Abstract
Detachment from biofilms is an important consideration in the dissemination of infection and the contamination of industrial systems but is the least-studied biofilm process. By using digital time-lapse microscopy and biofilm flow cells, we visualized localized growth and detachment of discrete cell clusters in mature mixed-species biofilms growing under steady conditions in turbulent flow in situ. The detaching biomass ranged from single cells to an aggregate with a diameter of approximately 500 μm. Direct evidence of local cell cluster detachment from the biofilms was supported by microscopic examination of filtered effluent. Single cells and small clusters detached more frequently, but larger aggregates contained a disproportionately high fraction of total detached biomass. These results have significance in the establishment of an infectious dose and public health risk assessment.
The detachment of bacterial cells from biofilms is of fundamental importance to the dissemination of infection and to contamination in both clinical (12) and public health (13, 18, 19) settings. However, detachment is the least-studied biofilm process and remains poorly understood (10, 14). The spontaneous detachment of cells from bacterial biofilms has been divided into two processes, erosion and sloughing, based on the magnitude and frequency of the detachment event (3, 4). Erosion is the continual detachment of single cells and “small portions of the biofilm,” whereas sloughing is the “rapid, massive loss of biofilm” (4). However, the size distribution and detachment frequency of biofilm particulates have not been quantified. Information on biofilm detachment is usually inferred from monitoring the cell concentration in the liquid phase after sample homogenization. This process provides a temporally and spatially averaged detachment rate, but information on the size distribution and detachment frequency is lost. A minimum infectious dose of cells concentrated in a biofilm particulate may be overlooked if such an aggregate is disrupted and effectively diluted in the test tube.
Digital time-lapse microscopy (DTLM) has been used to track single cells moving between biofilm cell clusters (5) and to quantify the dynamic viscoelastic behavior and movement of mature biofilms over solid surfaces in situ (15, 16). In previous investigations on the influence of hydrodynamics and nutrient concentration on the structure of a four-species laboratory biofilm, it was reported that an increase in C and N concentrations resulted in a change in architecture from a thin layer of migratory ripples (regularly spaced ridges running perpendicular to the flow direction) and filamentous streamers (cell clusters elongated in the downstream direction with “tails” rapidly oscillating in the flow) to one of mushroom- and mound-shaped cell clusters (17). DTLM suggested that these cell clusters were continually growing and detaching from the biofilm.
The goal of the work presented here was to examine localized growth and detachment events at the microscopic scale and to use image analysis to quantify the size distribution and detachment rate for discrete biofilm cell clusters. We then broadened the scope of our experiments to include an undefined biofilm developed from a tap water inoculum to determine if similar detachment patterns might occur in “natural” biofilms. To support results from direct observation of the biofilm, we also microscopically quantified the distribution of particulate biomass in the filtered effluent from the tap water biofilm.
MATERIALS AND METHODS
Biofilm reactor system.
Biofilms were grown in square glass tubing flow cells (20 cm of 3- by 3-mm tubing; Friedrich & Dimmock Inc.) that were incorporated into a recirculation loop of a chemostat (17). The volume of the mixing chamber and the recycle loop was 175 ml. The nutrient influent flow rate was 4.3 ml min−1, giving a resulting residence time of 40 min. Under these conditions, the reactor system would only support suspended populations with a doubling time of less than 28 min (maximum specific growth rate [μmax], 1.47 h−1). The average flow velocity through the flow cells was maintained at 1 m s−1 throughout the experiments. The corresponding Reynolds number (Re) was 3,600, and flow was turbulent (15). The flow cells were positioned on a microscope stage to allow the biofilms to be imaged in situ. Under operating conditions, the water temperature in the reactor system was 26 ± 2°C.
Inocula and nutrients.
