Abstract
The aim of this work was to evaluate if rivers could be used for SARS-CoV-2 surveillance. Five sampling points from three rivers (AR-1 and AR-2 in Arenales River, MR-1 and MR-2 in Mojotoro River, and CR in La Caldera River) from Salta (Argentina), two of them receiving discharges from wastewater plants (WWTP), were monitored from July to December 2020. Fifteen water samples from each point (75 in total) were collected and characterized physico-chemically and microbiologically and SARS-CoV-2 was quantified by RT-qPCR. Also, two targets linked to human contributions, human polyomavirus (HPyV) and RNase P, were quantified and used to normalize SARS-CoV-2 concentration, which was compared to reported COVID-19 cases. Statistical analyses allowed us to verify the correlation between SARS-CoV-2 and the concentration of fecal indicator bacteria (FIB), as well as to find similarities and differences between sampling points. La Caldera River showed the best water quality; FIBs were within acceptable limits for recreational activities. Mojotoro River's water quality was not affected by the northern WWTP of the city. Instead, Arenales River presented the poorest water quality; at AR-2 was negatively affected by the discharges of the southern WWTP, which contributed to significant increase of fecal contamination. SARS-CoV-2 was found in about half of samples in low concentrations in La Caldera and Mojotoro Rivers, while it was high and persistent in Arenales River. No human tracers were detected in CR, only HPyV was found in MR-1, MR-2 and AR-1, and both were quantified in AR-2. The experimental and normalized viral concentrations strongly correlated with reported COVID-19 cases; thus, Arenales River at AR-2 reflected the epidemiological situation of the city. This is the first study showing the dynamic of SARS-CoV-2 concentration in an urban river highly impacted by wastewater and proved that can be used for SARS-CoV-2 surveillance to support health authorities.
Keywords: SARS-CoV-2, Water quality, Human tracers, Normalization, Rivers impacted by wastewater, Epidemiological surveillance
Graphical abstract
1. Introduction
The Coronaviridae family includes a broad spectrum of viruses that cause infections in various vertebrate animals including humans. Human Coronavirus are generally responsible for common colds. However, severe acute respiratory syndrome coronavirus 1 (SARS-CoV1) and Middle East respiratory syndrome coronavirus (MERS-CoV) were responsible for global epidemics, with high rates of morbidity and mortality, in 2003 and 2012, respectively (Peiris et al., 2003; Zaki et al., 2012). On 31 December 2019, the World Health Organization was informed about pneumonia cases of unknown origin in Wuhan, China. On 7 January 2020 the new virus, called 2019-nCoV at that moment, was identified as the cause and on 11 February 2020, it was named as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Virus Taxonomy (Gorbalenya et al., 2020). The virus spread rapidly among the population in the globe and the pandemic was declared in mid-March 2020. Since then, it has infected almost 550 million people and there are >6.3 million confirmed deaths by July 3rd, 2022 (WHO, 2022).
SARS-CoV-2 is an enveloped single-stranded positive-sense RNA virus with an approximate diameter of 80 to 120 nm. The main transmission pathway is person-to-person, mainly airborne by aerosols, droplets, and sputum, formed during breathing, sneezing, coughing, or talking (Zhang et al., 2020a). Another way of transmission described initially was by contact with contaminated fomites; however, this hypothesis lost strength and is yet under discussion (Harrison et al., 2020; Kampf et al., 2020; Mukhra et al., 2020).
Although the 2019-nCoV infection (COVID-19) is mainly a respiratory disease, SARS-CoV-2 also replicates in the enterocytes from ileum and colon (Zhang et al., 2020b). Thus, a fraction of infected people, symptomatic or asymptomatic, excrete the virus in feces (Xiao et al., 2020b; Zhang et al., 2020c; Zhang et al., 2020d) and urine (Guan et al., 2020; Jeong et al., 2020) in variable concentrations. There is still controversy about the infectivity of the virus in feces and wastewater. Despite their efforts, Wölfel et al. (2020) were not able to isolate infective viruses from stool samples; conversely, Xiao et al. (2020a) were successful and propagated them in Vero E6 cells. In another study the virus was isolated from urine (Sun et al., 2020). The fact that viral particles remain infective after being excreted through fecal matter or urine, supports the hypothesis of a possible fecal-oral (Amirian, 2020; Jiang et al., 2020; Park et al., 2020) or a fecal-respiratory - by inhalation of aerosols - (Xiao et al., 2020a) route of transmission, although they have not been fully established yet.
The viral circulation in the population of a city, or a region, is reflected in the viral loads in their wastewater (Medema et al., 2020a; Randazzo et al., 2020a). In fact, the so-called wastewater-based epidemiology has been applied in many developed countries (Ahmed et al., 2020; Lodder and de Roda Husman, 2020; Betancourt et al., 2021; Gonzalez et al., 2020; Albastaki et al., 2021; Saththasivam et al., 2021) to help authorities to better understand the situation and to make decisions regarding lockdowns. In most places, surveillance has been done in wastewater treatment plants, analyzing therefore, samples that represent the population of a city or part of it (Ahmed et al., 2020; Gonzalez et al., 2020; Kumar et al., 2020; Randazzo et al., 2020b; Rimoldi et al., 2020; Wurtzer et al., 2020).
Rivers, as well as other water bodies, usually receive the discharges of treated wastewater (although sometimes with insufficient treatment) and, on some occasions, also (illegal) discharges of raw sewage. In this way, surface water can be impacted by wastewater as well as by non-point source fecal contamination. At the same time many of those aquatic environments are used as source water, for the irrigation of vegetables or for recreational purposes (Chávez-Díaz et al., 2020; Gutiérrez Cacciabue et al., 2014; Poma et al., 2012). Despite this, there is little evidence about the presence of SARS-CoV-2 in freshwater and the information reported in most of the studies is limited (Haramoto et al., 2020; La Rosa et al., 2020; Rimoldi et al., 2020; Kumar et al., 2020; Mahlknecht et al., 2021).
Previous studies about the microbiological quality of surface water in Salta, northwest of Argentina, demonstrated that fecal contamination was considerable in some rivers (Chávez-Díaz et al., 2020; Cruz et al., 2012; Gutiérrez Cacciabue et al., 2014), and multiple pathogens, including bacteria, enteric virus, and parasites, were found (Pisano et al., 2018; Poma et al., 2012; Prez et al., 2020), representing a risk for the population in contact with those waters (Poma et al., 2019).
The aims of this work were to assess the water quality and the presence of SARS-CoV-2 in urban and peri-urban rivers used for recreational purposes, and to evaluate if rivers could be used for SARS-CoV-2 surveillance to support health authorities. Three rivers from the province of Salta (northwest of Argentina), two of them receiving the discharges of the two wastewater plants of the city of Salta (main city in the province, 700,000 inhabitants), were monitored from July to December 2020 during the first wave of COVID-19 in Argentina. Water samples were physico-chemically and microbiologically characterized, SARS-CoV-2 was quantitatively detected, and all these information was used to perform statistical tests to assess for correlations between variables. In addition, two targets linked to human contributions were quantified and used to normalize the SARS-CoV-2 concentration, which was ultimately compared to the reported COVID-19 cases.
2. Materials and methods
2.1. Sampling sites
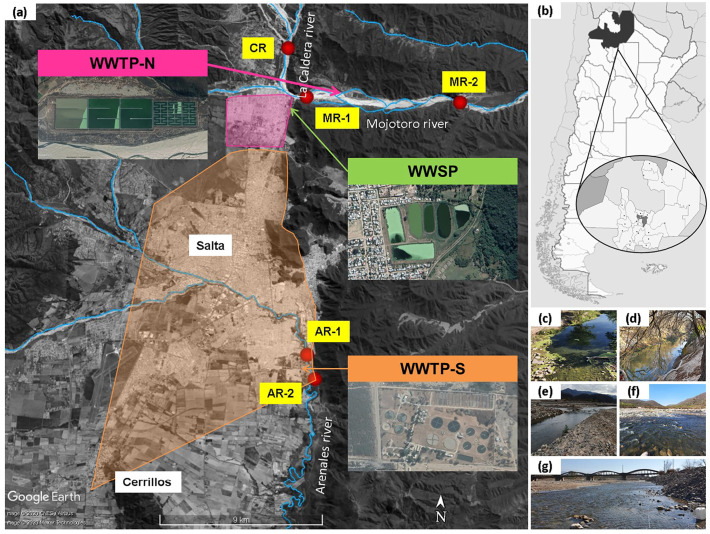
Three rivers from the province of Salta, northwest of Argentina, were selected for the study: Arenales, Mojotoro, and La Caldera Rivers (Fig. 1 ). The first two are impacted by the discharges of the two wastewater treatments plants (WWTP) of the city of Salta (capital of the province, with 700,000 inhabitants) and vicinities, while the third one is used as source water upstream and as a recreational ambient downstream.
Fig. 1.
Geographical representation of the sampling sites (a) in Salta city and vicinities at the Province of Salta, northwest of Argentina (b). Three rivers were monitored: Arenales River before (AR-1) and after (AR-2) the discharge of the southern wastewater treatment plant (WWTP-S); Mojotoro River before (MR-1) and after (MR-2) the discharge of the northern wastewater treatment plant (WWTP-N), and La Caldera River (CR). An old wastewater stabilization pond (WWSP), out of use, located between CR and MR-1is also indicated. Shaded pink area corresponds to the coverage of WWTP-N, while shaded orange area corresponds to the coverage of WWTP-S. Illustrative photographs of the sampling points are shown: AR-1 (c), AR-2 (d), MR-1 (e), MR-2 (f), and CR (g).
The Arenales River (AR) is the main river that crosses the city of Salta, from west to southeast, that belongs to the Juramento-Salado watershed in the province of Salta, northwest of Argentina. While passing across the city, the river receives stormwater, industrial effluents, and illegal raw sewage (Poma et al., 2012; Lomniczi et al., 2007). Two sampling points were selected (Fig. 1a): AR-1 (Fig. 1c) and AR-2 (Fig. 1d) located 0.7 km upstream and 0.7 km downstream the southern wastewater treatment plant (WWTP-S; serving approximately 615,000 people) and the municipal landfill, respectively. Despite the chemical and microbiological contamination of this river (Lomniczi et al., 2007; Poma et al., 2012; Prez et al., 2020), it is used for domestic irrigation of vegetables and for recreational purposes.
La Caldera River (CR) is located 13 km north from the city of Salta. It is used as source water and for recreational activities. Indeed, it is the most visited river in Salta city's vicinities because of its landscapes and accessibility. One sampling site (CR) in a recreational area was selected for this study (Fig. 1e).
