJ, Williams RW, et al. Cross fertility of Australian accessions of wild mungbean (Vigna radiata ssp sublobata) with green gram (V. radiata ssp. radiata) and black gram (V. mungo) Aust J Bot. 1999;47:601–610. doi: 10.1071/BT97068. [DOI] [Google Scholar]
- Jang S, Han J-H, Lee YK, Shin N-H, Kang YJ, Kim C-K, Chin JH. Mapping and validation of qtls for the amino acid and total protein content in brown rice. Front Genet. 2020;11:240. doi: 10.3389/fgene.2020.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajonphol T, Sangsiri C, Somta P, et al. SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek) SABRAO J Breed Genet. 2012;44:71–86. [Google Scholar]
- Kang YJ, Kim SK, Kim MY, et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun. 2014;5:5443. doi: 10.1038/ncomms6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasettranan W, Somta P, Srinives P. Genetics of the resistance to powdery mildew disease in mungbean (Vigna radiata (L.) Wilczek) J Crop Sci Biotechnol. 2009;12:37–42. doi: 10.1007/s12892-008-0074-4. [DOI] [Google Scholar]
- Kasettranan W, Somta P, Srinives P. Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean (Vigna radiata (L.) Wilczek) J Crop Sci Biotechnol. 2010;13:155–161. doi: 10.1007/s12892-010-0052-z. [DOI] [Google Scholar]
- Khajudparn P, Wongkew S, Thipyapong P. Mungbean powdery mildew resistance: identification of genes for resistance to powdery mildew in mungbean. Afr Crop Sci J. 2007;8:743–745. [Google Scholar]
- Khan MA, Han Y, Zhao YF, et al. A multi-population consensus genetic map reveals inconsistent marker order among maps likely attributed to structural variations in the apple genome. PLoS ONE. 2012;7:e47864. doi: 10.1371/journal.pone.0047864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak GSS, Haq MA, Ashraf M, et al. Detection of epistasis, and estimation of additive and dominance components of genetic variation for synchrony in pod maturity in mungbean (Vigna radiata (L.) Wilczek) Field Crops Res. 2001;72:211–219. doi: 10.1016/S0378-4290(01)00178-2. [DOI] [Google Scholar]
- Khattak GSS, Ashraf M, Zamir R. Gene action for synchrony in pod maturity and indeterminate growth habit in mungbean (Vigna radiata (L.) Wilczek) Pak J Bot. 2004;36:589–594. [Google Scholar]
- Khattak MK, Ali S, Chishti JI. Varietal resistance of mungbean (Vigna radiata L.) against whitefly (Bemisia tabaci Genn.), jassid (Amrasca devastans Dist.), and thrips (Thrips tabaci Lind.) Pak Entomology. 2004;26:9–12. doi: 10.18801/jbar.180118.183. [DOI] [Google Scholar]
- Kim SK, Nair RM, Lee J, Lee S-H. Genomic resources in mungbean for future breeding programs. Front Plant Sci. 2015;6:626. doi: 10.3389/fpls.2015.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrides CJ, Lawn RJ, Godwin ID, et al. Two genetic linkage maps of mungbean using RFLP and RAPD markers. Aust J Agric Res. 2000;51:415–425. doi: 10.1071/AR99052. [DOI] [Google Scholar]
- Lawn RJ. Agronomic studies on Vigna spp. in south-eastern Queensland I. Phenological response of cultivars to sowing date. Aust J Agric Res. 1979;30:855–870. doi: 10.1071/AR9790855. [DOI] [Google Scholar]
- Lawn RJ, Cottrell A. Wild mungbean and its relatives in Australia. Biologist. 1988;35:267–273. [Google Scholar]
- Lawn RJ, Rebetzke GJ. Variation among Australian accessions of the wild mungbean (Vigna radiata ssp. sublobata) for traits of agronomic, adaptive or taxonomic interest. Aust J Agric Res. 2006;57:119–132. doi: 10.1071/AR05215. [DOI] [Google Scholar]
- Lawn RJ, Williams RW, Imrie BC (1988) Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean. In Shanmugasundaram S, McLean BT (Ed): Proceedings of the second international symposium. Bangkok, Thailand, pp 136–145
- Lee E, Yang X, Ha J, et al. Identification of a locus controlling compound raceme inflorescence in mungbean [Vigna radiata (L.) R. Wilczek] Front Genet. 2021;12:642518–642518. doi: 10.3389/fgene.2021.642518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cui F, Ding A-M, Zhao C-H, Wang X-Q, Wang L, Bao Y-G, Qi X-L, Li X-F, Gao J-R, Feng D-S, Wang H-G. QTL detection of seven quality traits in wheat using two related recombinant inbred line populations. Euphytica. 2012;183:207–226. doi: 10.1007/s10681-011-0448-4. [DOI] [Google Scholar]
- Liu C, Wu J, Wang L, et al. Quantitative trait locus mapping under irrigated and drought treatments based on a novel genetic linkage map in mungbean (Vigna radiata L.) Theor Appl Genet. 2017;130:2375–2393. doi: 10.1007/s00122-017-2965-6. [DOI] [PubMed] [Google Scholar]