Two inocula were used, a defined four-species laboratory model and an undefined tap water consortium. The four-species biofilm was composed of the gram-negative environmental isolates Pseudomonas aeruginosa (ATTC 700829), Pseudomonas fluorescens (ATTC 700830), Klebsiella pneumoniae (ATTC 700831), and Stenotrophomonas maltophilia (identified by fatty acid analysis; similarity index, 0.727; MIDI, Newark, Del.). The μmax values obtained from batch culture growth curves for the first three species on minimal salts medium (MSM) were 0.50 ± 0.04 (n = 3), 0.22 ± 0.00 (n = 2), and 0.41 ± 0.09 (n = 3) h−1, respectively (mean and standard deviation [SD]). Because S. maltophilia requires growth factors, it could not be grown in pure culture on MSM alone and, in previous experiments with the four-species consortium, took between 7 and 10 days to reach detectable concentrations in the reactor (17). To simulate growth conditions in the reactor system, we grew S. maltophilia batch cultures on filter-sterilized spent MSM from a culture inoculated with the other three species and supplemented with fresh C and N sources (glucose and ammonium sulfate). The μmax was 0.41 ± 0.07 (n = 5) h−1.
The four-species biofilm was initially grown on MSM consisting of glucose (40 mg/liter), (NH4)2SO4 (10 mg/liter), KH2PO4 (70 mg/liter), K2HPO4 (30 mg/liter), MgSO4 · 7H2O (10 mg/liter), and trace elements: Na2EDTA · 2H2O (12 mg/liter), FeSO4 · 0.7H2O (2 mg/liter), CaCl2 (1 mg/liter), Na2SO4 (1 mg/liter), ZnSO4 · 0.7H2O (4 mg/liter), MnSO4 · 0.4H2O (4 mg/liter), CuSO4 · 0.5H2O (1 mg/liter), and Na2MoO4 · 0.2H2O (1 mg/liter) (pH 7.0 ± 0.2) (40 ppm glucose MSM). After 21 days, the glucose and (NH4)2SO4 concentrations were increased to 400 and 100 mg/liter, respectively (400 ppm glucose MSM). Detachment was monitored microscopically on days 24 and 26 at these nutrient concentrations. The C and N sources were then reduced back to the original concentrations for the remaining 3 days of the run to investigate the effect on biofilm structure. Detachment was not monitored during this period.
For inoculation, frozen stock cultures (0.5 ml of 3 × 108 to 4 × 109 CFU/ml) of each of the species were thawed and injected directly into the mixing chamber. The reactor was initially run as a recirculating batch culture for 24 h before it was switched to a continuous culture. The reactor was operated above washout at a dilution rate of 1.47 h−1 to favor biofilm growth and allow the assumption that cells in the effluent occurred predominantly from detachment.
For the tap water biofilm, tap water was recirculated through an autoclaved reactor for 24 h before the addition of sterile MSM with sodium succinate (1,000 mg/liter) as the carbon source. The biofilm was grown for 20 days.
Enumeration and identification of bacteria in the effluent and the biofilm. (i) Enumeration of cells in the effluent.
Effluent samples (0.5 ml) were periodically syringe drawn through an in-line septum and vortexed for 3 10-s periods before serial dilution and plating. For the four-species biofilm, we used King's B agar with added bromthymol blue (0.03 g/liter). Individual species were distinguished by colony morphology, acid production, and fluorescence (17). Plates were incubated at 26°C, and counts were determined after 48 h. For the tap water inoculum, we used nutrient agar (Difco). Nonvortexed samples were also enumerated to estimate aggregation in the effluent.
(ii) Enumeration of cells in the biofilm.
At the end of the experiments, the flow cells were removed and rinsed five times with 1 ml of sterile buffer (one-quarter-strength Ringer's solution). Three 1-cm sections were cut from each tube with a diamond knife and immersed in 5 ml of sterile buffer with 0.1% (wt/vol) Tween 20 in separate test tubes. The samples were sonicated for 3 min and vortexed for 10 s for five repeated cycles. Serial dilutions were plated on either King's B-bromthymol blue or nutrient agar. Plates were incubated at 26°C, and counts were determined after 48 h.
Microscopy and image analysis.
