Abstract
Biodegradation of methyl tert-butyl ether (MTBE) by the hydrogen-oxidizing bacterium Hydrogenophaga flava ENV735 was evaluated. ENV735 grew slowly on MTBE or tert-butyl alcohol (TBA) as sole sources of carbon and energy, but growth on these substrates was greatly enhanced by the addition of a small amount of yeast extract. The addition of H2 did not enhance or diminish MTBE degradation by the strain, and MTBE was only poorly degraded or not degraded by type strains of Hydrogenophaga or hydrogen-oxidizing enrichment cultures, respectively. MTBE degradation activity was constitutively expressed in ENV735 and was not greatly affected by formaldehyde, carbon monoxide, allyl thiourea, or acetylene. MTBE degradation was inhibited by 1-amino benzotriazole and butadiene monoepoxide. TBA degradation was inducible by TBA and was inhibited by formaldehyde at concentrations of >0.24 mM and by acetylene but not by the other inhibitors tested. These results demonstrate that separate, independently regulated genes encode MTBE and TBA metabolism in ENV735.
Methyl tert-butyl ether (MTBE) has been used as a gasoline additive since the late 1970s to replace lead and other toxic chemicals and as an oxygenate to meet the vehicle emissions requirements of the 1990 Clean Air Act Amendments (21). Reformulated gasoline presently contains approximately 11% (vol/vol) MTBE. The widespread use of MTBE in gasoline has led to accidental spills and its discharge into soils and groundwater. Because it is highly soluble in water (∼43,000 ppm) and has a low tendency to adsorb to soils, it moves rapidly in groundwater (25) and is now often found in groundwater near service stations, fuel storage facilities, and filling terminals throughout the United States. As little as 4 liters of reformulated gasoline can contaminate >106 liters of groundwater to above its odor and taste threshold of 40 μg/liter.
The full extent of MTBE contamination in groundwater in the United States has only recently been under careful assessment. A study performed as part of the U.S. Geological Survey's National Water-Quality Assessment Program revealed that MTBE is the second most commonly detected contaminant in urban groundwater (26). As an example of how widespread this problem has become, Buscheck et al. (5) reviewed groundwater monitoring data from 700 service station sites in the United States and observed that >80% of the active sites and 74% of the inactive sites had MTBE contamination. Approximately 96, 98, and 86% of the service station sites in Texas, Maryland, and California, respectively, where groundwater was analyzed for MTBE had significant MTBE contamination. Of these sites, 63, 82, and 47%, respectively, had MTBE concentrations greater than 1 mg/liter. This widespread contamination has led to increased public and regulatory scrutiny and a need to identify cost-effective remediation technologies.
Relatively little work has been done to address the biodegradability of MTBE. In an early study, an aerobic consortium isolated from acclimated sludge was maintained on MTBE as a sole source of carbon (23). MTBE was degraded to tert-butyl alcohol (TBA), which was also degraded by the enrichment culture. This culture has been the focus of a bioremediation demonstration where it was injected directly into an MTBE-contaminated aquifer at the Port Hueneme Naval Station in California (24). MTBE biodegradation has been reported in sewage sludge (20), soils (33), river sediments (3, 4), and a biofilter inoculated with groundwater (7, 8), although the responsible bacteria were not isolated or characterized. At least partial MTBE degradation has been observed in a few pure cultures of bacteria (9, 14, 15, 16, 17, 28) and fungi (12), and recent studies demonstrated growth of a pure culture (strain PM1) on MTBE as the sole carbon source (6, 11). Anaerobic degradation of MTBE has been observed in one aquifer (32), but it was not shown in anaerobic samples from several other sites (18, 30).
We previously reported that MTBE is mineralized by propane-oxidizing bacteria and proposed a pathway for MTBE degradation (28). Our initial studies suggested that MTBE is first oxidized to TBA, but more recent studies have demonstrated that the first oxidation product may be tert-butyl formate (16). TBA is subsequently degraded by the strains through the intermediate 2-hydroxy isobutyric acid (HIBA), which accumulates in the culture media. HIBA is not an effective growth substrate for the propane-oxidizing bacteria studied, but it is eventually metabolized to CO2 by the strains.
We recently isolated and described a new MTBE-degrading organism, Hydrogenophaga flava strain ENV735, which grows slowly on MTBE but can be grown rapidly on other substrates for research and bioremediation applications (29). In this report, we evaluate MTBE and TBA degradation by strain ENV735 more closely and attempt to identify factors that could account for the persistence of MTBE in the environment. The results of the study suggest that MTBE and TBA are oxidized by separate enzyme systems in this strain.
MATERIALS AND METHODS
Chemicals.
MTBE (98%) was purchased from Aldrich Chemical Co. (Milwaukee, Wis.). TBA (analytical reagent grade) was purchased from Mallinckrodt Specialty Chemical Co. (Paris, Ky.). R2A medium was from BBL, Inc. (Cockeysville, Md.), and Luria-Bertani (LB) medium was from Difco, Inc. (Sparks, Md.). Corn steep liquor (CSL) was from Grain Processing Corporation (Muscatine, Iowa). Uniformly labeled [14C]MTBE (10.1 mCi/mmol; lot no. 3048-175B) was purchased from Dupont New England Nuclear Products (Boston, Mass.). The chemical purity of the [14C]MTBE was >99%, as determined by gas chromatography, and the manufacturer's high-pressure liquid chromatography analysis indicated that it had a radiochemical purity of approximately 99%. Unless otherwise stated, all other chemicals were of the highest purity available and were purchased from either Aldrich Chemical Co., Mallinckrodt Specialty Chemical Co., J. T. Baker Inc. (Phillipsburg, N.J.), or Sigma Chemical Co. (St. Louis, Mo.).