The Mojotoro River (MR) is originated from the convergence of La Caldera and Vaqueros Rivers (Fig. 1a). It represents a natural border between Salta city and vicinities, going from urban to semi-rural areas, and it is mainly used for recreation and gravel extraction. Two sampling points, 10 km away from each other, were selected for monitoring: MR-1 (Fig. 1a, e) and MR-2 (Fig. 1a, f), located 5 km before and 5 km after the discharge of the northern wastewater treatment plant (WWTP-N; serving approximately 85,000 people) (Fig. 1a), respectively. There is also an old, out of use, wastewater stabilization pond (WWSP) located 3.5 km after CR and 0.7 km before MR-1 (Fig. 1a).
2.2. Sampling and characterization of water samples
A total of 15 monitoring campaigns were carried out weekly initially and then every other week, from the 13 July to 28 December 2020. Water samples were characterized physico-chemical and microbiologically. Physico-chemical variables: pH, temperature (°C), turbidity (NTU), and conductivity (mS/cm), were measured in situ using a multiparametric probe U10 Horiba (Japan). For microbiological characterization, one-liter water samples were collected in sterile bottles and refrigerated until fecal indicator bacteria (FIB) were analyzed in the laboratory. Also 20-liter water samples were collected in clean and rinsed plastic carboys at each point and brought to the laboratory for immediate concentration and molecular analysis.
Total (TC) and thermotolerant (TTC) coliforms were determined by the multiple-tube fermentation method, culturing them on MacConkey broth (Britania, Argentina) at 37 and 44.5 °C for 48 h, respectively (Eaton et al., 2005). Escherichia coli (EC) and enterococci (EN) were enumerated through the Membrane Filter technique (Eaton et al., 2005). E. coli was cultured in modified mTEC Agar (Fluka, USA) at 35 °C for 2 h and 44.5 °C for 22 h (Method 1603, USEPA, 2014), and enterococci in mE Agar (Difco, USA) at 41 °C for 48 h and confirmation in Esculin-Iron Agar at 41 °C for 20 min (Method 1106.1, USEPA, 2002). Results for TC and TTC were expressed as Most Probable Number (MPN) per 100 mL, while for EC and EN they were expressed as Colony Forming Unit (CFU) per 100 mL.
2.3. Water concentration and nucleic acid extraction
Twenty-liter water samples were concentrated down to 80 mL by ultrafiltration using a disposable hemodialysis hollow fiber cartridge (FX Classix 100, Fresenius Medical Care, Germany), following a previously optimized and validated protocol for the detection of enteric viruses (Poma et al., 2012; Rajal et al., 2007). A fraction of the final concentrated water sample (140 μL) was employed for nucleic acid extraction using a commercial kit (Puro Virus RNA, Productos Bio-Lógicos, Argentina), producing 50 μL of extract which was used for the quantitative detection of SARS-CoV-2, Polyomavirus (HPyV) and Ribonuclease P (RNase P).
2.4. Quantitative detection of SARS-CoV-2
The detection of SARS-CoV-2 was performed on the RNA extracts from each concentrated water sample by RT-qPCR based on the detection of a region of the N gene (N1 system) validated by the US Centers for Disease Control and Prevention (CDC, 2020). The determination and quantification of the gene N was performed in a StepOne Plus real time PCR system (Applied Biosystem). The final 20 μL reaction volume contained 5 μL of TaqMan Fast virus 1-Step Master Mix 4X (Applied Biosystems), primers and probe solutions, water (to complete 15 μL), and 5 μL of template. The final concentrations of forward and reverse primers were 400 nM each while it was 250 nM for the probe. The RT-qPCR was initiated with reverse transcription at 50 °C for 5 min followed by a step of 95 °C for 20 s and 45 cycles of 95 °C for 15 s and 59 °C for 1 min.
Dilutions of the RNA extract were performed when needed to overcome inhibition according to Rajal et al. (2007). All qPCR reactions were performed in duplicate, simultaneously with a non-template control (MiliQ water) and DNA plasmid (nCoV-ALL-Control Plasmid, Eurofins) as positive control. A protocol for rapid digestion with EcoR1 (Promega, USA) was used to linearize the plasmid, following the manufacturer's instructions.
The standard curve for SARS-CoV-2 quantification (y = −3.4700x + 43.1616; R2 = 0.9820; amplification efficiency: 94 %; dynamic range: 8) was performed with eight 10-fold serial dilutions, in triplicate, of the DNA plasmid that contains the interest target.
2.5. Normalization with human polyomaviruses (HPyV) and ribonuclease P (RNase P) as human input indicators
The viral concentration was expected to show variations along time. These variations could be due to changing viral discharges into the rivers but also due to fluctuations in the flow rate, i.e. because of stormwater. Thus, to accurately assess the evolution of SARS-CoV-2 concentration, without the impact of external factors, two human indicators (HPyV and RNaseP) were used as normalizers.
Human JC (JCPyV) and BK (BKPyV) polyomaviruses were quantitatively detected by qPCR using the oligonucleotides and reaction conditions described by Barrios et al. (2018). On the other hand, a fragment of the gene encoding for the human Ribonuclease P (RNase P) was quantitatively detected by qPCR using the system described by the US Centers for Disease Control and Prevention (CDC, 2020).
The final volume of each qPCR reaction contained 15 μL of reaction mix containing SensiFAST Probe HiRox kit 2X (Bioline) and specific primers and probes to reach the final concentrations established (a summary of all the oligonucleotides used for all the targets, including those for the detection of SARS-CoV-2 is shown in Table 1 ) and 5 μL of template. Cycling parameters were 95 °C for 5 min for polymerase activation and 45 cycles of 95 °C for 10 s for denaturation and 60 °C for 50 s for annealing/extension.
Table 1.
Oligonucleotides: forward (F) and reverse (R) primers and Taq-Man® probes (P) used for the detection of SARS-CoV-2 N gene (CDC, 2020), HPyV (Barrios et al., 2018) and RNase P gene (CDC, 2020).
| Target | Oligonucleotide | Sequence 5′ - 3′ | Final concentration (nM) |
|---|---|---|---|
| N1 system of SARS-CoV-2 gene N | 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | 400 |
| 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | 400 | |
| 2019-nCoV_N1-P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | 250 | |
| HPyV JC-BK |
S1-JC-BK (F) | CTGCTGCTGCCACAGGATT | 500 |
| AS2-JC-BK (R) | CCTCTACAGTAGCAAGGGATGCA | 500 | |
| P-JC-BK | FAM-AGCAGCAGCCTCYCCAGCAG CAATTTCAGC-BHQ | 250 | |
| RNase P | RP-F | AGATTTGGACCTGCGAGCG | 500 |
| RP-R | GAGCGGCTGTCTCCACAAGT | 500 | |
| RP-P | HEX-TTCTGACCTGAAGGCTCTGCGCG–BHQ-1 | 250 |
Standard curves for HPyV (y = −3.656x + 39.317; R2 = 0.996; amplification efficiency: 88 %; dynamic range 8) and RNase P (−3.71x + 42.8280; R2 = 0.9991; dynamic range 7) markers, were built using 10-fold dilutions of the corresponding plasmid DNA, containing the target sequences, in triplicate.
All the extracted nucleic acids from the concentrated water samples were analyzed for HPyV and RNaseP, by duplicate, using positive (plasmid) and negative (ultrapure water) controls. These results were used for the normalization of the SARS-CoV-2 concentrations; three alternative normalizations were assessed, as follows:
Alternative 1:
| (1) |
Alternative 2:
| (2) |
Alternative 3:
| (3) |
where C (gc/mL) is the concentration and subindexes CoV (as for SARS-CoV-2), HPyV and RNaseP indicate the targets; C a (gc/mL) is the average concentration and C N (gc/mL) is the normalized concentration. The super index (i) (i = 1, 2, 3) indicates the normalization alternative.
2.6. COVID-19 cases
Reported COVID-19 cases for the city of Salta were compiled and the prior two-weeks accumulated amount were assigned to each monitoring date to compare the epidemiological curve to the evolution of the viral concentration in the Arenales River. SARS-CoV-2 is shed by infected people for a variable period between one and three weeks (Wu et al., 2020a). Thus, considering that the excretion pattern is not constant and that when the excreted viral concentration is decreasing for some individuals it will be increasing for some others compensating such variations, we accounted for the two-weeks accumulated cases as an average regarding the viral contribution to wastewater, and also to synchronize this with the monitoring dates. The reason for choosing this river was that it crosses the city receiving the impact of various human activities (domestic and industrial) and also, it receives the discharges of the main wastewater treatment plant (WWTP-S) (covers 85 % of the population), which is outdated and not sufficient for treating the volume of a fast-growing city.
2.7. Statistical analyses
Physico-chemical variables (temperature, pH, conductivity, and turbidity) measured along the monitoring campaigns were compiled to show descriptive statistics: mean and standard deviation, median and range of variability (minimum–maximum).
To assess data variability during the monitoring campaign within each river and regarding the impact due to SARS-CoV-2, different statistical tests were run.
Most analyses were carried out using non-parametric tests, due to the non-normal distribution of the data (p < 0.001). Only the correlation between concentrations of HPyV and RNase P was carried out with a parametric test (Pearson) as both datasets showed normal distribution. Normality was tested by the Shapiro–Wilks W-test (Shapiro and Francia, 1972).
Kruskal-Wallis test (non-parametric) was performed with the physico-chemical and microbiological variables measured to compare all the sampling points. Different letters were assigned when there were significant differences within the variables between the sampling points with p-values < 0.05.
Correlations between all the physico-chemical variables and fecal indicator bacteria (describing water quality) and SARS-CoV-2 (as a descriptor of the level of viral circulation in the population) concentrations measured for all the sampling points were assessed by Spearman test (non-parametric). This test was also applied to all the same variables for each sampling point. Correlations were considered significant when the probabilities (p) were lower than 0.05 and they were arbitrary considered weak when coefficients were lower than 0.40, moderate when were between 0.40 and 0.75, and strong when were higher than 0.75.
A Cluster Analysis (CA) and a Linear Discriminant Analysis (DA) were applied using nine variables (TC, TTC, EC, EN, SARS-CoV-2, T, conductivity, pH and turbidity) to assess the behavior of the data in a multivariate sense. The CA aimed to search similarities between sampling points. This multivariate statistical test was performed by the Ward Method. Euclidean distances were calculated for each case. The DA was applied to distinguish between sampling points. The goal is to discriminate two or more groups of objects by maximizing their differences (Di Rienzo et al., 2016). All the variables were standardized before the multivariate performance, to eliminate the effect of scale.
Statistical analyses were run with the InfoStat software (Di Rienzo et al., 2016) and RStudio version 4.0.0 (Ihaka and Gentleman, 1996; R Development Core Team, 2005; RStudio Team, 2020; Wickham, 2009).