Biofilms were observed in situ by transmitted bright-field microscopy using an Olympus BH2 microscope. A COHU 4612-5000 charge-coupled-device camera (Cohu, Inc., San Diego, Calif.) and a VG-5 PCI framestore board (Scion Inc., Frederick, Md.) were used to capture images. Image capture, processing, and analysis were done using NIH-Image (rsb.info.nih.gov/nih-image/) on a Macintosh computer or Scion Image (www.scioncorp.com/) on a PC. Time-lapse sequences were made by capturing images at intervals ranging from 20 to 60 min over periods of up to 19 h using a ×10 or a ×20 objective. The objective resolution was approximately 1.1 or 0.7 μm, respectively. Distance and area measurements were calibrated using a 1-mm graticule with 10-μm divisions (CS990; Graticules Ltd., Tonbridge, Kent, United Kingdom).
Biofilm thickness and surface area coverage.
Biofilm thickness was measured by microscopic focusing (1), and surface coverage was measured by image analysis from digital images taken with either a ×10 or a ×20 objective (17). The average biofilm surface coverage and thickness were obtained from a minimum of five measurements taken at various days over the course of each experiment. Stable daily values in these structural parameters were used to ascertain biofilm maturity prior to DTLM imaging. Time-lapse sequences of the four-species biofilm were made on days 24 and 26. A time-lapse sequence of the tap water biofilm was made on day 14.
Frequency and size measurements for detaching cell clusters from direct observations.
The number and size of cell clusters that had detached between sequential frames were determined by subtracting each image from the preceding image with Scion Image. Cell clusters that had detached between frames appeared white, and areas of new growth appeared black. Similarly, cell clusters attaching from the effluent between frames appeared black. The gray-scale image was then inverted, and a threshold was applied so that the detached cell clusters appeared as black objects on a white background. The “Analyze Particles” command was used to quantify the number, area, and diameter of the detached cell clusters.
Size distribution of aggregates in the reactor effluent.
One milliliter of effluent from the tap water biofilm reactor was collected on days 9 and 14. The sample was stained with a Molecular Probes (www.probes.com) LIVE/DEAD BacLight bacterial viability kit. A dilution series was prepared in one-quarter-strength Ringer's buffer, and 1 ml of a 1:100 dilution was filtered onto a Nuclepore black polycarbonate 0.22-μm membrane. Samples were mixed by gently swirling to minimize aggregate disruption. Pipette tips were cut to increase orifice size and reduce shear, and a minimal vacuum was applied during filtering. Membranes were oil mounted and observed using either confocal (Leica TCSNT) or epifluorescence microscopy with a ×100 oil immersion objective. Fifty fields were captured for image analysis. Confocal microscopy showed that the aggregates flattened out on the membrane (data not shown), allowing a correlation to be made between the area of each aggregate on the filter and the number of cells in the aggregate. The number of cells in an aggregate was 1.23 × aggregate area (square micrometers) (r2, 0.85; n = 58). The area of 1 cell was 0.59 ± 0.37 μm2 (n = 113) (mean and SD). The “Analyze Particles” command was used to determine the number and area of aggregates on the membrane. Aggregates with areas in the range of 0.59 ± 0.37 μm2 were considered single cells, while the calibration curve was used to estimate cell number per aggregate for particles with areas of greater than 0.97 μm2
Statistics.
Regression analysis and analysis-of-variance comparisons were carried out using QuattroPro 9.0.0.528 (Corel Corp.) or MiniTab 13.20 (MiniTab). Differences were considered significant for P values of <0.05. Data were reported as mean and 1 SD.
RESULTS
Four-species inoculum. (i) Concentrations and proportions of bacteria in the effluent and biofilm.
The proportions of species in the effluent stabilized after 14 days and were dominated by K. pneumoniae (81.6%), with P. aeruginosa (11.5%), S. maltophilia (5.0%), and P. fluorescens (1.9%) in decreasing proportions. These proportions remained within a range of no more than ±6% over the course of the experiment. The concentration of viable cells in the bulk liquid was 2.1 × 107 ± 0.4 × 107 CFU/ml on day 21. After the glucose concentration was increased from 40 to 400 ppm, the effluent cell concentration increased by a factor of 10 to 2.2 × 108 ± 0.3 × 108 CFU/ml on day 23. There was a further increase to 8.3 × 108 ± 0.4 × 108 CFU/ml on day 26.