Bacterial strains.
H. flava ENV735 (ATCC PTA-2158) was isolated by enrichment culturing on MTBE (29). The strain is a gram-negative organism and was identified as H. flava by fatty acid analysis and 500-base 16S rRNA sequencing (Acculab, Newark, Del.). Fatty acid analysis indicated that the strain was most closely related to bacteria of the genus Hydrogenophaga (similarity index = 0.720), and 16S rRNA analysis indicated that the strain is most closely related to H. flava (0.58% difference from the library strain). The strain grew readily on hydrogen (H2) as a sole energy source. Because the cells constitutively expressed MTBE degradation activity (see Results), cells used for MTBE degradation assays could be grown at 30°C in either LB broth, basal salts medium (BSM) (13) with 0.4% yeast extract (YE), BSM with sucrose, or BSM with TBA. Because TBA degradation activity was inducible (see Results), cells used for TBA degradation assays were grown either on TBA or MTBE to ensure induction of TBA degradation activity or on the other media described above when noninducing conditions were required.
To isolate other hydrogen-oxidizing bacteria, approximately 5 g of turf soil or 5 ml of sludge from the Hamilton, N.J., wastewater treatment facility was added to 100 ml of 1246 medium (1) in a 250-ml Erlenmeyer flask fitted with a butyl rubber stopper. The rubber stopper was pierced with an 18-gauge needle onto which was fitted a two-way stopcock. The headspace of the flask was filled with a gas mixture designed for the culture of hydrogen oxidizers, which contained 60% H2, 10% CO2, 25% N2, and 5% O2 (1). The flasks were then placed on a shaker and incubated for several days or until the culture turbidity increased. The headspace of the flask was flushed daily with the gas mixture to ensure the availability of H2, CO2, and O2. The culture was then subcultured as above until an active hydrogen-oxidizing culture was selected.
The bacterial strains H. flava (ATCC 17724) and Hydrogenophaga palleronii (ATCC 33667) were purchased from the American Type Culture Collection (Rockville, Md.) and grown on rich media (YE or LB), 1246 medium (1), and hydrogen as recommended by the ATCC or on BSM with hydrogen as described above. Pure cultures of hydrogen-oxidizing bacteria were grown as described previously for enrichment cultures, except the gas mixture was passed through a sterile 0.2-μm-pore-size filter to prevent contamination.
MTBE and TBA degradation assays.
Biodegradation assays were performed as previously described (28). Cells were grown in shake flasks containing rich medium (LB or YE) or 1246 medium or containing BSM with the addition of MTBE (75 mg/liter), TBA (100 mg/liter), or sucrose (0.1 or 0.5% [wt/vol]). The bacteria were collected by centrifugation, washed, and suspended in BSM to an optical density at 550 nm (OD550) of 1, unless otherwise indicated. Subsamples of the cultures were placed in 60-ml serum vials, and MTBE was added to the culture as either undiluted compound or an aqueous solution depending on the desired final concentration. For assays utilizing high MTBE concentrations, cultures were placed in 160-ml serum vials with an aerobic headspace. The headspace gas was replaced as necessary to ensure oxygen availability. Vials were sealed with Teflon-lined septa and incubated on their side with shaking at 25°C.
To measure the amount of MTBE and TBA during culture assays, a portion of the culture liquid was removed, centrifuged, and analyzed by direct-liquid-injection gas chromatography (GC) with flame ionization detection as previously described (28). GC response with the samples was compared to the response of a three- to five-point standard curve for each analyte. This method has a detection limit of ∼300 μg/liter for MTBE and ∼500 μg/liter for TBA. When a lower detection limit for MTBE was desired, the samples were analyzed using GC coupled to mass spectroscopy (U.S. Environmental Protection Agency [EPA] method 8260 [31]) with sample preparation using the purge-and-trap method for aqueous samples (EPA method 5030B [31]). This method provides a detection limit for MTBE of 5 μg/liter but cannot be used for TBA detection. When lower detection limits for both MTBE and TBA were necessary, the samples were analyzed by GC-flame ionization detection using EPA method 8015b (31) and a heated purge-and-trap system.
Inhibitor assays.
To evaluate inhibition of MTBE and TBA degradation by known oxygenase inhibitors, ENV735 was grown in BSM with MTBE plus 0.01% YE. Cells were concentrated by centrifugation, washed with BSM, and resuspended in BSM to an OD550 of 2.0. Subsamples (5 ml) of the culture were placed in 25-ml serum vials, and the vials received one of the following: no addition, 0.05 mM 1-amino benzotriazole (ABT), carbon monoxide (CO; 30% [vol/vol] of headspace), 4 mM allyl thiourea (ATU), 0.5 mM butadiene monoepoxide, or acetylene (30% [vol/vol] of headspace). The vials were sealed with Teflon-lined septa and then shaken at room temperature for 20 min. MTBE or TBA was added to duplicate vials to a final concentration of 40 mg/liter, and the vials were incubated for 16 h with shaking at 25°C. Each sample was analyzed by direct-injection GC as described above.