Finally, the parametric Pearson's test was performed to evaluate is there was correlation between the concentrations of HPyV and RNaseP. In addition, Spearman test was performed to assess for correlation between the evolution of the concentration of SARS-CoV-2 in the Arenales River, at AR-1 and AR-2, and the number of active reported cases in Salta city. Then the test was applied for the normalized SARS-CoV-2 concentration at AR-2 and the number of cases.
3. Results
A total of 15 sampling events were carried out from July to December of 2020 in three rivers, monitoring a total of five points: AR-1 and AR-2 at the Arenales River, CR at La Caldera River, and MR-1 and MR-2 at Mojotoro River.
3.1. Physico-chemical characterization of water samples
Four physico-chemical variables (temperature, pH, conductivity, and turbidity) were measured in water (Table 2 ). Water temperature (7.2–23.8 °C) was directly affected by environmental temperature (5.0–25.0 °C) (July to middle September is winter and middle September to middle December is spring in the South Hemisphere), as expected. Although the maximum temperatures were similar for all the rivers, the minimum values were higher for AR-1 and AR-2 (Arenales River is in the middle of the city) compared to those from CR, MR-1 and MR-2 which are in the vicinities of Salta, in open space and more exposed to the wind and elements.
Table 2.
Physico-chemical variables measured in surface water, along 15 sampling events, in the Arenales River at AR-1 and AR-2 (before and after the discharges of the southern wastewater treatment plant, WWTP-S, respectively; distance between AR-1 and AR-2 is 1.4 km and WWTP-S is separated 0.7 km from each), in La Caldera River (CR), and in the Mojotoro River at MR-1 (after an old wastewater stabilization pond and before the discharges of the northern wastewater treatment plant, WWTP-N) and MR-2 (after the discharges of the WWTP-N; distance between MR-1 and MR-2 is 10 km and WWTP-N is 5 km away from each). K-W: Bonferroni Post Hoc test from Kruskal Wallis. Medians with different letters are significantly different (p < 0.05).
| Variables | Sampling points | Mean ± SD | Median | Range (min–max) | K-W |
|---|---|---|---|---|---|
| Temperature (°C) | AR-1 | 17.41 ± 3.99 | 16.30 | 12.10–23.50 | AB |
| AR-2 | 19.23 ± 3.19 | 18.40 | 14.70–23.80 | A | |
| CR | 15.83 ± 4.73 | 14.30 | 7.20–23.80 | C | |
| MR-1 | 15.69 ± 4.18 | 15.00 | 8.00–22.50 | A | |
| MR-2 | 16.28 ± 4.88 | 16.00 | 7.80–23.80 | BC | |
| pH | AR-1 | 8.07 ± 0.27 | 8.12 | 7.64–8.54 | E |
| AR-2 | 7.90 ± 0.12 | 7.91 | 7.70–8.09 | E | |
| CR | 8.45 ± 0.28 | 8.46 | 7.89–8.82 | D | |
| MR-1 | 7.96 ± 0.40 | 7.94 | 6.99–8.90 | D | |
| MR-2 | 8.24 ± 0.32 | 8.24 | 7.32–8.63 | D | |
| Conductivity (mS/cm) | AR-1 | 0.46 ± 0.07 | 0.48 | 0.31–0.54 | G |
| AR-2 | 0.58 ± 0.07 | 0.59 | 0.41–0.74 | G | |
| CR | 0.21 ± 0.03 | 0.20 | 0.17–0.27 | F | |
| MR-1 | 0.24 ± 0.01 | 0.24 | 0.21–0.27 | F | |
| MR-2 | 0.25 ± 0.02 | 0.24 | 0.20–0.28 | F | |
| Turbidity (NTU) | AR-1 | 17.07 ± 32.51 | 8.00 | 4.00–134.00 | H |
| AR-2 | 88.27 ± 28.44 | 80.00 | 55.00–159.00 | J | |
| CR | 16.07 ± 13.78 | 13.00 | 5.00–56.00 | HI | |
| MR-1 | 11.47 ± 5.57 | 10.00 | 5.00–26.00 | HI | |
| MR-2 | 16.27 ± 5.36 | 16.00 | 5.00–26.00 | I |
Kruskal-Wallis test was performed with physico-chemical variables measured in the five sampling points analyzed. The sampling points CR, MR-1 and MR-2 were similar except for the temperature that was different in MR-1. The two monitoring points at the Arenales River (AR-1 and AR-2; distance was 1.4 km, the WWTP-S was 0.7 km away from each) only showed significant differences in the turbidity, which was much higher in AR-2, probably due to the discharges of the WWTP-S, also reported in a previous work (Poma et al., 2012).
3.2. Bacterial characterization
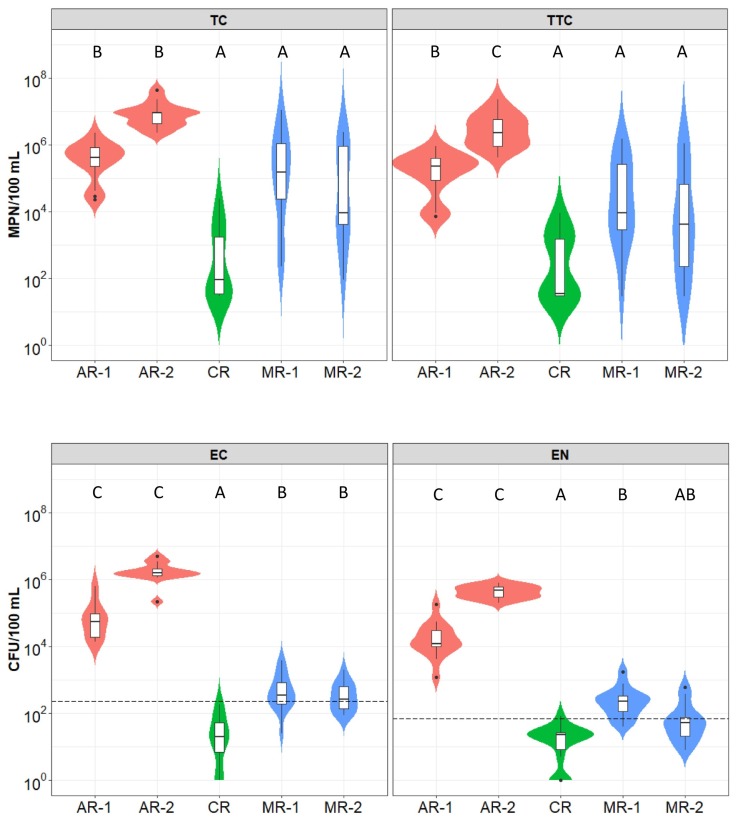
The highest concentrations of all FIBs were found in the Arenales River, followed by Mojotoro River and La Caldera River (Fig. 2 ). In general, variability in bacteria concentrations (within two orders of magnitude) was lower in AR-1 and AR-2 than in the other sampling points (Fig. 2). Conversely, the concentrations of total and thermotolerant coliforms were highly variable for MR-1, MR-2, and CR.
Fig. 2.
Violin plots representing indicator bacteria determined in 15 monitoring events in five different sampling points (75 samples in total): AR-1 and AR-2 in the Arenales River (before and after the discharges of the southern wastewater treatment plant, respectively), CR in La Caldera River, and MR-1 and MR-2 in the Mojotoro River (before and after the discharges of the northern wastewater treatment plant, respectively). The concentrations of total (TC) and thermotolerant coliforms (TTC) are expressed as the Most Probable Number per 100 mL, while those of E. coli (EC) and Enterococcus sp. (EN) are in Colony Forming Units (CFU) per 100 mL. Violin shapes show the data kernel probability with the boxplots embedded. Boxplots show the interquartile range (IQR) in boxes divided by median values (horizontal lines) with whiskers depicting ±1.5 IQR and outliers as points. The segmented horizontal lines represent the limit values accepted by legislation for recreational water (USEPA, 2012). Bonferroni Post Hoc test from Kruskal Wallis was performed. Medians with different letters for each variable are significantly different (p < 0.05).
Bacteria concentrations in Arenales River exceeded 104 CFU or MPN per 100 mL and furthermore, they surpassed the acceptable limits for E. coli and enterococci (235 CFU/100 mL and 71 CFU/100 mL, respectively, USEPA, 2012) in all the samples analyzed. The point AR-2, 700 m downstream from AR-1, was highly impacted by fecal contamination of untreated or insufficiently treated wastewater effluents from the WWTP-S (p < 0.05 for all the FBI) (Fig. 2).
Regarding Mojotoro River, bacteria concentrations were similar in both sampling sites (Fig. 2) showing that the effluents of the WWTP-N have no impact on the microbial quality of water. However, the concentration of E. coli and enterococci exceeded the recreational limits in most of the samples (67 and 80 % for MR-1, respectively, and 53 and 33 % for MR-2, respectively). In contrast, La Caldera River showed the best water quality, appropriate for recreational activities. In fact, all the samples analyzed (15 in total) were within the accepted values for E. coli; however, one of them exceeded the limit for enterococci.
3.3. Quantification of SARS-CoV-2, HPyV and RNase P
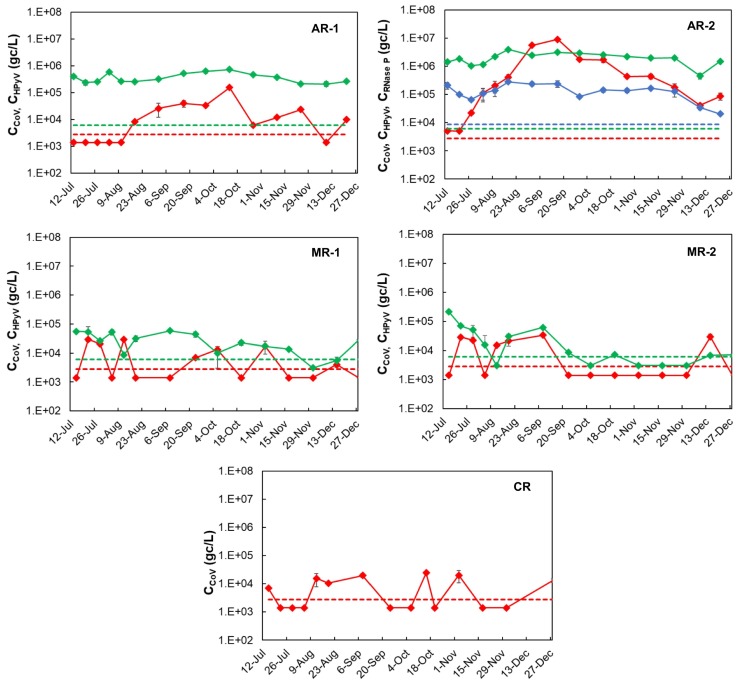
All the samples (75 in total) collected during the 15 sampling events (from July to December 2020, during the first wave of COVID-19) at five different points in three rivers, were concentrated, the nucleic acids were extracted, and then analyzed for SARS-CoV-2, HPyV and RNase P (Fig. 3 ). In La Caldera River, from the three targets evaluated only SARS-CoV-2 was found in around half of the samples (53.3 % samples were non-detects) and in those cases the viral concentrations were lower than 105 gc/L. The same occurred in the Mojotoro River where 7 and 6 out of 15 (46.6 and 40.0 %, respectively) samples were non detects at MR-1 and MR-2, respectively. However, HPyV was also detected in that river in both sampling points, in 14 and in 10 (out of 15) samples at MR-1 and MR-2, respectively. The HPyV was detected in all the samples in Arenales River, in both sampling points, showing the impact of human contamination. This effect was also seen through the detection of RNase P that was only found in AR-2 samples. While SARS-CoV-2 was not detected during the first five monitoring campaigns in AR-1, it was quantified in all samples collected at AR-2. The two targets selected for tracing human contamination showed a similar behavior (coefficient was 0.7228; p = 0.0021; positive strong correlation) along time. However, the concentrations of HPyV were around one order of magnitude higher than those of RNase P, which may be the reason for RNase P not being found consistently in other sites (average ratio HPyV/RNaseP was 15.8 ± 6.4, without considering one outlier on 27 July, where the ratio was 299.3).