The concentration of viable cells in the biofilm was 6.6 × 108 ± 0.5 × 108 CFU/cm2. The biofilm was dominated by K. pneumoniae (88.8% ± 4.0%; n = 3), with P. aeruginosa (7.2% ± 4.0%), P. fluorescens (2.2% ± 1.7%), and S. maltophilia (1.9% ± 0.5%) making up the remaining proportions. There was no significant difference (P > 0.5 for each species) between the proportions of species in the biofilm (n = 3) and those in the effluent (n = 10) (for data points between days 14 and 26).
(ii) Biofilm morphology.
When the nutrient concentration was increased to 400 ppm glucose MSM on day 21, there was a significant increase in biofilm thickness from 26 ± 6.0 μm (n = 5) to 57.8 ± 8.1 μm on day 23 (P < 0.01). The surface area coverage also significantly increased from 49.3% ± 4.3% to 83.8% ± 6.3% (P < 0.01) over the same period. The biofilm morphology stabilized after a further 24 h. The biofilm thickness and surface coverage on day 26 were 59.2 ± 9.7 μm and 81.0% ± 14.8%, respectively, values which were not significantly different from those measured on day 23 (P = 0.83 and P = 0.74, respectively). The DTLM sequence on day 24 showed that cell clusters were continually growing and detaching from the biofilm (Fig. 1; the complete time-lapse sequence “Growth and Detachment of Cell Clusters from a Four-Species Biofilm” has been submitted to the ASM MicrobeLibrary [www.microbelibrary.org]). On day 29, 3 days after the nutrient reduction, the surface coverage decreased to 63.2% ± 14.8% and the biofilm thickness was 48.7 ± 11.8 μm.
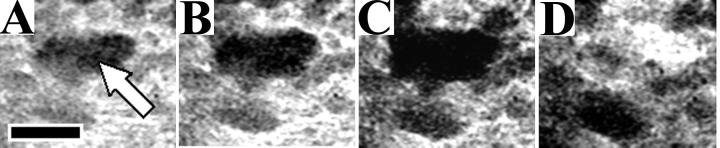
FIG. 1.
Growth and detachment of a representative cell cluster from the four-species biofilm. Panels A to D were taken at 2-h intervals on day 24. The arrow in panel A indicates the cell cluster. Underlying biofilm in the area beneath the detached cell cluster can be seen in panel D. Scale bar, 100 μm.
(iii) Size distribution and detachment rate for biofilm cell clusters.
The number and area of individual cell clusters that had detached from the biofilm between sequential images were determined by image subtraction (Fig. 2). Some biofilm was still visible in the areas underneath the detached clusters (Fig. 1). Because of the high surface area coverage (ca. 80%) and the use of low-power objectives, we estimated that we could confidently resolve only distinct cell clusters with areas as small as 140 μm2 (13-μm diameter). Attempts to use higher-power objectives to detect single cells detaching from the biofilm proved difficult due to the speckled nature of the background that resulted from the rapid twitching motion of the cells, which was presumably from motility and flow-induced biofilm vibration. In some instances, the cell clusters appeared to stretch and move slightly downstream prior to detaching. However, an absence of cell clusters entering or tracking across the field of view suggested that there was no downstream migration of the kind observed for the ripple-type architecture (16).
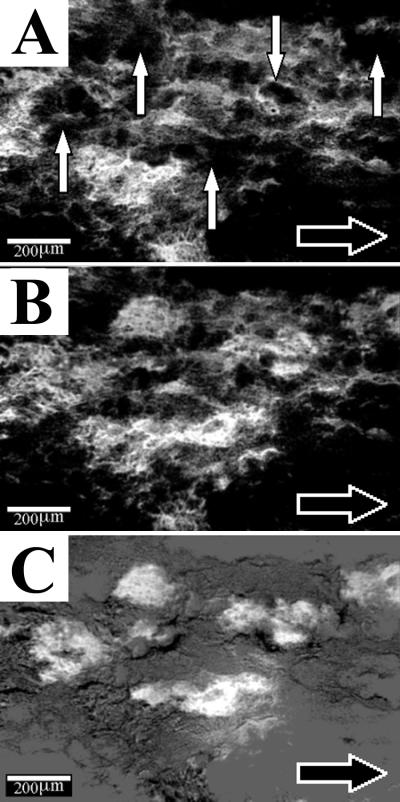
FIG. 2.