To assess inhibition by formaldehyde, a sample of TBA-grown ENV735 was harvested by centrifugation and resuspended in BSM to an OD550 of 1. Subsamples (5 ml) were added to 25-ml serum vials, and TBA was added to each vial to a final concentration of 2 mM. Formaldehyde (37% solution in methanol) was then added to duplicate vials to final concentrations ranging from 0 to 2.4 mM. The vials were sealed with Teflon-lined septa and incubated on a shaker (100 rpm) at 25°C for 20 h. Alternately, sucrose-grown ENV735 cultures (10 ml; OD550 = 1) were incubated with MTBE (25 mg/liter) and 0 to 2.6 mM formaldehyde and were analyzed periodically for MTBE and TBA. The concentration of TBA and MTBE remaining in each of the duplicate vials was determined by direct-injection GC analysis.
Protein labeling.
To label potential MTBE-degrading proteins, YE-grown ENV735 was harvested by centrifugation and suspended in BSM to an OD550 of 1. Subsamples (10 ml) of the cell suspension were placed in 3 separate 50-ml serum vials. Chloramphenicol was added to two of the vials to a final concentration of 200 μg/ml. Then, 5 to 10 μCi of uniformly labeled [14C]MTBE was added to each vial, and the vials were incubated for 3 h on an orbital shaker (100 rpm) at room temperature. After incubation, the vials were centrifuged to pellet the bacteria and the cells were suspended in 1 ml of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and were boiled to lyse cells and denature proteins. Subsamples (10 to 40 μl) of the lysates were loaded onto an 8% polyacrylamide gel and separated by electrophoresis. Broad-range protein molecular weight markers (catalog no. 7701S; New England Biolabs, Beverly, Mass.) were added to the gel to determine the size of the labeled fragments. The gels were stained with Coomassie blue, dried under vacuum, and then placed against a Bio-Rad standard phosphorimaging screen in a Molecular Dynamics PhosphorImager system (Sunnyvale, Calif.) for up to 4 weeks to generate phosphorimages of the labeled proteins. Images were generated and analyzed by using the system's Image Quantifier software.
Induction of TBA degradation.
To evaluate induction of TBA degradation, cells were grown on either LB broth or BSM with YE (0.01%) and MTBE or TBA. Total cellular proteins were analyzed by using SDS-PAGE analysis on an 8% polyacrylamide gel (2). Broad-range protein molecular weight markers (catalog no. 7701S; New England Biolabs) were used to determine the size of peptides in the gel.
RESULTS
Growth of strain ENV735.
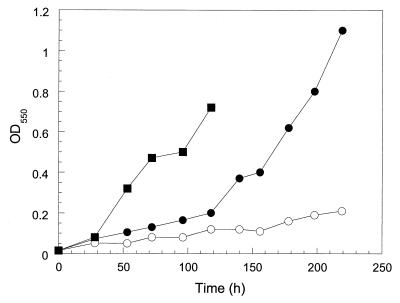
Growth of strain ENV735 on MTBE as a sole source of carbon was slow and resulted in the production of dense bacterial clumps and cells attached to the container surface at the air/water interface. This growth characteristic made it difficult to collect representative samples for measuring cell yield on MTBE. If a small amount of YE (0.01% [wt/vol]) was added to the media, however, the cells grew more rapidly and were dispersed throughout the media (Fig. 1). Biomass yield on MTBE in the presence of 0.01% YE was approximately 0.4 mg of biomass (dry weight) per mg of MTBE after subtraction of the amount of biomass produced in samples incubated in the absence of MTBE. The addition of the 20 essential amino acids, either in groups of five, individually, and/or in a complex vitamin solution did not enhance growth on MTBE.
FIG. 1.
Growth of ENV735 on TBA + 0.01% YE (▪), MTBE + 0.01% YE (●), and 0.01% YE (○). Cell growth on MTBE as a sole carbon source was slow and resulted in the formation of dense clumps that were difficult to sample and quantify.
To evaluate MTBE degradation by other hydrogen-oxidizing bacteria, a microbial enrichment with H2 was performed with turf soil and sewage sludge to grow indigenous hydrogen oxidizers and two type strains of hydrogen-oxidizing bacteria were purchased from ATCC. None of the H2 enrichment cultures degraded MTBE. In some experiments, however, small amounts of TBA were detected in cultures of H. flava (ATCC 17724) and H. palleronii (ATCC 33667) after growth in 0.3% YE in BSM. For example, in one experiment with H. flava (OD550 = 1.1; MTBE initial concentration = 25 mg/liter ), 0.9 mg of TBA/liter and 23.4 mg of MTBE/liter were present after 24 h of incubation at 30°C. A similarly incubated culture of H. palleronii (OD550 = 1.3) contained 0.25 mg of TBA/liter and 22 mg of MTBE/liter after 48 h of incubation at 30°C. In each case, there was no change in the MTBE concentration and no TBA production in poisoned samples. Furthermore, the addition of H2 as an energy source to the enrichment cultures did not enhance MTBE degradation, nor did it improve or prevent MTBE degradation by strain ENV735.
MTBE degradation assays.