Fig. 3.
Evolution of SARS-CoV-2 (CCoV, red solid line), human polyomavirus (CHPyV, green solid line), and RNase P (CRNase P, blue solid line) concentrations, determined in five sampling points: AR-1 and AR-2 in Arenales River, MR-1 and MR-2 in Mojotoro River, and CR in La Caldera River, from July to December 2020 (15 sampling events). Dashed lines represent the limit of detection (LOD) for SARS-CoV-2 (red), HPyV (green) and RNase P (blue). Non detects were arbitrarily represented as LOD/2. Values plotted are the average of duplicates and error bars correspond to the standard deviation (some bars are too small to be seen). Concentrations are expressed in gene copies per liter (gc/L).
3.4. Relationship between water quality and SARS-CoV-2 concentration
Spearman correlation was evaluated using all the physico-chemical, FIBs and SARS-CoV-2 concentration data, including all the sampling points (Supplementary Information, Table S1). Temperature was the only variable that did not show any correlation. Positive strong correlations were found between pH and COND, TURB and FIBs, and among FIBs. Regarding CoV, also positive strong correlations were confirmed with pH and COND, while they were moderate with TURB and all FIBs (Supplementary Information, Table 1).
When Spearman correlation was performed for each sampling point instead, positive strong or moderate correlations were found (p < 0.05) between COND and pH (and with temperature for MR-2 only) and also between TC, TTC and EC for all the sampling points (Table 3 ). In all cases, except for MR-2, there were positive moderate correlations (p < 0.05) between TURB and at least two FIBs. Only for AR-1 and AR-2 positive moderate correlations (p < 0.05) were found between CoV and pH, CoV and TURB, and EN and another FIB (Table 3).
Table 3.
Coefficients and probabilities (below and above the main diagonal, respectively) from Spearman correlation test using the physico-chemical variables, fecal indicator bacteria and SARS-CoV-2 concentration (CoV) for each sampling point (SP): AR-1 and AR-2 in Arenales River; CR in La Caldera River; MR-1 and MR-2 in Mojotoro River. Only significant correlations (p < 0.05) are shown and those that are strong are indicated in bold. Physico-chemical variables: temperature (T), pH, conductivity (COND), turbidity (TURB). Fecal indicator bacteria: total coliforms (TC), thermotolerant coliforms (TTC), E. coli (EC), enterococci (EN).
| SP | T | pH | COND | TURB | TC | TTC | EC | EN | CoV | |
|---|---|---|---|---|---|---|---|---|---|---|
| AR-1 | T | |||||||||
| pH | 1.8 × 10−4 | 4.6 × 10−3 | ||||||||
| COND | 1 | 4.6 × 10−3 | ||||||||
| TURB | 0.01 | |||||||||
| TC | 0.04 | 0.04 | ||||||||
| TTC | 0.53 | |||||||||
| EC | 0.65 | 0.54 | 0.03 | |||||||
| EN | 0.57 | |||||||||
| CoV | 0.69 | 0.69 | ||||||||
| AR-2 | T | 0.01 | ||||||||
| pH | 1.8 × 10−4 | 2.3 × 10−3 | ||||||||
| COND | 1 | 2.3 × 10−3 | ||||||||
| TURB | 0.05 | |||||||||
| TC | 0.02 | |||||||||
| TTC | 3.7 × 10−3 | |||||||||
| EC | 0.70 | |||||||||
| EN | 0.66 | 0.51 | 0.59 | |||||||
| CoV | 0.81 | 0.81 | ||||||||
| CR | T | |||||||||
| pH | 1.8 × 10−4 | |||||||||
| COND | 1 | |||||||||
| TURB | 0.01 | 0.04 | ||||||||
| TC | 0.63 | 1.1 × 10−6 | 0.02 | |||||||
| TTC | 0.55 | 0.92 | 4.7 × 10−3 | |||||||
| EC | 0.59 | 0.69 | ||||||||
| EN | ||||||||||
| CoV | ||||||||||
| MR-1 | T | |||||||||
| pH | 0 | |||||||||
| COND | 1 | |||||||||
| TURB | 0.01 | 0.02 | 0.05 | |||||||
| TC | 0.67 | 7.3 × 10−4 | 1.2 × 10−4 | |||||||
| TTC | 0.60 | 0.77 | 0.01 | |||||||
| EC | 0.51 | 0.83 | 0.64 | |||||||
| EN | ||||||||||
| CoV | ||||||||||
| MR-2 | T | 0.01 | 0.01 | |||||||
| pH | −0.66 | 0 | ||||||||
| COND | −0.66 | 1 | ||||||||
| TURB | ||||||||||
| TC | 9.4 × 10−6 | 1.7 × 10−3 | ||||||||
| TTC | 0.89 | 2.0 × 10−3 | ||||||||
| EC | 0.74 | 0.73 | ||||||||
| EN | ||||||||||
| CoV |
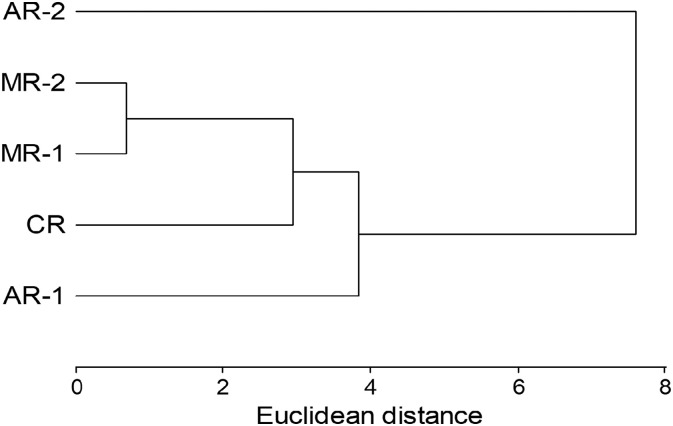
Cluster analysis including all the variables measured for a total of 75 water samples from five sampling points was performed and the Euclidean distances were determined (cophenetic correlation was 0.906) (Fig. 4 ). The most similar sampling points were MR-1 and MR-2 (Euclidean distance was 0.69), verifying once again that the Mojotoro River was not negatively impacted by the discharges of the WWTP-N. Conversely, the most different sampling points were CR and AR-2 (Euclidean distance was 6.99), in agreement with other observations, that CR was the point with the best water quality and AR-2 the most impacted one by the discharges of the WWTP-S.
Fig. 4.
Cluster analysis performed with nine variables (temperature, pH, conductivity, turbidity, and concentrations of total and thermotolerant coliforms, E. coli, enterococci and SARS-CoV-2) measured in a total of 75 water samples collected in five sampling points: AR-1 and AR-2 in the Arenales River, MR-1 and MR-2 in the Mojotoro River, and CR in La Caldera River, from July to December 2020 (15 sampling events).
The discriminant analysis performed with nine the variables measured in 75 water samples along 15 sampling events showed two main groups (Fig. 5 ). At the right side there were those samples from the Arenales River, where AR-1 was also discriminated from AR-2, due to the high microbial contamination. The second group, at the left side, included the samples from Mojotoro River, with MR-1 and MR-2 mingled together, slightly separated from those from Caldera River, which was the one with best microbial water quality.
Fig. 5.
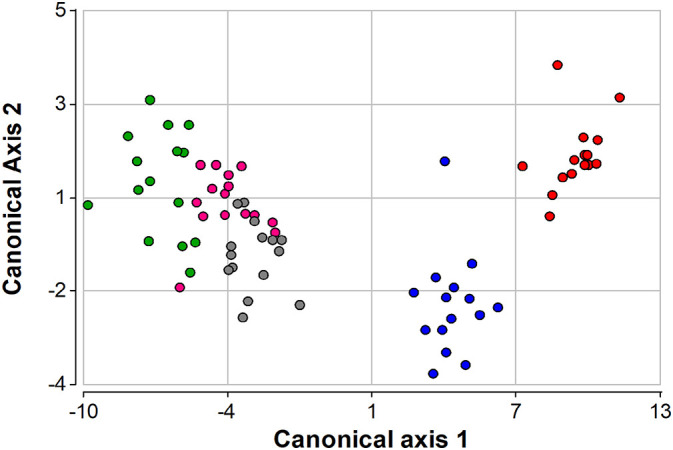
Plots of the first two axes (with eigenvalues higher than 1, explaining 97.7 % of the accumulated variance) obtained with a linear discriminant analysis conducted with nine variables: temperature, pH, conductivity, turbidity, and concentrations of total coliforms, thermotolerant coliforms, E. coli, enterococci, and SARS-CoV-2. A total of 75 water samples were collected in five sampling points: AR-1 (red circles) and AR-2 (blue circles) in the Arenales River, MR-1 (gray circles) and MR-2 (pink circles) in the Mojotoro River, and CR (green circles) in La Caldera River, from July to December 2020 (15 sampling events).
3.5. Comparison between SARS-CoV-2 concentration and reported COVID-19 cases
The total of COVID-19 reported cases in the city of Salta, accumulated for 14 days before each monitoring campaign, were assigned to that sampling date. The evolution of the number of cases in Salta city was confronted with the SARS-CoV-2 concentration found in the Arenales River (Fig. 6 ). This was not performed for the Mojotoro and La Caldera Rivers as SARS-CoV-2 was detected in around half of the samples, following no pattern, in low concentration, while the number of cases was constantly increasing.