- Mariyammal I, Seram D, Samyuktha SM, et al. QTL mapping in Vigna radiata × Vigna umbellata population uncovers major genomic regions associated with bruchid resistance. Mol Breed. 2019;39:110. doi: 10.1007/s11032-019-1006-3. [DOI] [Google Scholar]
- Mathivathana MK, Murukarthick J, Karthikeyan A, et al. Detection of QTLs associated with mungbean yellow mosaic virus (MYMV) resistance using the interspecific cross of Vigna radiata × Vigna umbellata. J Appl Genet. 2019;60:255–268. doi: 10.1007/s13353-019-00506-x. [DOI] [PubMed] [Google Scholar]
- Mogensen HL. The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot. 1996;83:383–404. doi: 10.1002/j.1537-2197.1996.tb12718.x. [DOI] [Google Scholar]
- Muchero W, Ehlers J, Roberts P. QTL analysis for resistance to foliar damage caused by Thrips tabaci and Frankliniella schultzei (Thysanoptera: Thripidae) feeding in cowpea [Vigna unguiculata (L.) Walp.] Mol Breed. 2010;25:47–56. doi: 10.1007/s11032-009-9307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RM, Pandey AK, War AR, et al. Biotic and abiotic constraints in mungbean production-progress in genetic improvement. Front Plant Sci. 2019;10:1340. doi: 10.3389/fpls.2019.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negeri AT, Coles ND, Holland JB, Balint-Kurti PJ. Mapping QTL controlling southern leaf blight resistance by joint analysis of three related recombinant inbred line populations. Crop Sci. 2011;51:1571–1579. doi: 10.2135/cropsci2010.12.0672. [DOI] [Google Scholar]
- Nguyen TD, Lawn RJ, Bielig LM. Expression and inheritance of perenniality and other qualitative traits in hybrids between mungbean cultivars and Australian wild accessions. Crop Pasture Sci. 2012;63:619–634. doi: 10.1071/CP12263. [DOI] [Google Scholar]
- Nguyen TD, Vu HTT, Lawn RJ, Bielig LM. Expression and heritability of late flowering and other quantitative traits in cultivated X Australian wild mungbean hybrids. Crop Pasture Sci. 2016;67:1235–1251. doi: 10.1071/CP16297. [DOI] [Google Scholar]
- Noble TJ, Tao Y, Mace ES, et al. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front Plant Sci. 2018;8:2102. doi: 10.3389/fpls.2017.02102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omo-Ikerodah EE, Fawole I, Fatokun CA. Genetic mapping of quantitative trait loci (QTLs) with effects on resistance to flower bud thrips (Megalurothrips sjostedti) identified in recombinant inbred lines of cowpea (Vigna unguiculata (L.) Walp) Afr J Biotechnol. 2008;7:263–270. [Google Scholar]
- Omo-Ikerodah EE, Fatokun CA, Fawole I. Genetic analysis of resistance to flower bud thrips (Megalurothrips sjostedti) in cowpea (Vigna unguiculata [L.] Walp.) Euphytica. 2009;165:145–154. doi: 10.1007/s10681-008-9776-4. [DOI] [Google Scholar]
- Pascual L, Ruiz M, López-Fernández M, et al. Genomic analysis of Spanish wheat landraces reveals their variability and potential for breeding. BMC Genomics. 2020;21:122–139. doi: 10.1186/s12864-020-6536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroli C, Sansaloni CP, Carling J, et al. Genomic characterization of DArT markers based on high-density linkage analysis and physical mapping to the Eucalyptus genome. PLoS ONE. 2012;7:e44684. doi: 10.1371/journal.pone.0044684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetzke GJ, Lawn RJ. Adaptive responses of wild mungbean (Vigna radiata ssp. sublobata) to photo-thermal environment I. Phenology. Aust J Agric Res. 2006;57:917–928. doi: 10.1071/AR05357. [DOI] [Google Scholar]
- Rebetzke GJ, Lawn RJ. Adaptive responses of wild mungbean (Vigna radiata ssp. sublobata) to photo-thermal environment. II. Growth, biomass, and seed yield. Aust J Agric Res. 2006;57:929–937. doi: 10.1071/AR05377. [DOI] [Google Scholar]
- Rehman A, Ali M, Saleem M, Tadesse W. Study of heritable variation and genetics of earliness in mungbean (Vigna radiata L. Wilczek) Euphytica. 2010;176:331–339. doi: 10.1007/s10681-010-0208-x. [DOI] [Google Scholar]
- Sandhya RC, Eshwari KB, Sudarshanam A. Field screening of greengram (Vigna radiata L.) entries against thrips (Thrips palmi) and spotted pod borer (Maruca vitrata) J Res ANGRAU. 2008;36:17–22. [Google Scholar]