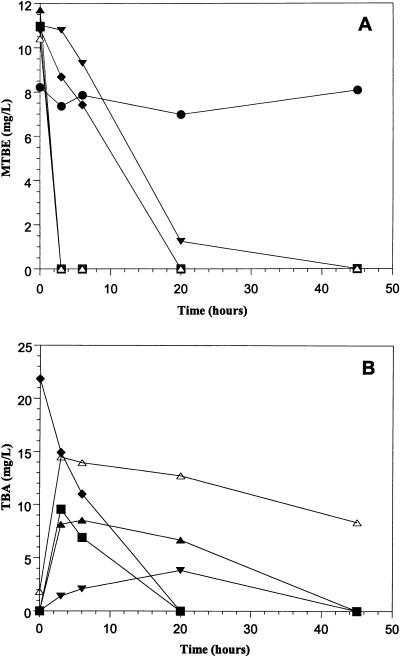
The ability of strain ENV735 to mineralize [14C]MTBE has been described (29). To evaluate induction of MTBE degradation activity in strain ENV735, the cells were grown in BSM + TBA (160 mg/liter) + 0.01% YE, BSM + 0.4% YE, BSM + sucrose (0.1%), LB, or BSM + CSL (0.1%) and incubated with MTBE. With the exception of LB-grown cells, MTBE was degraded without a lag period (Fig. 2A). The lag in MTBE degradation by LB-grown cells in this experiment may have been related to cell growth stage rather than gene induction, because the experiment was performed with overnight cultures and LB-grown cells may have entered early stationary phase. The other cultures were in the early to late log phase of growth. In other experiments, MTBE was degraded without a lag by both LB- and YE-grown cells. The highest rates of MTBE degradation with ENV735 were achieved with YE or sucrose-grown cells. The initial maximum MTBE oxidation rate at 30°C with YE-grown cells and 25 mg of MTBE/liter was 86 nmol of MTBE/min/mg of total cell protein.
FIG. 2.
Degradation of MTBE (A) and TBA (B) by ENV735. Cells were grown on either LB (▾), 0.4% YE (▪), TBA (⧫), CSL (▴), or sucrose (▵) and were assayed for MTBE and TBA biodegradation as described in Materials and Methods. Samples containing poisoned LB-grown cells are also represented (●). These samples are not shown in panel B because no TBA was produced in the samples.
To evaluate what concentrations of MTBE could be degraded by strain ENV735, YE-grown cells (OD550 = 2) were incubated for 72 h at room temperature with either 25, 100, 300, 500, 1,000 or 3,000 mg of MTBE/liter. No MTBE remained in the samples after incubation. TBA concentrations in the samples after incubation were 0, 0, 0.23, 0.85, 316, and 694 mg/liter, respectively.
Inhibitor studies.
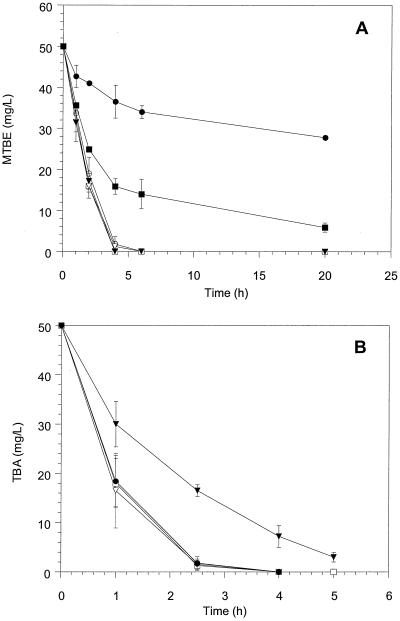
The influence of four metabolic inhibitors, an epoxide, and formaldehyde on MTBE and TBA degradation by ENV735 was tested. ABT and CO are known inhibitors of P450 monooxygenases, and they were previously observed to inhibit MTBE oxidation by propane-oxidizing bacteria (28). ATU chelates copper and irreversibly inhibits copper-containing monooxygenases, including some butane monooxygenases, and acetylene is a mechanism-based inactivator that binds tightly to specific monooxygenases (10). Butadiene monoepoxide was tested as an inhibitor because epoxides such as ethylene oxide have been shown to inactivate the butane oxidation system of Pseudomonas butanovora (10).
MTBE degradation was inhibited by ABT (0.1 mM) but not by CO (30% [vol/vol] of headspace) or acetylene (30% [vol/vol] of headspace) (Fig. 3A). MTBE degradation also was inhibited somewhat but not inactivated by butadiene monoepoxide, but the inhibition was not apparent until about 50% of the MTBE had been degraded. TBA degradation was slowed but not inactivated in the presence of acetylene (Fig. 3B). ABT, ATU, and CO did not inhibit TBA degradation. The influence of butadiene monoepoxide on TBA degradation was not tested.
FIG. 3.
Biodegradation of MTBE (A) and TBA (B) by strain ENV735 in the presence of known metabolic inhibitors and inactivators. ○, no inhibitor; □, CO; ▿, ATU; ●, ABT; ▪, butadiene monoepoxide; and ▾, acetylene. Values represent means of triplicate samples, and error bars represent 1 standard deviation of the mean.