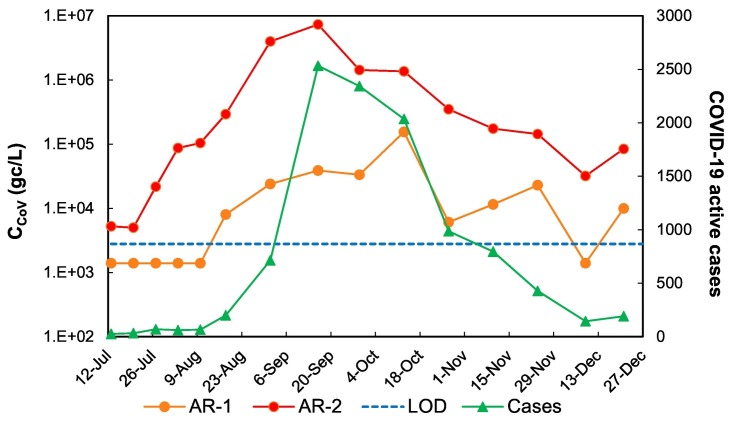
Fig. 6.
Evolution of SARS-CoV-2 concentration (CCoV) in the Arenales River at two sampling points: AR-1 and AR-2 before and after the southern wastewater treatment plant, respectively, and of the number of COVID-19 active cases accumulated for 14 days before sampling. LOD: limit of detection for SARS-CoV-2 in surface water. Non-detects were arbitrarily represented as LOD/2.
The viral concentration curve anticipated the epidemiological one. The ratio between the viral concentration and the number of active cases (14 days before) was variable, being 32 gc/mL/case the average for AR-1 and 1099 gc/mL/case the average for AR-2. The latest was a consequence of a higher fecal content in AR-2 due to the WWTP discharges in the river and was highly influenced by the extreme values. In fact, at the beginning and towards the end of the epidemiological curve the ratio was around 281 gc/mL/case while it was 2034 gc/mL/case during the two months (August–September) when the viral dissemination was higher.
Spearman correlation coefficient indicated a positive association between the number of cases and the viral concentrations in the Arenales River, at AR-1 (ρ = 0.89; p = 0.00001) and at AR-2 (ρ = 0.89; p = 0.00084).
3.6. Normalization of the SARS-CoV-2 concentration
The experimental concentration of SARS-CoV-2, although is calculated based on the detection of the gene of interest in water samples, is subject to fluctuations due to variations in the flow rate, i.e. from precipitations or from discharges of the wastewater plant. One strategy to compensate for that, thus, to help the data to get closer to the real situation, is to normalize the data, with some variable that accounts for the human input in this case. Two targets, T antigen for HPyV and RNase P, linked to human contamination were selected for that. Therefore, the concentration of SARS-CoV-2 in the Arenales River was normalized following three different alternatives, including the concentration of HPyV (Alternative 1), the concentration of RNase P (Alternative 2) and a combination of both (Alternative 3) (Fig. 7 ).
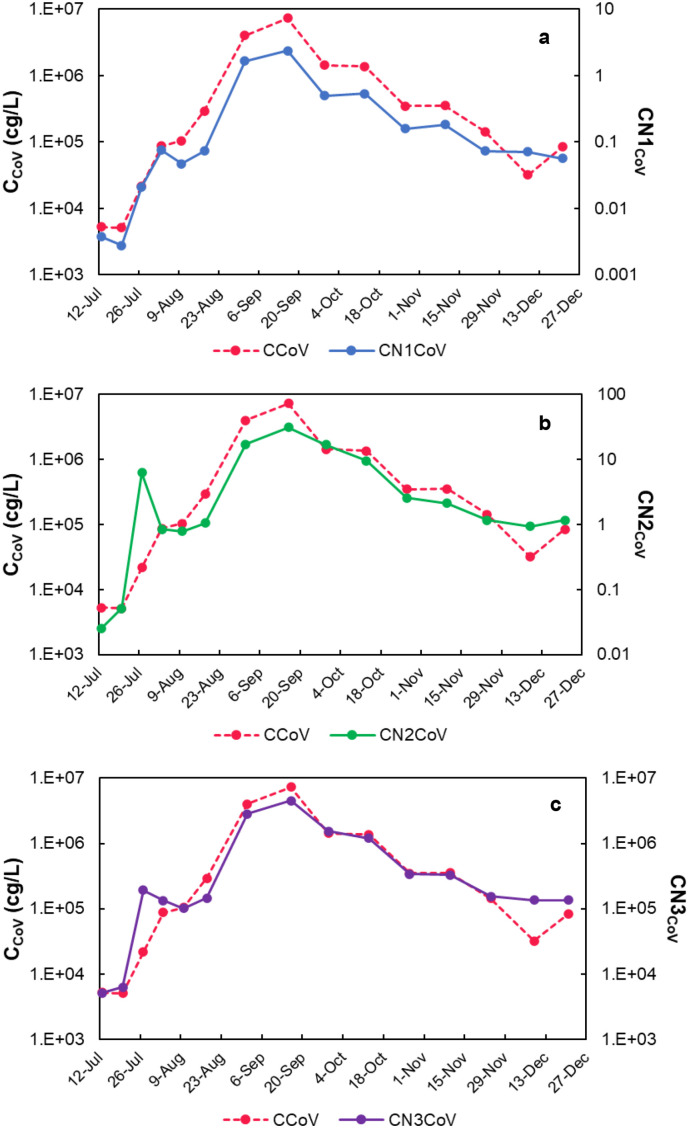
Fig. 7.
Concentration of SARS-CoV-2 experimentally determined (CCoV) at AR-2 in Arenales River and the normalized concentration using three different alternatives: (a) alternative 1 (CN1CoV), using the concentration of human polyomavirus (HPyV), according to Eq. (1), (b) alternative 2 (CN2CoV), using the concentration of RNase P, according to Eq. (2), and (c) alternative 3 (CN3CoV), using the concentration of HPyV and RNase P, according to Eq. (3).
Regarding the three alternatives for normalization of the SARS-CoV-2 concentration evaluated, the first one was as the ratio with the concentration of human polyomavirus (Eq. (1)), in which case the shape of the curve was not modified. However, the scale of the resulting number was too small, and the sense of the viral concentration magnitude was lost, and the units were (gc of SARS-CoV-2/gc of HPyV). The latest also happened when the ratio of SARS-CoV-2 was calculated with RNase P (Eq. (2)), being the units (gc of SARS-CoV-2/gc of RNase P); however, the shape of the resulting curve changed a bit due to the fluctuations of RNase P concentration. Instead, the third way of performing the normalization was using the two normalizers (HPyV and RNase P), according to Eq. (3). In this case, the shape of the experimental SARS-CoV-2 concentration remained constant, and the normalized values kept the sense of the concentration magnitude as well as the proper units. Strong positive correlations were found by Spearman test between the number of COVID-19 cases and the normalized viral concentrations at AR-2 using the three alternatives: CN1 CoV (ρ = 0.87; p = 0.0012), CN2 CoV (ρ = 0.88; p = 0.0011), and CN3 CoV (ρ = 0.93; p = 0.00054). Thus, the normalized concentration of SARS-CoV-2, obtained using both HPyV and RNase P, was selected and compared to the number of reported COVID-19 cases in the city (Fig. 8 ).
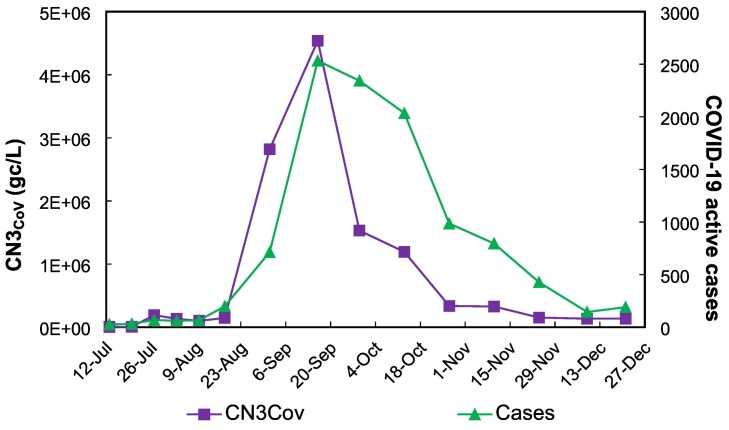
Fig. 8.
Evolution of the normalized concentration of SARS-CoV-2 (gc/L) (CN3CoV, according to Eq. (3)) in the Arenales River at AR-2 and the two-weeks accumulated reported COVID-19 cases, from the 12 July to 27 December 2020 during the first wave of COVID-19 in the city of Salta, Argentina.
4. Discussion
A dataset including experimental values for physico-chemical (four variables) and microbiological (four fecal indicator bacteria and SARS-CoV-2 concentrations) variables from a total of 75 grab water samples from five sampling points was used to perform various bi- and multi-variable analyses. These allowed us to compare among sampling points located in the same or different rivers, to evaluate correlations, and to find the most similar and different points.
The results of the monitoring from July to December 2020, allowed us to verify that from the three rivers analyzed La Caldera River has the best water quality, with low and sporadic fecal input, adequate for recreational activities, which was also described in previous work (Chávez-Díaz et al., 2020; Gutiérrez Cacciabue et al., 2014). The Mojotoro and the Arenales Rivers receive the impact of the discharges of the two wastewater treatment plants of the city, WWTP-N and WWTP-S, respectively. The WWTP-N, serving a small proportion (12 %) of the city population, is new and has capacity for treating efficiently the flow rate from the northern part of the city, thus, the discharges do not affect the water quality of the river at least between MR-1 and MR-2. Instead, the WWTP-S, serving the major proportion of the population, is an outdated treatment plant without enough capacity for a fast-growing city. The discharges of the WWTP-S significantly impacted the Arenales River, verified at AR-2, although the water quality was already poor at AR-1, 700 m upstream, due to domestic and industrial activities and to sporadic illegal discharges of raw sewage (Poma et al., 2012). The latest was evidenced by the high concentrations of fecal bacteria indicators.
Measured physicochemical and bacteriological variables gave us information about the rivers water quality at every monitoring event. Assessing for correlations between those variables and the SARS-CoV-2 concentrations allowed us to learn the behavior of the virus when entering into a particular aquatic environment with different characteristics, as it could eventually have implications for the population (these aquatic environments are used for recreation, consumption, irrigation). It is well known that the behavior (resistance, persistence, survival) of a microorganism once it reaches an aquatic environment depends on biotic and abiotic factors (that is, its quality measured by physicochemical, biological, and microbiological parameters). For example, the persistence of a microorganism in water is related to the presence of small solid particles. A high correlation between turbidity and SARS-CoV-2 may suggest a higher persistence of the virus in the environment (Gutiérrez-Cacciabue and Rajal, 2022). This could indicate that viral particles (infective or not) may sediment in the same environment (an eventual resuspension could bring the virus back to the water column becoming a risk) or be transported to other parts of the waterbody, generating a risk to people using the water there.