Detachment of biofilm cell clusters from the four-species biofilm on day 24. Image analysis was used to subtract the image made an hour earlier (A) from the subsequent image (B). The result is shown in panel C. Cell clusters that had detached (vertical white arrows in panel A) in the elapsed hour appeared white. Note that without using image subtraction, it would have been difficult to distinguish changes in the biofilm structure that had occurred between panels A and B. The horizontal black arrow indicates flow direction in the flow cell.
Cell clusters detached at a steady rate (14.9 detaching clusters mm−2 h−1; r2 = 0.97, n = 19) over the 19-h monitoring period on day 24. The histogram distributions from days 24 and 26 showed that although smaller clusters detached more frequently, larger clusters accounted for a disproportionately high fraction of the total detached biofilm area (Fig. 3). The mean diameters of the detached cell clusters were 69.8 ± 130 μm (n = 286) on day 24 and 61.2 ± 89 μm (n = 151) on day 26. The SDs reflect the large variability in the sizes of the cell clusters. The largest detached cluster had an area of 1.8 × 105 μm2 (diameter, 478 μm).
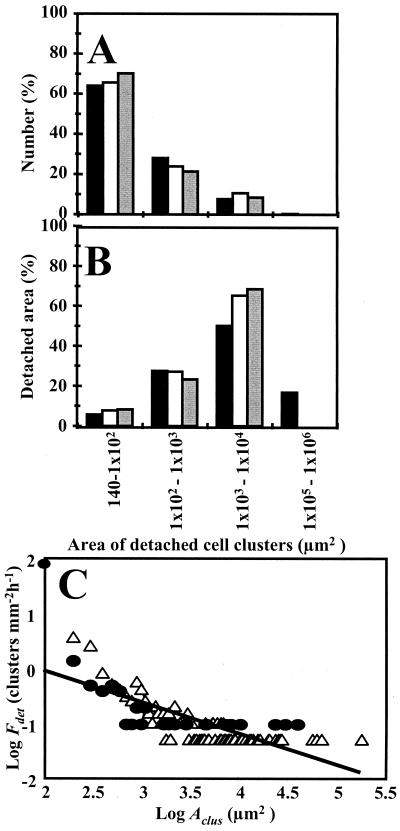
FIG. 3.
Histogram distributions of detached cell clusters in the four-species biofilm on day 24 (black bars) and day 26 (open bars) and the tap water biofilm on day 14 (gray bars). The size groups are based on the areas of the detached clusters. (A) Percentage of cell clusters detaching from each size group. (B) Proportion of detached biofilm area from each cluster size group. (C) Log-log plot of the calculated detachment frequency (Fdet) for cell clusters as a function of cell cluster area (Aclus) in the four-species biofilm on day 24 (▵) and the tap water biofilm on day 14 (●). The clusters were grouped by area into bin sizes of 100 μm2. The solid line is the linear regression of the log data and yields the empirical relationship Fdet = 14Aclus−0.58 (r2 = 0.57, n = 96).
Tap water inoculum. (i) Biofilm morphology.
The tap water biofilm formed discrete mound-shaped cell clusters, some of which became elongated in the downstream direction to form filamentous streamers. By day 11, the thickness of the biofilm had increased to 50.3 ± 19.9 μm (n = 10). The thickness subsequently did not change significantly between sequential daily measurements (P > 0.13) to the end of the experiment (day 18), when the thickness was 60.3 ± 33.1 μm. Similarly, the surface coverage had increased to 22.1% ± 7.3% (n = 10) by day 7, after which it stabilized, with no further significant change between sequential daily measurements (P > 0.09). At day 16, the surface coverage was 23% ± 7.5%.
(ii) Viable cells in the effluent and biofilm and species identification.