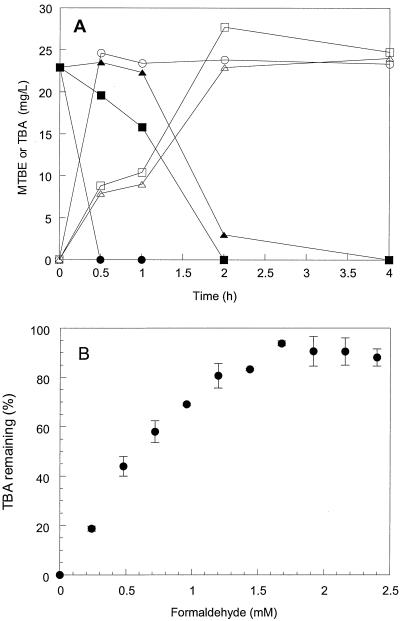
To evaluate formaldehyde toxicity in the culture, sucrose-grown ENV735 was incubated with 1.3 or 2.6 mM formaldehyde and 25 mg of MTBE/liter (Fig. 4A). All of the formaldehyde added to the cultures was degraded within 2 h of incubation. MTBE degradation was initiated only after the added formaldehyde was completely degraded by ENV735, but degradation rates were similar in cultures with and without initial formaldehyde addition. Likewise, when strain ENV735 was grown on formaldehyde as a sole carbon source, MTBE degradation was not inhibited (data not shown). TBA produced from MTBE degradation, however, was not degraded in formaldehyde-grown cultures during a 20-h incubation.
FIG. 4.
Effect of formaldehyde on MTBE (A) and TBA (B) degradation by ENV735. Symbols in panel A are as follows: ●, MTBE only; ○, TBA produced in MTBE-only samples; ▪, MTBE + 1.3 mM formaldehyde; □, TBA produced in MTBE + 1.3 mM formaldehyde samples; ▴, MTBE + 2.6 mM formaldehyde; and ▵, TBA produced in MTBE + 2.6 mM formaldehyde samples. Symbols in panel B represent mean ± standard deviation (n = 3) of TBA concentration after 20 h of incubation with different concentrations of formaldehyde.
In another experiment, TBA-grown ENV735 was incubated with TBA (2 mM) and different concentrations of formaldehyde (Fig. 4B). The amount of TBA remaining in the samples was determined after 20 h of incubation. TBA degradation was inhibited by as little as 0.24 mM (7.2-mg/liter) formaldehyde.
TBA degradation.
When YE-grown cells of ENV735 were incubated with MTBE, MTBE degradation was accompanied by a nearly stoichiometric accumulation of TBA and TBA degradation did not occur until after a lag period of about 5 h (29). A similar accumulation of TBA occurred in cells grown on YE, CSL, LB, or sucrose (Fig. 2B). There was no lag in TBA degradation, however, if cells were grown on TBA. The initial TBA concentration was somewhat higher than predicted from stoichiometric MTBE degradation, probably due to residual TBA in the cell suspensions after washing (Fig. 2B). TBA degradation was retarded in sucrose-, CSL-, and LB-grown cultures, and TBA accumulated to a lower level in LB-grown cultures than in cultures grown on other substrates. In another experiment, ENV735 was grown on either sucrose or sucrose + 0.01% YE and was incubated with 10 mg of MTBE/liter. TBA accumulated to 6 and 8 mg/liter within 4.5 h in the cultures without or with YE, respectively, but TBA was completely degraded within 20 h in cultures containing YE. No further TBA degradation occurred in sucrose-grown cultures without YE. Strain ENV735 did not degrade TBA in the absence of oxygen. HIBA was the only other water-soluble degradation intermediate detected in this study, and strain ENV735 was capable of growth on this metabolite (data not shown). No additional detailed analyses were performed to elucidate the entire MTBE degradation pathway of ENV735.
Induction of TBA degradation.
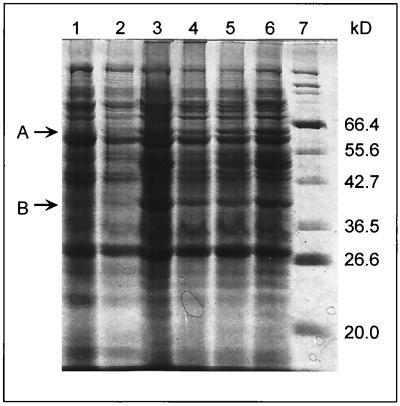
To further evaluate induction of TBA degradation, strain ENV735 was grown in BSM with either 0.4% YE (noninducing conditions) or with MTBE or TBA + 0.01% YE (inducing conditions). Alternately, cells were grown on YE and were then incubated for 5 h with 100 mg of TBA/liter. Cells were harvested, and total cell proteins were separated by PAGE (Fig. 5). When cells were grown with MTBE or TBA, they produced at least two polypeptides (∼60 and ∼40 kDa) that were not produced in abundance in YE-grown cells. The identity of these peptides is presently under investigation.
FIG. 5.
SDS-PAGE gel of ENV735 proteins after growth on different substrates. Lanes 1 and 2, LB medium; lanes 3 and 6, TBA + 0.01% YE; lanes 4 and 5, MTBE + 0.01% YE; and lane 7, protein size markers. Protein size in kilodaltons is shown to the right of the figure. Arrows A and B identify peptides produced during growth on MTBE and TBA but not during growth on LB.
Protein labeling.
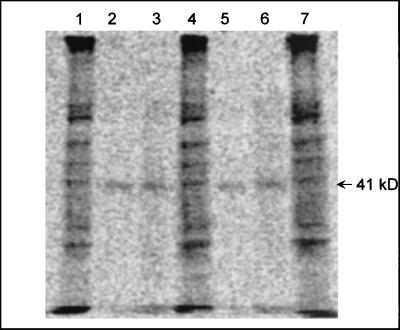
To identify peptides potentially involved in MTBE metabolism, YE-grown ENV735 cells were incubated with uniformly labeled [14C]MTBE in the presence or absence of the protein synthesis inhibitor chloramphenicol (Fig. 6). In the absence of chloramphenicol, many cellular proteins were labeled, thereby demonstrating incorporation of MTBE carbon into biomass. In the presence of chloramphenicol, however, a single peptide of approximately 41 kDa was labeled. Because the peptides were denatured by boiling in SDS prior to electrophoresis, it is suspected that an MTBE metabolite was covalently bound to the peptide. The identity of the labeled peptide is under investigation but has not yet been determined.