Although SARS-CoV-2, as an enveloped virus, was presumed to inactivate or degrade easily in water, different works have demonstrated their persistence for days and even weeks, mainly depending on abiotic factors. de Oliveira et al. (2021) found that infective SARS-CoV-2 spiked in sterile water and wastewater remained infective for a few days and the persistence time was strongly affected by temperature. Bivins et al. (2020) studied the persistence of SARS-CoV-2 in tap water and wastewater in microcosms in the laboratory verifying that the RNA persisted longer than the infective virus at 50 and 70 °C, and that was especially remarkable for high viral titers. The long persistence of the viral RNA in wastewater, especially at low temperatures, was also demonstrated by Yang et al. (2022).
Besides, there is not a direct relationship between water quality indicators and the epidemiology of the population. Knowing the water quality and its relationship with SARS-CoV-2 once it reaches the environment and the subsequent consequences (if any) that could occur there where it appears, is relevant data for modeling and for carrying out risk analysis (Kumar et al., 2020). Environmental surveillance as a tool to know about the circulation of viruses and other microorganisms causing disease in the population has been proposed for decades already. In fact, rivers that were impacted by wastewater have been monitored in multiple studies, like the analyses of circulation of human polio virus as part of the effort for eradication (Delogu et al., 2018; Iaconelli et al., 2017; WHO, 2003) and of Hepatitis A virus after vaccination campaigns (Blanco Fernández et al., 2012a), or of evolutionary relationships between norovirus (Blanco Fernández et al., 2012b).
In the case of environmental surveillance of SARS-CoV-2 almost all the efforts around the world were focused on monitoring wastewater (Medema et al., 2020a; Randazzo et al., 2020b; Wu et al., 2020b), most of the times analyzing the inlets (and sometimes also the outlets) of wastewater treatment plants (Gonzalez et al., 2020; Nasseri et al., 2021), but also at maintenance holes that are representative of defined areas of a city, or regions or neighborhoods (Larson et al., 2020). Rimoldi et al. (2020) in Italy conducted studies to assess the efficiency of wastewater treatment plants to remove SARS-CoV-2. Also, Wurtzer et al. (2020) evaluated the lockdown impact on SARS-CoV-2 dynamics in Paris wastewaters.
Wastewater samples are useful to learn about the true viral circulation in a population of geographical interest, especially to estimate the extension of virus dissemination due to asymptomatic people, that seems to play an important role in the COVID-19 pandemic (He et al., 2020). In addition, these samples present the advantages that they are anonymous and non-invasive, they do not depend on the population will (to be tested) under analysis and they can be used by the respective health authorities to make smart decisions regarding the pandemic's management (Thompson et al., 2020).
While monitoring wastewater is a useful tool to evaluate the situation in developed countries, this may not be the case in threshold or developing countries, where the sewage systems are insufficient or non-existent, or when the wastewater treatment is inefficient. In most of those places, rivers or other water bodies receive non-point source fecal contamination or the discharges of untreated (sometimes illegally) or insufficiently treated wastewater, impacting the water quality and thus, the use of the resource (Rimoldi et al., 2020). In such scenarios, surface water like natural or artificial ponds (Iglesias et al., 2020; Mahlknecht et al., 2021) or rivers could also be used as tools for assessing the epidemiological situation of the population. The first attempt in that direction was done by Guerrero-Latorre et al. (2020) that evaluated the presence of SARS-CoV-2 in a context of low sanitation countries. They sampled once in natural streams located in three different regions of Quito, Ecuador. Although they quantified SARS-CoV-2 and human adenoviruses, as well as the number of reported cases, the study is limited due to the small number of samples analyzed. The same was observed in another study performed in Monterrey, Mexico, where a total of 24 water samples were collected in two monitoring campaigns from three different rivers (Mahlknecht et al., 2021). In that case, groundwater and surface water from damns samples were also analyzed, as well as wastewater from different treatment plants. Although in that work no concentration procedure for surface water (they did not collect large volumes of water) was reported, they still found SARS-CoV-2 in 13 % of the river samples, and their results were expressed in terms of Ct. In a recently published study, Tandukar et al. (2022) collected 84 water samples, mainly from wastewater (71) (mostly inlets and outlets of two WWTPs), 13 of which were from an urban river of a densely populated area in Nepal. Although around half of the river samples were positive for SARS-CoV-2 with at least one of the detection methods used, only average concentrations were presented, without showing the dynamics of viral concentration. Issues like the presence, persistence, and potential infectivity of SARS-CoV-2 detected in the water environment have been under the discussion for the past years (Carducci et al., 2020; La Rosa et al., 2020). Until now all findings suggest that the transmission of SARS-CoV-2 is not via the oral-fecal route, i.e. in recreational activities; however, the presence of viral genetic material in surface water should not be ignored.
The concentrations of SARS-CoV-2 found in the Arenales River were high (peak measurements were between 106 and 107 gc/L), similar to (Randazzo et al., 2020a; Barrios et al., 2021) and higher than (Gonzalez et al., 2020; Giraud-Billoud et al., 2021; Wu et al., 2020b) the concentrations found (using a comparable detection system) in wastewater by other authors. After this work we verified that the concentrations of SARS-CoV-2 in AR-2 were approximately one order of magnitude lower than in raw wastewater from manholes in the city (data not shown). The fecal contamination observed in AR-2 (much higher than in AR-1), evidenced by SARS-CoV-2 concentrations but also through bacterial indicators (Fig. 2), showed that testing the Arenales River at AR-2 is in fact equivalent to testing a dilution of raw sewage, for SARS-CoV-2 surveillance.
According to the results of this and previous works, the Arenales River can properly play that role as it does reflect the pathogens circulating in the population. In fact, previous studies performed in the Arenales River revealed the circulation of multiples pathogens including bacteria, parasites, and various enteric viruses (Poma et al., 2012; Blanco Fernández et al., 2012a, Blanco Fernández et al., 2012b; Pisano et al., 2018; Prez et al., 2020). Furthermore, the microbiological risk for the population of being in contact with those water through recreational activities was estimated for enterovirus and norovirus (Poma et al., 2019).
The representation of the viral concentration in river water allows seeing the variation along time. It is like a succession of instant photos (or moments) showing the situation in that specific point under analysis. Although it allows understanding the magnitude of the viral circulation in the area, there are some limitations that should be considered, especially if the intention was to estimate the number of infected people. A water body like a river will show flow rate variations due to the input of stormwater, illegal raw sewage, and industrial effluents, among others. In other words, the viral concentration will depend on all those human or natural contributions; thus, those flow rates or dilution factors should be considered if the absolute number of viruses were to be calculated. On the other hand, there could be some discussion about the influence of population dynamic in the area of impact. One possibility to account for population dynamics and for other human and non-human inputs is to normalize the viral concentration using some other target or surrogate (Medema et al., 2020b). Some chemical compounds, like fecal sterols like coprostanol (Daughton, 2012; Chen et al., 2014), sucralose (Mahlknecht et al., 2021), other viruses common in human urinary tract, like human polyomavirus (HPyV) (McQuaig et al., 2009) or bacterial fecal indicators like human Bacteroidales (D’Aoust et al., 2020), just to mention some, have been suggested for normalization. The advantage of using any of them is that, as they are excreted by all the population all the time, then cultural or seasonal effects could be neglected. Although pepper mild mottle virus (PMMoV) has also been proposed as a normalizer (D’Aoust et al., 2020) the disadvantage is that is related to the population's diet (not related to the local diet in this particular case), thus the cultural effect cannot be neglected.
Measuring chemical tracers in river water could be interesting; however, their recovery would probably be highly affected by the concentration method used in this case. The ultrafiltration method applied here included a disposable hollow fiber unit that has a molecular weight cut-off of approximately 10,000 Da. Chemicals typically used for tracing human contamination, like coprostanol (C27H48O, MW 389), sucralose (C12H19Cl3O8, MW 398), caffeine (C8H10N4O2, MW 194), ibuprofen (C13H18O2, MW 206), to mention some, are much smaller than the membrane pore size; therefore, they would be part of the permeate rather than of the concentrate used for the detection of the target. Thus, if the aim were to use these chemical compounds as human tracers that should be planned from the beginning of the experiments to concentrate water in a different way.
Glassmeyer et al. (2005) analyzed 110 chemical compounds up- and down-stream 10 WWTP in an exhaustive study carried out in the United States to determine which one could be better as in indicator of human fecal contamination in environmental water. After discarding some chemicals, coprostanol (fecal sterol), proved to be the best candidate (Glassmeyer et al., 2005; Nakagawa et al., 2017) because it is of human origin, but the conversion of cholesterol to coprostanol is diet-dependent; thus, the diet of the population and their fluctuation over time should be studied jointly.
In addition to the detection of SARS-CoV-2, two other targets linked to human contamination were quantified in this work. One was the gene that encodes for human RNase P, used in many studies as an endogenous internal control (Arnold et al., 2020; Boddicker et al., 2007; Dahdouh et al., 2021), and it has been extensively used as a sample quality control in clinical tests in the current COVID-19 pandemic. The second one was the marker for human polyomaviruses, which was found ubiquitously in wastewater along the year, with concentrations raging 103 to 107 gc/L, in different geographical areas (Ahmed et al., 2010; Bofill-Mas et al., 2000, Bofill-Mas et al., 2006; Haramoto et al., 2010; McQuaig et al., 2009); thus, it has been proposed as a viral indicator of human fecal contamination and it is known to be stable in the environment. In addition, these viruses are excreted by feces and urine fluids (McQuaig et al., 2009; Bofill-Mas et al., 2000; Boothpur and Brennan, 2010; Dalianis and Hirsch, 2013), with the species JC and BK reported as the most frequent and prevalent HPyV detected in sewage samples of this geographical region (Barrios et al., 2018).
Neither RNase P nor HPyV were detected in La Caldera River, showing again that fecal human contamination was low and sporadic. In the case of Mojotoro River, RNase P was not detected and HPyV was present only in some of the samples, proving that those waters were only slightly impacted by wastewater. Conversely, in the case of the Arenales River, highly impacted by wastewater discharges, HPyV was quantified at both sampling points and the concentration was persistent and steady along the monitoring period. This was also the case for RNase P, which was only detected at AR-2.