The culturable cell concentration in the effluent of the tap water biofilm was stabilized at approximately 109 CFU/ml by day 7. The effluent concentration ranged between 2.7 × 108 ± 0.6 × 108 CFU/ml and 6.6 × 109 ± 1.9 × 109 CFU/ml until the end of the experiment. After 2 days of growth, two distinct colony types dominated the effluent plates (>99%). These were identified using API 20E test strips (BioMerieux, Vitek Inc., Hazelwood, Mo.) as S. maltophilia (API percent identification, 99.9; T = 0.92) and Ochrobacterium anthropi (API percent identification, 99.7; T = 0.55), two opportunistic pathogens. O. anthropi ranged between 90 and 92% of the cultured population while S. maltophilia ranged between 7 and 9% over the course of the experiment. The concentration of cells in the biofilm was 8.0 × 107 ± 1.0 × 107 CFU/cm2(n = 3). O. anthropi was the dominant species in the biofilm, making up 64% ± 5% of the total CFU. S. maltophilia made up 35% ± 5%, and other species made up 1%.
(iii) Growth and detachment of biofilm cell clusters.
A 19-h time-lapse sequence with a frame interval of 20 min on day 14 showed dynamic growth and detachment behaviors similar to those for the four-species biofilm (the complete time-lapse sequence “Growth and Detachment of Cell Clusters from a Tap Water Biofilm” has been submitted to the ASM MicrobeLibrary [www.microbelibrary.org]). Because the surface coverage was only ca. 20% (compared with 80% for the four-species biofilm), we estimated that we could resolve detached cell clusters with areas as small as 20 μm2 (diameter, 5 μm). Higher magnification with a ×40 objective showed that, after a period of growth, the cell clusters peeled away from the surface in the direction of fluid flow (“Growth and Detachment of Cell Cluster in Turbulent Flow”; www.microbelibrary.org). The calculated detachment rate was 3.7 clusters mm−2 h−1 (r2 = 0.86, n = 19) when frame intervals of 1 h were used. However, when frame intervals of 20 min were analyzed, the calculated detachment rate was 80.2 clusters mm−2 h−1 (r2 = 0.99, n = 53), demonstrating that a significant number of dynamic events were missed at the longer time interval. Generally, for sampling of periodic behavior, the sampling frequency should be at least three times the actual frequency, emphasizing the need to determine the temporal scales of dynamic behaviors in biofilms for sampling optimization. The size distribution of detached biofilm clusters was similar to that for the four-species biofilm (Fig. 3).
(iv) Aggregates in the effluent.
Epifluorescence microscopic examination revealed that the effluent sampled on day 14 contained both single cells and aggregates of cells. The number of cells in each particle ranged from single cells to a cluster containing an estimated 1.7 × 103 cells (Fig. 4). The size distribution of aggregates showed a trend similar to that seen from direct observation of cell clusters leaving the biofilm. Single cells and small cell clusters occurred most frequently, but larger aggregates contained a disproportionately high fraction of the total biomass (Fig. 4). On average, there were 5.9 ± 42.6 cells per aggregate. The CFU concentration of the vortexed samples was 2.4% ± 1.6% (n = 5) times greater than that of the nonvortexed samples, suggesting that conventional vortexing was not sufficient to totally disperse the detached aggregates.
FIG. 4.
Biomass particulates in the filtered tap water biofilm effluent collected on day 14. (A) Single cells and small aggregates of less than 10 cells. (B) Single cells and an aggregate containing approximately 80 cells. (C) Large aggregate containing approximately 900 cells. Scale bar, 20 μm. (D) Histogram showing the distribution of detached particles by size (white bars) and by the total number of cells in each size group (gray bars).
DISCUSSION
In this study, DTLM was used to quantify the size distribution and detachment frequency for biofilm particulates from two types of mature mixed-species biofilms growing in turbulent flow. In both biofilms, there was a steady detachment of cell clusters over the 16- to 19-h monitoring periods. The four-species biofilm produced an estimated detached population of up to 3.6 × 109 CFU/min, representing a significant transfer of biomass from the biofilm to the liquid phase. Our results agree with those of Baty et al. (2), who estimated that up to 97% of biofilm-generated biomass detached from a chitin-degrading marine biofilm over a 200-h period.