FIG. 6.
Phosphorimage of 14C-labeled proteins from ENV735. TBA-grown cells were incubated with uniformly labeled [14C]MTBE in the presence (lanes 2, 3, 5, and 6) or absence (lanes 1, 4, and 7) of chloramphenicol and were then denatured and separated on a PAGE gel. The gel was dried and placed on a phosphorimaging system to identify 14C-labeled proteins. Protein size (41 kDa) is indicated by arrow.
DISCUSSION
MTBE oxidation by strain ENV735 occurred constitutively at an initial rate of approximately 86 nmol/min/mg of cell protein at 25°C. This was approximately 5 to 10 times greater than the rate of MTBE oxidation by propane-oxidizing bacteria (28). The strain converted MTBE rapidly to CO2, especially if the TBA pathway was induced by growth on or exposure to TBA. Thus, unlike propane-oxidizing bacteria (28) or some other pure cultures described to date (9, 14, 17), strain ENV735 appears to have an efficient pathway for metabolizing TBA and its oxidation products, albeit at a slightly lower initial rate than MTBE oxidation.
To evaluate the possible role of hydrogen oxidation in MTBE degradation by ENV735 and to determine if the ability to metabolize MTBE is widespread among aerobic hydrogen-oxidizing bacteria, we tested two type strains of Hydrogenophaga and enriched for hydrogen oxidizers from soil and sludge. None of the organisms or enrichments tested metabolized MTBE well, and the presence of H2 did not appear to affect MTBE oxidation by strain ENV735. Thus, the ability of ENV735 to oxidize MTBE may be an acquired trait, but the extent of MTBE-degrading ability among indigenous hydrogen-oxidizing bacteria in MTBE-contaminated aquifers requires further investigation.
To date, few pure or mixed cultures of bacteria have been shown to grow on MTBE as a carbon source. The cultures that have been isolated grow relatively slowly on the compound and have low cell yields (11, 22, 23). These growth characteristics could result from slow initial oxidation of the oxygenate, poor energy yield, poor utilization of the compound and its metabolites in the biosynthetic process, specific nutritional requirements of the organisms, or a combination of these factors. Salanitro and colleagues (22) compared the heats of combustion of several compounds with the typical growth yields of bacteria on these substrates. With common bacterial growth substrates, the relationship between these parameters was linear. MTBE and its metabolite TBA, however, had heats of combustion similar to isopropanol, propane, and methanol, but cell yields of the mixed culture BC-1 on MTBE and TBA were only about 12 to 15% of that expected based on heat of combustion alone. Interestingly, many of the substrates that exhibited a typical linear relationship between growth yield and heat of combustion were suspected or identified MTBE metabolites (28). Thus, although MTBE had sufficient chemical energy to support bacterial growth and BC-1 could presumably degrade MTBE to produce good growth substrates, growth on MTBE was still poor. Similarly, in previous work (28), it was observed that propane-oxidizing bacteria could oxidize MTBE and TBA but that the inability to efficiently oxidize certain metabolites (e.g., HIBA) apparently limited growth of the organisms on MTBE. Thus, poor growth yield on MTBE may in part be due to a lack of necessary downstream metabolic pathways to efficiently mineralize MTBE. The observation that the growth yield of strain ENV735 on MTBE and TBA can be enhanced by adding YE (Fig. 1) suggests that certain cofactors or inducers may be necessary for efficient growth on MTBE.
Because of the possibility that product toxicity is caused by the binding of metabolites to cellular proteins, the labeling of cellular proteins during oxidation of uniformly labeled [14C]MTBE was evaluated. In the absence of chloramphenicol, MTBE metabolism led to the incorporation of [14C]MTBE metabolites into multiple cellular proteins (Fig. 6). In the presence of the protein synthesis inhibitor chloramphenicol, however, only a single peptide of approximately 41 kDa was labeled. It is possible that the protein labeled in the presence of chloramphenicol is associated with MTBE or TBA metabolism and that [14C]-labeled products produced from [14C]MTBE oxidation react immediately with the degradative enzyme. Similar enzyme-inactivating reactions have been observed during the monooxygenase-catalyzed epoxidation of alkenes (10) and chlorinated alkenes (19). Binding of degradation products to other similarly sized peptides cannot be ruled out, but persistence of the label after protein denaturation indicates a strong peptide-label interaction. These results suggest that the poor growth of microorganisms on MTBE may at least partially be related to the toxic effects of products produced during MTBE metabolism. Similar labeling experiments performed with methoxy-labeled [14C]MTBE and/or purified enzymes will help to better understand this finding.