Regarding normalization, although most of the times it has been done with only one target as reference, calculating the ratio between the gene of interest and the normalizer (McQuaig et al., 2009), it is highly recommended to use multiple genes for a more accurate normalization. For averaging of the control genes, it is recommended to use the geometric instead of the arithmetic mean, as the former controls are better for possible outlying values and abundance differences between the different genes (Vandesompele et al., 2002).
In this case the concentration of SARS-CoV-2 from AR-2 was normalized in three different ways. The first one was as the ratio with the concentration of human polyomavirus, in which case the shape of the curve was not modified, the scale of results became too small, and the sense of the viral concentration magnitude was lost, plus the units were gc of SARS-CoV-2 per gc of HPyV. A similar situation resulted when the ratio of SARS-CoV-2 was calculated with RNase P (gc of SARS-CoV-2/gc of RNase P), although the curved was slightly modified due to the fluctuations of RNase P concentration. Instead, the third way of normalizing, using both HPyV and RNase P according to Eq. (3), seemed more appropriate as the shape of the original concentration curved was not changed but mostly because the normalized values kept the sense of the magnitude.
In the case of the Arenales River the flow rate fluctuations were not measured; however, the difference in flow rate between the sampling points AR-1 and AR-2 (separated 700 m, no additional inputs verified) are only due to the discharges of the WWTP-S. What is interesting is that this discharge, far from producing a “dilution effect”, decreasing the viral concentration, has an “addition effect” increasing it due to the huge fecal contribution. This effect observed in the concentration of SARS-CoV-2 was also reflected in the concentration of the bacterial indicators measured. The magnitude of the discharge modified substantially, in a short distance, the microbiological and physico-chemical characteristics of the water river, differentiating this sampling point from all the others that were analyzed and minimizing any other effect related to the population dynamic or to seasonal effects. Finally, strong correlations were found between both the experimental and the normalized (using all three alternatives) SARS-CoV-2 concentrations and the curve of active COVID-19 cases in the city of Salta. In this way, the water from the Arenales River, at AR-2, reflected the population situation regarding viral circulation of COVID-19 cases, therefore, it could be well used as a surveillance tool.
This is, to the best of our knowledge, the first study that showed the dynamic of SARS-CoV-2 concentration in an urban river highly impacted by wastewater and proved that it can be used for SARS-CoV-2 surveillance to support health authorities´ decisions.
5. Conclusions
-
•
From the three rivers analyzed La Caldera River had the best water quality, and indicator bacteria were within acceptable limits for recreational activities.
-
•
Although Mojotoro River receives the discharges of an old wastewater stabilization pond before MR-1 and the discharges of the northern wastewater treatment plant of the city before MR-2, they did not have a negative effect on the water quality.
-
•
The Arenales River presented the poorest water quality. In addition to the contamination existing at AR-1 the discharges of the southern wastewater treatment plant contributed to the significant increase of fecal contamination at AR-2.
-
•
SARS-CoV-2 was found in about half of the samples analyzed in low concentrations in La Caldera and Mojotoro Rivers. Instead, the virus concentration in the Arenales River was persistently high.
-
•
Two human tracers, human polyomavirus (HPyV) and RNase P, were analyzed. None of them was detected in La Caldera River, only HPyV was found in Mojotoro River and at AR-1 in Arenales River, and both were quantified at AR-2 (Arenales River).
-
•
The Arenales River, at AR-2, was highly impacted by wastewater and the concentration of SARS-CoV-2 was normalized using both human tracers. The experimental and the normalized concentrations correlated with the curve of accumulated reported COVID-19 cases in the city.
-
•
Rivers highly impacted by wastewater, like the Arenales River at AR-2, can reflect the viral circulation of the population. Thus, environmental water monitoring is a useful tool for epidemiological surveillance as well as particularly important for quantitative microbial risk assessment regarding recreational activities and crop irrigation.
The following is the supplementary data related to this article.
Coefficients and probabilities (below and above the main diagonal, respectively) from Spearman correlation test using the physico-chemical variables, fecal indicator bacteria and SARS-CoV-2 concentration (CoV) of all sampling points. Significant correlations (p < 0.05) are indicated in bold. Physico-chemical variables: temperature (T), pH, conductivity (COND), turbidity (TURB). Fecal indicator bacteria: total coliforms (TC), thermotolerant coliforms (TTC), E. coli (EC), enterococci (EN).
CRediT authorship contribution statement
María Noel Maidana-Kulesza: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. Hugo Ramiro Poma: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Visualization. Diego Gastón Sanguino-Jorquera: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. Sarita Isabel Reyes: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. María del Milagro Said-Adamo: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. Juan Martín Mainardi-Remis: Formal analysis, Investigation, Writing – original draft. Dolores Gutiérrez-Cacciabue: Formal analysis, Investigation, Writing – review & editing. Héctor Antonio Cristóbal: Formal analysis, Investigation, Writing – original draft. Mercedes Cecilia Cruz: Methodology, Formal analysis, Visualization, Writing – original draft. Mónica Aparicio González: Methodology, Validation, Investigation, Writing – original draft. Verónica Beatriz Rajal: Conceptualization, Resources, Writing – review & editing, Visualization, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by Project COVID-19 233-785, from Fondo para la Investigación Científica y Tecnológica (FONCyT), Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación, Argentina. María Noel Maidana Kulesza, Diego Gastón Sanguino-Jorquera, Sarita Reyes, María del Milagro Said-Adamo, and Martín Mainardi Remis are recipients of doctoral fellowships from CONICET.
Editor: Damià Barceló
References
- Ahmed W., Wan C., Goonetilleke A., Gardner T. Evaluating sewage-associated JCV and BKV polyomaviruses for sourcing human fecal pollution in a coastal river in Southeast Queensland, Australia. J. Environ. Qual. 2010;39(5):1743–1750. doi: 10.2134/jeq2010.0062. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bibbins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albastaki A., Naji M., Lootah R., Almeheiri R., Almulla H., Almarri I., Alreyami A., Aden A., Alghafri R. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: the use of wastewater-based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M.T., Temte J.L., Barlow S.K., Bell C.J., Goss M.D., Temte E.G., Checovich M.M., Reisdorf E., Scott S., Guenther K., Wedig M., Shult P., Uzicanin A. Comparison of participant-collected nasal and staff-collected oropharyngeal specimens for human ribonuclease P detection with RT-PCR during a community-based study. PloS One. 2020;15(10) doi: 10.1371/journal.pone.0239000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios M.E., Fernández M.D.B., Cammarata R.V., Torres C., Mbayed V.A. Viral tools for detection of fecal contamination and microbial source tracking in wastewater from food industries and domestic sewage. J. Virol. Methods. 2018;262:79–88. doi: 10.1016/j.jviromet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Barrios M.E., Díaz S.M., Torres C., Costamagna D.M., Blanco Fernández M.D., Mbayed V.A. Dynamics of SARS-CoV-2 in wastewater in three districts of the Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: a pilot study. Sci. Total Environ. 2021;800 doi: 10.1016/j.scitotenv.2021.149578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Brown K.M.P., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco Fernández M.D., Torres C., Riviello-López G., Poma H.R., Rajal V.B., Nates S., Cisterna D.M., Campos R.H., Mbayed V.A. Analysis of the circulation of hepatitis A virus in Argentina since vaccine introduction. Clin. Microbiol. Infect. 2012;18(12):E548–E551. doi: 10.1111/1469-0691.12034. [DOI] [PubMed] [Google Scholar]
- Blanco Fernández M.D., Torres C., Poma H.R., Riviello-López G., Martínez L.C., Cisterna D.M., Rajal V.B., Nates S., Mbayed V.A. Environmental surveillance of norovirus in Argentina revealed distinct viral diversity patterns, seasonality and spatio-temporal diffusion processes. Sci. Total Environ. 2012;437:262–269. doi: 10.1016/j.scitotenv.2012.08.033. [DOI] [PubMed] [Google Scholar]
- Boddicker J.D., Rota P.A., Kreman T., Wangeman A., Lowe L., Hummel K.B., Thompson R., Bellini W.J., Pentella M., DesJardin L.E. Real-time reverse transcription-PCR assay for detection of mumps virus RNA in clinical specimens. J. Clin. Microbiol. 2007;45(9):2902–2908. doi: 10.1128/JCM.00614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Pina S., Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 2000;66(1):238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Albinana-Gimenez N., Clemente-Casares P., Hundesa A., Rodriguez-Manzano J., Allard A., Calvo M., Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006;72(12):7894–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothpur R., Brennan D.C. Human polyoma viruses and disease with emphasis on clinical BK and JC. J. Clin. Virol. 2010;47(4):306–312. doi: 10.1016/j.jcv.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Veran M. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179(2020):115907. doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Centers for Disease Control and Prevention; U.S: 2020. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcrpanel-primer-probes.html (Last accessed 16th June, 2020) [Google Scholar]
- Chávez-Díaz L.V., Gutiérrez-Cacciabue D., Poma H.R., Rajal V.B. Sediments quality must be considered when evaluating freshwater aquatic environments used for recreational activities. Int. J. Hyg. Environ. Health. 2020;223:159–170. doi: 10.1016/j.ijheh.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Chen C., Kostakis C., Gerber J.P., Tscharke B.J., Irvine R.J., White J.M. Towards finding a population biomarker for wastewater epidemiology studies. Sci. Total Environ. 2014;487:621–628. doi: 10.1016/j.scitotenv.2013.11.075. [DOI] [PubMed] [Google Scholar]
- Cruz M.C., Gutiérrez Cacciabue D., Gil J.F., Gamboni O., Vicente M.S., Wuertz S., Gonzo E.E., Rajal V.B. The impact of point source pollution on shallow groundwater used for human consumption in a threshold country. J. Environ. Monit. 2012;14:2338–2349. doi: 10.1039/c2em30322a. [DOI] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier É., Montpetit D., Jia J.J., Alexandrov I., Tariq Baig A., Mayne J., Zhang X., Alain T., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2020;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdouh E., Lázaro-Perona F., Romero-Gómez M.P., Mingorance J., García-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J. Infect. 2021;82(3):414–451. doi: 10.1016/j.jinf.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalianis T., Hirsch H.H. Human polyomaviruses in disease and cancer. Virology. 2013;437(2):63–72. doi: 10.1016/j.virol.2012.12.015. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Using biomarkers in sewage to monitor community-wide human health: isoprostanes as conceptual prototype. Sci. Total Environ. 2012;424:16–38. doi: 10.1016/j.scitotenv.2012.02.038. [DOI] [PubMed] [Google Scholar]