The size distributions of detached cell clusters were similar for both biofilms. Cursory statistical analysis showed that the distributions differed markedly from ordinary probability functions and appeared to resemble a Pareto distribution (6). We are currently investigating the applicability of applying a Pareto distribution to model the detachment of cell clusters. This approach may be useful for predicting the probability of detachment of large cell clusters from biofilms. The importance of modeling detachment as a discrete process in which local conditions may lead to local detachment events and nonuniform biofilm thickness has been recognized (11, 14).
Numerically, cell clusters with areas of greater than 1,000 μm2 made up only about 10% of the total number of detached cell clusters but contributed over 60% of the total detached biofilm area. This result suggests that, although the larger cell clusters detached less frequently, they contained a disproportionately large proportion of the total detached biomass. The largest cluster that detached from the four-species biofilm had a diameter of approximately 500 μm. From the biofilm thickness, we estimated that the volume of the cell cluster was 7.2 × 10−6 cm3 and that it could have contained up to 105 CFU (based on a cell density of 1010 CFU/cm3 for cell cluster material measured from individual cell clusters that had been selectively removed from similar biofilms by micropipette (P. Stoodley and D. deBeer, unpublished results). The frequency data (Fig. 3) suggest that a biofilm covering a 1-cm2 area could shed five such clusters every hour.
Microscopic analysis of the effluent from the tap water biofilm supported the conclusions made from direct microscopic observations of cell clusters leaving the biofilm. The effluent contained a wide range of biomass particulates from single cells to cell clusters containing over 103 cells. Although single cells and aggregates of 10 cells or less accounted for over 90% of the biomass particles, they contained only approximately 50% of the total effluent biomass concentration. Conversely, a disproportionately high percentage of the total cells occurred in the larger aggregates. The largest cell cluster that we detected in the effluent contained approximately 1.6 × 103 cells. This number is above the estimated infective dose for a number of pathogenic biofilm-forming organisms, such as Escherichia coli, Salmonella spp., and Listeria monocytogenes (9) which, depending on the strain and age and health of the host, is estimated to be as low as 10 cells (http://vm.cfsan.fda.gov /∼mow/intro.html). If cell clusters spontaneously detach from these types of biofilms, it is possible that an infective dose may be provided by the ingestion of a single detached cluster which, if not dispersed, may produce only 1 CFU during routine monitoring. It is also likely that detached biofilm clusters will possess the protective properties against antibiotics and host defense systems that are commonly attributed to attached biofilms (7, 8). Since the detachment frequency was inversely related to the cell cluster size, it is likely that larger clusters detached over the course of the experiment but did so too infrequently to be captured in our filtered sample. Further statistical analysis is required to more fully describe the distribution of detached biomass.
Finally, this work raises questions regarding current monitoring techniques. Liquid microbiological samples are usually vortexed prior to enumeration, and the impact to public health is based on averaged cell concentrations. Although homogenization may improve confidence in the mean, our data suggest that the distribution of biomass in liquid samples should also be of concern. The current study represents an important first step in characterizing detachment of cell clusters from biofilms. Future work will focus on the genetic and environmental regulation of detachment.
ACKNOWLEDGMENTS
Work in the laboratories of P.S. and H.M.L.-S. was supported by National Institutes of Health grant 5RO1GM60052-02.
From Montana State University we thank Peter Suci for discussion and insights, Gill Geesey for comments on the manuscript, and Marty Hamilton and Tom Oakberg for statistical input.