During the degradation of MTBE by propane-oxidizing bacteria, TBA metabolism proceeded at a lower rate than MTBE oxidation and TBA accumulated stoichiometrically during MTBE oxidation (28). This finding led us to suggest that the same enzyme was involved in oxidizing both substrates. Similar findings were observed and conclusions were drawn by others investigating MTBE degradation by alkane-oxidizing bacteria (16). With strain ENV735, MTBE was degraded without a lag period, regardless of the growth substrate tested, suggesting that MTBE oxidation activity is constitutively expressed in this strain. Like propane-oxidizing bacteria, TBA accumulated during MTBE degradation by rich medium-grown ENV735, and TBA was not degraded until after about 5 h of incubation (Fig. 2B). A similar 5-h lag in TBA degradation was observed when ENV735 was grown on rich media and then fed TBA in the absence of MTBE (data not shown). TBA did not accumulate, however, in cultures grown on TBA or MTBE, even in the presence of MTBE. These results suggest that TBA oxidation in ENV735 is inducible and that separate genes control MTBE and TBA degradation. These results are supported by experiments that demonstrated that MTBE was degraded rapidly by sucrose-grown cells, whereas TBA was not degraded well by cells grown on sucrose. Additionally, MTBE degradation was inhibited by ABT but not by acetylene, and TBA degradation was not inhibited by ABT but was slower in the presence of acetylene. Furthermore, when strain ENV735 was incubated with formaldehyde, MTBE degradation occurred at essentially the same rate as for cells that were not treated with formaldehyde, albeit not until the added formaldehyde had been degraded by the cells (Fig. 4A). Likewise, cells grown on formaldehyde as a sole carbon source readily degraded MTBE (data not shown). TBA degradation, however, was inhibited by formaldehyde at relatively low concentrations (Fig. 4B).
To further investigate the induction of TBA metabolic genes, the proteins produced by ENV735 before and after exposure to TBA were examined (Fig. 5). At least two polypeptides were expressed at much higher levels in cells exposed to TBA, either through direct addition or from MTBE oxidation, than in unexposed cells. Thus, it is likely that these polypeptides are involved in TBA metabolism and that TBA or TBA degradation products induce their production. The identity of these peptides is presently under investigation.
Although in situ MTBE degradation can occur in some aquifers without the addition of specialized organisms (3, 24), in other cases MTBE-degrading organisms may be needed as biocatalysts (27) or seed cultures (24) to facilitate degradation. In these cases, the use of pure bacterial cultures like ENV735 can have advantages over the use of consortia. The advantages include lower fermentation costs, the ability to manipulate or screen cultures to select variants with desirable phenotypic characteristics (27), and an increased certainty about the safety of the microbes that will be added to the environment. Thus, the isolation of pure cultures of degradative bacteria, like H. flava ENV735, is important both for studying the mechanisms and limitations of contaminant degradation and for providing biocatalysts that can be used to solve real pollution problems. The results of this study suggest that strain ENV735 can degrade MTBE regardless of the fermentation substrate used to grow it but that complete degradation of MTBE metabolites like TBA requires induction of additional degradative genes. Thus, TBA can be expected to transiently accumulate in MTBE-contaminated aquifers treated with ENV735 as a remediation biocatalyst, but the strain will degrade TBA and other metabolites upon induction of the downstream pathway(s).
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation through the Small Business Innovative Research program (grant no. DMI-9960886). Patents are pending on this technology.
REFERENCES
- 1.American Type Culture Collection. Catalogue of bacteria and bacteriophages. 18th ed. Rockville, Md: American Type Culture Collection; 1992. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 3.Bradley P M, Chapelle F H, Landmeyer J E. Methyl t-butyl ether mineralization in surface-water sediment microcosms under denitrifying conditions. Appl Environ Microbiol. 2001;67:1975–1978. doi: 10.1128/AEM.67.4.1975-1978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley P M, Landmeyer J E, Chapelle F H. Widespread potential for microbial MTBE degradation in surface-water sediments. Environ Sci Technol. 2001;35:658–662. doi: 10.1021/es0015489. [DOI] [PubMed] [Google Scholar]
- 5.Buscheck T E, Gallagher D J, Peargin T R, Kuehne D L, Zuspan C R. Proceedings of the National Ground Water Association Southwest Focused Ground Water Conference, Anaheim, Calif. 1998. Occurrence and behavior of MTBE in groundwater; pp. 2–3. [Google Scholar]
- 6.Deeb R A, Hu H-Y, Hanson J R, Skow K M, Alvarez-Cohen L. Substrate interactions in BTEX and MTBE mixtures by an MTBE-degrading isolate. Environ Sci Technol. 2001;35:312–317. doi: 10.1021/es001249j. [DOI] [PubMed] [Google Scholar]
- 7.Fortin N Y, Deshusses M A. Treatment of methyl tert-butyl ether vapors in biotrickling filters. 1. Reactor startup, steady-state performance, and culture characteristics. Environ Sci Technol. 1999;33:2980–2986. [Google Scholar]
- 8.Fortin N Y, Deshusses M A. Treatment of methyl tert-butyl ether vapors in biotrickling filters. 2. Analysis of the rate-limiting step and behavior under transient conditions. Environ Sci Technol. 1999;33:2987–2991. [Google Scholar]