- de Oliveira L.C., Torres-Franco A.F., Coelho Lopes B., da Silva Álvares, Santos B.S., Azevedo Costa E., Costa M.S., Reis M.T.P., Melo M.C., Polizzi R.B., Teixeira M.M., Rossas Mota C. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021;195 doi: 10.1016/j.watres.2021.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu R., Battistone A., Buttinelli G., Fiore S., Fontana S., Amato C., Cristiano K., Gamper S., Simeoni J., Frate R., Pellegrinelli L., Binda S., Veronesi L., Zoni R., Castiglia P., Cossu A., Triassi M., Pennino F., Germinario C., Balena V., Cicala A., Mercurio P., Fiore L., Pini C., Stefanelli P. Poliovirus and other enteroviruses from environmental surveillance in Italy, 2009–2015. Food Environ. Virol. 2018;10(4):333–342. doi: 10.1007/s12560-018-9350-8. [DOI] [PubMed] [Google Scholar]
- Di Rienzo J.A., Casanoves F., Balzarini M.G., Gonzalez L., Tablada M., Robledo C.W. Grupo InfoStat, FCA, Universidad Nacional de Córdoba; Argentina: 2016. InfoStat version 2016.http://www.infostat.com.ar URL. [Google Scholar]
- Eaton A.D., Clesceri L.S., Rice E.W., Greenberg A.E., Franson M.A.H. 21st edition. APHA. American Public Health Association; 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Giraud-Billoud M., Cuervo P., Altamirano J.C., Pizarro M., Aranibar J.N., Catapano A., Cuello H., Masachessi G., Vega I.A. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza,Argentina. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.148887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassmeyer S.T., Furlong E.T., Kolpin D.W., Cahill J.D., Zaugg S.D., Werner S.L., Meyer M.T., Kryak D.D. Transport of chemical and microbial compounds from known wastewater discharges: potential for use as indicators of human fecal contamination. Environ. Sci. Technol. 2005;39:5157–5169. doi: 10.1021/es048120k. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R., Groot R.J.D., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L., Samborskiy D., Sidorov I.A., Solá-Gurpegui I., Ziebuhr J. Severe acute respiratory syndrome-related coronavirus: the species and its viruses–a statement of the coronavirus study group. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.-L., Hui D., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Zhong C., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of 2019 novel coronavirus infection in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez Cacciabue D., Teich I., Poma H.R., Cruz M.C., Balzarini M., Rajal V.B. Strategies to optimize monitoring schemes of recreational waters using a multivariate approach. Environ. Monit. Assess. 2014;186(12):8359–8380. doi: 10.1007/s10661-014-4010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Cacciabue D., Rajal V.B. Estimating decay kinetic parameters and persistence of bacteria in water is essential for future modelling. Chem. Eng. Res. Des. 2022;179:175–187. [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ohgaki S. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res. 2010;44(6):1747–1752. doi: 10.1016/j.watres.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Zhao S., Lin Q., Zhuang Z., Cao P., Wang M.H., Yang L. The relative transmissibility of asymptomatic COVID-19 infections among close contacts. Int. J. Infect. Dis. 2020;94:145–147. doi: 10.1016/j.ijid.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconelli M., Muscillo M., Della Libera S., Fratini M., Meucci L., De Ceglia M., Giacosa D., La Rosa G. One-year surveillance of human enteric viruses in raw and treated wastewaters, downstream river waters, and drinking waters. Food Environ. Virol. 2017;9(1):79–88. doi: 10.1007/s12560-016-9263-3. [DOI] [PubMed] [Google Scholar]
- Iglesias N.G., Gebhard L.G., Carballeda J.M., Aiello I., Recalde E., Terny G., Ambrosolio S., L'Arco G., Konfino J., Brardinelli J.I. MedRxiv preprint; 2020. SARS-CoV-2 Surveillance in Untreated Wastewater: First Detection in a Low-resource Community in Buenos Aires, Argentina. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5(3):299–314. [Google Scholar]
- Jeong H.W., Kim S.M., Kim H.S., Kim Y.I., Kim J.H., Cho J.Y., Kim S.H., Kang H., Kim S.G., Park S.J., Kim E.H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26(11):1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020;92(10):1807–1809. doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R.C., Berman O., Nourinejad M. Sampling manholes to home in on SARS-CoV-2 infections. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0240007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi I., Boemo A., Musso H. Location and characterization of pollution sites by principal component analysis of trace contaminants in a slightly polluted seasonal river: a case study of the Arenales River (Salta, Argentina) Water SA. 2007;33(4) [Google Scholar]
- Mahlknecht J., Padilla Reyes D.A., Ramos E., Reyes L.M., Alvarez M.M. The presence of SARS-CoV-2 RNA in different freshwater environments in urban settings determined by RT-qPCR: implications for water safety. Sci. Total Environ. 2021;784 doi: 10.1016/j.scitotenv.2021.147183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S.M., Scott T.M., Lukasik J.O., Paul J.H., Harwood V.J. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 2009;75(11):3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petersen S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhra R., Krishan K., Kanchan T. Possible modes of transmission of novel coronavirus SARS-CoV-2: a review. Acta Bio-Med. 2020;91(3) doi: 10.23750/abm.v91i3.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Amano H., Takao Y., Hosono T., Berndtsson R. On the use of coprostanol to identify source of nitrate pollution in groundwater. J. Hydrol. 2017;550:663–668. [Google Scholar]
- Nasseri S., Yavarian J., Norouzian Baghani A., Mokhtari Azad T., Nejati A., Nabizadeh R., Hadi M., Shafiei Jandaghi N.Z., Vakili B., Azam Vaghefi S.K., Baghban M., Yousefi S., Nazmara S., Alimohammadi M. The presence of SARS-CoV-2 in raw and treated wastewater in 3 cities of Iran: Tehran, Qom and Anzali during coronavirus disease 2019 (COVID-19) outbreak. J. Environ. Health Sci. Eng. 2021 doi: 10.1007/s40201-021-00629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Lee C.W., Park D.I., Woo H.Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.J., Joo E.J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano M.B., Lugo B., Poma H.R., Cristóbal H.A., Raskovsky V., Martínez Wassaf M.G., Rajal V.B., Ré V.E. Environmental hepatitis E virus detection supported by serological evidence in the northwest of Argentina. Trans. R. Soc. Trop. Med. Hyg. 2018;112(4):181–187. doi: 10.1093/trstmh/try048. [DOI] [PubMed] [Google Scholar]
- Poma H.R., Cacciabue D.G., Garcé B., Gonzo E.E., Rajal V.B. Towards a rational strategy for monitoring of microbiological quality of ambient waters. Sci. Total Environ. 2012;433:98–109. doi: 10.1016/j.scitotenv.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poma H.R., Kundu A., Wuertz S., Rajal V.R. Data fitting approach more critical than exposure scenarios and treatment of censored data for quantitative microbial risk assessment. Water Res. 2019;154:45–53. doi: 10.1016/j.watres.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Prez V.E., Poma H.R., Giordano G.G., Victoria M., Nates S.V., Rajal V.B., Barril P.A. Rotavirus contamination of surface waters from the northwest of Argentina. J. Water Health. 2020;18(3):409–415. doi: 10.2166/wh.2020.005. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2005. R: A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Rajal V.B., McSwain B.S., Thompson D.E., Leutenegger C.M., Kildare B., Wuertz S. Validation of hollow fiber ultrafiltration and real-time PCR for the quantification of viruses from environmental water samples. Water Res. 2007;41:1411–1422. doi: 10.1016/j.watres.2006.12.034. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . RStudio, PBC; Boston, MA: 2020. RStudio: Integrated Development for R.http://www.rstudio.com/ URL. [Google Scholar]
- Saththasivam J., El-Malah S.S., Gomez T.A., Jabbar K.A., Remanan R., Krishnankutty A.K., Ogunbiyi O., Rasool K., Ashhab S., Rashkeeb S., Bensaad M., Ahmed A.A., Mohamoud Y.A., Malek J.A., Abu-Raddad L.J., Jeremijenko A., Abu-Halaweh H.A., Lawler J., Mahmoud K.A. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro S.S., Francia R.S. An approximate analysis of variance test for normality. J. Am. Stat. Assoc. 1972;67(337):215–216. [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S.B., Liu X., Dai J., Li X., Huang S., Luo L., Wen L., Zhuo J., Li Y., Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Y.M. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;9(1):991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S., Sthapit N., Thakali O., Malla B., Sherchan S.P., Shakya B.M., Shrestha L.P., Sherchand J.B., Joshi D.R., Lama B., Haramoto E. Detection of SARS-CoV-2 RNA in wastewater, river water, and hospital wastewater of Nepal. Sci. Total Environ. 2022;824 doi: 10.1016/j.scitotenv.2022.153816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lin Lee W., Rajal V.B., Haines M.B., Girones R., Ching Ng L., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA, United States Environmental Protection Agency . 2002. Method 1106.1: Enterococci in Water by Membrane Filtration Using Membrane-Enterococcus-Esculin Iron Agar (mE-EIA) EPA 600-4-85-076. [Google Scholar]
- USEPA, United States Environmental Protection Agency . Office of Water, United States Environmental Protection Agency (USEPA); Washington, DC: 2012. Recreational Water Quality Criteria.https://www.epa.gov/sites/production/.../rec-factsheet-2012.pdf Online: [Google Scholar]
- USEPA, United States Environmental Protection Agency . 2014. Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia coli Agar (Modified mTEC) EPA-82 1-R-14-010. [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation (No. WHO/V&B/03.03) [Google Scholar]
- WHO World Health Organization. 2022. https://covid19.who.int/ (Last accessed 10th April, 2022)
- Wickham H. Springer-Verlag; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvalle C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Rhode S.F., Wuertz S., Thompson J., Alm E.J. MedRxiv preprint; 2020. Wastewater Surveillance of SARS-CoV-2 Across 40 U.S. States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. medRxiv; 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics Through Viral Genome Quantification in Paris Wastewaters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao Jingxian, Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Dong Q., Li S., Cheng Z., Kang X., Ren D., Xu C., Zhou X., Liang P., Sun L., Zhao J., Jiao Y., Han T., Liu Y., Qian Y., Liu Y., Huang X., Qu J. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J. Hazard. Mater. 2022;429 doi: 10.1016/j.jhazmat.2022.128358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Zhixiu, Li Zifu, Cui X., Xiao J., Zhan J., Meng T., Zhou W., Liu J., Xu H. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69(6):1010–1018. [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. 2020;117(26):14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., Xu J., Xu W. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coefficients and probabilities (below and above the main diagonal, respectively) from Spearman correlation test using the physico-chemical variables, fecal indicator bacteria and SARS-CoV-2 concentration (CoV) of all sampling points. Significant correlations (p < 0.05) are indicated in bold. Physico-chemical variables: temperature (T), pH, conductivity (COND), turbidity (TURB). Fecal indicator bacteria: total coliforms (TC), thermotolerant coliforms (TTC), E. coli (EC), enterococci (EN).