REFERENCES
- 1.Bakke R, Olsson P Q. Biofilm thickness measurements by light-microscopy. J Microbiol Methods. 1986;5:93–98. [Google Scholar]
- 2.Baty A M, Eastburn C C, Techkarnjanaruk S, Goodman A E, Geesey G G. Spatial and temporal variations in chitinolytic gene expression and bacterial biomass production during chitin degradation. Appl Environ Microbiol. 2000;66:3574–3585. doi: 10.1128/aem.66.8.3574-3585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryers J D. Modeling biofilm accumulation. In: Bazin M J, Prosser J I, editors. Physiological models in microbiology. Vol. 2. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 109–144. [Google Scholar]
- 4.Characklis W G. Biofilm processes. In: Characklis W G, Marshall K C, editors. Biofilms. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 195–231. [Google Scholar]
- 5.Dalton H M, Goodman A E, Marshall K C. Diversity in surface colonization behavior in marine bacteria. J Ind Microbiol. 1996;17:228–234. [Google Scholar]
- 6.Ellis B D. Biomass distributions, activity, growth, and carbon utilization in heterotrophic bacterial communities. Ph.D. thesis. Bozeman: Montana State University; 1999. [Google Scholar]
- 7.Gilbert P, Brown M R W. Mechanisms of the protection of bacterial biofilms from antimicrobial agents. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms—plant and microbial biotechnology research series. Vol. 5. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 118–130. [Google Scholar]
- 8.Hoiby N, Fomsgaard A, Jensen E T, Johansen H K, Kronborg G, Pedersen S S, Pressler T, Kharazmi A. The immune response to bacterial biofilms. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms—plant and microbial biotechnology research series. Vol. 5. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 233–250. [Google Scholar]
- 9.Hood S K, Zottola E A. Adherence to stainless steel by foodborne microorganisms during growth in model food systems. Int J Food Microbiol. 1997;37:145–153. doi: 10.1016/s0168-1605(97)00071-8. [DOI] [PubMed] [Google Scholar]
- 10.Kwok W K, Picioreanu C, Ong S L, van Loosdrecht M C M, Ng W J, Heijnen J J. Influence of biomass production and detachment forces on biofilm structures in a biofilm airlift suspension reactor. Biotechnol Bioeng. 1998;58:400–407. doi: 10.1002/(sici)1097-0290(19980520)58:4<400::aid-bit7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Morgenroth E, Wilderer P A. Influence of detachment mechanisms on competition in biofilms. Water Res. 2000;34:417–426. [Google Scholar]
- 12.Nickel J C, Costerton J W, McLean R J C, Olson M. Bacterial biofilms—influence on the pathogenesis, diagnosis and treatment of urinary-tract infections. J Antimicrob Chemother. 1994;33(Suppl. A):31–41. doi: 10.1093/jac/33.suppl_a.31. [DOI] [PubMed] [Google Scholar]
- 13.Piriou P, Dukan S, Levi Y, Jarrige P A. Prevention of bacterial growth in drinking water distribution systems. Water Sci Technol. 1997;35:283–287. [Google Scholar]
- 14.Stewart P S. A model of biofilm detachment. Biotechnol Bioeng. 1993;41:111–117. doi: 10.1002/bit.260410115. [DOI] [PubMed] [Google Scholar]
- 15.Stoodley P, Lewandowski Z, Boyle J D, Lappin-Scott H M. Structural deformation of bacterial biofilms caused by short term fluctuations in flow velocity: an in-situ demonstration of biofilm viscoelasticity. Biotechnol Bioeng. 1999;65:83–92. [PubMed] [Google Scholar]
- 16.Stoodley P, Lewandowski Z, Boyle J D, Lappin-Scott H M. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ Microbiol. 1999;1:447–457. doi: 10.1046/j.1462-2920.1999.00055.x. [DOI] [PubMed] [Google Scholar]
- 17.Stoodley P, Dodds I, Boyle J D, Lappin-Scott H M. Influence of hydrodynamics and nutrients on biofilm structure. J Appl Microbiol. 1999;85:19S–28S. doi: 10.1111/j.1365-2672.1998.tb05279.x. [DOI] [PubMed] [Google Scholar]
- 18.Walker J T, Mackerness C W, Mallon D, Makin T, Williets T, Keevil C W. Control of legionella-pneumophila in a hospital water-system by chlorine dioxide. J Ind Microbiol. 1995;15:384–390. doi: 10.1007/BF01569995. [DOI] [PubMed] [Google Scholar]
- 19.Zottola E A, Sasahara K C. Microbial biofilms in the food industry—should they be a concern? Int J Food Microbiol. 1994;23:125–148. doi: 10.1016/0168-1605(94)90047-7. [DOI] [PubMed] [Google Scholar]