- Sansaloni C, Petroli C, Carling J, et al. A high-density diversity arrays technology (DArT) microarray for genome-wide genotyping in Eucalyptus. Plant Methods. 2010;6:16–27. doi: 10.1186/1746-4811-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariatipour N, Heidari B, Tahmasebi A, Richards C. Comparative genomic analysis of quantitative trait loci associated with micronutrient contents, grain quality, and agronomic traits in wheat (Triticum aestivum L.) Front Plant Sci. 2021;12:709817. doi: 10.3389/fpls.2021.709817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekmann D, Jansen G, Zaar A, et al. A genome-wide association study pinpoints quantitative trait genes for plant height, heading date, grain quality, and yield in rye (Secale cereale L.) Front Plant Sci. 2021;12:718081. doi: 10.3389/fpls.2021.718081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tah PR. Induced synchrony in pod maturity in mungbean (Vigna radiata (L.) Wilczek) J Agric Bio Sci. 2009;4:41–44. [Google Scholar]
- Tinker N, Kilian A, Wight C, et al. New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genomics. 2009;10:39–61. doi: 10.1186/1471-2164-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomooka N, Vaughan DA, Moss H, Maxted N. The Asian Vigna: Genus Vigna subgenus Ceratotropis genetic resources. Netherlands: Kluwer Academic Publisher; 2002. [Google Scholar]
- Tyrka M, Bednarek PT, Kilian A, et al. Genetic map of triticale compiling DArT, SSR, and AFLP markers. Genome. 2011;54:391–401. doi: 10.1371/journal.pone.0145714. [DOI] [PubMed] [Google Scholar]
- Uga Y, Siangliw M, Nagamine T, Ohsawa R, Fujimura T, Fukuta Y. Comparative mapping of QTLs determining glume, pistil and stamen sizes in cultivated rice (Oryza sativa L.) Plant Breed. 2010;129:657–669. doi: 10.1111/j.1439-0523.2009.01765. [DOI] [Google Scholar]
- Vu HTT, Lawn RJ, Bielig LM, et al. Development and initial evaluation of diversity array technology for soybean and mungbean. Euphytica. 2012;186:741–754. doi: 10.1007/s10681-011-0573-0. [DOI] [Google Scholar]
- Vu HTT, Kilian A, James AT, et al. Use of DArT molecular markers for QTL analysis of drought-stress responses in soybean. II. Marker identification and QTL analyses. Crop Pasture Sci. 2015;66:817–830. doi: 10.1071/CP14304. [DOI] [Google Scholar]
- Vu HTT (2013) Use of Diversity Array Technology (DArT) to identify QTLs for physiological traits in mungbean (Vigna radiata) and soybean (Glycine max). Dissertation, James Cook University. https://researchonline.jcu.edu.au/40601/
- Wang G, He QQ, Xu ZK, Song RT. High segregation distortion in maize B73 x teosinte crosses. Genet Mol Res. 2012;11:693–706. doi: 10.4238/2012. [DOI] [PubMed] [Google Scholar]
- Wang L, Wu C, Zhong M, et al. Construction of an integrated map and location of a bruchid resistance gene in mung bean. Crop J. 2016;4:360–366. doi: 10.1016/j.cj.2016.06.010. [DOI] [Google Scholar]
- Wang J, Li J, Liu Z, et al. Construction of a high-density genetic map and its application for QTL mapping of leaflet shapes in mung bean (Vigna radiata L.) Front Genet. 2020;11:1032. doi: 10.3389/fgene.2020.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Zhang L et al (2011) Users' Manual of QTL IciMapping (Institution of Crop Science, Chinese Academy of Agricultural Sciences (CAAS) and Crop Research Informatics Lab, International Maize and Wheat Improvement Center (CIMMYT))
- Weller JL, Ortega R. Genetic control of flowering time in legumes. Front Plant Sci. 2015;6:207. doi: 10.3389/fpls.2015.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg AHJ, van der Lee T, Cayla C, et al. Validation of the high-throughput marker technology DArT using the model plant Arabidopsis thaliana. Mol Genet Genom. 2005;274:30–39. doi: 10.1007/s00438-005-1145-6. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bhat PR, Close TJ, Lonardi S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008;4:e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe JM, Poehlman JM. Genetic variability in the mungbean, Vigna radiata (L.) Wilczek. Crop Sci. 1972;12:461–464. doi: 10.2135/cropsci1972.0011183X001200040019x. [DOI] [Google Scholar]
- Young ND, Danesh D, Menancio-Hautea D. Mapping oligogenic resistance to powdery mildew in mungbean with RFLPs. Theoret Appl Genetics. 1993;87:243–249. doi: 10.1007/BF00223772. [DOI] [PubMed] [Google Scholar]
- Zhao D, Cheng X-Z, Wang L-X, et al. Construction of mungbean genetic linkage map. Acta Agron Sin. 2010;36:932–939. doi: 10.1016/S1875-2780(09)60054-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.