- 9.Garnier P M, Auria R, Augur C, Revah S. Cometabolic biodegradation of methyl t-butyl ether by Pseudomonas aeruginosa grown on pentane. Appl Microbiol Biotechnol. 1999;51:498–503. doi: 10.1007/s002530051423. [DOI] [PubMed] [Google Scholar]
- 10.Hamamura N, Storfa R T, Semprini L, Arp D J. Diversity in butane monooxygenases among butane-grown bacteria. Appl Environ Microbiol. 1999;65:4586–4593. doi: 10.1128/aem.65.10.4586-4593.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson J R, Ackerman C E, Scow K M. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl Environ Microbiol. 1999;65:4788–4792. doi: 10.1128/aem.65.11.4788-4792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardison L K, Curry S S, Ciuffetti L M, Hyman M R. Metabolism of diethyl ether and cometabolism of methyl tert-butyl ether by a filamentous fungus, a Graphium sp. Appl Environ Microbiol. 1997;63:3059–3067. doi: 10.1128/aem.63.8.3059-3067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hareland W, Crawford R L, Chapman P J, Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975;121:272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez-Perez G, Fayolle F, Vandecasteele J-P. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl Microbiol Biotechnol. 2001;55:117–121. doi: 10.1007/s002530000482. [DOI] [PubMed] [Google Scholar]
- 15.Hyman M, Taylor C, O'Reilly K. Cometabolic degradation of MTBE by iso-alkane-utilizing bacteria from gasoline-impacted soils. In: Wickramanayake G B, Gavaskar A R, Alleman B C, Magar V S, editors. Bioremediation and phytoremediation of chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 2000. pp. 149–155. [Google Scholar]
- 16.Hyman M, Kwon P, Williamson K, O'Reilly K. Cometabolism of MTBE by alkane-utilizing microorganisms. In: Wickramanayake G B, Hinchee R E, editors. Natural attenuation of chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 1998. pp. 321–326. [Google Scholar]
- 17.Mo K, Lora C O, Wanken A E, Javanmardian M, Yang X, Kulpa C F. Biodegradation of methyl t-butyl ether by pure bacterial cultures. Appl Microbiol Biotechnol. 1997;47:69–72. doi: 10.1007/s002530050890. [DOI] [PubMed] [Google Scholar]
- 18.Mormile M R, Liu S, Suflita J M. Anaerobic biodegradation of gasoline oxygenates: extrapolation of information to multiple sites and redox conditions. Environ Sci Technol. 1994;28:1727–1732. doi: 10.1021/es00058a026. [DOI] [PubMed] [Google Scholar]
- 19.Newman L M, Wackett L P. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J Bacteriol. 1997;179:90–96. doi: 10.1128/jb.179.1.90-96.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park K, Cowan R M. Proceedings of the 213th American Chemical Society National Meeting. Washington, D.C.: American Chemical Society; 1997. Effects of oxygen and temperature on the biodegradation of MTBE; pp. 421–423. [Google Scholar]
- 21.Peaff G. Court ruling spurs continued debate over gasoline oxygenates. Chem Eng News. 1994;72:8–13. [Google Scholar]
- 22.Salanitro J P, Chou C S, Wisniewski H L, Vipond T E. Proceedings of the National Ground Water Association Southwest Focused Ground Water Conference, Anaheim, Calif. 1998. Perspectives on MTBE biodegradation and the potential for in situ aquifer bioremediation; pp. 40–54. [Google Scholar]
- 23.Salanitro J P, Diaz L A, Williams M P, Wisniewski H L. Isolation of a bacterial culture that degrades methyl t-butyl ether. Appl Environ Microbiol. 1994;60:2593–2596. doi: 10.1128/aem.60.7.2593-2596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salanitro J P, Johnson P C, Spinnler G E, Maner P M, Wisniewski H L, Bruce C. Field-scale demonstration of enhanced MTBE bioremediation through aquifer bioaugmentation and oxygenation. Environ Sci Technol. 2000;34:4152–4162. [Google Scholar]
- 25.Squillace P J, Pankow J F, Korte N E, Zogorski J S. Review of the environmental behavior and fate of methyl tert-butyl ether. Environ Toxicol Chem. 1997;16:1836–1844. [Google Scholar]
- 26.Squillace P J, Zogorski J S, Wilber W G, Price C V. Preliminary assessment of the occurrence and possible sources of MTBE in groundwater in the United States, 1993–1994. Environ Sci Technol. 1996;30:1721–1730. [Google Scholar]
- 27.Steffan R J, Sperry K L, Walsh M T, Vainberg S, Condee C W. Field-scale evaluation of in situ bioaugmentation for remediation of chlorinated solvents in groundwater. Environ Sci Technol. 1999;33:2771–2781. [Google Scholar]
- 28.Steffan R J, McClay K, Vainberg S, Condee C W, Zhang D. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl Environ Microbiol. 1997;63:4216–4222. doi: 10.1128/aem.63.11.4216-4222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steffan R J, Vainberg S, Condee C W, McClay K, Hatzinger P. Biotreatment of MTBE with a new bacterial isolate. In: Wickramanayake G B, Gavaskar A R, Alleman B C, Magar V S, editors. Bioremediation and phytoremediation of chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 2000. pp. 165–173. [Google Scholar]
- 30.Suflita J M, Mormile M R. Anaerobic biodegradation of known and potential gasoline oxygenates in the terrestrial subsurface. Environ Sci Technol. 1993;27:976–978. [Google Scholar]
- 31.U.S. Environmental Protection Agency. Measurement of purgeable organic compounds in water by capillary-column gas chromatography/mass spectroscopy, method 524.2, revision 4.0. Cincinnati, Ohio: Environmental Monitoring Systems Laboratory; 1992. [Google Scholar]
- 32.Wilson J T, Cho J S, Wilson B H, Vardy J A. Natural attenuation of MTBE in the subsurface under methanogenic conditions. EPA/600/R-00/006. U.S. Washington, D.C.: Environmental Protection Agency; 2000. [Google Scholar]
- 33.Yeh C K, Novak J T. Anaerobic biodegradation of gasoline oxygenates in soils. Water Environ Res. 1994;66:744–752. [Google Scholar]